Abstract
Infectious Human respiratory syncytial virus (HRSV) with an aberrant RNA synthesis pattern was recovered from a cDNA clone. The virus displayed increased levels of polycistronic readthrough mRNAs resulting from failure of the polymerase to terminate transcription efficiently at the gene ends. An asparagine (N) to aspartic acid (D) change at amino acid 1049 in the large (L) polymerase protein was found to be responsible for the readthrough phenotype. Virus encoding N at position 1049 displayed an RNA synthesis pattern indistinguishable from the A2 strain of HRSV. We compared the transcriptional activities of polymerases that encoded an N or D at position 1049 (L1049N or L1049D) by using dicistronic subgenomic replicons and found that the alteration of amino acid 1049 specifically affected transcriptional termination but had no effect on genome replication. L1049N recognized and terminated transcription at each of the naturally occurring gene junctions with differing efficiencies but at significantly higher efficiency than L1049D. The abilities of the polymerases to recognize the cis-acting gene end signals required for termination were compared by examining the effect of single nucleotide changes at positions 1 to 8 of the M gene end and were found to be similar. This work identifies a single amino acid position in the polymerase that specifically affects the ability of the polymerase to terminate transcription, but which does not affect genome replication or interaction with the M2-1 protein. This work shows the critical importance of the previously defined cis-acting signals for termination, confirms differential termination at the varied gene junctions, and shows that the polymerase responds to the cis-acting sequences similarly, whether it has N or D at position 1049.
Human respiratory syncytial virus (HRSV) is a nonsegmented negative-strand (NNS) RNA virus belonging to the Paramyxoviridae family in the order Mononegavirales. The HRSV genome contains 10 genes, in the order 3′-NS1-NS2-N-P-M-SH-G-F-M2-L-5′, encoding at least 11 proteins (10, 11). The genes are flanked by noncoding leader (Le) and trailer (Tr) sequences at the 3′ and 5′ ends of the genome, respectively. The virally encoded RNA-dependent RNA polymerase (RdRp), containing the large (L) protein and the phosphoprotein (P), carries out genome replication and transcription (17, 47). Genome replication additionally requires the synthesis of nucleocapsid (N) protein for encapsidation of progeny genome RNA, which is essential for recognition by the RdRp (17, 47). In addition to the L and P proteins, efficient transcription of HRSV requires a further viral component, the M2-1 protein. Produced from the first open reading frame of the M2 gene, the M2-1 protein is a transcription factor that decreases intragenic and intergenic termination, allowing synthesis of full-length mRNAs (9, 15, 18, 20).
Available evidence supports a stop-start model of transcription for the Mononegavirales (5, 14, 24) in which the RdRp enters the genome at a single 3′ site and transcribes each gene in an obligatorily sequential manner from 3′ to 5′, producing capped and polyadenylated mRNAs (13). The RdRp must terminate synthesis of each upstream mRNA to initiate synthesis of the next downstream mRNA (21, 32). The sequential mode of transcription, combined with the process of transcriptional attenuation at each gene junction, results in a gradient of mRNA synthesis such that genes nearest the 3′ end are transcribed more abundantly than 3′-distal genes (2, 13). If the RdRp fails to terminate at a gene end sequence, it reads through the gene junction and a polycistronic readthrough product is generated that contains both the upstream and downstream RNAs (18, 21). If readthrough occurs, the RdRp cannot return to the gene start of the downstream gene and synthesize a monocistronic mRNA, and there is no evidence for internal entry by the RdRp other than at the 3′ entry site. Thus, transcriptional termination is a key step at which gene expression can be controlled in the NNS RNA viruses. Analysis of RNAs produced in HRSV-infected cells has shown that in addition to the expected monocistronic mRNAs, numerous polycistronic mRNAs are observed. These polycistronic mRNAs are produced when the polymerase fails to terminate at a gene end signal, and such RNAs are detected that correspond to most of the gene junctions with the exception of the SH/G gene junction (11).
Each HRSV gene has a well-conserved gene start sequence, but the gene end sequences are semiconserved and the genes are separated by nonconserved intergenic regions of 1 to 52 nucleotides (nt) (8). The semiconserved gene end sequences of HRSV are 13 to 14 nt in length and comprised of a semiconserved A (in 8 of the 10 gene ends) followed by the conserved pentanucleotide 3′-UCAAU-5′, an A/U-rich central region of one to four nucleotides, and a U tract of four to seven residues (8, 22). Thus, an HRSV gene junction is 3′-…AUCAAUN1-4U4-7N1-52CCCCGUUUA…-5′ (negative sense). The numbering system used to identify the residues in the gene end specifies the first U of the UCAAU as position 1. Previous work examined the termination efficiencies of the various HRSV genes by constructing a series of dicistronic subgenomic replicons containing each of the gene junction sequences of HRSV in their authentic sequence context and found that the termination efficiencies of the different gene ends varied (18). The sequence requirements for efficient termination at a gene end have been examined at the M and F gene ends (21, 44). Any change at positions 1 through 8 of the M gene end led to a decrease in termination efficiency, although the extent varied (21). The spacing between the conserved UCAAU and U tract and the length of the U tract were both found to be important for efficient termination (21, 44). In addition to the gene end sequence, the composition of the intergenic region was important when the U tract was only four residues (22, 44). This finding was similar to results with Simian parainfluenza virus 5 (37, 38).
While the effects on termination of the cis-acting genomic sequence elements as well as the trans-acting M2-1 protein have been previously examined, the role of the trans-acting HRSV L protein component of the RdRp complex in transcriptional termination has not been well studied (15, 18, 19, 21, 22, 32, 44). The sequences of L proteins from NNS RNA viruses have been aligned, and six conserved sequence blocks, I to VI, have been identified (35, 43, 45). Mutagenic analyses of the NNS virus L proteins have been done to investigate the possible functional significance of these domains (2, 7, 12, 16, 29, 41, 42, 46). Domain III contains a set of conserved motifs, A to D, found in all RdRp (30, 36). The predicted active site of the polymerase, the GDNQ motif, lies in domain III, and mutation of any of these conserved residues inactivated the polymerase (28, 39, 40). Domain II is proposed to be involved in template recognition due to the presence of a conserved area of highly charged residues (35), and mutations in this domain inhibited RNA synthesis (42). Domain VI has a putative nucleotide binding motif (35). Domains I, IV, and V, while conserved among the NNS viruses, have no sequence elements that indicate a specific function. Although much work has focused on determining the precise function of each domain, it has become clear that the individual domains may contribute to several of the distinct functions of the polymerase, supported by recent evidence showing that polymerases inactivated by mutations in separate conserved domains can complement one another (41).
