Abstract
The replicase polyprotein of murine coronavirus is extensively processed by three proteinases, two papain-like proteinases (PLPs), termed PLP1 and PLP2, and a picornavirus 3C-like proteinase (3CLpro). Previously, we established a trans-cleavage assay and showed that PLP2 cleaves the replicase polyprotein between p210 and membrane protein 1 (MP1) (A. Kanjanahaluethai and S. C. Baker, J. Virol. 74:7911-7921, 2000). Here, we report the results of our studies identifying and characterizing this cleavage site. To determine the approximate position of the cleavage site, we expressed constructs that extended various distances upstream from the previously defined C-terminal end of MP1. We found that the construct extending from the putative PLP2 cleavage site at glycine 2840-alanine 2841 was most similar in size to the processed MP1 replicase product generated in a trans-cleavage assay. To determine which amino acids are critical for PLP2 recognition and processing, we generated 14 constructs with amino acid substitutions upstream and downstream of the putative cleavage site and assessed the effects of the mutations in the PLP2 trans-cleavage assay. We found that substitutions at phenylalanine 2835, glycine 2839, or glycine 2840 resulted in a reduction in cleavage of MP1. Finally, to unequivocally identify this cleavage site, we isolated radiolabeled MP1 protein and determined the position of [35S]methionine residues released by Edman degradation reaction. We found that the amino-terminal residue of MP1 corresponds to alanine 2841. Therefore, murine coronavirus PLP2 cleaves the replicase polyprotein between glycine 2840 and alanine 2841, and the critical determinants for PLP2 recognition and processing occupy the P6, P2, and P1 positions of the cleavage site. This study is the first report of the identification and characterization of a cleavage site recognized by murine coronavirus PLP2 activity.
Mouse hepatitis virus (MHV) is a member of the coronavirus family of positive-strand RNA, enveloped viruses (23). The name coronavirus refers to the fact that these viruses have spike glycoproteins that project from the surface of the virion, and these spikes give the appearance of a corona or crown-like structure when virus particles are visualized under an electron microscope (5). Coronaviruses generally cause species-specific respiratory and gastrointestinal infections in animals and birds. The well-documented human coronaviruses (HCoVs) 229E and OC43 are responsible for respiratory infections that are similar to the common cold (19). Recently, a new coronavirus has been implicated as a possible etiologic agent of severe acute respiratory syndrome (SARS) in humans (11, 35). Our understanding of the molecular mechanisms that regulate coronavirus replication will aid the development of antiviral agents and vaccines for this emerging infection.
Coronaviruses were recently merged with the arteriviruses into the order Nidovirales because the viruses in these two families exhibit a discontinuous mechanism of transcription to generate a “nested set” (nido is Latin for nest) of mRNAs (10, 22, 38). In addition, these viruses exhibit similarities in the translation and subsequent proteolytic processing of the polyprotein containing the viral RNA-dependent RNA polymerase, termed the replicase polyprotein (reviewed in reference 42). The Nidovirus replicase proteins have also been shown to generate novel cytoplasmic vesicles, termed double-membrane vesicles, where arterivirus and coronavirus RNA synthesis occurs (16, 32). Elucidation of the mechanisms used to process and assemble the membrane-associated viral replication complex is the focus of our current studies.
