Abstract
Simian immunodeficiency virus of chimpanzees (SIVcpz) is the immediate precursor to human immunodeficiency virus type 1 (HIV-1), yet remarkably, the distribution and prevalence of SIVcpz in wild ape populations are unknown. Studies of SIVcpz infection rates in wild chimpanzees are complicated by the species' endangered status and by its geographic location in remote areas of sub-Saharan Africa. We have developed sensitive and specific urine and fecal tests for SIVcpz antibody and virion RNA (vRNA) detection and describe herein the first comprehensive prevalence study of SIVcpz infection in five wild Pan troglodytes schweinfurthii communities in east Africa. In Kibale National Park in Uganda, 31 (of 52) members of the Kanyawara community and 39 (of ∼145) members of the Ngogo community were studied; none were found to be positive for SIVcpz infection. In Gombe National Park in Tanzania, 15 (of 20) members of the Mitumba community, 51 (of 55) members of the Kasekela community, and at least 10 (of ∼20) members of the Kalande community were studied. Seven individuals were SIVcpz antibody and/or vRNA positive, and two others had indeterminate antibody results. Based on assay sensitivities and the numbers and types of specimens analyzed, we estimated the prevalence of SIVcpz infection to be 17% in Mitumba (95% confidence interval, 10 to 40%), 5% in Kasekela (95% confidence interval, 4 to 7%), and 30% in Kalande (95% confidence interval, 15 to 60%). For Gombe as a whole, the SIVcpz prevalence was estimated to be 13% (95% confidence interval, 7 to 25%). SIVcpz infection was confirmed in five chimpanzees by PCR amplification of partial pol and gp41/nef sequences which revealed a diverse group of viruses that formed a monophyletic lineage within the SIVcpzPts radiation. Although none of the 70 Kibale chimpanzees tested SIVcpz positive, we estimated the likelihood that a 10% or higher prevalence existed but went undetected because of sampling and assay limitations; this possibility was ruled out with 95% certainty. These results indicate that SIVcpz is unevenly distributed among P. t. schweinfurthii in east Africa, with foci or “hot spots” of SIVcpz endemicity in some communities and rare or absent infection in others. This situation contrasts with that for smaller monkey species, in which infection rates by related SIVs are generally much higher and more uniform among different groups and populations. The basis for the wide variability in SIVcpz infection rates in east African apes and the important question of SIVcpz prevalence in west central African chimpanzees (Pan troglodytes troglodytes) remain to be elucidated.
The origin and evolutionary history of human immunodeficiency virus type 1 (HIV-1) and the circumstances leading to the AIDS pandemic remain important questions for our understanding of emerging infectious diseases. Chimpanzees (Pan troglodytes) are widely regarded as a natural host for simian immunodeficiency virus SIVcpz and as the simian source of HIV-1, but the prevalence of SIVcpz in wild chimpanzee populations is completely unknown (2, 25). Only eight naturally infected chimpanzees have thus far been identified, and of those, seven were studied in captivity (13, 18, 41, 42; E. Nerrienet, unpublished data). One wild chimpanzee from Gombe National Park in Tanzania was recently found to harbor SIVcpz (49). This seeming scarcity of SIVcpz in captive and wild ape populations has raised questions as to whether chimpanzees represent a true SIVcpz reservoir (59) or whether a third as yet unidentified primate species might be a source for both human and chimpanzee infections (10, 58).
Chimpanzees are widely dispersed across sub-Saharan Africa. Four subspecies with distinct, nonoverlapping ranges are currently recognized on the basis of mitochondrial DNA (mtDNA) analyses (21, 33). These include the western Pan troglodytes verus, the Nigerian P. t. vellerosus, the central P. t. troglodytes, and the eastern P. t. schweinfurthii (Fig. 1A). The great majority of captive chimpanzees in US and European primate centers are of the P. t. verus subspecies because they were imported from west Africa (51). Although more than 1,500 of these apes have been screened, none have been found to harbor SIVcpz (44; W. M. Switzer, B. Parekh, V. Shanmugam, V. Bhullar, S. Phillips, T. M. Folks, J. Ely, and W. Heneine, 9th International Workshop on HIV Dynamics and Evolution, Lake Arrowhead, Calif., 2002). Members of the P. t. vellerosus subspecies occupy a smaller region in Nigeria and Cameroon north of the Sanaga River (Fig. 1A). Their infection status is unknown because very few have been tested (E. Nerrienet, unpublished data). Only P. t. troglodytes and P. t. schweinfurthii are known to be naturally infected with SIVcpz (13, 42, 49). We have explained the absence of SIVcpz from P. t. verus by postulating that chimpanzees as a species acquired SIVcpz infection after the geographic separation and isolation of P. t. verus from the other subspecies (2, 26, 48, 49). Recent data concerning the origin of SIVcpz are consistent with this interpretation, indicating that SIVcpz is actually a recombinant of ancestral viruses that infected west central African monkey species upon which chimpanzees prey (3). Under this scenario, P. t. troglodytes would have been the initial host of this recombinant virus and P. t. schweinfurthii would have become infected subsequently by the eastward spread of the virus. Whether SIVcpz might also have spread to P. t. vellerosus is not known.
FIG.1.
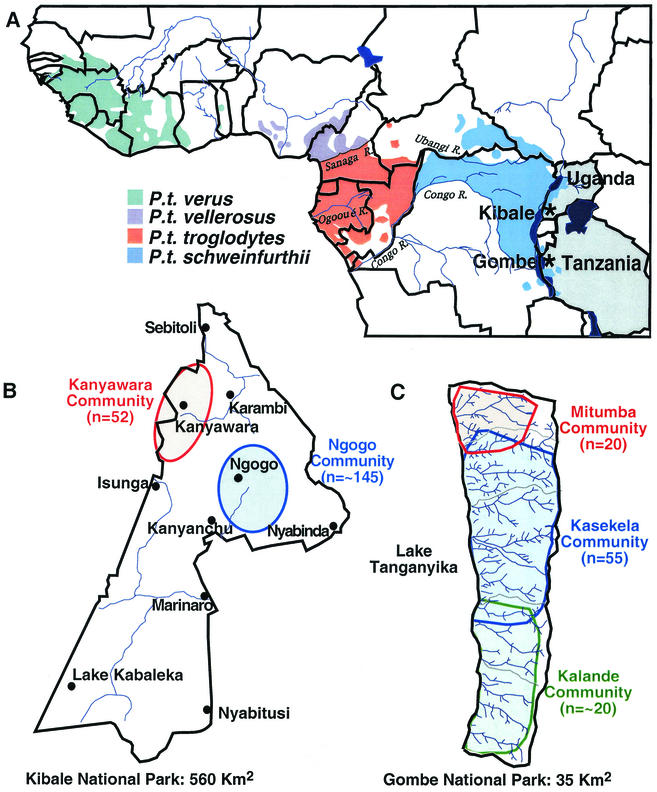
Location of wild chimpanzee study sites. (A) The locations of Kibale (Uganda) and Gombe (Tanzania) National Parks (indicated by asterisks) are shown in relation to the range of the common chimpanzees in equatorial Africa (Copyright © 2001, Smithsonian Institution. Adapted from reference 8 with permission). The four recognized chimpanzee subspecies are color coded. International borders (black lines) and major rivers (blue lines) are shown. Uganda and Tanzania are highlighted. (B) Map of Kibale National Park, indicating the approximate ranges of the Kanyawara (red) and Ngogo (blue) communities. The two communities are not adjacent, thus limiting direct contact between members. Black dots denote ranger outposts. (C) Map of Gombe National Park, indicating the approximate ranges of the northern Mitumba (red), the main Kasekela (blue), and the nonhabituated southern Kalande community (green). Gombe National Park is bordered by Lake Tanganyika to the east, and the rift escarpment (>1,500 m) to the west. Interactions between members of the different communities have been observed in regions of overlap.
Although isolated instances of SIVcpz infection have been documented in P. t. troglodytes and P. t. schweinfurthii, neither chimpanzee subspecies has been studied systematically in its natural habitat. Wild chimpanzees live in communities of 5 to 150 individuals in the forests of tropical Africa. They avoid human contact, except in the rare instances in which they have been habituated to the presence of human observers (60). Wild chimpanzees are also highly endangered. Deforestation, habitat destruction, and relentless poaching have reduced the population size from over 2 million to less than 120,000 individuals in just 50 years (38). Invasive studies, such as sampling of blood or other tissues, are thus neither practical nor ethical. As an alternative, we have recently developed urine- and fecal-based assays for detecting SIVcpz antibodies and virion RNA (vRNA), and we have used these to screen a small number of habituated chimpanzees in east Africa (49). Analysis of urine samples from 24 wild chimpanzees from the Kanyawara community in Kibale National Park (Uganda) failed to detect evidence of SIVcpz infection, whereas testing of only six apes from the Kasekela community in Gombe National Park identified one chimpanzee (Ch-06) to be both urine antibody and fecal vRNA positive. Subsequent molecular analyses confirmed SIVcpz infection in this individual (49) and even permitted the amplification of a complete viral genome from fecal samples (48). While these studies established the utility of the noninvasive assays for the detection and characterization of SIVcpz under field conditions, they did not examine the sensitivity and specificity of the assays, nor did they address the frequency of SIVcpz infection in these wild chimpanzee communities.
In this paper, we describe a comprehensive field survey of 146 wild chimpanzees from five different communities in east Africa. By sampling individuals from two national parks in Uganda and Tanzania, we could examine the prevalence, genetic diversity, and transmission patterns of SIVcpz in both habituated and nonhabituated settings. Our data reveal foci of SIVcpz endemicity in some but not all wild communities—an unexpected finding that is distinctly different from comparable studies of SIV infection in other nonhuman primate species for which infection rates are generally high and uniform. The reasons for the scattered distribution of SIVcpz and for its overall low prevalence in east African chimpanzee populations remain to be determined. Nonetheless, the seven new cases of SIVcpz infection in Gombe double the number of known natural infections and formally establish chimpanzees as a natural host and reservoir for SIVcpz.
MATERIALS AND METHODS
Captive chimpanzees.
Fecal and/or urine samples were collected from 47 captive chimpanzees of known HIV-1 or SIVcpz infection status housed at primate facilities in the United States (Yerkes National Primate Research Center, Atlanta, Ga.; n = 35), The Netherlands (Biomedical Primate Research Centre, Rijswijk; n = 9), and Cameroon (wildlife rescue centers; n = 3). Fifteen of these had been experimentally infected with HIV-1 more than a decade earlier as part of AIDS research studies (Table 1). Although five had significantly reduced CD4 counts (less than 200/mm3; normal range 1,000 to 2,500/mm3), none exhibited signs of clinical illness (35, 36, 39). Fecal and urine samples were also obtained from four SIVcpz-infected chimpanzees. Of these, Ch-No and CAM13 were wild-caught in the Democratic Republic of Congo (DRC) and Cameroon, respectively, and thus likely acquired SIVcpz infection naturally (E. Nerrienet, unpublished data) (41); Ch-Ni was infected experimentally in captivity by using blood from Ch-No (28); and CAM4 is believed to have been infected from a naturally infected cage mate, CAM3 (13). Twenty-eight uninfected chimpanzees served as negative controls (Table 1). A subset of the experimentally infected apes was examined for microhematuria and proteinuria as well as occult blood in stool, but all tested negative. All studies were carried out in strict accordance with international guidelines for the ethical scientific use and humane care of primates in research (the Yerkes National Primate Research Center is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International).
TABLE 1.
