Abstract
Virus-specific cytotoxic T lymphocytes (CTL) are critical for control of human immunodeficiency virus type 1 replication. However, viral escape from CTL recognition can undermine this immune control. Here we demonstrate the high frequency and pattern of viral escape from dominant epitope-specific CTL in SIV gag DNA-vaccinated rhesus monkeys following a heterologous simian immunodeficiency virus (SIV) challenge. DNA-vaccinated monkeys exhibited initial effective control of the SIV challenge, but this early control was lost by serial breakthroughs of viral replication over a 3-year follow-up period. Increases in plasma viral RNA correlated temporally with declines of dominant SIV epitope-specific CD8+ T-lymphocyte responses and the emergence of viral mutations that escaped recognition by dominant epitope-specific CTL. Viral escape from CTL occurred in a total of seven of nine vaccinated and control monkeys, including three animals that initially controlled viral replication to undetectable levels of plasma viral RNA. These data suggest that CTL exert selective pressure on viral replication and that viral escape from CTL may be a limitation of CTL-based AIDS vaccine strategies.
Virus-specific cytotoxic T-lymphocyte (CTL) responses have been shown to be critical for the control of human immunodeficiency virus type 1 (HIV-1) replication in humans and simian immunodeficiency virus (SIV) replication in rhesus monkeys (17, 23, 28). It is therefore widely believed that an effective AIDS vaccine should elicit potent CTL responses (20). Recent studies have shown that plasmid DNA, recombinant live vectors, and a number of combined modality vaccines can elicit virus-specific CTL responses and afford significant control of viral replication following pathogenic viral challenges in rhesus monkeys (5, 6, 27, 29). Human trials with these AIDS vaccine strategies are in progress.
It has also been shown that viral mutations can develop in CTL epitopes and result in escape of these viruses from CTL recognition. This phenomenon has been observed in both HIV-1-infected humans (8, 15, 16, 18, 25, 26) and SIV-infected rhesus monkeys (1, 9, 12, 24). Moreover, HIV-1 mutations have been shown to cluster in HLA-restricted epitopes at a population level, suggesting the importance of CTL responses in shaping viral evolution in human populations (22).
We recently demonstrated that a rhesus monkey immunized with cytokine-augmented DNA vaccines and subsequently infected with simian-human immunodeficiency virus (SHIV) strain 89.6P eventually lost control of viral replication as a result of viral escape from CTL recognition (7). The frequency and pattern of viral escape from CTL in cohorts of vaccinated animals, however, have not previously been investigated. In this study, we performed a longitudinal analysis of the durability of immunologic control and viral sequence evolution in a cohort of SIVmac239 gag DNA-vaccinated rhesus monkeys following a heterologous SIVsmE660 challenge.
MATERIALS AND METHODS
Rhesus monkeys.
Nine Mamu-A∗01-positive rhesus monkeys were selected for inclusion in this study. The monkeys were immunized intramuscularly with 5 mg of SIVmac239 gag DNA vaccine or a sham plasmid at weeks 0, 4, 8, and 24 and challenged intravenously at week 28 with 50 monkey-infectious doses of SIVsmE660 as described previously (10). Plasma viral RNA levels were measured by an ultrasensitive branched DNA amplification assay with a detection limit of 500 copies/ml (Bayer Diagnostics, Emeryville, Calif.). Monkeys were maintained in accordance with the Committee on Animals of Harvard Medical School and the Guide for the Care and Use of Laboratory Animals (23a).
Tetramer staining.
A 0.2-μg sample of phycoerythrin-labeled tetrameric Mamu-A∗01-peptide complex was used in conjunction with fluorescein isothiocyanate-labeled anti-human CD8α (Leu2a; Becton Dickinson, San Jose, Calif.), Texas Red-labeled anti-human CD8αβ (2ST8-5H7; Beckman Coulter, Miami, Fla.), and allophycocyanin-labeled anti-rhesus monkey CD3 (FN18) monoclonal antibodies to stain epitope-specific CD8+ T cells as described previously (4, 19). Whole blood (100 μl) from the monkeys was stained with these reagents, lysed, washed, and fixed. Samples were analyzed by four-color flow cytometry on a Becton Dickinson FACScalibur, and gated CD3+ CD8+ T cells were examined for staining with tetrameric Mamu-A∗01-peptide complexes.
Viral sequencing.