In this work we recovered from cDNAs infectious HRSVs with two distinctly different RNA synthesis patterns. The phenotypic difference was mapped to a single amino acid change in the HRSV L protein that specifically affected transcriptional termination, such that one of the viruses exhibited elevated levels of readthrough at gene ends rather than efficient termination. The ability of polymerases to recognize and respond to the cis-acting termination signals at each of the HRSV gene junctions was examined using subgenomic replicons to determine how a site that affects termination in the trans-acting polymerase interacted with the cis-acting sequence elements previously identified as important for termination. The effect of the M2-1 protein on the ability of the two polymerases to terminate transcription was also examined.
MATERIALS AND METHODS
cDNA constructs.
Plasmids expressing the HRSV proteins N, P, and M2-1 (pN, pP, and pM2ORF-1) have been described previously (20, 47). Our laboratory also previously generated a cDNA plasmid that expressed the HRSV L protein (pL) (47). The L protein expressed from this plasmid was active in both replication and transcription (20, 21, 47). After examining the effect of the nucleotide change at 11642 in the full-length cDNA of HRSV, we reexamined the sequence of pL and found that it also had this mutation; therefore, it was renamed pL1049D. To construct pL1049N, nt 11642 (HRSV genome) in pL1049D was altered by replacing a NheI-PmlI fragment (nt 11002 to 12254) derived from a reverse transcription-PCR (RT-PCR) of the A2 strain of HRSV. An additional mutation at nt 8546 (amino acid S17G; pL) was repaired by QuikChange (Stratagene) using complementary mutagenic primers. The dicistronic subgenomic replicons containing each of the nine authentic HRSV gene junctions, the dicistronic subgenomic replicons containing the M/SH gene junction with the M gene end mutations, and the dicistronic subgenomic replicons with U4A, U4G, and U4C at the M gene end have been described previously (18, 20-22, 47).
HRSV cDNAs were created using standard cloning techniques. Briefly, they were constructed from RT-PCRs of viral RNAs and cloned into pBR322 (New England Biolabs) and were derived independently of cDNAs described previously (33). These cDNAs have a T7 promoter followed by three guanosine residues and then the sequence of the A2 strain of HRSV inserted, such that a positive-sense antigenome is transcribed by T7 RNA polymerase. A hepatitis delta virus ribozyme, which cleaves the RNA to create a perfect 3′ end (34), and a T7 terminator sequence follow the trailer sequence of HRSV. To generate prsL1049D, the SH coding region (nt 4303 to 4497) was replaced with that of human recombinant green fluorescent protein (GFP) without altering the existing gene start, gene end, or the noncoding regions of the SH gene. Briefly, PCR was used to introduce remote cutting BsmBI sites flanking the SH coding regions into the full-length cDNA. The GFP coding sequence with flanking BsmBI sites was cloned into pCR-BluntII-Topo, using the Zero Blunt TOPO PCR cloning kit (Invitrogen). Digestion of both with the BsmBI remote cutting enzyme allowed the insertion of the GFP coding sequence in the place of the SH open reading frame without the introduction of restriction site sequences or alterations in the genome. The L gene sequence in prsL1049D was derived from pL1049D (formerly pL [47]), except that the mutation at nt 8546 was repaired in the full-length cDNA. The prsL1049N described here was constructed by replacing a SphI fragment (nt 10742 to 13933) in prsL1049D with a SphI fragment from the pL1049N plasmid. The only difference between the two full-length cDNAs was the nucleotide change at position 11642.
Constructs were verified by sequence analysis carried out by fluorescent dye termination on ABI Genetic Analyzer 3100 and ABI model 377 DNA sequencers (DNA Sequencing Facility, Center for AIDS Research, University of Alabama at Birmingham).
Cells and virus.
HEp-2 cells were maintained in minimal essential medium (Invitrogen) supplemented with 5% fetal calf serum (FCS) (MEM5). Vero cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% FCS. The A2 strain of HRSV was used. Viral infections were carried out in HEp-2 cells at 33°C. Virus was allowed to adsorb for 2 h, and then MEM with 2% FCS was added. Viruses were recovered from cDNAs essentially as previously reported (33).
RT-PCR and sequencing.
The identity of nt 11642 of the recovered viruses was examined by RT-PCR and sequencing to determine whether the viruses retained the sequence of the input cDNA. RT-PCR was performed using the Qiagen One-Step RT-PCR kit according to the manufacturer's instructions. HEp-2 cells were infected with one of the engineered rsL1049D viruses, and RNAs were extracted at 48 h postinfection. RNAs were resuspended in 30 μl of water, and 2 μl was used per RT-PCR. The primers amplified a 1.2-kb fragment of the L gene of the recovered viruses corresponding to nt 10800 to 12000 of the HRSV sequence. The RT-PCR products were then sequenced.
RNA synthesis and RNase H analysis.
Synthesis of RNAs in HEp-2 cells transfected with HRSV subgenomic replicons was analyzed as described previously (18, 21). Viral RNAs were characterized by cleavage with RNase H following annealing to specific oligonucleotides. Oligo(dT) in combination with RNase H was used to remove the poly(A) tails of the mRNAs, causing sharpening of the bands and faster migration. An oligonucleotide was designed that was specific for the conserved gene start sequence found at the beginning of all HRSV genes, 5′-TTATTTGCCCCAT-3′. Binding of this oligonucleotide to the gene start sequence in polycistronic messages would result in cleavage by RNase H to yield messages corresponding to the size of monocistronic RNAs. RNAs were digested as previously described (18) by incubating with 1 μg of oligo(dT) (15-mer) alone, or in combination with 1 μg of the “gene start” oligonucleotide, and 1 U of RNase H in 1× RNase H buffer for 20 min at 37°C. RNAs were ethanol precipitated and separated on an agarose urea gel.
RESULTS
Recovery of HRSV with an altered RNA pattern.