Fecal and urine test results from captive chimpanzees of known HIV-1 and SIVcpz infection status
| Chimpanzee | Primate center | Type of infectiona | Infecting virus | CD4 countsb | Date of sample collection | Fecal samples
|
Urine samples
|
||
|---|---|---|---|---|---|---|---|---|---|
| Ab pos/no. testedc | RNA pos/no. testedd | Ab pos/no. testedc | RNA pos/no. testedd | ||||||
| HIV-1/(HIV-2) infected | |||||||||
| C455 | Yerkes | Exp | JC | 0 | 4/99 | 0/4 | 0/2 | — | — |
| C487 | Yerkes | Exp | LAV-Ib | 698 | 4/99 | 3/4 | 0/2 | — | — |
| C463 | Yerkes | Exp | LAV-I | 123 | 4/99 | 2/4 | 1/2 | 1/1 | 0/1 |
| C544 | Yerkes | Exp | LAV-Ib/(EHO) | 953 | 4/99 | 4/4 | 0/2 | 1/1 | 0/1 |
| C550 | Yerkes | Exp | LAV-Ib | 95 | 4/99 | 2/2 | 1/2 | 1/1 | 1/1 |
| C560 | Yerkes | Exp | LAV-Ib/SF2 | 182 | 4/99 | 2/2 | 0/2 | 1/1 | 0/1 |
| C459 | Yerkes | Exp | LAV-I/NDK/NC | 186 | 4/99 | 2/2 | 0/1 | 1/1 | 0/1 |
| C552 | Yerkes | Exp | LAV-Ib | 1100 | 4/99 | 0/2 | 0/2 | — | — |
| C497 | Yerkes | Exp | NDK | 850 | 4/99 | 0/2 | 0/2 | — | — |
| C542 | Yerkes | Exp | LAV-Ib | 1241 | 4/99 | 1/2 | 0/2 | — | — |
| C534 | Yerkes | Exp | NC/(ROD) | 19 | 4/99 | 2/2 | 0/2 | — | — |
| Ch-La | Rijswijk | Exp | LAI/HAN2 | 1700 | 7/99 | 3/3 | 0/3 | 3/3 | 0/3 |
| Ch-On | Rijswijk | Exp | HAN2 | 2300 | 7/99 | 1/3 | 0/3 | 3/3 | 0/3 |
| Ch-Ma | Rijswijk | Exp | LAI/HAN2 | 1300 | 7/99 | 2/3 | 0/3 | 3/3 | 0/3 |
| Ch-Bu | Rijswijk | Exp | LAI/HAN2 | 1000 | 7/99 | 2/3 | 0/3 | 3/3 | 0/3 |
| SIVcpz infected | |||||||||
| Ch-No | Rijswijk | Nat | ANT | 1400 | 7/99 | 7/9 | 7/9 | 3/3 | 1/3 |
| Ch-Ni | Rijswijk | Exp | ANT | — | 7/99 | 3/3 | 3/3 | 3/3 | 0/3 |
| CAM4 | Cameroon | Cage | CAM4 | — | 11/99 | 2/7 | 1/10 | 4/4 | 0/4 |
| CAM13 | Cameroon | Nat | CAM13 | — | 2/01 | — | 3/3 | — | — |
| Uninfected | |||||||||
| C575 | Yerkes | None | n/a | 1330 | 4/99 | 0/4 | — | — | — |
| C369 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C431 | Yerkes | None | n/a | 2552 | 4/99 | 0/2 | — | — | — |
| C452 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C382 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C425 | Yerkes | None | n/a | 1035 | 4/99 | 0/2 | — | — | — |
| C520 | Yerkes | None | n/a | 1220 | 4/99 | 0/2 | — | — | — |
| C507 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C536 | Yerkes | None | n/a | 1850 | 4/99 | 0/2 | — | — | — |
| C408 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C475 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C471 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C447 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C505 | Yerkes | None | n/a | 1600 | 4/99 | 0/2 | — | — | — |
| C548 | Yerkes | None | n/a | 977 | 4/99 | 0/2 | — | — | — |
| C400 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C407 | Yerkes | None | n/a | 772 | 4/99 | 0/2 | — | 0/1 | — |
| C421 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C433 | Yerkes | None | n/a | 932 | 4/99 | 0/2 | — | — | — |
| C202 | Yerkes | None | n/a | — | 4/99 | 0/2 | — | — | — |
| C494 | Yerkes | None | n/a | 2362 | 4/99 | — | — | 0/1 | — |
| C586 | Yerkes | None | n/a | 1815 | 4/99 | — | — | 0/1 | — |
| C222 | Yerkes | None | n/a | — | 4/99 | — | — | 0/1 | — |
| C694 | Yerkes | None | n/a | — | 5/01 | — | — | 0/1 | — |
| Ch-Ph | Rijswijk | None | n/a | — | 7/99 | 0/3 | — | 0/3 | — |
| Ch-Pe | Rijswijk | None | n/a | — | 7/99 | 0/3 | — | 0/3 | — |
| Ch-To | Rijswijk | None | n/a | — | 7/99 | 0/3 | — | 0/3 | — |
| CAM10 | Cameroon | None | n/a | — | 11/99 | — | — | 0/1 | — |
Exp, experimental infection; Nat, natural infection; Cage, cage transmission; none, not infected with HIV-1 or SIVcpz; n/a, not applicable.
CD4 counts per mm3 (normal range 1,000-2,500/mm3); —, specimens not available.
Ab, antibody. Number of antibody-positive fecal and urine samples per total number tested as determined by ECL Western blot analysis (samples which yielded a gp160 band, alone or in combination with other virus specific bands, or any three structural proteins exclusive of gp160, were scored antibody positive).
Number of vRNA-positive fecal and urine samples per total number tested as determined by diagnostic RT PCR analysis (authenticity of amplification products was confirmed by sequence analysis).
Wild chimpanzees.
Fecal and urine samples were collected from wild chimpanzee communities in Kibale and Gombe National Parks (Fig. 1). Kibale National Park is located in western Uganda and has two major research sites, Kanyawara and Ngogo (Fig. 1B). The Kanyawara community has been studied by Wrangham and colleagues since 1987 and comprised 52 individuals at the time of sampling (61). The Ngogo community has been studied continuously since 1995 and comprised about 145 individuals at the time of sampling (32, 57). Both Kanyawara and Ngogo chimpanzees tolerate the presence of human observers at close distances (5 to 10 m). All group members are individually known and named, allowing for sample collection under direct observation. Field studies in Kibale National Park were authorized by the Makerere University Biological Field Station and the Uganda Wildlife authority.
Gombe National Park is located on the eastern shores of Lake Tanganyika in Tanzania and is home to three chimpanzee communities (Fig. 1C). The largest, Kasekela, is habituated and has been studied by Goodall since 1960 (22). Habituation of northern Mitumba community began in the 1980s, with most individuals identified by the mid-1990s. The southern Kalande community is not habituated and thus is the least studied of all Gombe communities. The three Gombe communities comprise a total of 95 individuals and have partially overlapping ranges (Fig. 1C). The Kasekela and Mitumba chimpanzees are monitored daily, with particular individuals selected for all-day observation, and their reproductive states and social interactions are recorded. For some of the more recent Kasekela offspring, paternity has been determined by microsatellite analysis (12). Fieldwork in Gombe National Park was authorized by the Tanzania Commission for Science and Technology, the Tanzania Wildlife Research Institute, and the Tanzania National Parks.
Sample collection and storage.
Captive chimpanzees were sampled at the time of cage cleaning, with fecal and urine samples frozen immediately and stored at −20°C until analysis. Wild chimpanzees from habituated communities were sampled under direct observation, with date, time, and name of the particular chimpanzee recorded. Urine (1 to 3 ml) was collected by placing plastic bags under night nests or by pipetting from leaves. Aliquots were placed into sterile tubes without additives and frozen within hours of collection. Stool samples (20 to 50 g) were also collected from known individuals under direct observation and placed in 50-ml tubes with or without 20 ml of RNAlater (Ambion, Austin, Tex.). RNAlater preserves nucleic acids (RNA and DNA), allowing storage and shipment at room temperature without vRNA degradation, but precludes immunoblot analysis. Samples from the nonhabituated Kalande community, some of which were estimated to be more than a day old at the time of collection, were all preserved in RNAlater. Fecal samples not preserved in RNAlater were frozen. All samples were analyzed under code, with the investigator performing immunoblot or RNA analyses unaware of the infection status of the study subject.
Detection of urine and fecal antibodies by Western immunoblot analysis.
Enhanced chemiluminescent Western blotting was performed as previously described (49) by using commercially available HIV-1 nitrocellulose test strips (Calypte Biomedical, Rockville, Md.). Fecal samples (∼2.5 g, frozen without additives) were resuspended (33% [wt/vol]) in 1× immunoblot buffer (phosphate-buffered saline [PBS; pH 7.4], 5 mM EDTA, 0.05% [wt/vol] Tween 20, 0.15 mM NaN3, 1% [wt/vol] bovine serum albumin, and 0.01% [wt/vol] IGEPAL detergent [Sigma Biochemicals, St. Louis, Mo.]), vortexed, and centrifuged at 13,800 × g for 25 min at 4°C to pellet solid debris. Supernatants were transferred to a new tube and centrifuged a second time at 4,000 × g for 10 min at 4°C to remove additional solid matter. Urine samples were tested without further processing. Western blot strips were hydrated for 10 min in PBS-Tween containing 138 mM NaCl, 2.7 mM KCl, and 0.05% (wt/vol) Tween 20 (pH 7.4), blocked with 5% skim milk in PBS-Tween for 1 h at room temperature, incubated with 1 ml of undiluted urine or clarified fecal extracts in 1× immunoblot buffer overnight at 4°C on a rocker platform, reacted with goat anti-human immunoglobulin G (IgG) (1:1,000) conjugated to horseradish peroxidase (Southern Biotechnology, Birmingham, Ala.) for 1 h at room temperature, and developed by using an ECL detection system (Amersham/Pharmacia Biotech, Piscataway, N.J.) on Hyperfilm-ECL. A subset of Western blots was also probed with goat anti-human Ig or goat anti-human IgA secondary antibodies. A plasma sample from an HIV-1-infected human was used as a positive control at dilutions of 1:10,000 and 1:100,000.
Nucleic acid extraction from chimpanzee fecal and urine samples.
Fecal RNA was extracted by using the RNAqueous-Midi kit (Ambion). Fecal aliquots (0.5 g or 1 ml of a 1:1 suspension in RNAlater) were resuspended in 6 ml of lysis buffer, vortexed, and centrifuged (16,000 × g, 3 min) to remove solid debris. Clarified supernatants were mixed with an equal volume of 64% ethanol and passed through a glass fiber filter unit. Bound nucleic acids were eluted (1 ml), precipitated with LiCl2 to enrich for RNA and to remove potential PCR inhibitors, pelleted, washed with 70% ethanol, air dried, and resuspended in 50 μl of RNA storage solution (Ambion).
Fecal DNA was extracted by using the QIAamp DNA Stool Mini Kit (QIAGEN, Valencia, Calif.). Briefly, 400 μl of fecal sample (in RNAlater) was resuspended in stool lysis buffer, treated with InhibitEx to remove possible PCR inhibitors, and spun. Supernatants were subjected to proteinase K digestion and then loaded on microspin columns for DNA purification and concentration. DNA was eluted in 100 μl of elution buffer (10 mM Tris-Cl, pH 8.5), and 5-μl aliquots were used for mitochondrial DNA amplification and sex determinations (see below).
Urine aliquots (1 ml) were ultracentrifuged (23,500 × g, 1 h at 4°C) to pellet virions and cellular components. Total nucleic acids (RNA and DNA) were extracted from the pellets by using the Boom method (7), as adapted in the NucliSens HIV-1 QT Kit (Organon-Teknika, Boxtel, The Netherlands), and the final eluate was resuspended in 50 μl of RNA storage solution (Ambion).
Amplification of SIVcpz vRNA by RT PCR.
Diagnostic reverse transcriptase PCR (RT PCR) was performed by using two sets of primer pairs based on consensus HIV-1 and SIVcpz sequences (29). The pol primer set amplifies a 330-bp fragment in the conserved integrase region and consists of first-round primers pol-F1 (5′-CCAGCNCACAAAGGNATAGGAGG-3′) and pol-R1 (5′-ACBACYGCNCCTTCHCCTTTC-3′) and second-round primers pol-F2 (5′-GGAAGTGGATACTTAGAAGCAGAAGT-3′) and pol-R2 (5′-CCCAATCCCCCCTTTTCTTTTAAAATT-3′). The gp41/nef primer set amplifies a 740- to 790-bp fragment at the junction of gp41 and nef and consists of first-round primers gp41/nef-F1 (5′-AAATGGCTGTGGTATATAAAAAT-3′) and gp41/nef-R1 (5′-CCCWTCCAGTCCCCCCTTTTC-3′) and second-round primers gp41/nef-F2 (5′-GCTTAAGAAAGGTTAGGCAGGG-3′) and gp41/nef-R2 (5′-TCCCCCCTTTTCTTTTAAAAA-3′). For cDNA synthesis, 10 μl of the fecal- or urine-derived nucleic acid solution was added to an RT PCR master mix consisting of 1× Buffer II (Perkin-Elmer, La Jolla, Calif.), 5 mM MgCl2, 1 mM dNTP, 5 mM dithiothreitol, 20 pmol of cDNA synthesis primer (either pol-R1 or gp41/nef-R1), 20 U of RNase inhibitor (Promega, Madison, Wis.), and 100 U of Superscript RT II (Gibco-BRL, Rockville, Md.); the mixture was then incubated for 1 h at 42°C. Ten microliters of the cDNA was then added to a PCR mix consisting of 1× Expand Buffer II (Roche Molecular Biochemicals, Indianapolis, Ind.), 0.35 mM dNTP, 10 pmol of the corresponding primers for the first-round PCR (F1/R1), 0.1 μg of bovine serum albumin per ml, and 2.5 U of Expand High-Fidelity Taq polymerase (Roche Molecular Biochemicals). PCR amplifications included 45 cycles of denaturation (94°C, 30 s), annealing (50°C, 30 s), and elongation (68°C, 1 min) in a Perkin-Elmer 2400 thermocycler. Two microliters of the first-round reaction was used for the nested PCR with the second-round primers, F2/R2, by using the same thermocycling conditions. Amplified products were gel purified (QIAGEN) and directly sequenced with an ABI automated DNA sequencer by using the nested primers. Sequences were analyzed with the Sequencher program (Gene Codes Corporation, Ann Arbor, Mich.), and deposited at GenBank: SIVcpzTAN2 (AY181990, AY181991), SIVcpzTAN3 (AY181988, AY181992), SIVcpzTAN4 (AY181989), and SIVcpzTAN5 (AY181993). The full-length SIVcpzTAN1 sequence (AF447763) has been reported previously (48).