For MHV, the assembly and membrane association of the viral replication complex involve extensive proteolytic processing of an ∼800-kDa replicase polyprotein. The replicase polyprotein is encoded by gene 1, which encompassed the 5′-most 22 kb of the ∼31.2-kb genomic RNA (6, 24). Gene 1 contains two large open reading frames (ORFs), ORF1a and ORF1b, which are joined by a ribosomal frameshifting region to allow production of the replicase polyprotein (8). ORF1b encodes the core polymerase, helicase, and zinc finger domains (24). ORF1a encodes three proteinase domains, two papain-like proteinases (PLP1 and PLP2), and a picornavirus 3C-like proteinase domain (3CLpro); integral membrane protein 1 (MP1); and several additional products of unknown function (15, 24). The MHV replicase polyprotein is processed both in cis and in trans by the three ORF1a-encoded proteinases, and the PLP1 and 3CLpro enzymes and cleavage sites have been extensively characterized. PLP1 is responsible for the amino-terminal processing events to generate p28 (3, 4, 12, 20) and the adjacent protein, p65 (7). For the p28 cleavage site, PLP1 cleaves between Gly-248 and Val-249, and mutagenesis studies have shown that a basic residue (arginine or lysine) in the P5 position, arginine in the P2 position, and glycine in the P1 position are the critical determinants for PLP1 recognition and processing (12, 20). For the p65 cleavage site, in vitro studies have shown that PLP1-mediated processing occurs between Ala-832 and Gly-833. Mutagenesis studies have indicated that the critical determinants for the p65 cleavage site are P5-Arg, P1-Ala, and P1′-Gly and that PLP1 processing is retained even when the p65 and p28 cleavage sites are interchanged (7). MHV 3CLpro recognizes seven cleavage sites in the C-terminal region of ORF1a and four sites in ORF1b to ultimately yield 12 proteolytic products (16, 27-30, 33, 34, 36, 37). We showed that 3CLpro is part of the p150 intermediate generated during the cascade of proteolytic processing events (16, 21, 36). Pinon and coworkers showed that 3CLpro functions most efficiently when associated with cellular membranes (33), which is consistent with the idea that 3CLpro is part of a membrane-associated replication complex. Cleavage sites recognized by 3CLpro are generally well conserved, with P2-Leu, P1-Gln, and P1′-(Ser/Ala) being the consensus cleavage sites (42). The crystal structure of the porcine coronavirus transmissible gastroenteritis virus 3CLpro has recently been determined, revealing that the enzyme has a chymotrypsin fold with an extra alpha-helical domain (2). Regarding PLP2, we showed that PLP2 cleaves the central region of MHV ORF1a between the C-terminal region of p210 and the amino terminus of p150, which is further processed by 3CLpro to generate the amino-terminal product MP1 and several additional cleavage products (21) (see Fig. 1). We also showed that PLP2 can act in trans to cleave a substrate encoding the cleavage site to release p44 (MP1) (21). However, the cleavage site recognized by MHV PLP2 had not yet been determined.
FIG. 1.
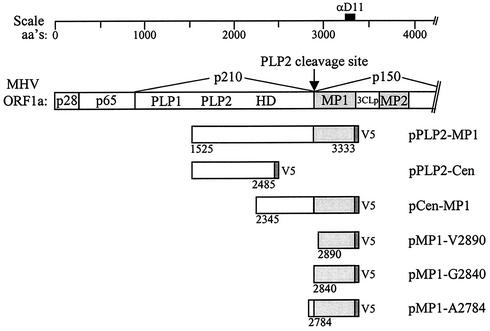
Schematic diagram of MHV ORF1a and PLP2-MP1 expression constructs. Papain-like proteinase domains 1 (PLP1) and 2 (PLP2), the hydrophobic domain (HD), membrane protein 1 (MP1), the poliovirus 3C-like proteinase domain (3CLpro), and membrane protein 2 (MP2), which are encoded by MHV ORF1a, are designated. Constructs generated for expression of the PLP2-MP1 region, the Cen-MP1 region, and the putative MP1 region alone are indicated. aa's, amino acids.
The aim of this study was to identify and characterize the MP1 cleavage site recognized by MHV PLP2. By expressing MHV ORF1a constructs extending for various lengths from the previously defined C-terminal end of MP1, we found a putative PLP2 cleavage site, glycine 2839-glycine 2840-alanine 2841, at the amino terminus of MP1. By using a trans-cleavage assay to assess the impact of specific substitutions in this region, we found that PLP2 recognizes phenylalanine 2835, glycine 2839, and glycine 2840 as the critical determinants for cleavage of the precursor polyprotein. Amino-terminal sequencing of radiolabeled MP1 revealed that PLP2 cleaves between glycine 2840 and alanine 2841. Thus, the critical determinants occupy the P6, P2, and P1 positions of the PLP2 cleavage site.
MATERIALS AND METHODS
Virus and cells.