Analyses of 500-bp regions of viral gag, env, and pol sequences were performed essentially as described previously (7, 30). Virus from frozen plasma samples was isolated by centrifugation at 25,000 × g for 1 h and lysed with 48% guanidine thiocyanate, 1.4% dithiothreitol, 1% N-laurolylsarcosine, and 1% sodium citrate. RNA was precipitated with isopropanol and solubilized. First-strand cDNA was synthesized with reverse transcriptase by using the SIV gag primer 5′-TGTTTGTTCTGCTCTTAAGCTTTTGTAG-3′ or the SIV pol primer 5′-ATGCCATGAGAAATGCTTCCA-3′. First-strand cDNA was synthesized during PCR amplification for HIV-1 env. Initial PCR amplification used the primers gag-fwd (5′-ACCTAGTGGTGGAAACAGGAACAG-3′), gag-rev (5′-TGTTTGTTCTGCTCTTAAGCTTTTGTAG-3′), env-fwd (5′-ATCTGTGAGCAGTCAGCGAATG-3′), env-rev (5′-TTGCCTCTAGGAAGTCAACCTGTC-3′), pol-fwd (5′-TTCTCAGTCAGGAACAAGAAGGATG-3′), and pol-rev (5′-ATGCCATGAGAAATGCTTCCA-3′). Secondary nested PCR amplification used the primers gag-fwd (5′-AGCACCATCTAGTGGCAGAGGA-3′), gag-rev (5′-GAAATGGCTCTTTTGGCCCTT-3′), env-fwd (5′-AGAATCCTGTGACAAGCATTATTGG-3′), env-rev (5′-CTGTCTTTCCCACTCTTGCCA-3′), pol-fwd (5′-CAAGAAGGCAAGCCATTAGAAGC-3′), and pol-rev (5′-TCTGTGATATATCCTGCTTTCCCTTC-3′). The amplified fragments were cloned into pCR4-TOPO (Invitrogen, Carlsbad, Calif.) or pAMPI (Stratagene, La Jolla, Calif.), and individual transformed colonies were subjected to T3 or T7 dideoxy sequencing.
Peptide binding assays.
Peptide binding assays were performed by measuring inhibition of the iodinated p11C analog peptide (ATPYDINQM) binding to 721.221 cells that express the major histocompatibility complex (MHC) class I molecule Mamu-A∗01. Cells (2 × 106) were incubated overnight at 26°C with 3 μg of human β2 microglobulin per ml. Iodinated p11C peptide (1.5 × 105 cpm) and serial half-log dilutions of unlabeled test peptides were then incubated with washed cells for 4 h at 20°C. Cells were then washed three times, and the radioactivity of the cell pellet was measured with a scintillation counter. The concentration of test peptide required for inhibition of binding of the index peptide by 50% was designated the 50% inhibitory concentration (IC50).
ELISPOT assays.
Ninety-six-well multiscreen plates were coated overnight with 100 μl of 10 μg/ml anti-human gamma interferon (B27; Becton Dickinson) per well in endotoxin-free Dulbecco's phosphate-buffered saline (D-PBS; Life Technologies, Gaithersburg, Md.). The plates were then washed three times with D-PBS containing 0.25% Tween 20 (D-PBS/Tween), blocked for 2 h with D-PBS containing 5% fetal bovine serum at 37°C, washed three times with D-PBS/Tween, rinsed with RPMI 1640 medium containing 10% fetal bovine serum to remove the Tween 20, and incubated with 8, 0.8, or 0.08 μg of peptide per ml and 2 × 105 peripheral blood mononuclear cells (PBMC) in triplicate in 100-μl reaction volumes. Following an 18-h incubation at 37°C, the plates were washed nine times with D-PBS/Tween and once with distilled water. The plates were then incubated with 2 μg of biotinylated rabbit anti-human gamma interferon (Biosource, Camarillo, Calif.) per ml for 2 h at room temperature, washed six times with Coulter Wash (Beckman Coulter, Miami, Fla.), and incubated for 2.5 h with a 1:500 dilution of streptavidin-alkaline phosphatase (Southern Biotechnology, Birmingham, Ala.). Following five washes with Coulter Wash and one with PBS, the plates were developed with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate chromogen (Pierce, Rockford, Ill.), stopped by washing with tap water, air dried, and read with an ELISPOT reader (Hitech Instruments, Edgemont, Pa.). Numbers of spot-forming cells (SFC) per 106 PBMC were calculated. Backgrounds of media were consistently <15 SFC per 106 PBMC.
RESULTS
Breakthroughs of SIV replication in DNA-vaccinated rhesus monkeys.
Nine rhesus monkeys expressing the MHC class I allele Mamu-A∗01 were included in this vaccine trial. At weeks 0, 4, 8, and 24, these animals were immunized intramuscularly with either 5 mg of a sham plasmid (n = 5) or 5 mg of SIVmac239 gag DNA vaccine (n = 4). At week 28, these animals were infected intravenously with 50 monkey-infectious doses of the heterologous, pathogenic virus SIVsmE660. We have previously reported that the vaccine-elicited Gag-specific CTL responses in these animals afforded partial control of early set point plasma viral RNA levels (10). In the present study, we investigated the durability of immunologic control by assessing plasma viral RNA levels, virus-specific cellular immune responses, viral sequence evolution, and clinical disease progression for 3 years following the challenge.