Sensitivity and specificity calculations.
Assay sensitivities and specificities were calculated by using test results obtained under code from captive chimpanzees of known HIV-1 or SIVcpz infection status (Table 1). The sensitivity values of antibody and vRNA detection in urine and fecal samples were determined as the fraction of positive tests per total number of samples analyzed (rather than number of individuals analyzed), with confidence limits determined given the assumption of binomial sampling (Table 2). For these calculations, it was assumed that successive test results from the same individual were not correlated. The specificities of fecal and urine antibody detection were calculated by using test results from uninfected chimpanzees. The specificity of vRNA detection in both fecal and urine samples was deduced to be 100% since all amplification products were subsequently sequence confirmed.
TABLE 2.
Sensitivity and specificity of antibody and vRNA detection in feces and urine, respectivelya
| Detection method | HIV-1-infected chimpanzees
|
SIVcpz-infected chimpanzees
|
Uninfected chimpanzees
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Individuals | Samples | Sensitivity (95% CI) | Individuals | Samples | Sensitivity (95% CI) | Individuals | Samples | Specificity (95% CI)c | |
| ECL immunoblot (antibodies) | |||||||||
| Urine | 9/9 | 17/17 | 1.00 (0.84-1.00) | 3/3 | 10/10 | 1.00 (0.74-1.00) | 0/9 | 0/15 | 1.00 (0.82-1.00) |
| Feces | 12/5 | 26/42 | 0.62 (0.50-0.74) | 3/3 | 12/19 | 0.63 (0.47-0.83) | 0/23 | 0/51 | 1.00 (0.94-1.00) |
| RT PCR (vRNA) | |||||||||
| Urine | 1/9 | 1/17 | 0.06 (0.02-0.25) | 1/3 | 1/10 | 0.10 (0.04-0.39) | NDb | ND | 1.00 |
| Feces | 2/15 | 2/33 | 0.06 (0.02-0.18) | 4/4 | 14/25 | 0.56 (0.42-0.73) | ND | ND | 1.00 |
Sensitivity and specificity estimates are based on the number of samples rather than the number of individuals tested (each with 95% confidence interval [CI]).
ND, not done.
Specificity of vRNA detection by RT PCR is 100%, since all amplification products are sequence confirmed.
Although the sensitivity of urine antibody detection in the captive population was 100%, subsequent field studies identified, from an SIVcpz-infected wild chimpanzee (Ch-37), one urine sample that exhibited an indeterminant Western blot result. Thus, of 21 urine samples from a total of six SIVcpz-infected chimpanzees (Ch-No, Ch-Ni, CAM4, Ch-06, Ch-30, and Ch-37), only 20 met the criteria for Western blot positivity. For the prevalence calculations, we therefore used a more conservative estimate of 0.95 (for 20 of 21 samples) for the sensitivity of the urine antibody test.
Prevalence estimations.
The prevalence rates of SIVcpz infection in the five wild communities were estimated by using the test sensitivities determined for SIVcpz (not HIV-1-)-infected apes. For each chimpanzee, the probability of infection being detected, if it was truly infected, was calculated, taking into consideration the different types and numbers of specimens analyzed. The prevalence for the community was then estimated as the number of chimpanzees that tested positive divided by the number of chimpanzees expected (given the assay sensitivities) to test positive, assuming that all individuals were truly infected. For these calculations, it was again assumed that the probabilities of a positive test result in successive samples from an infected chimpanzee were not correlated.
For the Kanyawara and Ngogo communities, where no infected individuals were identified, the upper confidence limits on the rate of infection were calculated as the prevalences that would give only a 5% chance of having missed infection, given the number and types of samples analyzed and the particular tests used. For these communities, it was assumed that the total Kibale population size was large and unknown. For the more isolated Gombe chimpanzees (31), the actual number of individuals per community was used, which was 55 for Kasekela, 20 for Mitumba, and 20 for Kalande at the time of sampling. For these populations, the calculations of upper and lower confidence limits were based on the number of individuals per community who could be infected and yet would have a greater than 5% chance of showing as few or as many infected individuals as were detected, respectively.
For the nonhabituated Kalande community, 25 fecal samples from an unknown number of chimpanzees were available. It was thus assumed that each of the 20 chimpanzees in that community was sampled a Poisson number of times, with a mean number of samples of 1.25 per chimpanzee. The probability that an infected chimpanzee would be detected as being positive was estimated, and the prevalence was then calculated as the number of infected chimpanzees detected divided by the number of chimpanzees expected to be positive, assuming that all were truly infected. The upper and lower bounds of the possible numbers of chimpanzees infected (out of 20) in that community were also calculated. These were the largest, or smallest, number of infected chimpanzees that would have a greater than 5% chance of showing as few or as many infected chimpanzees as were detected, respectively.
Subspecies and sex determinations.
To confirm the subspecies origin of fecal samples from nonhabituated communities, a 450-bp region of the mtDNA genome (D loop) was amplified as previously described (33) by using primers L15997 (5′-CACCATTAGCACCCAAAGCT-3′) and H16498 (5′-CCTGAAGTAGGAACCAGATG-3′). The resulting products were sequenced directly with L15997 used as the sequencing primer. For sex determination, a region of the amelogenin gene was targeted which is known to contain a deletion near the centromere of the Y, but not of the X, chromosome (16). Primers AMEL-F (5′-CTGATGGTTGGCCTCAAGCCTGTG-3′) and AMEL-R (5′-TAAAGAGATTCATTAACTTGACTG-3′) amplify a 977-bp fragment from the X chromosome and amplify a 788-bp fragment from the Y chromosome (16). As a consequence, genomic DNA from a male chimpanzee yields two amplification products (977 and 788 bp, respectively), while genomic DNA from a female chimpanzee only yields one fragment (977 bp). PCR conditions were the same as for mtDNA amplification (33), except that an annealing temperature of 55°C was used and 55 amplification cycles were performed.
Phylogenetic methods.
Viral and mtDNA sequences were aligned by using CLUSTAL W (viral sequences were aligned bearing in mind their predicted protein sequences) (53). Gaps in any of the sequences or ambiguous areas were excluded from all comparisons. Phylogenies were inferred by the neighbor-joining method (47) and by the maximum likelihood method with a gamma distribution used to allow for variation in the substitution rate between sites (Phylogenetic Analysis by Maximum Likelihood [PAML], software version 3.0b; University College London, London, United Kingdom).
RESULTS
Fecal- and urine-based methods for SIVcpz antibody and vRNA detection.
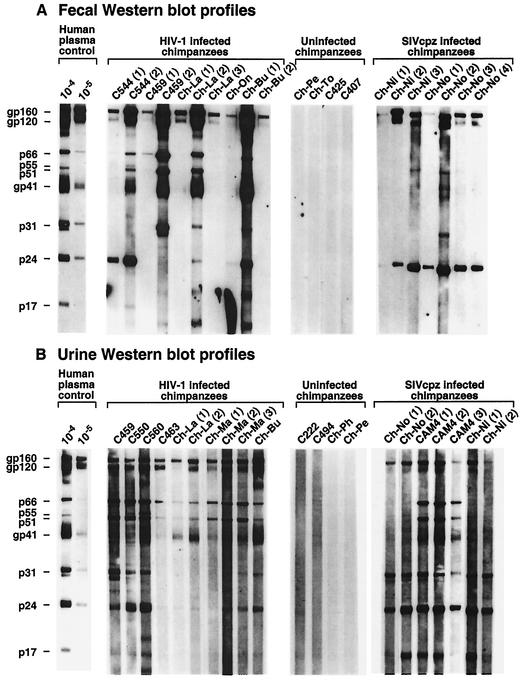
To validate the urine- and fecal-based SIVcpz detection approaches, assay sensitivities and specificities were determined in captive chimpanzees of known infection status. Figure 2 shows representative enhanced chemiluminescent (ECL) Western blots of fecal suspensions and urine samples from captive chimpanzees chronically infected with HIV-1 or SIVcpz and from uninfected controls (see Table 1 for characteristics of captive chimpanzees and summary of test results). HIV-1 was used as the test antigen because of its commercial availability and its close genetic and antigenic relatedness to SIVcpz (2, 42). Fecal and urine specimens from both HIV-1- and SIVcpz-infected chimpanzees contained readily detectable antibodies which were of the IgG class and were directed against multiple HIV-1 proteins. However, fecal antibodies were generally lower in titer and produced a more variable banding pattern (Fig. 2A) than urine antibodies (Fig. 2B). Moreover, there was substantially more variability among different individuals and between different samples from the same individual in fecal blots than in urine blots, again likely as a consequence of differences in antibody titers. The use of anti-Ig secondary antibodies did not enhance HIV-1 or SIVcpz antibody detection, and anti-IgA secondary antibodies were generally nonreactive (data not shown). Importantly, specimens from uninfected chimpanzees did not react with any viral antigens.
FIG. 2.
Detection of HIV-1- and SIVcpz-specific antibodies in chimpanzee fecal and urine samples. Fecal (A) and urine (B) specimens from captive chimpanzees chronically infected with either HIV-1 or SIVcpz, and specimens from uninfected controls, were tested by ECL Western immunoblot analysis for the presence of virus-specific antibodies. Chimpanzees are identified by code number (additional information for each individual is provided in Table 1); Ch-No and Ch-Ni are infected with the highly divergent SIVcpzANT strain (28, 55). Serial samples from the same individual are identified in parentheses. Molecular weights of HIV-1-specific proteins are indicated. The banding pattern of plasma from an HIV-1-infected human (analyzed at dilutions of 1:10,000 and 1:100,000) is shown for positive control.
A total of 112 fecal samples and 42 urine samples from 46 captive chimpanzees of known infection status were analyzed by ECL immunoblotting in order to develop criteria for positive, negative, and indeterminant reactivity and to assess the sensitivity and specificity of the assays (all analyses were performed under code). Western blots exhibiting a band corresponding to the HIV-1 envelope glycoprotein gp160 alone or in combination with any other viral band, or reactivity to any three structural proteins exclusive of gp160, were scored as positive, similar to the interpretive guidelines of commercial HIV-1 urine and serum immunoassays (9, 54). The absence of viral bands was scored negative, and samples which did not meet either criterion were considered indeterminant. By using these criteria, the sensitivity of fecal antibody detection was 62% for specimens from HIV-1-infected chimpanzees and 63% for specimens from SIVcpz-infected chimpanzees (Table 2). For urine antibody detection, the sensitivity was 100% for both HIV-1- and SIVcpz-infected chimpanzees. Specificity was 100% in all instances. None of the fecal or urine samples from infected or uninfected chimpanzees exhibited indeterminant or false-positive banding patterns.
We also investigated whether HIV-1 or SIVcpz nucleic acids could be detected in urine and fecal specimens from infected chimpanzees by nested PCR amplification. A genomic region in the viral pol gene (330 bp), highly conserved among divergent SIVcpz and HIV-1 strains, was chosen for analysis. Viral RNA, but not DNA, was detected in both feces and urine from a proportion of infected individuals (Tables 1 and 2), suggesting that viral nucleic acids were present in the form of virions and not in infected cells or as free nucleic acid. However, viral RNA was more frequently detected in feces (6 of 19 individuals positive) than in urine (only 2 of 12 individuals positive) and much more frequently in feces of SIVcpz-infected chimpanzees (4 of 4) than in HIV-1-infected chimpanzees (2 of 15). In fact, 14 of 25 fecal samples from SIVcpz-infected chimpanzees were found to be vRNA positive, compared with only 2 of 33 samples from HIV-1-infected apes. This difference was statistically significant (P = 0.002) and suggested a sensitivity of 56% for fecal vRNA detection in naturally infected chimpanzees. Although this sensitivity was lower than the sensitivity of both urine and fecal antibody detection (Table 2), the fact that all four SIVcpz-infected chimpanzees had at least one fecal sample that was positive by RT PCR indicated to us that vRNA analyses would be useful for confirmation and molecular identification of SIVcpz infection as well as for screening, if serial specimens could be collected and tested.
Noninvasive survey of wild chimpanzees in Kibale National Park.