MHV strain JHM-x was propagated as previously described (36). HeLa cells expressing the MHV receptor, HeLa-MHVR cells (14), were used for all infection and transfection experiments. The cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 0.5% penicillin-streptomycin, 2% glutamine, and 5 mM sodium HEPES (pH 7.4).
Generation of pMP1 expression constructs.
Constructs expressing the MP1 coding region were generated by using specific primers to amplify the designated region from the parental plasmid pCen-MP1 (21). See Table 1 for a list of the primers used to generate each clone. The region of interest was generated by PCR with LA-Taq polymerase in accordance with the manufacturer's (Clontech, Palo Alto, Calif.) instructions. The amplified region was then digested with restriction enzymes BamHI and XbaI and ligated into the corresponding sites in the pcDNA 3.1/V5-His expression vector (Stratagene, La Jolla, Calif.). The ligated DNA was transformed into XL-1 Blue competent cells in accordance with the manufacturer's (Stratagene) instructions, except that the bacteria were grown at 25°C.
TABLE 1.
Primers used for amplification or mutagenesis of MHV-JHM sequences
| Construct and primer | Oligonucleotide sequence (5′ to 3′)a | Nucleotidesb | First MHV amino acid or change(s) |
|---|---|---|---|
| MP1 expression | |||
| B242 | TTGGATCCAGAATGGTGCTAAGGGATGTGT | 8882-8897 | V2890 |
| B273 | TAGGATCCAAAATGGGCGCTGTGTTTAGTA | 8732-8747 | G2840 |
| B274 | TTGGATCCGGGATGGCTAATGTGGCTTGC | 8564-8578 | A2784 |
| B207 | AATCTAGAAATGATGTAGTAACAGAGGCA | 10195-10216 | 3′ primer |
| pCen-MP1c | |||
| B347 | CCTATTTTAACTACACCGGCCTCCCTTAAAGGG | 8699-8731 | F2835A |
| B349 | CTATTTTAACTACACCGTTCGCCCTTAAAGGG | 8700-8731 | S2836A |
| B351 | CTACACCGTTCTCCGCTAAAGGGGGCGCTGTG | 8709-8740 | L2837A |
| B353 | CTACACCGTTCTCCCTTGCAGGGGGCGCTGTGTTTAG | 8709-8745 | K2838A |
| B275 | GTTCTCCCTTAACGGGGGCGCTGTG | 8716-8740 | K2838N |
| B277 | GTTCTCCCTTAAAAACGGCGCTGTGTTTAG | 8716-8745 | G2839N |
| B301 | GTTCTCCCTTAAAGCGGGCGCTGTGTTTAG | 8716-8745 | G2839A |
| B303 | GTTCTCCCTTAAAGTGGGCGCTGTGTTTAG | 8716-8745 | G2839V |
| B279 | CTCCCTTAAAGGGAACGCTGTGTTTAG | 8719-8745 | G2840N |
| B305 | CCCTTAAAGGGGCCGCTGTGTTTAGTAG | 8721-8748 | G2840A |
| B307 | CCCTTAAAGGGGTCGCTGTGTTTAGTAG | 8721-8748 | G2840V |
| B281 | CCTTAAAGGGGGCAATGTGTTTAGTAGAG | 8722-8750 | A2841N |
| B283 | TAAAGGGGGCGCTAACTTTAGTAGAGTT | 8725-8752 | V2842N |
| pPLP2-MP1 | |||
| B384c | GGGCGCTATGTTTAGTAGAATGTTGCAATGG | 8731-8761 | V2842M, V2846M |
Transfection and vaccinia virus T7-mediated expression of MHV ORF1a products.
MHV ORF1a regions cloned into plasmids under T7 promoter control were transfected into HeLa-MHVR cells infected with recombinant vaccinia virus expressing the bacteriophage T7 polymerase as previously described (13, 21). Newly synthesized proteins were metabolically labeled with 50 μCi of Tran35S-label (ICN, Costa Mesa, Calif.) per ml from 4.5 to 9.5 h postinfection. Cells were washed in phosphate-buffered saline, and cell lysates were prepared by scraping the cells in lysis buffer A (4% sodium dodecyl sulfate [SDS], 3% dithiothreitol, 40% glycerol, 0.065 M Tris, pH 6.8) (36). The cell lysates were diluted in 1.0 ml of radioimmunoprecipitation assay buffer and subjected to immunoprecipitation with the designated antibody and protein A-Sepharose beads (Amersham Bioscience, Piscataway, N.J.). The antibodies used in this study included rabbit polyclonal anti-D11 serum (21) and mouse anti-V5 monoclonal antibody (Invitrogen, Carlsbad, Calif.).