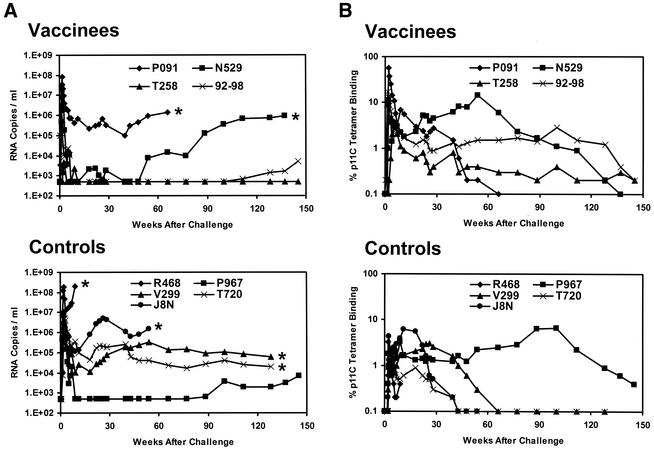
As shown in Fig. 1A, despite the high stringency of this heterologous SIV challenge (13), three of four vaccinated animals exhibited low or undetectable levels of plasma viral RNA at the set point of viral replication between weeks 10 and 50 following the challenge. In contrast, only one of five sham-vaccinated animals controlled viral replication. Over a 3-year follow-up period, however, serial breakthroughs of viral replication resulted in loss of this early control. Of the four monkeys that had undetectable initial set point viral RNA levels (vaccinees N529, 92-98, and T258 and control P967), three eventually developed breakthroughs of viral replication and clinical disease progression. Vaccinated monkey N529 exhibited a breakthrough of viral replication 1 year following challenge, with a dramatic rise in plasma viral RNA levels. This animal developed a clinical AIDS-like syndrome and was euthanized 2.5 years following challenge. Pathological examination revealed generalized wasting and opportunistic infections consistent with advanced SIV infection (data not shown). Vaccinated monkey 92-98 had undetectable plasma viral RNA levels for >2 years prior to a breakthrough of viral replication. Similarly, control monkey P967 appeared to be a clinical long-term nonprogressor for nearly 2 years following the challenge prior to a breakthrough of viral replication. In contrast, vaccinated monkey T258 maintained persistently undetectable plasma viral RNA levels for the entire 3-year follow-up period.
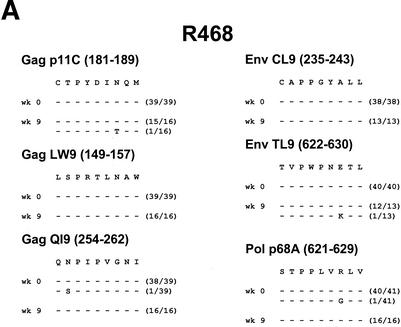
FIG. 1.
Plasma viral RNA levels and CD8+ T-lymphocyte responses following SIVsmE660 challenge. (A) Plasma viral RNA levels were determined for 3 years following challenge in the SIV gag DNA-vaccinated and control rhesus monkeys. The limit of detection of this assay was 500 RNA copies/ml. Asterisks denote animal deaths. (B) Gag p11C-specific CD8+ T-lymphocyte responses were determined for 3 years following challenge in the SIV gag DNA-vaccinated and control rhesus monkeys. The tetramer responses represent the percentages of gated CD3+ CD8+ T lymphocytes that bound tetramer. Functional CTL activity was confirmed by chromium release assays (data not shown).
All of the monkeys with initially high set point viral RNA levels (vaccinee P091 and controls R468, J8N, V299, and T720) developed clinical AIDS-like syndromes and were euthanized during this period. Monkey R468 had 107 to 108 copies of viral RNA per ml of plasma and died shortly after the primary infection. Monkeys J8N and P091 had approximately 106 copies of viral RNA per ml of plasma and died 1.5 years after the challenge. Monkeys V299 and T720 had 104 to 105 copies of viral RNA per ml of plasma and survived for 2.5 years. Pathological examination of these animals revealed opportunistic infections and lymphomas consistent with advanced SIV infection (data not shown).
Declines of dominant epitope-specific CD8+ T-cell responses.
We next studied longitudinal virus-specific CD8+ T-cell responses specific for the Mamu-A∗01-restricted immunodominant Gag p11C epitope (181 to 189; CTPYDINQM) (2, 21). As shown in Fig. 1B, CD8+ T-cell responses were monitored by tetramer binding (4, 19) throughout the 3-year follow-up period. Functional chromium release cytotoxicity assays were also performed at various time points following challenge and confirmed the results of these tetramer assays (data not shown).