In a previous study, we failed to identify SIVcpz infection among 24 members of the Kanyawara community in Kibale National Park (49). To examine whether this was due to insufficient sampling, we obtained additional specimens from Kanyawara and expanded our survey to a second, larger community, termed Ngogo (Fig. 1B). Importantly, the ranges of the Kanyawara and Ngogo communities are not adjoining, thus limiting direct interactions between their members. Ngogo is also unusual in that it comprises the largest community with the highest number of male members known to exist in the wild (32, 57). This is believed to be responsible for the social and behavioral differences that distinguish Ngogo chimpanzees from other wild ape communities (57). Screening both Ngogo and Kanyawara chimpanzees thus ensured a more representative sampling of the larger Kibale population, which includes still other communities. Both Ngogo and Kanyawara chimpanzees are habituated, permitting repeated sampling from known individuals under direct observation (34, 57, 61).
Study subjects, sex, sample collection dates, numbers of samples tested from each chimpanzee, and the results of tests for SIVcpz antibodies and for vRNA are summarized in Table 3. For Kanyawara, a total of 80 urine specimens were obtained from 31 chimpanzees, representing all of the adult and adolescent males as well as 82% of the adult and adolescent females (the community comprised a total of 52 members at the time of study). All tested negative for SIVcpz antibodies (Table 3). Moreover, at least one urine sample from each individual was also subjected to RT PCR analysis and found to be negative for vRNA. Similarly, none of 39 chimpanzees from the Ngogo community tested positive for SIVcpz infection (the community comprised a total of ∼145 members at the time of study). A total of 45 urine and 58 fecal samples from 32 adult and adolescent males and 7 adult and adolescent females were negative for SIVcpz antibodies and vRNA, respectively (Table 3). This was also true for an additional set of 16 urine and 7 fecal samples for which the respective donors could not be determined with certainty (data not shown). Thus, screening more than 70 wild chimpanzees from two different communities, including 61 individuals with the highly sensitive urine antibody test, failed to yield evidence for SIVcpz infection in Kibale National Park.
TABLE 3.
Noninvasive survey of two wild chimpanzee communities in Kibale National Park, Uganda
| Chimpanzee | Sexa | Date of sample collection | Urine samples
|
Fecal samples
|
Chimpanzee | Sexa | Date of sample collection | Urine samples
|
Fecal samples | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ab pos/ no. testedb | vRNA pos/ no. testedc | vRNA pos/ no. testedc | Ab pos/ no. testedb | vRNA pos/no. testedc | vRNA pos/ no. testedc | |||||||
| Kanyawara community | ||||||||||||
| ARd | F | 2/98-10/98 | 0/4 | 0/2 | ||||||||
| BLd | F | 2/99 | 0/1 | 0/1 | ||||||||
| EKd | F | 4/99 | 0/1 | 0/1 | ||||||||
| KLd | F | 3/98-6/98 | 0/2 | 0/1 | ||||||||
| ALd | F | 9/98-11/98 | 0/4 | 0/1 | ||||||||
| LPd | F | 12/97-2/98 | 0/3 | 0/1 | ||||||||
| MUd | F | 6/99 | 0/1 | 0/1 | ||||||||
| NLd | F | 4/98 | 0/1 | 0/1 | ||||||||
| OUd | F | 4/98-9/98 | 0/4 | 0/1 | ||||||||
| PUd | F | 6/99 | 0/1 | 0/1 | ||||||||
| TGd | F | 4/98-8/98 | 0/3 | 0/1 | ||||||||
| AS | F | 2/98 | 0/1 | 0/1 | ||||||||
| NE | F | 10/98 | 0/1 | 0/1 | ||||||||
| LR | F | 4/98 | 0/1 | 0/1 | ||||||||
| FG | F | 2/98 | 0/1 | 0/1 | ||||||||
| FN | F | 2/98 | 0/1 | 0/1 | ||||||||
| GO | F | 12/98 | 0/1 | 0/1 | ||||||||
| UM | F | 2/00 | 0/1 | 0/1 | ||||||||
| BFd | M | 3/98-3/98 | 0/4 | 0/1 | ||||||||
| BBd | M | 12/97-7/98 | 0/4 | 0/3 | ||||||||
| MSd | M | 1/98-10/98 | 0/3 | 0/1 | ||||||||
| AJd | M | 1/98-9/98 | 0/4 | 0/1 | ||||||||
| KKd | M | 1/98-8/98 | 0/5 | 0/1 | ||||||||
| LBd | M | 3/98-9/98 | 0/4 | 0/1 | ||||||||
| LKd | M | 3/98-11/98 | 0/3 | 0/2 | ||||||||
| SLd | M | 8/98-11/98 | 0/3 | 0/1 | ||||||||
| SYd | M | 2/98-11/98 | 0/3 | 0/1 | ||||||||
| STd | M | 2/98-11/98 | 0/5 | 0/2 | ||||||||
| TUd | M | 2/98-11/98 | 0/6 | 0/1 | ||||||||
| PGd | M | 2/99 | 0/1 | 0/1 | ||||||||
| YBd | M | 3/98-11/98 | 0/3 | 0/2 | ||||||||
| Total | 18F/13M | 12/97-2/00 | 0/80 | 0/37 | ||||||||
| Ngogo community | ||||||||||||
| NG-01 | F | 3/02 | 0/1 | 0/1 | ||||||||
| NG-02 | F | 3/02 | — | 0/1 | ||||||||
| NG-03 | F | 4/02 | 0/1 | 0/1 | ||||||||
| NG-04 | F | 4/02 | — | 0/2 | ||||||||
| NG-05 | F | 4/02 | — | 0/1 | ||||||||
| NG-06 | M | 3/02-4/02 | 0/2 | 0/1 | ||||||||
| NG-07 | M | 3/02 | 0/1 | 0/3 | ||||||||
| NG-08 | M | 3/02 | 0/1 | 0/3 | ||||||||
| NG-09 | M | 3/02-4/02 | 0/1 | 0/3 | ||||||||
| NG-10 | M | 3/02 | 0/1 | — | ||||||||
| NG-11 | M | 3/02 | 0/1 | 0/1 | ||||||||
| NG-12 | M | 3/02-4/02 | 0/2 | 0/3 | ||||||||
| NG-13 | M | 3/02-4/02 | 0/1 | 0/3 | ||||||||
| NG-14 | M | 3/02 | 0/1 | — | ||||||||
| NG-15 | M | 3/02-4/02 | 0/2 | 0/2 | ||||||||
| NG-16 | M | 3/02 | 0/4 | — | ||||||||
| NG-17 | M | 3/02 | — | 0/2 | ||||||||
| NG-18 | M | 3/02-4/02 | 0/1 | 0/3 | ||||||||
| NG-19 | M | 4/02 | 0/1 | 0/1 | ||||||||
| NG-20 | M | 3/02-4/02 | 0/1 | 0/3 | ||||||||
| NG-21 | M | 3/02-4/02 | 0/2 | 0/3 | ||||||||
| NG-22 | M | 3/02-4/02 | 0/2 | — | ||||||||
| NG-23 | M | 3/02 | 0/1 | 0/3 | ||||||||
| NG-24 | M | 3/02 | — | 0/1 | ||||||||
| NG-25 | M | 3/02 | 0/1 | 0/2 | ||||||||
| NG-26 | M | 3/02-4/02 | 0/2 | 0/2 | ||||||||
| NG-27 | M | 3/02 | 0/4 | 0/3 | ||||||||
| NG-28 | M | 3/02 | — | 0/2 | ||||||||
| NG-29 | M | 3/02-4/02 | 0/1 | 0/2 | ||||||||
| NG-30 | M | 3/02 | 0/1 | — | ||||||||
| NG-31 | M | 3/02-4/02 | 0/4 | — | ||||||||
| NG-32 | F | 3/02 | 0/1 | — | ||||||||
| NG-33 | F | 4/02 | 0/1 | — | ||||||||
| NG-34 | M | 3/02 | — | 0/1 | ||||||||
| NG-35 | M | 3/02 | — | 0/2 | ||||||||
| NG-36 | M | 3/02-4/02 | 0/1 | 0/1 | ||||||||
| NG-37 | M | 4/02 | 0/1 | — | ||||||||
| NG-38 | M | 3/02-4/02 | 0/1 | 0/1 | ||||||||
| NG-39 | M | 4/02 | — | 0/1 | ||||||||
| Total | 7F/32M | 3/02-4/02 | 0/45 | 0/58 | ||||||||
M, male; F, female.
Number of antibody-positive (Ab pos) urine samples per total number tested as determined by ECL-based immunoblot analysis (see Materials and Methods). —, specimens not available.
Number of vRNA-positive urine or fecal samples per total number tested as determined by diagnostic RT PCR.
Individuals included in a previous study (49).
Noninvasive survey of wild chimpanzees in Gombe National Park.
In contrast to Kibale, our previous survey had identified a single SIVcpz-infected chimpanzee in Gombe National Park (49). This individual was one of six tested from the Kasekela community, which is the largest in Gombe and currently comprises 55 individuals (Fig. 1C). We thus targeted the remaining members of this community for additional fecal and urine collections but also screened individuals from the neighboring Mitumba and Kalande communities (Fig. 1C). Both of these are much smaller than Kasekela, each comprising only 20 individuals, and the southern Kalande community is not habituated. The three communities have adjoining ranges, thus providing an opportunity for direct interaction among chimpanzees mainly in the form of border disputes and the migration of adolescent and adult females. Because of extensive deforestation and habitat destruction surrounding the National Park, the Gombe chimpanzees are almost completely isolated from other east African chimpanzee communities (31).
Analysis of 118 fecal and 81 urine samples from 51 of the 55 Kasekela chimpanzees confirmed infection of the known index case, Ch-06, and identified one additional SIVcpz-infected chimpanzee (Table 4). This chimpanzee (Ch-30) is a 32-year-old, healthy, and sexually active female who has no offspring. Two urine samples collected in March and April 2002 were strongly antibody positive, with reactivity to multiple HIV-1 proteins, including core, RT, and envelope glycoproteins and an overall Western blot profile virtually identical to that of Ch-06 (Fig. 3). Although two fecal samples collected in December 2001 and July 2002 were vRNA negative, detection of high titers of antibodies in urine on two independent occasions identified Ch-30 as being SIVcpz infected (Table 5).
TABLE 4.