Site-directed mutagenesis of pCen-MP1 and pPLP2-MP1 expression constructs.
Plasmid DNAs pCen-MP1 and pPLP2-MP1 were subjected to site-directed mutagenesis with synthetic oligonucleotides with mismatches encoding specific nucleotide changes, as shown in Table 1. QuikChange site-directed mutagenesis was performed in accordance with the manufacturer's (Stratagene, La Jolla, Calif.) instructions and as previously described (21). Mutations were confirmed by DNA sequencing.
Protein microsequencing.
Radiolabeled lysates were generated from vTF7.3-infected, pPLP2-MP1v2842,46m-transfected cells as described above. The V5 epitope-tagged MP1 protein was immunoprecipitated with anti-V5 antibody and separated by electrophoresis on an SDS-7.5% polyacrylamide gel. Following electrophoresis, the MP1 cleavage product was transferred to ProBlot polyvinylidene difluoride (PVDF) membrane (Applied Biosystems) at 250 mA for 2.5 h at 4°C in buffer containing 10 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid], pH 11.0, and 10% methanol. After transfer, the PVDF membrane was air dried and exposed to Kodak X-ray film at −70°C. The 44-kDa MP1 protein was identified by autoradiography and excised from the membrane. The excised sample was subjected to an Applied Biosystems sequencer for partial N-terminal amino acid sequence analysis based on the Edman degradation reaction (31). The radioactivity released from each cycle was quantitated by scintillation counting.
RESULTS
Characterization of the cleavage site recognized by MHV PLP2.
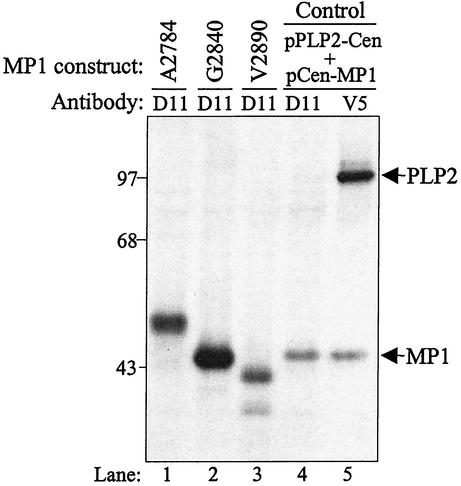
The MHV replicase polyprotein is processed by three distinct proteinases, PLP1, PLP2, and 3CLpro. Cleavage sites recognized by PLP1 and 3CLpro have been identified and characterized (reviewed in reference 42), but the site recognized by PLP2 was not known. We showed that MHV PLP2 cleaves the replicase polyprotein to separate the p210 and MP1 (p44) products (21). To further identify and characterize this cleavage site, we scanned the replicase polyprotein coding sequence between amino acids 2780 and 2895 and identified three possible cleavage sites for papain-like proteinase recognition and processing. Characterization of cleavage sites recognized by other viral papain-like proteinases suggested that the cleavage site was likely to have a series of small, uncharged amino acids (glycine, alanine or valine), sometimes flanked by a charged residue. Thus, the putative PLP2 cleavage sites we identified were as follows: site 1, alanine 2783-alanine 2784; site 2, glycine 2839-glycine 2840-alanine 2841; site 3, glycine 2889-valine-2890. To determine which of these sites is the probable amino terminus of MP1, we generated expression constructs that began at either alanine 2784, glycine 2840, or valine 2890 and extended to the carboxy-terminal region of p44, with an added V5 epitope tag (Fig. 1). The encoded proteins were expressed via vaccinia virus T7-mediated expression, and radiolabeled products were immunoprecipitated with anti-D11 or anti-V5 antibody. The products were then analyzed by electrophoresis on an SDS-polyacrylamide gel (Fig. 2). The protein products generated by expression of the pPLP2-Cen and pCen-MP1 cotransfection served as a control for the size of the MP1 cleavage product (p44). As seen in lane 2, the product generated from the pMP1-G2840 construct migrated with an apparent molecular mass of 44 kDa, very similar to that of the control (lanes 4 and 5). The pMP1-A2784 product migrated more slowly than p44, and the pMP1-V2890 product migrated more quickly than p44. Thus, the pMP1-G2840 construct likely encodes the entire p44 sequence, and we hypothesized that MP1 extends from alanine 2841 to the 3CLpro cleavage site at glutamine 3336. However, we noted that the calculated molecular mass of this 496-amino-acid region is 56.5 kDa, much larger than the observed molecular mass of 44 kDa determined by SDS-polyacrylamide gel electrophoresis analysis. This aberrant migration of MP1 is likely due to the compact nature of this hydrophobic protein (24).