As expected, early and potent anamnestic p11C-specific CD8+ T-cell responses were observed in the vaccinated monkeys following the challenge, compared with weaker primary responses in the control animals. Interestingly, nearly all of the vaccinated and control animals exhibited initial stable p11C tetramer+ CD8+ T-cell responses that were followed by profound declines in the numbers of these cells. Monkeys with high set point plasma viral RNA levels (vaccinee P091 and controls J8N, V299, and T720) had short periods of stable p11C tetramer+ CD8+ T-cell responses, followed by marked declines in these responses between weeks 20 and 60 following the challenge. In contrast, monkeys with low set point plasma viral RNA levels (vaccinees N529, 92-98, and T258 and control P967) had longer periods of stable p11C tetramer+ CD8+ T-cell responses. However, declines eventually occurred in monkeys N529 (week 54), 92-98 (week 100), and P967 (week 100). These declines correlated temporally with breakthroughs of viral replication in monkeys N529 (week 54), 92-98 (week 112), and P967 (week 89). Monkey T258 had low but stable p11C tetramer+ CD8+ T-cell responses during the 3-year study period.
Viral sequence evolution.
We reasoned that the declines in the p11C tetramer+ CD8+ T-cell populations in the setting of high levels of plasma viral RNA may have reflected viral sequence evolution in CTL epitopes in these animals. To assess this possibility, we isolated plasma virus samples from each monkey at various time points; amplified 500-bp regions of gag, env, and pol; and sequenced multiple clones of each region. We also analyzed multiple clones of the SIVsmE660 challenge stock. Viral sequence evolution was evaluated in the Mamu-A∗01-restricted CTL epitopes Gag p11C (181 to 189; CTPYDINQM), Gag LW9 (149 to 157; LSPRTLNAW), Gag QI9 (254 to 262; QNPIPVGNI), Env CL9 (235 to 243; CAPPGYALL), Env TL9 (622 to 630; TVPWPNETL), and Pol p68A (621 to 629; STPPLVRLV) (2, 3, 11, 14, 21). The Gag p11C and Env TL9 epitopes have previously been reported to be high-frequency dominant epitopes in SIV-infected Mamu-A∗01-positive rhesus monkeys (2, 14, 21), whereas the Gag LW9, Gag QI9, Env CL9, and Pol p68A epitopes have been reported to be low-frequency subdominant epitopes. Consistent with these observations, tetramer+ CD8+ T-cell responses specific for the Gag p11C and Env TL9 epitopes were readily detected in freshly isolated PBMC of these animals, whereas tetramer+ CD8+ T-cell responses for the subdominant epitopes were below the detection limit of this assay (data not shown).
As demonstrated in Fig. 2, viral sequence evolution was observed in the dominant Gag p11C and Env TL9 CTL epitopes in three of the four vaccinated monkeys. Monkey P091 was the only vaccinated monkey that failed to control primary viremia. A viral variant with a mutation in the Gag p11C epitope emerged by week 30 following the challenge in this animal. This mutation encoded a threonine to serine substitution at position 2 of the epitope and was found in the majority of viral clones at week 30 and in all clones at subsequent time points. A mutation in the Env TL9 epitope encoding a proline to threonine change at position 3 was also detected in the majority of viral clones by week 30. In contrast, little viral sequence evolution was observed in the subdominant Gag LW9, Gag QI9, Env CL9, and Pol p68A epitopes in this animal.
FIG. 2.
Viral sequence evolution in SIV gag DNA-vaccinated animals. Viral sequence evolution over time in the Mamu-A∗01-restricted Gag p11C, Gag LW9, Gag QI9, Env CL9, Env TL9, and Pol p68A CTL epitopes. Sequence analyses are shown for SIV gag DNA-vaccinated rhesus monkeys P091 (A), N529 (B), 92-98 (C), and T258 (D) at various time points following the challenge. The numbers of viral clones analyzed per time point are shown in parentheses.
Vaccinated monkeys N529 and 92-98 both exhibited initial immune control of viral replication with later breakthroughs of viral replication. In monkey N529, a viral variant with a mutation in the Gag p11C epitope emerged by week 66, coincident with the breakthrough of viral replication. This mutation also encoded a position 2 threonine to serine substitution and was found in half of the viral clones at the time of viral breakthrough and in all clones at subsequent time points. A mutation in the Env TL9 epitope developed at later time points and encoded aspartate substitutions at position 6 or 7. In monkey 92-98, a viral variant with a mutation in the Gag p11C epitope emerged by week 100, just prior to the breakthrough of viral replication. This mutation encoded a position 2 threonine-to-alanine substitution and was found in the majority of the viral clones at week 100 and in all of the clones at a subsequent time point. A late position 3 proline to leucine mutation also developed in the Env TL9 epitope. No major mutations were found in the subdominant epitopes in these animals.