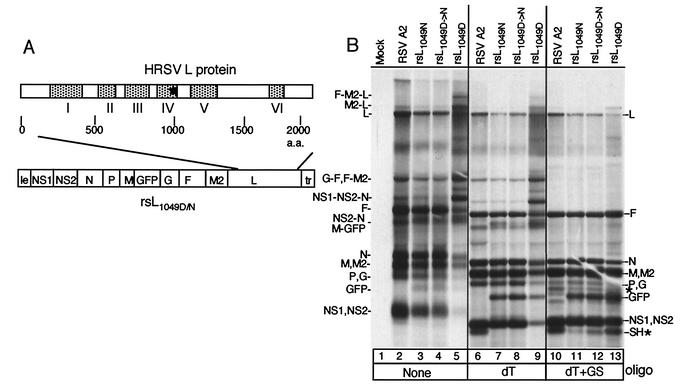
Infectious HRSVs were recovered from several independent full-length cDNA clones in which the SH coding region was replaced with that of GFP. Most of these viruses displayed an RNA synthesis pattern that was identical to that of the A2 strain of HRSV (33). However, one of the viruses, produced from a cDNA in which the L gene came from a preexisting cDNA, pL (47), showed an aberrant RNA synthesis pattern. We analyzed the sequence of the L gene in this cDNA clone and found a base change from the A2 strain sequence at nt 11642, which resulted in an asparagine-to-aspartic acid change at amino acid 1049. We named this virus rsL1049D. To examine whether this single amino acid change in the L protein was responsible for the altered RNA phenotype, a full-length viral cDNA was engineered which was identical to rsL1049D except that it encoded an asparagine at amino acid 1049 in the L protein, and virus was recovered (rsL1049N). HEp-2 cells were infected with the recombinant viruses rsL1049N or rsL1049D, or the A2 strain of HRSV for comparison, and RNAs were examined by metabolic labeling with [3H]uridine. HRSV A2 strain infection resulted in the replication of the viral genome and transcription of monocistronic RNAs for each gene as expected (Fig. 1B, lane 2). In addition, several polycistronic readthrough transcripts were produced which were typical for an HRSV infection (Fig. 1B, lane 2; e.g., NS2-N, G-F, F-M2, and NS1-NS2-N polycistronic RNAs) (11). In contrast, the rsL1049D virus showed an RNA synthesis pattern with a substantial decrease in the faster-migrating RNA products comigrating with monocistronic RNAs (Fig. 1B, lane 5; e.g., NS1, NS2, and N) and increased amounts of slower-migrating polycistronic RNAs (e.g., NS1-NS2-N and NS2-N) (Fig. 1B, compare lanes 2 and 5; see below for characterization of viral RNAs). The rsL1049N virus displayed an RNA synthesis pattern indistinguishable from that of the HRSV A2 virus, except for the expected loss of the SH mRNA and the gain of the GFP mRNA (Fig. 1B, compare lanes 2 and 3, or see below for lanes 6 and 7). These data indicated that the asparagine-to-aspartic acid change at amino acid 1049 was responsible for the altered RNA pattern of the rsL1049D virus. Furthermore, four independent virus stocks were recovered from the rsL1049D cDNA and, interestingly, one showed an RNA synthesis pattern that did not have the enhanced readthrough phenotype, such that it was indistinguishable from rsL1049N and HRSV A2 (Fig. 1B, lane 4). Therefore, we analyzed the sequences of the recovered viruses and found that the three virus stocks that had an altered RNA synthesis pattern had retained the input sequence of the rsL1049D cDNA, while the fourth had changed from a G to an A (positive sense) at nt 11642, which caused a reversion to asparagine at position 1049, rsL1049D→N (Fig. 1B, lane 4). We conclude that during the passaging to recover the rsL1049D virus (43 days compared to 21 days for rsL1049N), one of the virus stocks, rsL1049D→N, reverted at position 11642 to the consensus A2 strain sequence and recovered a normal RNA synthesis pattern (Fig. 1B, lane 4).
FIG. 1.
RNA synthesis of HRSV A2 and recovered engineered viruses. (A) Schematic of the rsL1049D or rsL1049N cDNA in which the SH coding region was replaced with that of GFP. To create rsL1049N, an SphI fragment in the L gene was replaced. Schematic of L protein illustrating the positions of the conserved domains is shown above the cDNA. An asterisk shows the location of amino acid 1049. Viruses were recovered by transfecting appropriate cDNAs along with the HRSV support plasmids pN, pP, pM2ORFI, and L1049D or L1049N into HEp-2 cells infected with MVA-T7. (B) HEp-2 cells were infected with the A2 strain of HRSV or individual engineered viruses, and viral RNAs were labeled 16 h postinfection with [3H]uridine in the presence of actinomycin D. Cells were infected at a multiplicity of infection of approximately 0.5 for each virus, except the rsL1049D virus, for which a multiplicity of infection of approximately 3 was used. RNAs were either undigested (lanes 1 to 5), digested with RNase H and 1 μg of oligo(dT) (lanes 6 to 9), or digested with RNase H, 1 μg of oligo(dT), and 1 μg of gene start oligonucleotide (lanes 10 to 13). Use of the gene start oligonucleotide resulted in some nonspecific cleavage, indicated by new bands (*) not present in the undigested samples or samples digested with oligo(dT) alone. The positions of monocistronic and polycistronic mRNAs are shown to the right and left side of the gel. Those separated by commas are monocistronic or polycistronic RNAs that comigrate. Those separated by hyphens denote polycistronic RNAs. dT, oligo(dT); GS, gene start oligonucleotide.
Characterization of viral RNAs.
The larger RNAs observed in the rsL1049D virus comigrated with polycistronic RNAs that occur naturally in HRSV-infected cells (Fig. 1B, lanes 2 and 5). To determine if the larger RNAs were indeed readthrough RNAs, we designed an oligonucleotide that was complementary to the highly conserved HRSV gene start sequence to use in an RNase H cleavage assay. Following annealing of the gene start oligonucleotide to the gene start present in a polycistronic RNA, RNase H would cleave the oligonucleotide-RNA hybrid. This cleavage would convert any polycistronic readthrough products to monocistronic RNAs. Separately, we also used oligo(dT) to anneal to poly(A) tracts which, when followed by RNase H cleavage, removed the poly(A) tails and resolved mRNAs that were similar in size (Fig. 1B, lanes 6 to 9). When oligo(dT) alone was used (Fig. 1B, lanes 6 to 9), the RNA pattern sharpened and RNAs migrated faster because the poly(A) tails had been removed. RNAs from the engineered viruses did not include the SH mRNA and instead expressed the GFP mRNA (Fig. 1B, compare lane 6 with lanes 7 to 9). When viral RNAs were digested with RNase H in the presence of the gene start oligonucleotide and oligo(dT), there was a loss of the polycistronic RNAs and a concomitant increase in the monocistronic RNAs in all samples, indicating that the slower-migrating RNAs observed with the rsL1049D virus were polycistronic readthrough RNAs that were converted to monocistronic RNAs upon digestion (Fig. 1B, lanes 10 to 13). For example, RNAs corresponding to NS1-NS2-N, NS2-N, G-F, F-M2, and M2-L readthrough products were clearly observed in the untreated or oligo(dT)-treated rsL1049D virus (Fig. 1B, lanes 5 and 9) but were nearly undetectable following digestion with RNase H in the presence of the gene start oligonucleotide (Fig. 1B, lane 13). Some nonspecific cleavage occurred with the gene start oligonucleotide, such that a band was observed migrating at a position similar to that of the SH mRNA in the samples which did not have the SH coding sequence (Fig. 1B, lanes 11 to 13); however, it was clear that these viruses did not have the SH mRNA, as shown by the samples treated with RNase H in the presence of oligo(dT) alone (Fig. 1B, lanes 7 to 9). These data suggested that the asparagine-to-aspartic acid change in the rsL1049D virus decreased termination efficiency, as shown by the increased synthesis of polycistronic readthrough RNAs.