FIG. 2.
Analysis of pMP1 expression constructs. HeLa-MHVR cells were infected with vTF7.3 and transfected with plasmid DNAs encoding MHV ORF1a regions. Newly synthesized proteins were labeled with Trans35S-label from 4.5 to 9.5 h postinfection, and lysates were prepared and subjected to immunoprecipitation with anti-D11 or anti-V5 antibody. Immunoprecipitated proteins were analyzed by electrophoresis on 5 to 10% polyacrylamide gels, processed, and subjected to autoradiography. Molecular size markers (in kilodaltons) are indicated to the left.
Identification of the determinants critical for PLP2 recognition and processing.
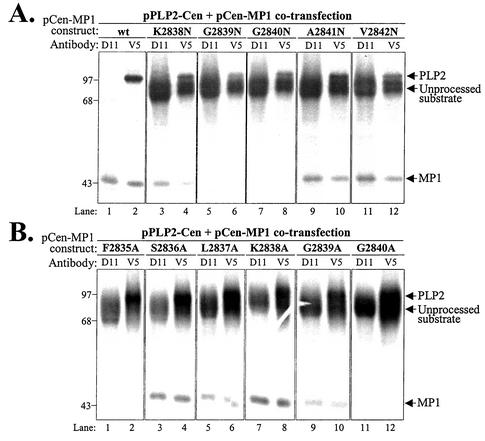
To determine which amino acid residues are critical for recognition and processing by PLP2, we performed site-directed mutagenesis of the sequences encoding ORF1a amino acid residues 2835 to 2842 in the pCen-MP1 substrate. The effects of the mutations on PLP2 recognition and processing were assessed with a trans-cleavage assay (21). The trans-cleavage assay was performed by cotransfection of pPLP2-Cen (proteinase) and either wild-type or mutant pCen-MP1 (substrate) DNA into vTF7.3-infected cells. Newly synthesized proteins were radiolabeled with Trans35S-label for 5 h, cells were lysed, and the lysates were subjected to immunoprecipitation with anti-D11 serum, which recognizes p44 and the uncleaved substrate, and anti-V5 antibody, which recognizes the epitope-tagged proteinase, the substrate, and the cleavage product, p44. Immunoprecipitated products were analyzed by SDS-polyacrylamide gel electrophoresis, and representative results are shown in Fig. 3. The wild-type substrate was processed efficiently by PLP2, and p44 was detected by anti-D11 and anti-V5 antibodies (Fig. 3A, lanes 1 and 2). In contrast, replacement of either glycine 2839 or glycine 2840 with asparagine (Fig. 3A, lanes 5 to 8) or alanine (Fig. 3B, lanes 9 to 12) resulted in reduction or complete loss of processing of the substrate. Replacement of these amino acids with valine gave results very similar to the results of the asparagine substitutions, with complete loss of processing when either glycine 2840 or glycine 2839 was changed to valine (data not shown). Interestingly, we also detected an inhibition of processing when phenylalanine 2835 was changed to alanine (Fig. 3B, lanes 1 and 2). These results indicate that a site upstream of the cleavage site is also important for recognition and processing by PLP2. In contrast, replacement of amino acids 2836, 2837, 2838, 2841, and 2842 had little or no effect on PLP2 recognition and processing (Fig. 3A, lanes 3, 4, and 9 to 12, and B, lanes 3 to 8). Overall, these results indicate that phenylalanine 2835, glycine 2839, and glycine 2840 are critical for PLP2 recognition and processing.