Noninvasive survey of three wild chimpanzee communities in Gombe National Park, Tanzania
| Chimpanzee | Sexa | Date of sample collection | Urine samples
|
Fecal samples | ||
|---|---|---|---|---|---|---|
| Ab pos/no. testedc | Ab ID/no. testedd | vRNA pos/no. testedb | vRNA pos/no. testedb | |||
| Kasekela community | ||||||
| Ch-01e | F | 7/00-4/02 | 0/3 | 0/3 | 0/1 | 0/4 |
| Ch-02e | F | 7/00-12/01 | 0/1 | 0/1 | 0/1 | 0/2 |
| Ch-03e | M | 7/00-3/02 | 0/3 | 0/3 | 0/1 | 0/4 |
| Ch-04e | M | 7/00-12/01 | 0/2 | 0/2 | 0/2 | 0/3 |
| Ch-05e | M | 7/00-5/02 | 0/5 | 0/5 | 0/1 | 0/4 |
| Ch-06e | M | 7/00-4/02 | 4/4 | 0/4 | 0/2 | 6/8 |
| Ch-07 | M | 10/00-4/02 | 0/3 | 0/3 | 0/1 | 0/3 |
| Ch-08 | M | 1/01-5/02 | 0/3 | 0/3 | 0/1 | 0/4 |
| Ch-09 | F | 7/00-3/02 | 0/3 | 0/3 | 0/1 | 0/4 |
| Ch-10 | M | 1/01-3/02 | 0/2 | 0/2 | 0/1 | 0/2 |
| Ch-11 | F | 7/00-4/02 | 0/3 | 0/3 | 0/1 | 0/4 |
| Ch-12 | F | 11/00-10/01 | — | — | — | 0/2 |
| Ch-13 | M | 8/00-4/02 | 0/3 | 0/3 | 0/1 | 0/2 |
| Ch-14 | M | 11/01 | — | — | — | 0/1 |
| Ch-15 | F | 11/01-3/02 | 0/1 | 0/1 | — | 0/3 |
| Ch-16 | M | 11/00-7/02 | 0/7 | 0/7 | 0/1 | 0/7 |
| Ch-17 | F | 11/01 | — | — | — | 0/1 |
| Ch-18 | F | 11/01 | — | — | — | 0/1 |
| Ch-19 | F | 9/01-11/01 | 0/2 | 0/2 | — | 0/1 |
| Ch-20 | M | 11/00-4/02 | 0/1 | 0/1 | — | 0/3 |
| Ch-21 | F | 11/01-1/02 | — | — | — | 0/2 |
| Ch-22 | F | 12/01-6/02 | 0/1 | 1/1 | — | 0/4 |
| Ch-23 | M | 10/00-5/02 | 0/2 | 0/2 | 0/1 | 0/2 |
| Ch-24 | F | 12/01-4/02 | 0/2 | 0/2 | — | 0/2 |
| Ch-25 | F | 11/00-4/02 | 0/3 | 0/3 | — | 0/4 |
| Ch-26 | F | 12/01 | — | — | — | 0/2 |
| Ch-27 | M | 11/00-4/02 | 0/3 | 0/3 | 0/1 | 0/3 |
| Ch-28 | M | 12/01-4/02 | 0/2 | 0/2 | — | 0/1 |
| Ch-29 | F | 12/01-3/02 | 0/3 | 1/3 | — | 0/4 |
| Ch-30 | F | 12/01-4/02 | 2/2 | 0/2 | — | 0/2 |
| Ch-31 | F | 10/00-4/02 | 0/3 | 0/3 | 0/1 | 0/4 |
| Ch-32 | F | 12/01-1/02 | 0/1 | 0/1 | — | 0/2 |
| Ch-33 | F | 10/01-2/02 | 0/1 | 0/1 | — | 0/2 |
| Ch-34 | M | 1/02; 4/02 | 0/1 | 0/1 | — | 0/1 |
| Ch-35 | M | 1/02-4/02 | 0/2 | 0/2 | — | 0/2 |
| Ch-36 | F | 1/02-2/02 | 0/1 | 0/1 | — | 0/2 |
| Ch-40 | F | 12/01 | — | — | — | 0/2 |
| Ch-41 | F | 1/02 | 0/1 | 0/1 | — | 0/1 |
| Ch-43 | F | 1/02-4/02 | 0/3 | 0/3 | — | 0/1 |
| Ch-50 | F | 1/02-4/02 | 0/2 | 0/2 | — | 0/1 |
| Ch-51 | M | 12/01 | — | — | — | 0/1 |
| Ch-52 | M | 1/02 | — | — | — | 0/1 |
| Ch-53 | F | 12/01 | — | — | — | 0/1 |
| Ch-54 | M | 1/02 | — | — | — | 0/1 |
| Ch-55 | F | 1/02 | — | — | — | 0/1 |
| Ch-56 | M | 1/02 | — | — | — | 0/1 |
| Ch-57 | F | 4/02-7/02 | 0/2 | 0/2 | — | 0/2 |
| Ch-58 | M | 4/02 | — | — | — | 0/1 |
| Ch-60 | M | 8/02 | — | — | — | 0/1 |
| Ch-61 | F | 4/02 | — | — | — | 0/1 |
| Ch-62 | M | 8/02 | — | — | — | 0/1 |
| Total | 23M/28F | 7/00-8/02 | 6/81 | 2/81 | 0/18 | 6/119 |
| Mitumba community | ||||||
| Ch-37 | M | 9/00 | 4/5 | 1/5 | 0/5 | — |
| Ch-38 | F | 11/01-12/01 | — | — | — | 0/2 |
| Ch-39 | F | 9/00-12/01 | 0/1 | 0/1 | 0/1 | 0/1 |
| Ch-42 | F | 10/00-4/02 | 0/2 | 0/2 | — | 0/1 |
| Ch-44 | F | 10/00-1/02 | — | — | — | 0/3 |
| Ch-45 | M | 1/02 | — | — | — | 2/2 |
| Ch-46 | F | 11/00 | — | — | — | 0/1 |
| Ch-47 | F | 9/00-12/01 | 0/1 | 0/1 | 0/1 | 0/2 |
| Ch-48 | M | 12/01; 1/02 | — | — | — | 0/2 |
| Ch-49 | F | 8/02 | — | — | — | 0/2 |
| Ch-65 | M | 3/02 | — | — | — | 0/1 |
| Ch-66 | F | 5/02 | — | — | — | 0/2 |
| Ch-67 | M | 8/02 | — | — | — | 0/1 |
| Ch-68 | M | 8/02 | — | — | — | 0/1 |
| Ch-69 | M | 8/02 | — | — | — | 0/1 |
| Total | 7M/8F | 9/00-8/02 | 4/9 | 1/9 | 0/7 | 2/22/PICK> |
| Kalande community | ||||||
| Ch-64g | F | 11/01 | — | — | — | 1/1 |
| KA-02 | F | 8/02 | — | — | — | 0/1 |
| KA-03 | M | 6/02 | — | — | — | 0/1 |
| KA-04 | M | 6/02 | — | — | — | 0/1 |
| KA-05 | M | 3/02 | — | — | — | 0/1 |
| KA-06 | U | 4/02 | — | — | — | 0/1 |
| KA-07 | F | 6/02 | — | — | — | 0/1 |
| KA-08 | U | 3/02 | — | — | — | 0/1 |
| KA-09 | U | 8/02 | — | — | — | 0/1 |
| KA-10 | U | 8/02 | — | — | — | 0/1 |
| KA-11 | U | 6/02 | — | — | — | 0/1 |
| KA-12 | U | 4/02 | — | — | — | 0/1 |
| KA-13 | F | 8/02 | — | — | — | 0/1 |
| KA-14 | U | 7/02 | — | — | — | 0/1 |
| Ch-70 | Ff | 3/02 | — | — | — | 1/1 |
| KA-16 | U | 8/02 | — | — | — | 0/1 |
| KA-17 | U | 3/02 | — | — | — | 0/1 |
| Ch-71 | Ff | 6/02 | — | — | — | 1/1 |
| KA-19 | U | 7/02 | — | — | — | 0/1 |
| KA-20 | M | 8/02 | — | — | — | 0/1 |
| KA-21 | U | 5/02 | — | — | — | 0/1 |
| KA-22 | F | 8/02 | — | — | — | 0/1 |
| KA-23 | U | 6/02 | — | — | — | 0/1 |
| KA-24 | U | 6/02 | — | — | — | 0/1 |
| KA-25 | U | 4/02 | — | — | — | 0/1 |
| Total | ≥ 10 | 11/01-8/02 | — | — | — | 3/25 |
M, male; F, female; U, unknown.
Number of vRNA-positive fecal and urine samples per total number tested as determined by diagnostic RT PCR (authenticity of amplification products was confirmed by sequence analysis). —, specimens not available.
Number of antibody-positive (Ab pos) urine samples per total number tested as determined by ECL-based immunoblot analysis (see Materials and Methods).
Number of antibody indeterminant (Ab ID) urine samples per total number tested.
Individuals included in a previous study (49).
Sex was determined by PCR amplification of the amelogenin gene (see Materials and Methods).
Boldface rows indicate chimpanzees confirmed or suspected to be SIVcpz infected.
FIG. 3.
Western blot profiles of urine samples from wild-living chimpanzees. Representative Western immunoblots of urine samples collected in Kanyawara, Kasekela, and Mitumba are shown; collection dates are indicated (Tables 3 to 5 show details on the particular chimpanzees analyzed). Molecular weights of HIV-1-specific proteins are indicated. The banding pattern of plasma from an HIV-1-infected human (analyzed at dilutions of 1:100,000) is shown for positive control.
TABLE 5.
Gombe chimpanzees with confirmed or suspected SIVcpz infection
| Chimpanzeea | Birth date | Urine collection date | WBb | Feces collection date | vRNAc | Virus | Region amplified | Comments |
|---|---|---|---|---|---|---|---|---|
| Confirmed | ||||||||
| Ch-06 (KK) | 10/77 | 07/24/00 | + | 11/09/00 | + | TAN1(a) | pol | Mid-ranking male |
| 07/25/00 | + | 11/13/00 | + | TAN1(b) | pol/gp41 | |||
| 04/11/02 | + | 11/18/00 | + | TAN1(c) | pol | |||
| 04/28/02 | + | 11/24/01 | − | |||||
| 12/18/01 | − | |||||||
| 12/23/01 | + | |||||||
| 01/03/02 | + | |||||||
| 01/11/02 | + | TAN1(d) | gp41 | |||||
| Ch-30 (KK) | 03/70 | 03/31/02 | + | 12/13/01 | − | Sterile adult female | ||
| 04/08/02 | + | 07/02/02 | − | |||||
| Ch-37 (MI) | 07/71 | 09/09/00 | + | Low-ranking male, lost to follow-up 9/01 | ||||
| 09/09/00 | + | |||||||
| 09/09/00 | + | |||||||
| 09/09/00 | + | |||||||
| 09/18/00 | ID | |||||||
| Ch-45 (MI) | 07/76 | 01/03/02 | + | TAN3 | pol/gp41 | High-ranking male | ||
| 01/10/02 | + | |||||||
| Ch-64 (KA) | Unknown | 03/11/01 | + | TAN2 | pol/gp41 | Female | ||
| Ch-70 (KA) | Unknown | 03/13/02 | + | TAN4 | pol | Adolescent female | ||
| Ch-71 (KA) | Unknown | 06/22/02 | + | TAN5 | gp41 | Female | ||
| Suspected | ||||||||
| Ch-22 (KK) | Unknown | 02/10/02 | ID | 12/14/01 | − | Adolescent female, joined KK 10/00, presumably from Kalande | ||
| 01/06/02 | − | |||||||
| 06/18/02 | − | |||||||
| 06/20/02 | − | |||||||
| Ch-29 (KK) | Unknown | 01/27/02 | ID | [no date] | − | Adult female joined KK 9/00, presumably from Kalande | ||
| 03/10/02 | − | 12/13/01 | − | |||||
| 03/18/02 | − | 03/06/02 | − | |||||
| 06/25/02 | − |
KK, Kasekela; MI, Mitumba; KA, Kalande.
WB, Western blot of urine samples; +, positive; −, negative; ID, indeterminant.
vRNA, detection of SIVcpz vRNA in fecal samples.
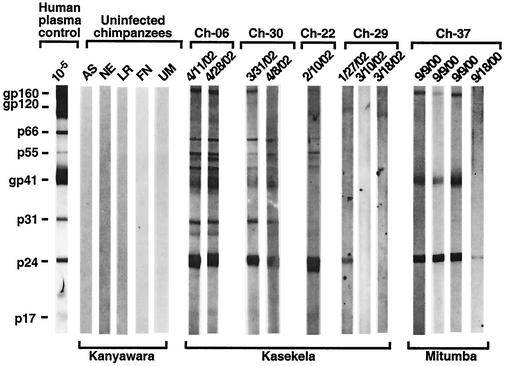
Two other Kasekela chimpanzees exhibited indeterminant urine Western blot profiles (Fig. 3). One of these is a healthy adolescent female, Ch-22, who is believed to have joined the Kasekela community from Kalande in September 2000 (Table 5). Figure 3 depicts the Western blot profile of a single available urine sample from Ch-22 which exhibits a very strong p24 reactivity as well as reactivity to p55. Although there was no detectable reactivity with envelope glycoproteins, this profile suggests infection, possibly with a divergent SIVcpz exhibiting little antigenic cross-reactivity in envelope. The second chimpanzee, Ch-29, is a healthy adult female believed to have joined Kasekela from Kalande in October 2000 (Table 5). One of her urine samples also exhibited a distinct p24 band, but this reactivity was not as strong as the p24 reactivity observed for Ch-22, and two subsequent urine samples were Western blot negative (Fig. 3). Thus, the infection status of both Ch-22 and Ch-29 is uncertain and additional samples will have to be analyzed.
The documentation of SIVcpz in Kasekela prompted us to examine members of the neighboring Mitumba community. Analysis of 22 fecal and nine urine samples from 15 Mitumba chimpanzees identified two additional SIVcpz-infected apes (Table 4). Ch-37, a 29-year-old low-ranking male at the time of study, was repeatedly positive for virus-specific antibodies in urine (Fig. 3), and Ch-45, a high-ranking 26-year-old male, was positive for fecal vRNA (Table 5). Ch-37 became chronically ill and was eventually lost to follow-up in September 2001, thus precluding confirmation of his infection by amplification of viral sequences from fecal RNA. By contrast, Ch-45 has remained healthy with no signs of wasting or of other disease. Two different fecal samples from Ch-45 yielded pol (330 bp) and gp41/nef (750 bp) fragments, and sequence analysis confirmed a new SIVcpz strain, termed TAN3 (Table 5).