Transcriptional activity of L1049D and L1049N.
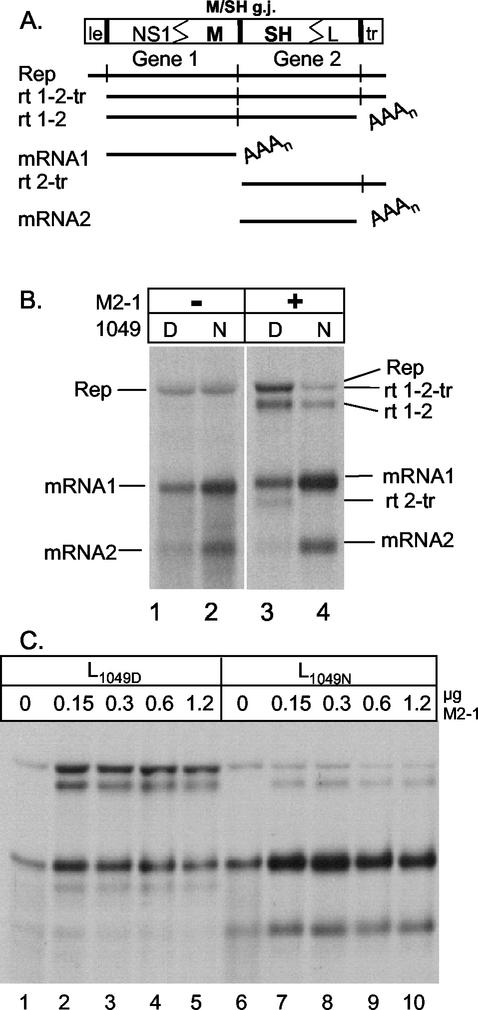
To further examine the transcriptional activity of these two polymerases, we moved our analysis to the subgenomic replicon system. The ability of the HRSV L proteins L1049D (formerly pL) and L1049N to carry out transcription and replication was examined using a subgenomic replicon containing the M/SH gene junction, pM/SH-B, whose RNA synthesis products have been extensively characterized in previous work (Fig. 2A) (18, 20, 21). RNA synthesis by the two polymerases was compared in the absence and presence of the M2-1 protein. Subgenomic RNA replication was indistinguishable when supported by either of the two polymerases in the absence of the M2-1 protein (Fig. 2B, lanes 1 and 2). Transcription from the M/SH-B replicon (shown schematically in Fig. 2A) resulted in the synthesis of several RNA species, and these products differed, depending on the presence of the M2-1 protein. In the presence of N, P, and L, but no M2-1, mRNA1 and mRNA2 produced from gene 1 and gene 2, respectively, were the predominant products of transcription (Fig. 2B, lanes 1 and 2). Upon addition of M2-1, mRNA1 and mRNA2 as well as three readthrough products were produced when the polymerase failed to terminate at the first gene end sequence (rt 1-2), the second gene end sequence (rt 2-tr), or both (rt 1-2-tr, which comigrates with the replication product) (Fig. 2B, lanes 3 and 4). In the presence of the M2-1 protein and L1049D, we observed synthesis of mRNA1, mRNA2, and three readthrough products (rt 1-2, rt 1-2-tr, and rt 2-tr), consistent with previous results (Fig. 2B, lane 3) (18, 20). With L1049N, mRNA1 and mRNA2 were synthesized in increased amounts, concurrent with a low level of rt 1-2 and undetectable levels of rt 2-tr and rt 1-2-tr (Fig. 2B, lane 4), showing that L1049N terminated transcription at gene ends more efficiently than L1049D. L1049D had a high rate of readthrough at the L/tr junction (Fig. 2B, lane 3) (18), while L1049N terminated efficiently at the L/tr junction, as rt 2-tr or rt 1-2-tr were not detected (Fig. 2B, lane 4). The termination efficiency, determined by densitometric analysis of fluorographs, at the M/SH gene junction with the L1049D polymerase was approximately 60% (40% readthrough), consistent with previous results (18, 21), whereas with L1049N the termination efficiency was increased to 95% (5% readthrough), showing that the two polymerases differed significantly in their ability to terminate. Concomitant with more efficient termination at the end of gene 1, more monocistronic mRNA2 was synthesized by L1049N (Fig. 2B), consistent with the stop-start model of transcription of negative-strand RNA viruses in which termination must occur at the end of the upstream gene in order to initiate at the downstream gene.
FIG. 2.
Effect of L1049D or L1049N on RNA synthesis from M/SH-B subgenomic replicon. (A) Diagram of the dicistronic subgenomic replicon containing the authentic M/SH gene junction, pM/SH-B, and the predicted products of RNA synthesis. le, leader; tr, trailer; g.j., gene junction; rt, readthrough; Rep, replication product; AAAn, polyadenylate. (B) RNA synthesis by L1049D and L1049N. HEp-2 cells, infected with MVA-T7, were transfected with pM/SH-B and plasmids pN, pP, pL1049D (lanes 1 and 3), or pL1049N (lanes 2 and 4) and 0 or 0.3 μg of pM2ORF1. Cells were labeled 16 h posttransfection with [3H]uridine in the presence of actinomycin D. 3H-labeled RNAs were visualized by fluorography after separation on an agarose urea gel. RNA products are indicated to the left and right of the gel. (C) Effect of increasing concentration of M2-1 on RNA synthesis. HEp-2 cells were transfected and labeled as for panel B, except that increasing amounts (0 to 1.2 μg) of pM2ORF1 were used as indicated above each lane.
When cells were cotransfected with various amounts of the plasmids encoding each of the two polymerases, an intermediate readthrough phenotype was observed. This finding suggested that neither polymerase had a dominant effect (data not shown).