FIG. 3.
Analysis of trans-acting PLP2 activity on wild-type and mutant forms of the MP1 cleavage site. Protein products detected after cotransfection of DNAs encoding the MHV PLP2-Cen region and the Cen-MP1 wild-type (wt) or mutant substrate. The specific amino acid substitution present in each mutant substrate is indicated above the gel, with the change to asparagine shown in panel A and the change to alanine shown in panel B. Products were analyzed as described in the legend to Fig. 2. Molecular size markers (in kilodaltons) are indicated to the left.
Identification of the cleavage site recognized by MHV PLP2.
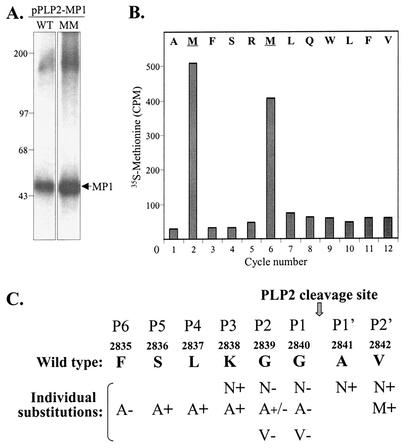
To unequivocally determine the cleavage site recognized by MHV PLP2, we wanted to identify the amino-terminal residues of MP1. However, the purification and subsequent amino acid analysis of the very hydrophobic MP1 protein proved to be problematic. By using lysates prepared from pPLP2-MP1-transfected cells, we were unable to solubilize and affinity purify sufficient quantities of MP1 for direct amino acid microsequencing. Another approach, which we previously used to identify the p28 cleavage site, is the microsequencing of radiolabeled in vitro-translated protein (12). For this approach, we performed in vitro transcription-translation with pPLP2-MP1 and analyzed the translation products. Surprisingly, we found that the PLP2-MP1 polyprotein is not processed efficiently in vitro, even in the presence of canine microsomal membranes (D. Jukneliene and S. C. Baker, unpublished data). Therefore, we decided to affinity purify radiolabeled protein from pPLP2-MP1-transfected cells and to identify the position of the radiolabeled amino acid by Edman degradation and scintillation counting of the product generated from each cycle. To facilitate this analysis, we generated a construct encoding valine-to-methionine substitutions at amino acid positions 2842 and 2846 (designated pPLP2-MP1v2842,46m). We had already shown that a substitution at valine 2842 did not affect proteolytic process (Fig. 3A, lanes 11 and 12), and we knew that MHV strain A59 contains a methionine at position 2846. Therefore, these substitutions were likely to be tolerated during processing of the polyprotein. We verified that the PLP2-MP1 polyprotein with the methionine substitutions was expressed and processed as efficiently as the wild-type polyprotein (Fig. 4A). The radiolabeled MP1 protein was transferred to PVDF membrane and subjected to Edman degradation microsequencing as described in Materials and Methods. Methionine residues were released in cycles 2 and 6 of the Edman degradation reaction (Fig. 4B). By aligning the methionine residues with the known amino acid sequence in this region, we determined that PLP2 processing takes place between glycine 2840 and alanine 2841, with alanine 2841 as the amino-terminal residue of MP1. A summary of the cleavage site analysis and the results of the mutagenesis study is shown in Fig. 4C. Thus, the critical determinants identified in the mutagenesis study occupy positions P6, P2, and P1 of the MP1 cleavage site recognized by PLP2.
FIG. 4.