Vaccinated monkey T258, in contrast, had undetectable plasma viral RNA for the entire 3-year follow-up period. In fact, plasma virus could not be amplified by nested PCR in multiple samples despite numerous attempts, indicating extraordinarily low levels of plasma viral RNA in this animal. We thus were unable to generate viral sequence data after primary infection. While we can assume that functionally significant mutations in dominant CTL epitopes likely did not occur in this animal, we cannot demonstrate this explicitly.
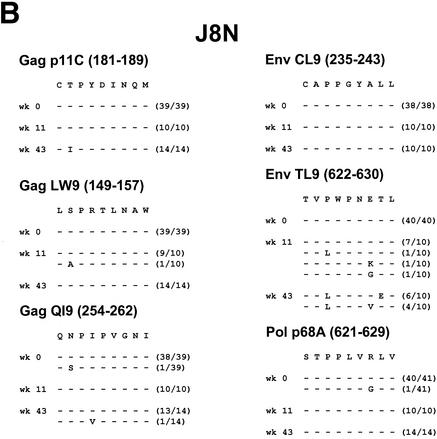
As demonstrated in Fig. 3, viral sequence evolution in these same two dominant Gag p11C and Env TL9 epitopes also occurred in four of the five control monkeys. Control monkey R468 died shortly after primary infection with uncontrolled viral replication. This animal had high set point plasma viral RNA levels (107 to 108 copies/ml) but very low p11C tetramer+ CD8+ T-cell responses. Interestingly, this monkey was the only control animal whose virus did not develop mutations in CTL epitopes, suggesting that very high levels of viral replication in the absence of potent immune pressure do not select for CTL escape mutations.
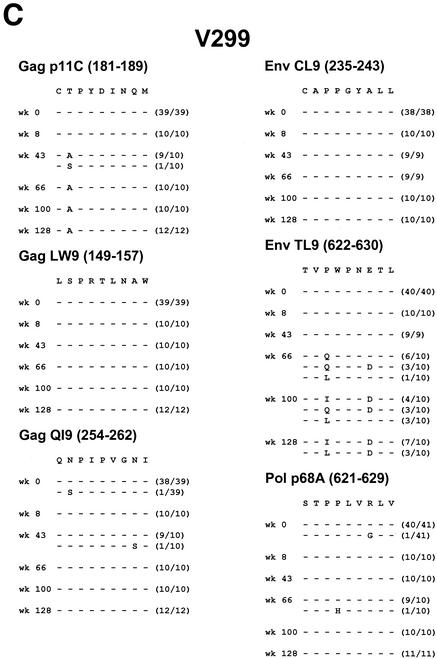
FIG. 3.
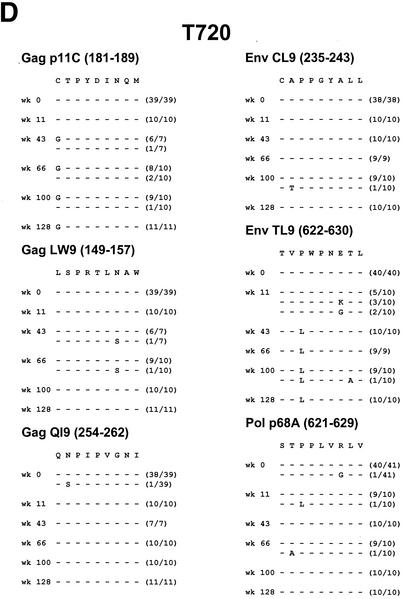
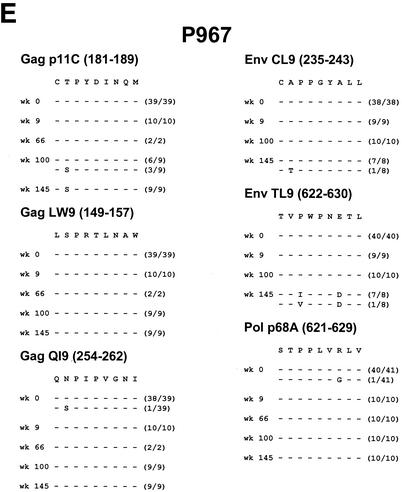
Viral sequence evolution in control animals. Viral sequence evolution over time in the Mamu-A∗01-restricted Gag p11C, Gag LW9, Gag QI9, Env CL9, Env TL9, and Pol p68A CTL epitopes is shown. Sequence analyses are shown for control rhesus monkeys R468 (A), J8N (B), V299 (C), T720 (D), and P967 (E) at various time points following the challenge. The numbers of viral clones analyzed per time point are shown in parentheses.