Effect of M2-1 on termination by L1049D and L1049N.
The activities of the two polymerases were compared as a function of increasing amounts of M2-1 protein, using the M/SH-B replicon as described above. Upon addition of increasing amounts of the M2-1 plasmid (0 to 1.2 μg), we observed an increase in the amount of rt 1-2 with both polymerases, although the increase was much less with L1049N than with L1049D. Increased levels of rt 1-2-tr and rt 2-tr were observed only with L1049D, reiterating the high termination at the L gene end with the L1049N polymerase. Both polymerases reached peak levels of readthrough products with 0.15 to 0.3 μg of input M2-1 plasmid (Fig. 2C, lanes 3 and 8). Increasing the input M2-1 plasmid from 0.3 to 1.2 μg did not increase the level of readthrough observed with L1049N, indicating that the reduced readthrough phenotype of L1049N could not be compensated for by increased M2-1 protein. Both polymerases responded similarly when increasing amounts of M2-1 were added, showing increased readthrough synthesis at equivalent M2-1 concentrations (Fig. 2C). Since the interaction of the two polymerases with the M2-1 protein did not differ, it is unlikely that the M2-1 protein caused the increased readthrough observed with the L1049D polymerase.
Termination at HRSV gene junctions.
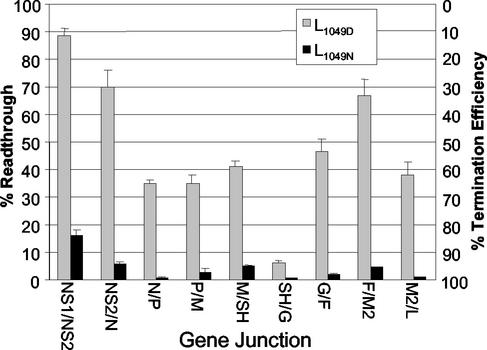
The HRSV gene junctions vary in sequence and termination efficiency; therefore, we compared termination by the two polymerases at each of the HRSV gene junctions. Our laboratory previously made a series of dicistronic subgenomic replicons similar to the M/SH-B replicon (Fig. 2A) containing each of the nine HRSV gene junctions in their natural sequence context (18, 20). Plasmids encoding each of these replicons were individually cotransfected with the pN, pP, pM2-ORF1, and pL1049D or pL1049N support plasmids. The RNAs were harvested, purified, and analyzed. The results in Fig. 3 are presented as both percent readthrough and percent termination for ease of comparison to previously published data (18). However, for consistency, data throughout the rest of the paper are calculated and based on termination efficiency. As with the M/SH junction (Fig. 2B), there was a striking difference between the activities of the two polymerases at each of the HRSV gene junctions. The L1049N polymerase consistently terminated more efficiently and produced lower levels of readthrough products than the L1049D polymerase (Fig. 3). For example, 67% readthrough was observed when the F/M2 junction was assayed with the L1049D polymerase, whereas with L1049N readthrough was reduced to 4.5% (Fig. 3). Readthrough at each of the gene ends was reduced with the L1049N polymerase, consistent with its more efficient termination ability; however, substantial differences were still observed in the extent of readthrough at the different gene junctions, which ranged from 17% at the NS1/NS2 junction to less than 2% at the SH/G, G/F, N/P, and M2/L junctions (Fig. 3). rt 1-2-tr and rt 2-tr were undetectable for all of the gene junctions when assayed with L1049N (data not shown), similar to the results at the M/SH junction (Fig. 2B and C), suggesting that termination at the L/tr junction was very efficient. The increased synthesis of readthrough RNAs by L1049D in the subgenomic replicon system was consistent with the increased presence of polycistronic readthrough RNAs observed with the rsL1049D virus. High levels of readthrough were observed in both systems at the NS1, NS2, G, F, and M2 gene ends.
FIG. 3.
Termination efficiency of the nine natural HRSV gene junctions with L1049D or L1049N. HEp-2 cells, infected with MVA-T7, were transfected with dicistronic subgenomic replicons containing each of the naturally occurring HRSV gene junctions as indicated and support plasmids pP, pN, pM2ORF1, and pL1049D or pL1049N. Cells were labeled at 16 h posttransfection with [3H]uridine in the presence of actinomycin D. RNAs were purified, separated on an agarose urea gel, and visualized by fluorography. Percent readthrough at each gene end (left side) and percent termination efficiency (right side) were calculated for each replicon by densitometric analysis of the fluorographs. The average percent readthrough and standard deviation were calculated from at least three experiments.
Effect of U tract length and first intergenic nucleotide.
The U tracts at the HRSV gene ends vary in length from four to seven residues. HRSV gene ends that have a U4 tract (NS1, NS2, F, and M2) terminated less efficiently when assayed with L1049D (Fig. 3) (18). The gene junctions with short U4 tracts also exhibited the lowest termination efficiency with L1049N, with the exception of the M2/L junction, which is unique in that it has an overlap. The presence of U tracts of four, five, six, and seven residues at the natural gene ends which terminate with varying efficiency shows that HRSV does not have as strict of a U tract length requirement as vesicular stomatitis virus, Indiana (VSIV), which requires a minimum U tract length of seven (4).
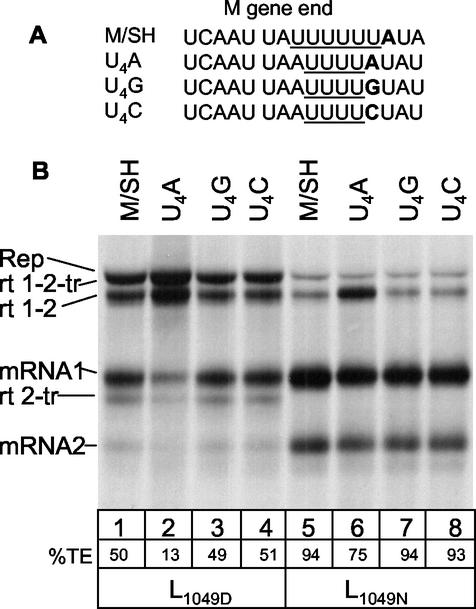
Recent work has shown that the first intergenic nucleotide is important for termination when the U tract is short (22, 44); therefore, we examined the effect of U tract length and the first intergenic nucleotide on termination efficiency with L1049N and L1049D using a series of replicons in which the U tract at the M gene end was shortened from U6 to U4 and the first intergenic nucleotide was altered from an A to a G or C (Fig. 4A) (22). Consistent with the previous report with L1049D (22), an A located downstream of a U4 tract resulted in decreased termination efficiency (Fig. 4B, lane 2), while a G or C at this position did not have an effect (Fig. 4B, lanes 3 and 4). The situation was similar with L1049N, where the M gene end having a U4 tract followed by a G or C terminated with 94% efficiency (Fig. 4B, lanes 5, 7, and 8), whereas when the first intergenic nucleotide was an A following the U4 tract, the termination efficiency dropped to 75% (Fig. 4B, lane 6). Thus, the first intergenic nucleotide following a U4 tract is important for termination by both polymerases.