N-terminal sequence analysis of the MHV MP1 cleavage product. (A) Products immunoprecipitated from cells transfected with either pPLP2-MP1 (WT) or pPLP2-MP1v2842,46m (MM) were generated and analyzed as described in the legend to Fig. 2. Molecular size markers (in kilodaltons) are indicated to the left. (B) Detection of [35S]methionine during Edman degradation cycle sequencing of the MP1 protein isolated from pPLP2-MP1v2842,46m-transfected cells. (C) Summary of mutagenesis data characterizing the cleavage site recognized by MHV PLP2. The results of each substitution are indicated, with + meaning PLP2 recognition and processing of the substrate, +/− meaning partial processing, and − meaning that no processing was detected.
DISCUSSION
In this study, we identified and characterized the cleavage site recognized by MHV PLP2 to generate replicase product MP1 (p44). MHV PLP2 cleaves between glycine 2840 and alanine 2841, with phenylalanine 2835 (P6), glycine 2839 (P2), and glycine 2840 (P1) acting as critical determinants for efficient PLP2 recognition and processing. This is the first identification of a cleavage site recognized by MHV PLP2 activity.
Site-directed mutagenesis studies of the MHV PLP2 cleavage sites revealed that the cleavage site is similar to, but distinct from, cleavage sites recognized by other viral papain-like proteinases (Table 2). Viral papain-like proteinases generally recognize a region of small, uncharged amino acids, such as glycine, alanine, or valine. For MHV PLP1, we and others showed that, in addition to a glycine in the P1 position, the arginine in the P2 position, and a basic amino acid (either arginine or lysine) in the P5 position are important for PLP1 recognition and processing (12, 20). For MHV PLP2, we found that glycine 2840 in the P1 position of the cleavage site is essential for processing. Even conservative changes to alanine (Fig. 3B, lanes 11 and 12) or valine (data not shown), resulted in complete loss of processing at the cleavage site. Our results are in agreement with other studies of coronavirus PLP cleavage sites, which showed that glycine is most commonly found in the P1 position and that the glycine residue is a critical determinant for efficient recognition and processing (7, 12, 17, 18, 20, 25, 26, 43). Our studies also indicated that glycine 2839, in the P2 position, is important for PLP2 recognition and processing. A conservative change to alanine results in a reduction in processing (Fig. 3B, lanes 9 and 10), but a more drastic change to asparagine results in complete loss of processing by PLP2 (Fig. 3A, lanes 5 and 6). Thus, the P2 position is also a critical determinant for PLP2 recognition and processing. Finally, we noted that changing the phenylalanine in the P6 position to alanine also resulted in loss of cleavage at the PLP2 cleavage site. This indicates that an upstream sequence can also play a role in the substrate-proteinase interaction required for efficient proteolytic processing.
TABLE 2.
Comparison of cleavage sites recognized by coronavirus PLP1 and PLP2 activitiesd
| Cleavage products | Amino acid at cleavage site position:
|
Reference(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P6 | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | |||
| Cleaved by PLP1 | ↓
|
|||||||||
| MHV p28/p65 | L | K/Ra | G | Y | Ra | Ga | V | K | 12, 20 | |
| MHV p65/p210 | W | Ra | F | P | C | Aa | Ga | K | 7 | |
| HCoV p9/p87 | G | Ka | R | G | G | Ga | N | V | 17 | |
| Cleaved by PLP1 and PLP2 | ||||||||||
| HCoV p87/p210b | F | T | K | A | A | G | G | K | 43 | |
| Cleaved by PLP2 | ||||||||||
| IBV p87/p195c | V | V | C | K | A | Ga | G | K | 25 | |
| IBV p195/p41c | V | E | K | K | Aa | Ga | G | I | 26 | |
| MHV p210/p44 | Fa | S | L | K | Ga | Ga | A | V | This paper | |
A critical determinant for PLP processing as determined by mutagenesis studies.
In vitro transcription-translation studies indicate that both PLP1 and PLP2 can cleave at this site. Critical determinants have not been identified.
A revised alignment study indicates that the domain previously referred to as IBV PLPD has an inactive PLP1 domain and an active PLP2 domain (43).
The arrow indicates where the polyprotein is cleaved.