Control monkeys J8N, V299, and T720 had more typical courses of SIV infection, with persistent moderate to high plasma viral RNA levels (104 to 106 copies/ml). These animals all developed early mutations in the Gag p11C and Env TL9 epitopes. In monkey J8N, a position 2 threonine to isoleucine mutation developed in the Gag p11C epitope and a position 3 proline to leucine mutation with additional position 7 and 8 changes developed in the Env TL9 epitope. In monkey V299, a position 2 threonine to alanine mutation developed in the Gag p11C epitope and a position 3 proline to isoleucine or leucine mutation with an additional position 7 aspartate substitution developed in the Env TL9 epitope. In monkey T720, a position 1 cysteine to glycine mutation developed in the Gag p11C epitope and a position 3 proline to leucine mutation developed in the Env TL9 epitope.
Control monkey P967 initially appeared to be a clinical long-term nonprogressor, with undetectable plasma viral RNA levels for nearly 2 years following the challenge. However, a viral variant with a mutation in the Gag p11C epitope emerged coincident with the breakthrough of viral replication. This mutation encoded a position 2 threonine to serine substitution and was found in a fraction of the clones at the time of viral breakthrough at week 100 and in all of the clones at a subsequent time point. Late mutations also developed in the Env TL9 epitope encoding position 3 proline to isoleucine or valine mutations with an additional position 7 aspartate substitution.
Mutations in both the dominant Gag p11C and Env TL9 epitopes thus emerged in seven of nine vaccinated and control monkeys. The emergence of mutations in the Gag p11C epitope correlated in all cases with a decline in p11C tetramer+ CD8+ T-cell responses and with breakthroughs of viral replication in the animals that initially controlled viral replication. No major mutations in the subdominant Gag LW9, Gag QI9, Env CL9, and Pol p68A epitopes were observed.
Mutant peptides effectively escape CTL recognition.
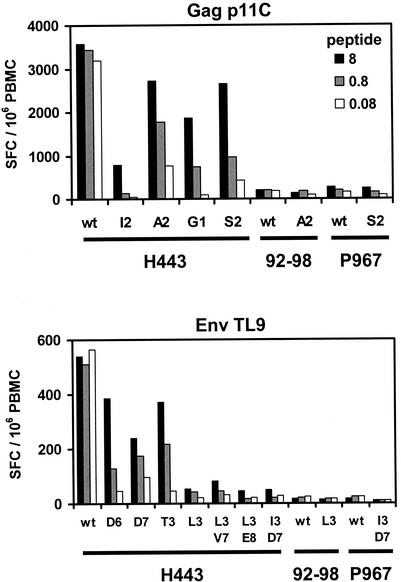
We next synthesized the mutant Gag p11C and Env TL9 peptides in vitro and assessed their abilities to bind the MHC class I molecule Mamu-A∗01 and to be recognized by epitope-specific CTL. In competitive peptide binding assays, whose results are summarized in Table 1, all of the mutant peptides exhibited 1- to 3-log decreases in the ability to bind Mamu-A∗01, compared with the wild-type p11C and TL9 peptides. Moreover, as shown in Fig. 4, the mutant peptides were not efficiently recognized by PBMC in functional gamma interferon ELISPOT assays, although several of these peptides were still inefficiently recognized by PBMC at high peptide concentrations. These data demonstrate that the viruses containing these mutations in dominant CTL epitopes effectively escaped from p11C-specific and TL9-specific CTL responses. Interestingly, PBMC from monkeys 92-98 and P967 were unable to recognize both the wild-type and mutant peptides following viral escape from CTL.
TABLE 1.
Wild-type and mutant Gag p11C and Env TL9 peptidesa
| Epitope and peptide | Sequence | Monkey(s) | Binding IC50 (μm) |
|---|---|---|---|
| Gag p11C | |||
| p11C-wt | C T P Y D I N Q M | T258, R468 | 10 |
| p11C-I2 | C I P Y D I N Q M | J8N | 1,000 |
| p11C-A2 | C A P Y D I N Q M | 92-98, V299 | 1,000 |
| p11C-G1 | G T P Y D I N Q M | T720 | 1,000 |
| p11C-S2 | C S P Y D I N Q M | P091, N529, P967 | 3,000 |
| Env TL9 | |||
| TL9-wt | T V P W P N E T L | T258, R468 | 30 |
| TL9-D6 | T V P W P D E T L | N529 | 300 |
| TL9-D7 | T V P W P N D T L | N529 | 300 |
| TL9-T3 | T V T W P N E T L | P091 | 3,000 |
| TL9-L3 | T V L W P N E T L | 92-98, T720 | 10,000 |
| TL9-L3V7 | T V L W P N V T L | J8N | 10,000 |
| TL9-L3E8 | T V L W P N E E L | J8N | 30,000 |
| TL9-I3D7 | T V I W P N D T L | P967, V299 | 30,000 |
Mutations are shown in bold and underlined. The monkeys in which these sequences were detected are noted. Binding affinities for the MHC class I molecule Mamu-A∗01 were determined by measuring IC50 in competitive peptide binding assays.