FIG. 4.
Effect of gene end U tract length and the first intergenic nucleotide on transcriptional termination. (A) Sequence of the M/SH gene junction aligned with the sequence from replicons having shortened U tracts and variation in the first intergenic nucleotide. The U tracts are underlined, and the first intergenic nucleotide is in bold. (B) RNA synthesized from wild-type M/SH junction compared to that from altered M/SH junctions. HEp-2 cells, infected with MVA-T7, were transfected with pM/SH-B or one of the mutant replicons along with the HRSV support plasmids pN, pP, pM2ORF1, and L1049D or L1049N. RNAs were labeled and visualized as described for Fig. 2B. Quantitation of percent termination efficiency (TE) was calculated by densitometric analysis of fluorographs from three experiments and is shown below each lane.
cis-acting sequences critical for termination.
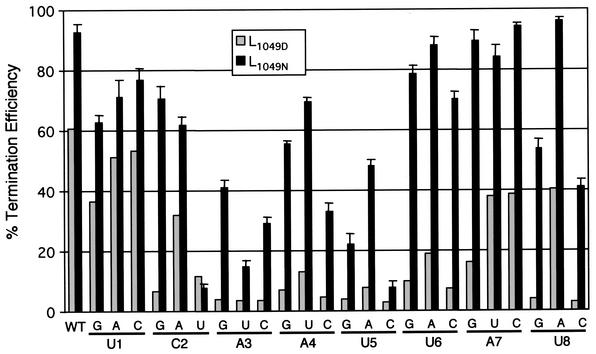
We further compared the cis-acting sequence requirements for termination to determine whether the two polymerases recognized the gene end sequences similarly. The residues of the gene end most critical for termination were examined previously by changing each nucleotide at positions 1 through 8 of the M gene end (UCAAUUAU) to all other possible nucleotides (21). The gene end mutants were used to compare the termination efficiencies of the two polymerases. The gene end mutations were named by their position and the nucleotide change, such that A3U corresponds to an A-to-U change at position 3 in the M gene end. Mutations in the gene end sequence led to decreased termination efficiency with either polymerase; however, the effect was more dramatic with L1049D (Fig. 5). Positions 1 and 7 in the M gene end were the most tolerant of change with either polymerase, with changes at position 7 showing only a small decrease in termination efficiency with L1049N. Positions 3, 4, and 5 were the most sensitive with either polymerase; all changes at positions 3 and 5 reduced termination efficiency to less than 50% with L1049N and to less than 15% with L1049D. The C2U, A3U, and U5C mutations were the most debilitating, reducing the termination efficiency to less than 15% with either polymerase. Positions 4, 5, 6, and 8 tolerated changes to A or U more readily than to G or C, with position 8 showing the most intolerance for a G or C, suggesting that the conserved A/U-rich nature of the gene end was critical for efficient termination. Positions 2 and 6 showed the greatest disparity in termination efficiency between the two polymerases. All changes at position 6 resulted in less than 20% termination efficiency with L1049D, while the termination efficiency was 70% or greater with L1049N. At position 2, the C2U mutation debilitated both polymerases. However, the C2G mutation severely impaired the ability of L1049D to terminate, while having only a moderate effect on termination by L1049N. The results showed that the polymerases differed in their ability to recognize the cis-acting signals for termination at positions 2 and 6. However, the polymerases behaved similarly overall, despite their differences in termination efficiency.
FIG. 5.
Importance of gene end sequences in signaling termination by L1049D and L1049N. Positions 1 through 8 of the M gene end (3′…UCAAUUAU…5′) were individually changed to all other possible nucleotides in the pM/SH-B replicon shown in Fig. 2A. HEp-2 cells, infected with MVA-T7, were transfected with pN, pP, pM2ORF1, pL1049D, or pL1049N and a subgenomic replicon with the indicated mutation in the M gene end. RNAs were labeled and visualized as described for Fig. 2B. The percent termination efficiency was calculated from densitometric analysis of the fluorographs. The black bars (L1049N) represent the average termination efficiency and standard deviation from at least two experiments. The gray bars represent a single experiment with L1049D, which was used as a control, and can be compared to previously published results (21).
DISCUSSION
The presence of numerous polycistronic readthrough transcripts, due to inefficient transcriptional termination, is common in HRSV infection. There are two main mechanisms by which the efficiency of termination could vary in a virus infection: either the gene end sequences are inefficient at signaling termination, or the polymerase may be inherently poor at terminating. Previous work and the work presented here show that the HRSV gene ends, which vary in sequence, do signal termination with different efficiencies (18, 31). Other viruses within the Mononegavirales, such as Subacute sclerosing panencephalitis virus or Human parainfluenza virus type 1, have been shown to have significant variation at gene ends which results in different termination efficiencies at certain gene ends (1, 6). In contrast, viruses such as VSIV have gene ends that are identical in sequence and all terminate with approximately 99% efficiency, such that transcriptional attenuation is the predominant factor in control of gene expression (4, 5, 27). The data presented here demonstrate that in addition to control of termination by response to variation in cis-acting signals, there is also a region of the polymerase that is involved in determining termination efficiency. We identified a single amino acid change in the polymerase that affects its ability to terminate transcription and when introduced into the viral genome causes increased synthesis of polycistronic readthrough RNAs. Further, the work presented here shows that these two factors, the cis-acting gene end sequences and the trans-acting polymerase, function together to determine efficiency of termination.
The recovery of the rsL1049D virus showed that while L1049D was functional in replication and transcription and produced viable virus from a full-length cDNA, it gave increased synthesis of polycistronic readthrough RNAs indicative of lower termination efficiency compared to the prototypic HRSV A2 virus. The decreased termination at the gene ends of this virus was detrimental as evidenced by the longer recovery time and the reversion of one of the viruses to encode an asparagine at position 1049. The rsL1049N virus, however, had an RNA synthesis pattern that was indistinguishable from the A2 strain of HRSV, indicating that this polymerase was representative of the wild-type HRSV polymerase.