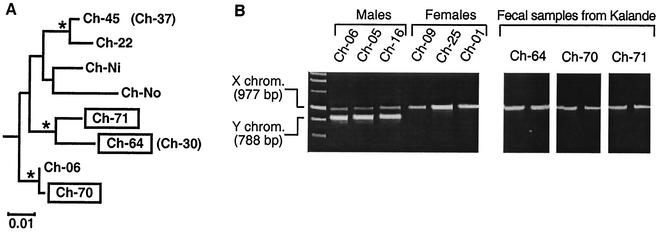
Finally, we extended our survey to the southern Kalande community, screening a total of 25 fecal samples. Although these were collected from an unknown number of chimpanzees, observational data combined with mitochondrial DNA analysis allowed us to infer that they were derived from at least 10 different individuals. Surprisingly, 3 of the 25 fecal samples (including one indicating severe diarrhea) were SIVcpz vRNA positive (Table 4). Since they could have been derived from the same individual, we amplified and sequenced a 498-bp mitochondrial control region (D loop) fragment and compared the newly derived sequences with other mtDNA sequences from Gombe as well as reference sequences from the database by phylogenetic tree analysis (Fig. 4A). This analysis revealed three distinct mtDNA haplotypes, one for each of the three SIVcpz-positive fecal samples, indicating that they were indeed collected from three different individuals (termed Ch-64, Ch-70, and Ch-71). To determine the sex of these three chimpanzees, we amplified a portion of the (nuclear) amelogenin gene. This gene is known to contain a 189-bp deletion near the centromere of the Y, but not the X, chromosome (16). Thus, PCR amplification of a fragment spanning this deletion yields two PCR products for males (one from the X chromosome and one from the Y chromosome), but only one product for females (from the X chromosome). Amplification of such a fragment from fecal DNA extracted from the three SIVcpz-infected Kalande chimpanzees (performed in duplicate) identified Ch-64, Ch-70, and Ch-71 as females (Fig. 4B).
FIG. 4.
Subspecies and sex determination of three SIVcpz-infected, nonhabituated chimpanzees from Kalande. (A) Phylogenetic tree of mitochondrial DNA sequences. D-loop sequences (498 bp) amplified from three SIVcpz vRNA-positive fecal specimens (boxed) from Kalande were compared with mtDNA sequences from other eastern chimpanzees (P. t. schweinfurthii). The mtDNA sequence for Ch-No has been reported previously (18). Sequences for Ch-06, Ch-22, Ch-45, Ch-37, Ch-30, and Ch-Ni have been determined in this study. The tree was obtained by maximum likelihood analysis of 377 gap-stripped sites by using the REV + Gamma substitution model implemented in BASEML from the PAML package (PAML, software version 3.0b; University College London, London, United Kingdom) testing 48 tree topologies obtained from NUCML from the MOLPHY package (program package for MOLecular PHYlogenetics, software version 2.2; Institute of Statistical Mathematics, 4-6-7 Minami-Azabu, Minato-ku, Tokyo 106, Japan). Asterisks denote clades to the right found in at least 70% of bootstrapped replicates analyzed by the neighbor-joining method implemented in CLUSTAL W (53) using Kimura's two-parameter correction (27). The three Kalande sequences (boxed) represent different haplotypes within the P. t. schweinfurthii radiation, indicating that the samples came from three different individuals. Ch-30 and Ch-37 (in parentheses) have haplotypes identical to those of Ch-64 and Ch-45, respectively. The tree is rooted on sequences from the subspecies P. t. verus and P. t. vellerosus. (B) Sex determination of Ch-64, Ch-70, and Ch-71 from Kalande. A region of the amelogenin gene, known to contain a 189-bp deletion near the centromere of the Y, but not the X, chromosome, was amplified (16). Fecal DNA from known male (Ch-06, Ch-05, and Ch-16) and female (Ch-09, Ch-25, and Ch-01) chimpanzees from Kasekela was analyzed for control. Sex determination for Ch-64, Ch-70, and Ch-71 was performed in duplicate.
Taken together, of 76 Gombe chimpanzees analyzed, seven (Ch-06 and Ch-30 from Kasekela; Ch-37 and Ch-45 from Mitumba; and Ch-64, Ch-70, and Ch-71 from Kalande) were SIVcpz infected, and two (Ch-22 and Ch-29 from Kasekela) had indeterminant urine antibody results suggestive of SIVcpz infection.
Prevalence of SIVcpz in Gombe.
Using conservative estimates of assay sensitivities determined for SIVcpz-infected chimpanzees and the numbers and types of specimens actually analyzed, we calculated the prevalence of SIVcpz infection for each of the three Gombe communities. These values were 5% for Kasekela, 17% for Mitumba, and 30% for Kalande. Estimates of the 95% confidence limits showed that for Kasekela, a total of two infected individuals in the population resulted in a greater than 5% chance of detecting two or more infected individuals in the sample set, making 2/55 (4%) the lower confidence limit on the prevalence in this population. Any number greater than four infected chimpanzees in the population gave a lower than 5% chance of two or fewer infected chimpanzees being detected, thus making 4/55 (7%) the 95% upper confidence limit on the prevalence. Corresponding values for Mitumba were two and eight infected chimpanzees in the population of 20 individuals, corresponding to confidence limits of 10 and 40% as the lower and upper bounds, respectively. In Kalande, the range of possible numbers in the community was from 3 to 12, corresponding to a range of prevalences from 15 to 60%.
For the Gombe population as a whole, the mean SIVcpz prevalence (weighted by population size) was 13%, and the range indicated by the lower and higher bounds of the individual populations was from 7/95 (7%) to 24/95 (25%).
Molecular epidemiology of SIVcpz in Gombe.
In addition to the index chimpanzee Ch-06, four other chimpanzees (Ch-45, Ch-64, Ch-70, and Ch-71) were found to be fecal vRNA positive. We thus used RT-PCR approaches to amplify viral sequences of sufficient length for phylogenetic analyses. Two regions of the SIVcpz genome were targeted, a 330-bp pol fragment in the highly conserved integrase region and a 750-bp env fragment spanning the junction of gp41 and nef. For Ch-64 and Ch-45, amplification was successful for both regions, and the corresponding viruses were designated TAN2 and TAN3, respectively. For the remaining two chimpanzees, only one of the two genomic regions could be amplified: a pol fragment for Ch-70 (TAN4) and a gp41/nef fragment for Ch-71 (TAN5). All newly derived sequences were compared to each other as well as to the previously reported SIVcpz TAN1 strain (48).
In pairwise sequence comparisons, the nucleotide sequences of all five Gombe SIVcpz strains (TAN1 to TAN5) differed by 5.9 to 9.6% in the pol region and by 9.5 to 14.6% in the gp41/nef region (Table 6), while pol sequences obtained from Ch-06 at different time points (TAN1a to TAN1c) differed by only 0.7 to 1.4% (not shown). When compared to the only other P. t. schweinfurthii strain, SIVcpzANT, TAN1 to TAN5 differed from by 16 to 21% in the pol and by 40.6% to 43.5% in the gp41/nef regions, respectively. Thus, the Gombe strains were much more similar to one another than any one of them was to SIVcpzANT, but no two were so similar as to suggest that they represented a direct transmission pair. Phylogenetic tree analyses confirmed these conclusions, revealing that all Gombe viruses formed a monophyletic cluster within the SIVcpzPts radiation (Fig. 5). The extent of divergence among the Gombe viruses was similar to that seen among HIV-1 group M subtypes (Fig. 5). Since HIV-1 and SIVcpz appear to have evolved at similar rates (50), this suggests that the common ancestor of the Gombe SIVcpz strains existed within the last 100 years. This common ancestor could have resulted from a single introduction of SIVcpz into the larger Gombe community. In each tree, it is one of the Kalande strains (TAN4 for pol, TAN5 for gp41/nef) that forms the outgroup. These relationships are consistent with an initial introduction into Kalande, followed by onward transmission of the virus to the other communities. However, additional sequence data are necessary to confirm this.
TABLE 6.
Percent nucleotide sequence differences between pol and gp41/nef regions of Gombe SIVcpz strains
| Virus strain | % nucleotide sequence difference, pol and gp41/nef regions
|
|||||
|---|---|---|---|---|---|---|
| TAN1 | TAN2 | TAN3 | TAN4a | TAN5a | ANT | |
| TAN1 | 6.6 | 9.1 | 8.7 | — | 21.0 | |
| TAN2 | 13.0 | 5.9 | 9.6 | — | 19.9 | |
| TAN3 | 13.9 | 9.5 | 8.7 | — | 19.2 | |
| TAN4a | — | — | — | — | 16.1 | |
| TAN5a | 13.8 | 14.6 | 14.4 | — | — | |
| ANT | 41.6 | 40.6 | 43.3 | — | 43.5 | |
—, sequences not available.
FIG. 5.
Phylogenetic analyses of SIVcpz in Gombe. Five different SIVcpz strains identified in Mitumba, Kasekela, and Kalande were analyzed in two different genomic regions, a 286-bp pol and 564-bp gp41/nef fragment (after gap stripping). The newly derived sequences TAN2, TAN3, TAN4, and TAN5 were compared with TAN1 sequences from different time points (Table 5) as well as reference sequences for HIV-1 M subtype A; U455 (GenBank accession number M62320), subtype B; LAI (K02013), subtype D; ELI (K03454), HIV-1 N; YBF30 (AJ006022), HIV-1 O; MVP5180 (L20771), and ANT70C (L20587), SIVcpz; CAM3 (AF115393), CAM5 (AJ271369), US (AF103818), GAB1 (X52154), and GAB2 (F. Bibollet-Ruche and B. H. Hahn, unpublished data), and ANT (U42720). The trees were obtained by the neighbor-joining method (47) implemented in CLUSTAL W (53) by using Kimura's two-parameter correction (27) and 1,000 bootstrapped replicates. Asterisks indicate that the clades to the right were found in at least 80% of bootstrapped replicates.
DISCUSSION
Noninvasive assay development.
Detection of infectious agents in wild-living, endangered primates is a particular challenge. Nowhere is this more evident than with SIVcpz infection of chimpanzees. Field studies of chimpanzees requiring genetic, hormonal, or biochemical analyses have relied on noninvasively collected hair, urine, or feces (12, 33, 34). We thus sought to develop urine- and fecal-based methods for SIVcpz antibody and vRNA detection that would be suitable for viral testing under field conditions. To accomplish this, assay sensitivities and specificities were first determined in captive chimpanzees of known infection status. Assay performance was then evaluated in habituated chimpanzee communities, which permitted repeated sampling of the same individuals and thus confirmation of test results. Finally, assays were tested in nonhabituated apes who represent the vast majority of chimpanzees in the wild. The combined results from these studies demonstrate that (i) fecal- and urine-based assays provide sufficient sensitivity and specificity for viral testing under field conditions; (ii) noninvasive prevalence studies are feasible in habituated as well as nonhabituated settings; (iii) urine antibody screening combined with fecal vRNA amplification and sequence determination represents the most effective strategy to detect with certainty and characterize SIVcpz infection in the wild; and (iv) these approaches are applicable to other wild-living endangered primate species (30) (M. Santiago and B. H. Hahn, unpublished data).
Of the noninvasive methods that we evaluated, the ECL-based Western blot analysis of urine was by far the most sensitive (>95%) in detecting SIVcpz infection. Antibodies in urine were readily detectable and directed against multiple viral proteins, with little variation between repeat samples from the same individual. This was true not only for urine obtained from captive apes (Fig. 2) but also for samples collected in the wild (Fig. 3). Analysis of antibodies in urine was also highly specific (100%), since samples from uninfected chimpanzees did not react with any HIV-1 protein, including p24 (Fig. 2 and 3). On a practical level, urine antibodies were stable in the absence of refrigeration. Three urine specimens from Ch-37 exhibited strong gp160, gp41, and p24 reactivities despite storage at room temperatures for several weeks after a freezer breakdown at the Gombe field station (Fig. 3). Finally, urine contained sufficient quantities of cellular nucleic acids for host genetic analyses (not shown). Analysis of antibody in urine thus represents a highly sensitive, specific, and practical diagnostic tool that can be used in habituated as well as nonhabituated settings. However, insufficient quantities (<1 ml) can yield false-negative test results. For example, a 200-μl urine aliquot (taken 18 September 2000) from Ch-37 produced only a single p24 band and thus an indeterminant Western blot profile (Fig. 3).
In contrast to urine antibody testing, fecal antibody analysis was found to be only moderately sensitive (60%). Fecal antibodies were generally of lower titer than urine antibodies and produced more variable banding patterns even when samples were collected under optimal conditions (Fig. 2). Moreover, fecal samples required immediate storage at −20°C and shipment on dry ice, thus posing logistical problems at most field sites. The addition of antibiotics and protease inhibitors at the time of collection did not improve the sensitivity or reproducibility of antibody detection (data not shown). Finally, storage of fecal samples in RNAlater, a preservative of nucleic acids (see below), precluded antibody analysis. Taken together, these reasons render fecal antibody testing impractical for most field settings.