FIG. 4.
CD8+ T-lymphocyte recognition of mutant peptides. Results of functional gamma interferon ELISPOT assays using PBMC from long-term nonprogressor monkey H443 and from study monkeys 92-98 and P967 at week 150 following challenge are shown. PBMC from these Mamu-A∗01-positive animals were stimulated with either the wild-type (wt) or the mutant p11C or TL9 peptide at 8, 0.8, or 0.08 μg/ml. Mean numbers of SFC per 106 PBMC are shown. Mean responses determined by assays performed in triplicate are shown. Control media were <25 SFC per 106 PBMC.
DISCUSSION
The observation that vaccine-elicited CTL responses can provide a degree of control of viral replication following pathogenic viral challenges in rhesus monkeys has supported the advancement of CTL-based AIDS vaccine candidates into large-scale human trials. In the present study, we demonstrate that the initial partial protection afforded by DNA vaccination against a heterologous SIV challenge waned over a 3-year follow-up period. Breakthroughs of viral replication were observed in these monkeys and correlated temporally with viral escape from dominant epitope-specific CTL.
We observed viral escape from CTL in seven of the nine vaccinated and control monkeys, including three animals that initially controlled viral replication to undetectable levels of plasma viral RNA for 1 to 2 years. Interestingly, viral escape from CTL recognition appeared to occur in a predictable pattern. In all seven monkeys, viral mutations developed in the same two dominant Gag p11C and Env TL9 epitopes, whereas no significant mutations were observed at four subdominant epitopes. The majority of viral mutations were position 2 mutations in the Gag p11C epitope and position 3 mutations in the Env TL9 epitope. In addition, the Gag p11C mutations appeared to precede or to occur concurrently with the Env TL9 mutations.
The temporal correlation between the emergence of mutations in the Gag p11C epitope and the breakthroughs of viral replication in monkeys N529, 92-98, and P967 suggests the functional importance of this epitope-specific CTL response in the control of SIV replication. Although the present study was limited to analyses of Mamu-A∗01-restricted CTL epitopes in Mamu-A∗01-positive rhesus monkeys, we predict that responses restricted by alleles other than Mamu-A∗01 will likely exert similar immune selection pressure on the virus. Thus, viral escape from dominant epitope-specific CTL may prove to be an important and general strategy of immune evasion.
Rapid viral escape from CTL occurred in the monkeys with moderate set point viral RNA levels (P091, J8N, V299, and T720). In contrast, slower viral escape from CTL occurred in the monkeys with low initial set point viral RNA levels (N529, 92-98, and P967), suggesting that effective blunting of viral replication may reduce the stochastic generation of mutant viruses. The absence of CTL escape mutations in monkey R468 suggests that very high levels of viral RNA in the absence of potent immune pressure may be insufficient to select for CTL escape mutations (30). We have also analyzed viral sequences in two additional Mamu-A∗01-positive monkeys that had uncontrolled primary SIVsmE660 replication and have similarly observed no mutations within the Gag p11C and Env TL9 epitopes (data not shown).
We recently demonstrated that one vaccinated rhesus monkey infected with SHIV-89.6P eventually lost immune control of the virus as a result of viral escape from CTL recognition (7). This report did not, however, assess the frequency or pattern of viral escape from CTL. In the present study, we observed a high frequency of viral escape from dominant epitope-specific CTL in vaccinated and control monkeys over a 3-year study period. The higher frequency of viral escape from CTL observed here, compared with our previous study, likely reflects the differences between the study designs. In particular, the present study assessed a less potent DNA vaccine (gag alone versus gag plus env with a cytokine adjuvant) and a more stringent challenge virus (heterologous SIV versus homologous SHIV).
Previous studies have demonstrated frequent viral escape from CTL at a Tat epitope during acute SIV infection (1, 24) and at Env and Nef epitopes during chronic SIV infection (12). Since the Tat SL8 epitope is not present in SIVsmE660, we did not study tat sequences in the present study and focused instead on gag, pol, and env sequences. We did not find consistent viral escape from CTL during acute infection, although we cannot exclude the possibility that mutations occurred in subdominant epitopes in other regions of the viral genome or in epitopes restricted by alleles other than Mamu-A∗01. In contrast, we found that clinically significant viral escape mutations occurred at dominant CTL epitopes during chronic infection.