The identification of this mutation presented the opportunity to investigate the relationship between the trans-acting polymerase and the cis-acting genomic sequences specifically shown to be involved in signaling termination. We first investigated the ability of both polymerases to terminate at each of the nine distinct HRSV gene junctions by using dicistronic subgenomic replicons and found that in all cases L1049N terminated with higher efficiency, although variability was observed in the efficiency of termination at the different gene junctions, as reported previously with L1049D (18). Thus, the polymerases showed different levels of termination on the same sequence, but the different cis-acting gene end sequences directed different levels of termination with either polymerase, confirming that these two factors, one cis and one trans, work in concert to terminate transcription. These results differ slightly from those of Kuo et al., who concluded that except for the NS1 and NS2 gene ends there were similar levels of termination at the HRSV gene ends (32). We found that termination at the NS1 gene end was the lowest, termination at the NS2 gene end was similar to that of the M and F gene ends, and termination at the N, SH, and M2 gene ends was very high. The differences can be attributed to several factors. The gene end sequences of HRSV were assayed out of their natural sequence context by Kuo et al. and included neither the natural nucleotide at the −1 position of the gene end nor the naturally occurring first intergenic nucleotide for each of the genes, both of which play a role in determining termination efficiency (22, 44). In addition, there may be differences in the sensitivity of the reporter gene assay used by Kuo et al. and the direct metabolic labeling of RNAs used in this study.
We further examined the ability of the polymerases to recognize the gene end sequence by introducing mutations within the M gene end and determining the effect on termination. The first intergenic nucleotide following a U4 tract at the gene end was important for termination by either polymerase, similar to results observed at the F gene of HRSV and with simian parainfluenza virus 5 (22, 37, 38, 44). Analysis of the first eight nucleotides of the M gene end showed that at most positions within the gene end, the polymerases responded similarly to the mutations, although the extent of termination varied (Fig. 5). Position 6, which is not well conserved, showed the greatest disparity between the two polymerases: any change resulted in less than 20% termination with L1049D but greater than 70% termination efficiency with L1049N. Positions 4, 5, 6, and 8 showed a preference toward an A or U (Fig. 5), as replacement with a G or C at these positions was more detrimental. In a separate study of the F gene end, which has a U4 tract, positions 6, 7, and 8 showed greater sensitivity to a G or C change than what we observed at the M gene end (44), suggesting that there may be interplay between the composition of the central region and the length of the U tract. It is interesting to speculate that the preference for an A/U-rich sequence in the central region and particularly at position 8 may be important to allow proper slippage on the U tract. It is possible that the presence of G or C increases the strength of interaction between the template and nascent strands and hinders the ability of the polymerase to slip. In a recent study, increasing the G/C content of the VSIV gene ends prevented slippage on the U tract, while A/U-rich sequences prompted slippage, suggesting that the strength of the interaction between the template and nascent strands was important for slippage by the VSIV polymerase (3). A similar situation was reported for the process of editing in Sendai virus (23).
We examined the effect of the M2-1 protein, a processivity factor and an antitermination factor, on termination by both polymerases. The synthesis of readthrough products by L1049N and L1049D was dependent on the presence of the M2-1 protein, and both polymerases responded to the M2-1 protein at the same concentrations. These data indicated that the effect of the M2-1 protein on both polymerases was similar, suggesting that the elevated synthesis of readthrough products observed with L1049D was not due to an altered interaction with the M2-1 protein that increased M2-1 protein-mediated antitermination, but was due solely to the aspartic acid at position 1049 in the L protein.
Very few mutations in NNS virus RNA polymerases have been identified that specifically affect transcriptional termination. An interesting mutation was found in the L protein of a temperature-sensitive mutant of VSIV, which exhibited increased polyadenylation (25). The mutation was a phenylalanine-to-serine change at amino acid 1488, which lies between conserved domains V and VI. Further examination of this mutant revealed that it also exhibited increased synthesis of polycistronic readthrough RNAs, indicative of a defect in termination. This is not surprising, since polyadenylation has been shown to be an integral part of termination in the case of VSIV (26). Another mutation that increased readthrough was found in a cold-passaged, chemically mutagenized HRSV vaccine candidate. The mutation was a methionine-to-valine change at position 1169 in the L protein, which lies between conserved domains IV and V, and conferred at most a fourfold increase in readthrough in both the replicon system and in the virus (29). The HRSV mutation identified in this work, N1049D, lies in domain IV and confers an increase in readthrough ranging from 5-fold (NS1 gene end) to greater than 20-fold (N, G, and M2 gene ends) when assayed using the subgenomic replicon system. The mechanistic cause of the readthrough phenotype of the L1049D polymerase is unknown. Transcriptional termination in the NNS RNA viruses is complex and not only involves conserved sequences essential to signaling termination but also requires mRNA polyadenylation by reiterative stuttering on a U tract (3, 4). It is possible that the 1049 mutation may affect a domain of the polymerase that is specifically involved in termination. The three mutations identified in HRSV and VSIV thus far that affect termination are between domains IV and VI; however, they are separated in primary amino acid sequence, suggesting that multiple areas of the protein may contribute to the termination process. It is possible that these areas are brought together in the tertiary structure of the protein or that they represent independent domains that affect different steps in the termination process, as with the VSIV mutation that affects termination by altering polyadenylation (25). Clearly, further work needs to be done to dissect the areas of the polymerase that control termination.
Acknowledgments
We thank past and present members of the Wertz and L. A. Ball laboratories, especially Shawn Harmon, for helpful suggestions on this work and critical reading of the manuscript.
This work was supported by Public Health Service grant AI20181 from the NIH to G. W. Wertz.