Fecal and urine samples from HIV-1- and SIVcpz-infected chimpanzees also provided a source of viral RNA (Table 1). However, the frequency of vRNA detection differed greatly depending on the types of specimens analyzed (Table 2). Amplification of viral sequences from urine was rare for both HIV-1- and SIVcpz-infected chimpanzees (<10%), suggesting that virus particles are not normally passed through (or produced in) intact kidneys. Amplification of viral sequences from fecal samples of chimpanzees experimentally infected with HIV-1 was also rare (6%); however, the frequency of vRNA detection in fecal samples of naturally SIVcpz-infected (captive) chimpanzees was surprisingly high (56%). Spiking of fecal samples with known copy numbers of HIV-1 virions revealed that 104 to 105 virus particles per gram of fecal matter were required for fecal vRNA detection (data not shown). These data indicated that most virions were lost during the extraction and purification step and that high fecal viral loads were required for successful amplification. Moreover, noninvasive studies of SIVsm in sooty mangabeys revealed that recovery of viral RNA sequences from fecal samples depended on high systemic viral loads and that mangabeys with plasma viral loads of less than 3,500 particles per ml have a greater than 95% chance of being fecal vRNA negative (30). These findings explained the low frequency of fecal vRNA recovery from HIV-1-infected chimpanzees (Table 2), since most of these experimentally infected apes had very low or undetectable plasma virus loads (6, 39, 52). However, it also raised the question of to what extent SIVcpz-infected wild chimpanzees could be expected to be fecal vRNA positive. To address this, we examined the frequency of fecal vRNA detection in the chimpanzees from Gombe who were either known or suspected to be SIVcpz infected (Table 5). Including all individuals with positive and indeterminant urine antibody results, this frequency was 11 vRNA-positive fecal samples of 23 tested (from eight apes) or 49%. Including only the two urine antibody-positive individuals, this frequency was 6 of 10 samples, or 60%. Thus, the frequency of vRNA detection in the feces of SIVcpz-infected wild chimpanzees appears to be similar to that of captive SIVcpz-infected chimpanzees as well as SIVsm-infected sooty mangabeys (30).
Given the importance of fecal samples as a source of viral sequences, we tested different methods of fecal preservation previously reported to permit host genetic analyses (12, 15, 56). These studies showed that fecal treatment with 70% ethanol, guanidine thiocyanate, or silica did not preserve SIVcpz sequences (not shown); however, resuspension in RNAlater (Ambion), maintained viral as well as cellular nucleic acids, even in the absence of refrigeration. For example, fecal samples from Ch-06 shipped at ambient temperature from Tanzania to the United States yielded nucleic acids of sufficient quality to amplify a complete SIVcpzTAN1 genome (48). RNAlater also preserved fecal DNA, permitting mtDNA amplification (Fig. 4A), microsatellite analyses (not shown), and amplification of nuclear gene fragments of up to 2,000 bp, such as portions of the genes encoding amelogenin (Fig. 4B), CCR5, or glucose-6 phosphate dehydrogenase (not shown). This allowed us to independently confirm the species origin (P. t. schweinfurthii) of three SIVcpz-positive fecal samples from the nonhabituated Kalande community and to infer that these samples were derived from three different females. Unfortunately, RNAlater precludes Western blot analysis of fecal samples, and attempts to circumvent this problem have thus far remained unsuccessful.
Endemic SIVcpz infection in Gombe.
Our previous finding of a single SIVcpz-positive chimpanzee (Ch-06) in Kasekela (49) raised the question of to what extent SIVcpz was prevalent in this and other Gombe communities. Remarkably, after screening almost the entire Kasekela community (51 of 55 individuals, 93%), only one other SIVcpz-infected chimpanzee, Ch-30, was identified. Prevalence calculation based on the types and numbers of samples tested predicted a 5% infection rate for this community, with a 95% confidence interval of 4 to 7%. However, the actual prevalence may be closer to the upper limit since two chimpanzees with indeterminant urine Western blot results were considered negative in these calculations (Fig. 3). One of these, an adolescent female, Ch-22, believed to have joined Kasekela from the southern Kalande community, is likely to be SIVcpz infected since her urine Western blot profile showed reactivity with two structural proteins (p24 and p55). The second chimpanzee, an adult female, Ch-29, also believed to have originated from Kalande, is more difficult to assess since only one of three urine samples exhibited a single p24 band (Fig. 3). Analyses of additional urine (and fecal) samples will be required to determine whether this p24 reactivity is unspecific, indicative of a low-titer antibody response, or the result of infection with a more divergent (antigenically less cross-reactive) virus. Urine antibody testing using Western blot strips containing SIVcpz antigens derived from a cognate P. t. schweinfurthii virus should aid in distinguishing between these possibilities.
We also screened the neighboring Mitumba and Kalande chimpanzees. In the northern Mitumba community, we identified two males as harboring SIVcpz. With 15 of the 20 individuals tested, this accounts for a prevalence estimate of 17%. Since most Mitumba chimpanzees were tested for fecal vRNA only, the confidence interval is wide, ranging from 10 to 40%. In the southern Kalande community, three SIVcpz-infected females were identified, accounting for a prevalence of 30%, again with a wide confidence interval (15 to 60%). This prevalence is the highest of all Gombe communities and suggests a hot spot of SIVcpz infection in Kalande. The fact that two Kasekela chimpanzees with indeterminant Western blot results (Ch-22 and Ch-29) originated in Kalande, along with the phylogenetic data suggesting Kalande as the point of origin for the Gombe infections (Fig. 5), are consistent with this interpretation.
Although the phylogenetic relationships of the five Gombe strains indicate passage of virus among the three communities (Fig. 5), the routes of SIVcpz transmission remain unknown. To date, we have not been able to document sexual transmission. Similarly, we have not been able to assess vertical transmission because the females known or suspected to be infected are either infertile (Ch-30), adolescent (Ch-22), recent immigrants (Ch-29), or not habituated (Ch-64, Ch-70, and Ch-71). Finally, there has been no information to link the observed infections to border fights which are sometimes violent and thus represent a plausible route of transmission. An extreme example occurred in 1974, when border fights between Kasekela and neighboring Kahama males led to the dissolution of the entire Kahama community and the death of all Kahama males (23). Analysis of all chimpanzees who have not yet been tested, especially those in Mitumba and Kalande, as well as the molecular characterization of all circulating viruses, will be necessary to decipher the transmission patterns of SIVcpz within and between the Gombe communities and to elucidate why the two smallest communities seem to have a disproportionate number of infected individuals.
Absence of SIVcpz infection from Kibale?
In light of the seven documented and two suspected cases of SIVcpz infection in Gombe chimpanzees, the failure to find infection among the 70 chimpanzees from Ngogo and Kanyawara came as a surprise. We therefore examined the possibility that SIVcpz infection existed in Kibale but went undetected. We asked three questions. (i) What is the likelihood that any particular chimpanzee could have been infected despite negative test results, given the numbers and types of specimens analyzed and the sensitivities of the respective assays? (ii) What is the likelihood that one chimpanzee in each of the two groups studied was infected? (iii) What inferences can be drawn about the likely prevalence of SIVcpz in the larger Kibale communities surrounding and including the Ngogo and Kanyawara study groups?
The likelihood of a negative test result despite true infection in any one chimpanzee studied in Kanyawara was very low, ranging from 0.043 for chimpanzees for which only a single urine sample was tested to <0.001 for chimpanzees for which four or more urine samples were analyzed. In Ngogo, the likelihood of a falsely negative test ranged from 0.44 for chimpanzees for which only a single fecal sample was tested to <0.001 for individuals for which four urine samples were analyzed. Assuming there actually had been a single infected chimpanzee among those tested in Kibale, the chance of having missed this individual was 1.8% in Kanyawara and 8.2% in Ngogo, respectively. These values reflect the fact that a conservative estimate of urine antibody detection sensitivity (0.95) was used, and that in Ngogo, 8 of 39 individuals had only fecal samples available for testing. Finally, within the larger Kibale population (assuming an unlimited population size), we could exclude the possibility of a 10% or higher prevalence rate with 95% certainty. However, the possibility of a 1% or lower prevalence could not be ruled out. In fact, the probability of having missed a 1% prevalence was estimated to be 49%. Thus, a low-level endemic infection of the larger Kibale population of chimpanzees certainly remains a possibility.
P. t. schweinfurthii represents an atypical SIV reservoir.
The finding of endemic infection in Gombe identifies P. t. schweinfurthii as a natural host and reservoir for SIVcpz; however, the observed frequencies and distribution of infection indicate that naturally infected apes differ from SIV-infected monkey species in fundamental ways. First, the great majority of these monkey species harbor SIV at much higher frequencies (5, 11, 14, 25, 40, 43). Surveys of primate bushmeat as well as wild-living populations have yielded infection rates of greater than 40% for most of these species. Second, SIV infection is more uniformly distributed among members of these various species. Random sampling at any site within the natural range seems to be sufficient to identify SIV-infected animals (1, 4, 5, 14, 40, 43). Third, field studies of wild grivets and sabaeus monkeys have shown that SIV infection rates increase with age and that >90% of sexually active females can be infected. This has been taken to indicate that SIV is transmitted in these communities predominantly by sexual routes (5, 43). SIVcpz infection in Gombe does not increase with age, nor is it preferentially found in multiparous females—and this is not due to a lack of sexual activity. For example, Ch-06 has mated at least 333 times with 25 different females since puberty. More importantly, since July 2000, the earliest date of his SIVcpz diagnosis, Ch-06 has mated at least 33 times with eight females, and none have become infected. These data suggest that transmission of SIVcpz by sexual and/or other routes is much less efficient than transmission of SIV in smaller monkey species. Although the reasons for this remain unknown, it is of interest that heterosexual transmission of HIV-1 in humans is also very inefficient, ranging from 1 in 250 to 1 in 3,000 per unprotected coital act (17, 24, 45, 46). Thus, chimpanzees seem to resemble humans more than monkeys in the frequency with which they transmit SIV.
Assuming that natural transmission of SIVcpz is rare, how can the observed differences in SIVcpz prevalence in Kibale and Gombe be reconciled? Studies of HIV-1 infection rates in remote rural human populations may provide an answer. In 1986, Nzilambi and colleagues found an HIV-1 prevalence of 0.8% in a rural population of the Equateur province of the Democratic Republic of the Congo (formerly Zaire) (37). Moreover, retrospective analysis of blood samples obtained in 1976 from this same population revealed that the frequency of HIV-1 infection had not changed over a 10-year time period (37). If HIV-1 infection can be sustained in a heterosexual human population at low frequencies (<1%), the same may be true for SIVcpz infection of chimpanzees. Under this scenario, a low-level rate of SIVcpz endemicity would be the rule, while much higher prevalence rates like those seen in Gombe would be the exception, possibly resulting from higher transmission rates in an increasingly isolated population. Screening of additional chimpanzees in Kibale in sufficient numbers to detect a 1% or lower prevalence rate will be necessary to address this possibility.
Alternatively, Kibale chimpanzees may have never been exposed to SIVcpz because of biogeographic divisions of the eastern chimpanzee population. Kibale National Park is approximately 500 km north of Gombe National Park (Fig. 1A). Intervening lacustrine and forest gap barriers may have limited gene flow between these populations in the past (19, 20). It is possible that SIVcpz infection may not have spread to Kibale if it was introduced into eastern chimpanzee populations subsequent to the establishment of such gene flow barriers. This hypothesis can be tested by screening chimpanzee communities on either side of putative biogeographic barriers, such as populations north (Budongo, Semliki, and Bugoma) and south (Nyungwe and Mahale) of the Kazinga Channel (between Lake George and Lake Edward in Uganda).
Still another possibility is that SIVcpz may have been present in the Kibale chimpanzees at some point in time but subsequently became extinct. Although a combination of biological, ecological, and epidemiologic factors, including stochastic events, could have been responsible for such an event, the fact that the Kibale population is much larger than the one in Gombe makes this possibility somewhat less likely. Finally, the unexpected irregular pattern of SIVcpz infection described here for Kibale and Gombe may be unique to P. t. schweinfurthii at the easternmost limits of their range, possibly reflecting a particularly fragmented habitat. Studies of P. t. schweinfurthii communities elsewhere, for example in the Democratic Republic of the Congo, will be necessary to address this possibility.
P. t. schweinfurthii is not the source of pandemic HIV-1.
Phylogenetic analyses of all known SIVcpz and HIV-1 strains, including the newly identified Gombe viruses, show quite clearly that SIVcpz from P. t. schweinfurthii was not responsible for the human AIDS pandemic (Fig. 5) (48, 49, 50). Instead, several lines of evidence implicate west central African P. t. troglodytes apes as the source of HIV-1 groups M, N, and O (2, 13, 18, 25). The findings of this report point to the importance of field studies in west central Africa to determine the molecular epidemiology and prevalence of SIVcpz in this area. The present study demonstrates the feasibility of such an undertaking and provides a frame of reference for future studies of SIVcpz natural history, epidemiology, and evolution.
Acknowledgments
We thank the staff at the Rijswijk and Yerkes Primate Research Centers and the Cameroonian wildlife sanctuaries for collecting fecal and urine samples from captive chimpanzees; we thank field researchers from the Makerere University Biological Field Station and the Gombe Stream Research Centre for collecting fecal and urine samples from wild chimpanzees. We also thank Michael A. Huffman, Tony Goldberg, and Michael L. Wilson for helpful discussions, Joann Schumacher-Stankey (University of Minnesota) for information on mating frequencies of the Gombe chimpanzees, Julie Decker and Maria Salazar for technical assistance, and W. J. Abbott for artwork and manuscript preparation.