A computer-generated sequence alignment study indicates that all coronaviruses likely encode an active PLP2 domain and a corresponding cleavage site in the central region of ORF1a (43). This revised computer alignment of the coronavirus PLP domains suggested that the PLP1 domain of the avian coronavirus infectious bronchitis virus (IBV) is inactive, and that the IBV PLP2 domain mediates cleavages between ORF1a p87/p195 and p195/p41 (25, 26, 43). Therefore, in Table 2, we indicate that the IBV ORF1a cleavages are mediated by PLP2 activity. Interestingly, it was recently shown that the HCoV PLP1 and PLP2 domains are both able to recognize and process the ORF1a p87/p210 cleavage site (43). This study assessed the ability of each proteinase domain to cleave the substrate in an in vitro transcription-translation reaction. However, it is not clear if the dual recognition and processing activities detected in vitro accurately reflect the processing pathway in virus-infected cells.
Overall, the studies with IBV, HCoV, and MHV all provide evidence that PLP2 plays an important role in the processing of the replicase polyprotein. Future studies will be directed toward determining the role of PLP2 processing in coronavirus replication by making mutations that introduce amino acid substitutions into the recently developed full-length clones for IBV (9), HCoV (39), transmissible gastroenteritis virus (1, 40), and MHV (41). Analysis of PLP2 cleavage site mutations in the context of the replicating RNA will help us determine the role of PLP2-mediated proteolytic processing in the replication of coronavirus. In addition, increased understanding of the interaction of proteinases and their cleavage sites has important implications for the development of antiviral drugs, such as proteinase inhibitors, that may be important for controlling emerging coronavirus infections, such as SARS.
Acknowledgments
We thank J. Myron Crawford and the staff at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for helpful advice and for the microsequencing of MP1. We thank Shi-Kang Zhang at Loyola University Chicago for his dedication and for all the DNA sequencing work.
This work was supported by Public Health Service research grant AI 45798 to S.C.B.
REFERENCES
- 1.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, S. C., C. K. Shieh, L. H. Soe, M. F. Chang, D. M. Vannier, and M. M. Lai. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 63:3693-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 67:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry, D. M., J. G. Cruickshank, H. P. Chu, and R. J. H. Wells. 1964. The structure of infectious bronchitis virus. Virology 23:403-407. [DOI] [PubMed] [Google Scholar]
- 6.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla, P. J., S. A. Hughes, and S. R. Weiss. 1997. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 71:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley, I., M. E. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2003. Update: outbreak of severe acute respiratory syndrome—worldwide, 2003. Morb. Mortal. Wkly. Rep. 52:241-246. [PubMed] [Google Scholar]
- 12.Dong, S., and S. C. Baker. 1994. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology 204:541-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher, T. M. 1996. Murine coronavirus membrane fusion is blocked by modification of thiols buried within the spike protein. J. Virol. 70:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold, J., A. E. Gorbalenya, V. Thiel, B. Schelle, and S. G. Siddell. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 72:910-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herold, J., S. G. Siddell, and A. E. Gorbalenya. 1999. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 274:14918-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, K. V. 2001. Coronaviruses, p. 1187-1203. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 20.Hughes, S. A., P. J. Bonilla, and S. R. Weiss. 1995. Identification of the murine coronavirus p28 cleavage site. J. Virol. 69:809-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 74:7911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Lee, H. J., C. K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. La Monica, J. Tuler, A. Bagdzhadzhyan, and M. M. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, K. P., and D. X. Liu. 1998. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology 245:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, K. P., L. F. Ng, and D. X. Liu. 2000. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 74:1674-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, X., Y. Lu, and M. R. Denison. 1996. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, Y., and M. R. Denison. 1997. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology 230:335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, Y., X. Lu, and M. R. Denison. 1995. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 69:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 32.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinon, J. D., R. R. Mayreddy, J. D. Turner, F. S. Khan, P. J. Bonilla, and S. R. Weiss. 1997. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology 230:309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinon, J. D., H. Teng, and S. R. Weiss. 1999. Further requirements for cleavage by the murine coronavirus 3C-like proteinase: identification of a cleavage site within ORF1b. Virology 263:471-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 31 March 2003, posting date. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. [Online.] www.nejm.org [DOI] [PubMed]
- 36.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 39.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 40.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 43.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]