REFERENCES
- 1.Ayata, M., K. Komase, M. Shingai, I. Matsunaga, Y. Katayama, and H. Ogura. 2002. Mutations affecting transcriptional termination in the P gene end of subacute sclerosing panencephalitis viruses. J. Virol. 76:13062-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barik, S. 1992. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s). J. Virol. 66:6813-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 75:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., S. P. Whelan, and G. W. Wertz. 1997. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 71:8718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, J. N., S. P. Whelan, and G. W. Wertz. 2002. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim. Biophys. Acta 1577:337-353. [DOI] [PubMed] [Google Scholar]
- 6.Bousse, T., T. Matrosovich, A. Portner, A. Kato, Y. Nagai, and T. Takimoto. 2002. The long noncoding region of the human parainfluenza virus type 1 F gene contributes to the read-through transcription at the M-F gene junction. J. Virol. 76:8244-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrika, R., S. M. Horikami, S. Smallwood, and S. A. Moyer. 1995. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213:352-363. [DOI] [PubMed] [Google Scholar]
- 8.Collins, P. L., L. E. Dickens, A. Buckler-White, R. A. Olmsted, M. K. Spriggs, E. Camargo, and K. V. Coelingh. 1986. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc. Natl. Acad. Sci. USA 83:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, P. L., Y. T. Huang, and G. W. Wertz. 1984. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J. Virol. 49:572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, P. L., and G. W. Wertz. 1983. cDNA cloning and transcriptional mapping of nine polyadenylated RNAs encoded by the genome of human respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 80:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortese, C. K., J. A. Feller, and S. A. Moyer. 2000. Mutations in domain V of the Sendai virus L polymerase protein uncouple transcription and replication and differentially affect replication in vitro and in vivo. Virology 277:387-396. [DOI] [PubMed] [Google Scholar]
- 13.Dickens, L. E., P. L. Collins, and G. W. Wertz. 1984. Transcriptional mapping of human respiratory syncytial virus. J. Virol. 52:364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerson, S. 1987. Transcription of vesicular stomatitis viruses, p. 245-269. In R. R. Wagner (ed.), The rhabdoviruses. Plenum Press, New York, N.Y.
- 15.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feller, J. A., S. Smallwood, S. M. Horikami, and S. A. Moyer. 2000. Mutations in conserved domains IV and VI of the large (L) subunit of the Sendai virus RNA polymerase give a spectrum of defective RNA synthesis phenotypes. Virology 269:426-439. [DOI] [PubMed] [Google Scholar]
- 17.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmon, S. B., A. G. Megaw, and G. W. Wertz. 2001. RNA sequences involved in transcriptional termination of respiratory syncytial virus. J. Virol. 75:36-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmon, S. B., and G. W. Wertz. 2002. Transcriptional termination modulated by nucleotides outside the characterized gene end sequence of respiratory syncytial virus. Virology 300:304-315. [DOI] [PubMed] [Google Scholar]
- 23.Hausmann, S., D. Garcin, C. Delenda, and D. Kolakofsky. 1999. The versatility of paramyxovirus RNA polymerase stuttering. J. Virol. 73:5568-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt, D. M., and K. L. Hutchinson. 1993. Amino acid changes in the L polymerase protein of vesicular stomatitis virus which confer aberrant polyadenylation and temperature-sensitive phenotypes. Virology 193:786-793. [DOI] [PubMed] [Google Scholar]
- 25.Hunt, D. M., E. F. Smith, and D. W. Buckley. 1984. Aberrant polyadenylation by a vesicular stomatitis virus mutant is due to an altered L protein. J. Virol. 52:515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchinson, K. L., R. C. Herman, and D. M. Hunt. 1992. Increased synthesis of polycistronic mRNA associated with increased polyadenylation by vesicular stomatitis virus. Virology 189:67-78. [DOI] [PubMed] [Google Scholar]
- 27.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477-484. [DOI] [PubMed] [Google Scholar]
- 28.Jin, H., and R. Elliott. 1992. Mutagenesis of the L protein encoded by Bunyamwera virus and production of monospecific antibodies. J. Gen. Virol. 73:2235-2244. [DOI] [PubMed] [Google Scholar]
- 29.Juhasz, K., B. R. Murphy, and P. L. Collins. 1999. The major attenuating mutations of the respiratory syncytial virus vaccine candidate cpts530/1009 specify temperature-sensitive defects in transcription and replication and a non-temperature-sensitive alteration in mRNA termination. J. Virol. 73:5176-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamer, G., and P. Argos. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12:7269-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo, L., R. Fearns, and P. L. Collins. 1997. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J. Virol. 71:4944-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, L., H. Grosfeld, J. Cristina, M. G. Hill, and P. L. Collins. 1996. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J. Virol. 70:6892-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oomens, A. G. P., A. G. Megaw, and G. W. Wertz. 2003. Infectivity of a human respiratory syncytial virus lacking the SH, G, and F proteins is efficiently mediated by the vesicular stomatitis virus G protein. J. Virol. 77:3785-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pattnaik, A. K., and G. W. Wertz. 1990. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J. Virol. 64:2948-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71:1153-1162. [DOI] [PubMed] [Google Scholar]
- 36.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rassa, J. C., and G. D. Parks. 1999. Highly diverse intergenic regions of the paramyxovirus simian virus 5 cooperate with the gene end U tract in viral transcription termination and can influence reinitiation at a downstream gene. J. Virol. 73:3904-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rassa, J. C., G. M. Wilson, G. A. Brewer, and G. D. Parks. 2000. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology 274:438-449. [DOI] [PubMed] [Google Scholar]
- 39.Schnell, M., and K. Conzelmann. 1995. Polymerase activity of in vitro mutated rabies virus L protein. Virology 214:522-530. [DOI] [PubMed] [Google Scholar]
- 40.Sleat, D. E., and A. K. Banerjee. 1993. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J. Virol. 67:1334-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smallwood, S., B. Cevik, and S. A. Moyer. 2002. Intragenic complementation and oligomerization of the L subunit of the Sendai virus RNA polymerase. Virology 304:235-245. [DOI] [PubMed] [Google Scholar]
- 42.Smallwood, S., C. D. Easson, J. A. Feller, S. M. Horikami, and S. A. Moyer. 1999. Mutations in conserved domain II of the large (L) subunit of the Sendai virus RNA polymerase abolish RNA synthesis. Virology 262:375-383. [DOI] [PubMed] [Google Scholar]
- 43.Stec, D. S., M. G. Hill, and P. L. Collins. 1991. Sequence analysis of the polymerase L gene of human respiratory syncytial virus and predicted phylogeny of nonsegmented negative-strand viruses. Virology 183:273-287. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland, K. A., P. L. Collins, and M. E. Peeples. 2001. Synergistic effects of gene-end signal mutations and the M2-1 protein on transcription termination by respiratory syncytial virus. Virology 288:295-307. [DOI] [PubMed] [Google Scholar]
- 45.Svenda, M., M. Berg, J. Moreno-Lopez, and T. Linne. 1997. Analysis of the large (L) protein gene of the porcine rubulavirus LPMV: identification of possible functional domains. Virus Res. 48:57-70. [DOI] [PubMed] [Google Scholar]
- 46.Tang, R. S., N. Nguyen, H. Zhou, and H. Jin. 2002. Clustered charge-to-alanine mutagenesis of human respiratory syncytial virus L polymerase generates temperature-sensitive viruses. Virology 302:207-216. [DOI] [PubMed] [Google Scholar]
- 47.Yu, Q., R. W. Hardy, and G. W. Wertz. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]