This work was supported in part by grants from the National Institutes of Health (B.H.H., H.M.M., and P.M.S.), the Institute Pasteur (A.A. and E.N.), Yale University (D.P.W.), the Leakey Foundation and the National Science Foundation (J.S.L., M.N.M., and R.W.W.), the Wenner Gren Foundation for Anthropological Research and Harvard University (M.E.), the Jane Goodall Institute (M.L., S.K., A.E.P., and D.A.C.), the University of Minnesota (A.E.P.), and the Howard Hughes Medical Institute (G.M.S.).
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailes, E., R. R. Chaudhuri, M. L. Santiago, F. Bibollet-Ruche, B. H. Hahn, and P. M. Sharp. 2002. The evolution of primate lentiviruses and the origins of AIDS, p. 65-96. In T. Leitner (ed.), The molecular epidemiology of human viruses. Kluwer Academic Publishers, Norwell, Mass.
- 3.Bailes, E., F. Gao, F. Bibollet-Ruche, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science, in press. [DOI] [PubMed]
- 4.Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77:773-781. [DOI] [PubMed] [Google Scholar]
- 6.Bogers, W. M., W. H. Koornstra, R. H. Dubbes, P. J. ten Haaft, B. E. Verstrepen, S. S. Jhagjhoorsingh, A. G. Haaksma, H. Niphuis, J. D. Laman, S. Norley, H. Schuitemaker, J. Goudsmit, G. Hunsmann, J. L. Heeney, and H. Wigzell. 1998. Characteristics of primary infection of a European human immunodeficiency virus type 1 clade B isolate in chimpanzees. J. Gen. Virol. 79:2895-2903. [DOI] [PubMed] [Google Scholar]
- 7.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butynski, T. M. 2001. Africa's great apes: an overview of current taxonomy, distribution, numbers, conservation status, and threats, p. 3-56. In A. Arluke (ed.), Great apes and humans: the ethics of coexistence. Smithsonian Institution Press, Washington, D.C.
- 9.Centers for Disease Control and Prevention. 1989. Interpretation and use of the Western Blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. Morb. Mortal. Wkly. Rep. 38:S1-S7.
- 10.Chakrabarti, L. 2002. Natural history of simian immunodeficiency viruses: clues to the emergence and virulence of AIDS viruses, p. 61-63. In J. Lederberg (ed.), The emergence of zoonotic diseases: understanding the impact on animal and human health (workshop summary). National Academy Press, Washington, D.C. [PubMed]
- 11.Chen, Z., P. Telfer, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constable, J. L., M. V. Ashley, J. Goodall, and A. E. Pusey. 2001. Noninvasive paternity assignment in Gombe chimpanzees. Mol. Ecol. 10:1279-1300. [DOI] [PubMed] [Google Scholar]
- 13.Corbet, S., M. C. Muller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. Env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courgnaud, V., P. Formenty, C. Akoua-Koffi, R. Noe, C. Boesch, E. Delaporte, and M. Peeters. 2003. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African Colobids: SIVwrc from Western Red Colobus (Piliocolobus badius) and SIVolc from Olive Colobus (Procolobus verus). J. Virol. 77:744-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damen, M., P. Sillekens, M. Sjerps, R. Melsert, I. Frantzen, H. W. Reesink, P. N. Lelie, and H. T. Cuypers. 1998. Stability of hepatitis C virus RNA during specimen handling and storage prior to NASBA amplification. J. Virol. Methods 72:175-184. [DOI] [PubMed] [Google Scholar]
- 16.Eng, B., P. Ainsworth, and J. S. Waye. 1994. Anomalous migration of PCR products using nondenaturing polyacrylamide gel electrophoresis: the amelogenin sex-typing system. J. Forensic Sci. 39:1356-1359. [PubMed] [Google Scholar]
- 17.Fideli, U. S., S. A. Allen, R. Musonda, S. Trask, B. H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S. H. Vermund, and G. M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retrovir. 17:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg, T. L. 1996. Genetics and biogeography of East African chimpanzees. Ph.D. dissertation. Harvard University, Boston, Mass.
- 20.Goldberg, T. L., and M. Ruvolo. 1997. The geographic apportionment of mitochondrial genetic diversity in east African chimpanzees, Pan troglodytes schweinfurthii. Mol. Biol. Evol. 14.:976-984. [DOI] [PubMed]
- 21.Gonder, M. K., J. F. Oates, T. R. Disotell, M. R. Forstner, J. C. Morales, and D. J. Melnick. 1997. A new west African chimpanzee subspecies? Nature 388:337. [DOI] [PubMed] [Google Scholar]
- 22.Goodall, J. 1968. The behaviour of free-ranging chimpanzees in the Gombe Stream Reserve. Anim. Behav. Monogr. 163-311.
- 23.Goodall, J. 1986. The Chimpanzees of Gombe: patterns of behavior. Belknap Press, Cambridge, Mass.
- 24.Gray, R. H., M. J. Wawer, R. Brookmeyer, N. K. Sewankambo, D. Serwadda, F. Wabwire-Mangen, T. Lutalo, X. Li, T. vanCott, and T. C. Quinn. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149-1153. [DOI] [PubMed] [Google Scholar]
- 25.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 26.Kaessmann, H., V. Wiebe, and S. Paabo. 1999. Extensive nuclear DNA sequence diversity among chimpanzees. Science 286:1159-1162. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 28.Koopman, G., A. G. Haaksma, J. ten Velden, C. E. Hack, and J. L. Heeney. 1999. The relative resistance of HIV type 1-infected chimpanzees to AIDS correlates with the maintenance of follicular architecture and the absence of infiltration by CD8+ cytotoxic T lymphocytes. AIDS Res. Hum. Retrovir. 15:365-373. [DOI] [PubMed] [Google Scholar]
- 29.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2001. HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 30.Ling, B., M. L. Santiago, S. Meleth, B. Gormus, H. M. McClure, C. Apetrei, B. H. Hahn, and P. M. Marx. 2003. Noninvasive detection of new simian immunodeficiency virus lineages in captive sooty mangabeys: ability to amplify virion RNA from fecal samples correlates with viral load in plasma. J. Virol. 77:2214-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massawe, E. T. 1992. Assessment of the status of chimpanzee populations in Western Tanzania. Afr. Study Monogr. 13:35-55. [Google Scholar]
- 32.Mitani, J. C., and D. P. Watts. 1999. Demographic influences on the hunting behavior of chimpanzees. Am. J. Phys. Anthropol. 109:439-454. [DOI] [PubMed] [Google Scholar]
- 33.Morin, P. A., J. J. Moore, R. Chakraborty, L. Jin, J. Goodall, and D. S. Woodruff. 1994. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science 265:1193-1201. [DOI] [PubMed] [Google Scholar]
- 34.Muller, M. N., and R. W. Wrangham. 2001. The reproductive ecology of male hominoids. In P. T. Ellison (ed.), Reproductive ecology and human evolution. Aldine, New York, N.Y.
- 35.Novembre, F. J., J. de Rosayro, S. Nidtha, S. P. O'Neil, T. R. Gibson, T. Evans-Strickfaden, C. E. Hart, and H. M. McClure. 2001. Rapid CD4+ T-cell loss induced by human immunodeficiency virus type 1NC in uninfected and previously infected chimpanzees. J. Virol. 75:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novembre, F. J., M. Saucier, D. C. Anderson, S. A. Klumpp, S. P. O'Neil, C. R. Brown II, C. E. Hart, P. C. Guenthner, R. B. Swenson, and H. M. McClure. 1997. Development of AIDS in a chimpanzee infected with human immunodeficiency virus type1. J. Virol. 71:4086-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nzilambi, N., K. M. De Cock, D. N. Forthal, H. Francis, R. W. Ryder, I. Malebe, J. Getchell, M. Laga, P. Piot, and J. B. McCormick. 1988. The prevalence of infection with human immunodeficiency virus over a 10-year period in rural Zaire. N. Engl. J. Med. 318:276-279. [DOI] [PubMed] [Google Scholar]
- 38.Oates, J. F. 1996. African Primates, Status Survey and Conservation Action Plan. Gland: IUCN/SSC Primate Specialist Group.
- 39.O'Neil, S. P., F. J. Novembre, A. B. Hill, C. Suwyn, C. E. Hart, T. Evans-Strickfaden, D. C. Anderson, J. deRosayro, J. G. Herndon, M. Saucier, and H. M. McClure. 2000. Progressive infection in a subset of HIV-1-positive chimpanzees. J. Infect. Dis. 182:1051-1062. [DOI] [PubMed] [Google Scholar]
- 40.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peeters, M., K. Fransen, E. Delaporte, M. Van den Haesevelde, G. M. Gershy-Damet, L. Kestens, G. van der Groen, and P. Piot. 1992. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS 6:447-451. [DOI] [PubMed] [Google Scholar]
- 42.Peeters, M., C. Honore, T. Huet, L. Bedjabaga, S. Ossari, P. Bussi, R. W. Cooper, and E. Delaporte. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625-630. [DOI] [PubMed] [Google Scholar]
- 43.Phillips-Conroy, J. E., C. J. Jolly, B. Petros, J. S. Allan, and R. C. Desrosiers. 1994. Sexual transmission of SIVagm in wild grivet monkeys. J. Med. Primatol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 44.Prince, A. M., B. Brotman, D. H. Lee, L. Andrus, J. Valinsky, and P. Marx. 2002. Lack of evidence for HIV type 1-related SIVcpz infection in captive and wild chimpanzees (Pan troglodytes verus) in West Africa. AIDS Res. Hum. Retrovir. 18:657-660. [DOI] [PubMed] [Google Scholar]
- 45.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, and R. H. Gray. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 342:921-929. [DOI] [PubMed] [Google Scholar]
- 46.Royce, R. A., A. Sena, W. Cates, Jr., and M. S. Cohen. 1997. Sexual transmission of HIV. N. Engl. J. Med. 336:1072-1078. [DOI] [PubMed] [Google Scholar]
- 47.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 48.Santiago, M. L., F. Bibollet-Ruche, E. Bailes, S. Kamenya, M. N. Muller, M. Lukasik, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2003. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J. Virol. 77:2233-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 50.Sharp, P. M., E. Bailes, R. R. Chaudhuri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of acquired immune deficiency syndrome viruses: where and when? Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teleki, G. 1989. Population status of wild chimpanzees (Pan troglodytes) and threats to survival. In L. A. Marquardt (ed.), Understanding chimpanzees. Harvard University Press, Cambridge, Mass.
- 52.ten Haaft, P., M. Cornelissen, J. Goudsmit, W. Koornstra, R. Dubbes, H. Niphuis, M. Peeters, C. Thiriart, C. Bruck, and J. L. Heeney. 1995. Virus load in chimpanzees infected with human immunodeficiency virus type 1: effect of pre-exposure vaccination. J. Gen. Virol. 76:1015-1020. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urnovitz, H. B., J. C. Sturge, T. D. Gottfried, and W. H. Murphy. 1999. Urine antibody tests: new insights into the dynamics of HIV-1 infection. Clin. Chem. 45:1602-1613. [PubMed] [Google Scholar]
- 55.Vanden Haesevelde, M. M., M. Peeters, G. Jannes, W. Janssens, G. van der Groen, P. M. Sharp, and E. Saman. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346-350. [DOI] [PubMed] [Google Scholar]
- 56.Vigilant, L., M. Hofreiter, H. Siedel, and C. Boesch. 2001. Paternity and relatedness in wild chimpanzee communities. Proc. Natl. Acad. Sci. USA 98:12890-12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watts, D. P. 1998. Coalitionary mate guarding by male chimpanzees at Ngogo, Kibale National Park, Uganda. Behav. Ecol. Sociobiol. 44:43-55. [Google Scholar]
- 58.Weiss, R. A. 2001. Natural and iatrogenic factors in human immunodeficiency virus transmission. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:947-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, R. A., and R. W. Wrangham. 1999. From Pan to pandemic. Nature 397:385-386. [DOI] [PubMed] [Google Scholar]
- 60.Whiten, A., J. Goodall, W. C. McGrew, T. Nishida, V. Reynolds, Y. Sugiyama, C. E. Tutin, R. W. Wrangham, and C. Boesch. 1999. Cultures in chimpanzees. Nature 399:682-685. [DOI] [PubMed] [Google Scholar]
- 61.Wrangham, R. W., C. A. Chapman, A. P. Clark-Arcadi, and G. Isabirye-Basuta. 1996. Social ecology of Kanyawara chimpanzees: implications for understanding the costs of great ape groups. In T. Nishida (ed.), Great ape societies. Cambridge University Press, Cambridge, United Kingdom.