These data have clear implications for both HIV-1 pathogenesis and AIDS vaccine development strategies. Viral escape from dominant epitope-specific CTL appears to be a frequent and important problem in both vaccinated and control monkeys. The high frequency of viral escape from CTL observed in this study likely reflects the low potency and narrow breadth of the SIV gag DNA vaccine that was used, and it is expected that more potent and broader spectrum vaccines will be associated with a lower frequency of viral escape. In fact, the development of viral mutations that evade CTL may be analogous to the development of drug resistance mutations following suboptimal antiretroviral therapy. HIV-1 vaccine candidates should therefore be designed to elicit both broad and potent cellular immune responses against multiple viral antigens in order to minimize the likelihood of viral escape from CTL recognition. However, viral escape from CTL clearly can occur even after more than 2 years of effective immune control. These data suggest that vaccines that control rather than prevent immunodeficiency virus infections will always be subject to viral immune evasion strategies.
Acknowledgments
We are grateful to T. Fu, J. Shiver, N. Miller, K. Reimann, C. Lord, V. Hirsch, H. Bielefeldt-Ohmann, and A. Schmidt for generous advice, assistance, and reagents.
We acknowledge support from National Institutes of Health grants AI-51223 (D.H.B), CA-50139 (N.L.L.), AI-20729 (N.L.L.), AI-85343 (N.L.L.), CFAR P30 AI-28691 (N.L.L.), and CFAR P30 CA-79458 (S.M.W.). D.H.B. is a recipient of a Doris Duke Clinical Scientist Development Award.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viremia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. M., J. Sidney, M.-F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A∗01) that binds an immunodominant CTL epitope from SIV. J. Immunol. 160:6062-6071. [PubMed] [Google Scholar]
- 3.Allen, T. M., B. R. Mothé, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A∗01: implications for vaccine design and testing. J. Virol. 75:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altman, J. D., P. A. H. Moss, P. J. R. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 5.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-K. Ma, B. D. Grimm, M. L. Hulsey, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of clinical AIDS in rhesus monkeys by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 6.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T.-M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M.-E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 8.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. A. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z. W., A. Craiu, L. Shen, M. J. Kuroda, U. C. Iroku, D. I. Watkins, G. Voss, and N. L. Letvin. 2000. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J. Immunol. 164:6474-6479. [DOI] [PubMed] [Google Scholar]
- 10.Egan, M. A., W. A. Charini, M. J. Kuroda, G. Voss, J. E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M. A. Lifton, C. E. Nickerson, T. M. Fu, J. W. Shiver, and N. L. Letvin. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74:7485-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan, M. A., M. J. Kuroda, G. Voss, J. E. Schmitz, W. A. Charini, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Use of a major histocompatibility complex class I/peptide/β2M tetramers to quantitate CD8+ cytotoxic T lymphocytes specific for dominant and nondominant viral epitopes in simian-human immunodeficiency virus-infected rhesus monkeys. J. Virol. 73:5466-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg, M. B., and J. P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207-210. [DOI] [PubMed] [Google Scholar]
- 14.Furchner, M., A. L. Erickson, T. Allen, D. I. Watkins, A. Sette, P. R. Johnson, and C. M. Walker. 1999. The simian immunodeficiency virus envelope glycoprotein contains two epitopes presented by the Mamu-A∗01 class I molecule. J. Virol. 73:8035-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulder, P. J. R., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 16.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 17.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, and C. Carter. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda, M. J., J. E. Schmitz, D. H. Barouch, A. Criau, T. M. Allen, A. Sette, D. I. Watkins, M. A. Forman, and N. L. Letvin. 1998. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J. Exp. Med. 187:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letvin, N. L., D. H. Barouch, and D. C. Montefiori. 2002. Prospects for vaccine protection against HIV-1 infection and AIDS. Annu. Rev. Immunol. 20:73-99. [DOI] [PubMed] [Google Scholar]
- 21.Miller, M. D., H. Yamamoto, A. H. Hughes, D. I. Watkins, and N. L. Letvin. 1991. Definition of an epitope and MHC class I molecule recognized by Gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J. Immunol. 147:320-329. [PubMed] [Google Scholar]
- 22.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 23.Musey, L., J. Hughes, T. Schacker, T. Shea, L. Corey, and M. J. McElrath. 1997. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337:1267-1274. [DOI] [PubMed] [Google Scholar]
- 23a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 24.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 25.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. M. Bangham, C. R. Rizza, and A. J. McMichael. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 26.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 29.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 30.Wolinksy, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]