Abstract
The XC cell line undergoes extensive syncytium formation after infection with ecotropic murine leukemia viruses (MLVs) and is frequently used to titrate these viruses. This cell line is unique in its response to the ecotropic MLV envelope protein (Env) in that it undergoes syncytium formation with cells expressing Env protein containing R peptide (R+ Env), which is known to suppress the fusogenic potential of the Env protein in other susceptible cells. To analyze the ecotropic receptor, CAT1, in XC cells, a mouse CAT1 tagged with the influenza virus hemagglutinin epitope (mCAT1-HA)-expressing retroviral vector was inoculated into XC and NIH 3T3 cells. The molecular size of the mCAT1-HA protein expressed in XC cells was smaller than that in NIH 3T3 cells due to altered N glycosylation in XC cells. Treatment of XC cells with tunicamycin significantly suppressed the formation of XC cell syncytia induced by the R+ Env protein but not that induced by the R− Env protein. This result indicates that N glycosylation is required for XC cell-specific syncytium formation by the R+ Env protein. The R+ Env protein induced syncytia in XC cells expressing a mutant mCAT1 lacking both of two N glycosylation sites, and tunicamycin treatment suppressed syncytium formation by R+ Env in those cells. This suggests that N glycosylation of a molecule(s) other than the receptor is required for the induction of XC cell syncytia by the R+ Env protein.
Ecotropic murine leukemia viruses (MLVs) enter host cells mediated by fusion between the viral envelope and the cellular membrane following the recognition of the viral envelope glycoprotein (Env) by the cellular receptor, CAT1 (1). The Env protein of the MLVs is synthesized as a precursor polyprotein and is cleaved to surface (SU) and transmembrane (TM) subunits in infected cells by a cellular protease (25). The C-terminal 16 amino acids of the TM protein (R peptide) are further cleaved off during virion maturation by the viral protease (5, 7). The R peptide-truncated Env protein (R− Env) has membrane fusion activity to induce syncytium formation when it is expressed in susceptible cells, but the R peptide-containing Env protein (R+ Env) does not. This result indicates that the R peptide inhibits membrane fusion (14, 23, 24). Because cells expressing the R− Env protein will form syncytia and die, the selective truncation of the R peptide may be an adaptive mechanism that allows ecotropic MLVs to delay membrane fusion until the viruses leave the cells.
Rat XC cells, which are widely used to measure ecotropic MLV titers (12), are more susceptible to ecotropic MLV infection than other rat cells, such as F10 cells (13, 28). In addition to the R− Env protein, R+ Env can induce syncytia in XC cells but not in other susceptible cells (9). Recently, it was found that some CAT1 cDNA clones isolated from XC cells have an amino acid substitution of aspartic acid for asparagine at one of two N-linked glycosylation sites, and the XC cell receptor was designated xcCAT1 (13). CHO cells expressing xcCAT1 were more susceptible to Moloney MLV (Mo-MLV) vector transduction than those expressing the wild-type rat CAT1 (rCAT1). In addition, F10 cells treated with tunicamycin, an inhibitor of protein N glycosylation, were more susceptible to Mo-MLV vector transduction than untreated F10 cells. These results indicate that the N glycosylation of rCAT1 inhibits Mo-MLV vector infection. In CHO cells expressing xcCAT1, however, the R+ Env protein does not induce syncytia, but R− Env does. This result indicates that the amino acid substitution detected in xcCAT1 is not sufficient for XC cell-specific syncytium formation by the R+ Env protein and that another mechanism is necessary.
To analyze the ecotropic MLV receptor in XC cells, a retroviral vector encoding a mouse CAT1 (mCAT1) protein C-terminally tagged with the influenza virus hemagglutinin (HA) epitope was inoculated into XC and NIH 3T3 cells. The molecular size of the HA-tagged mCAT1 (mCAT1-HA) expressed in XC cells was smaller than that in NIH 3T3 cells due to altered N glycosylation in XC cells. Tunicamycin treatment of XC cells significantly inhibited XC cell-specific syncytium formation by the R+ Env protein but not that by the R− Env protein. This result indicates that N glycosylation in XC cells is required for XC cell-specific syncytium formation by the R+ Env protein. N glycosylation inhibits Mo-MLV infection in rat and hamster cells (15, 29, 32) but is required for the formation of XC cell syncytia by the R+ Env protein.
MATERIALS AND METHODS
Expression plasmids.
cDNA clones of mCAT1 (1) and rCAT1 (28) were kindly provided by J. M. Cunningham (Harvard Medical School) and by R. Watanabe (Soka University), respectively. Two cDNA clones of CAT1 have been isolated from XC cells in our laboratory (13). One of them had an amino acid substitution at an N-linked glycosylation site compared to rCAT1, and we designated it xcCAT1. A retrovirus vector DNA (M3) encoding a mutant mCAT1 lacking both of two N glycosylation sites was kindly provided by D. Kabat (Oregon Health Science University) (31). A glucosidase I expression plasmid was kindly provided by E. Bause (Rheinische Friedrich-Wilhelms-Universitat) (10).
Expression plasmids of mCAT1, rCAT1, and xcCAT1 C-terminally tagged with the influenza virus HA epitope (mCAT1-HA, rCAT1-HA, and xcCAT1-HA) were constructed by PCR using the following primers: 5′-TTACTGCAGACAGATTTGCTCAGCGCGATG-3′ and 5′-TCATGCGTAATCCGGAACATCGTACGGGTATTTGCACTGGTCCAAGTTGCT-3′. The latter (antisense) primer contains the HA epitope sequence. The PCR product was ligated into the pTargeT expression vector (Plomega) by TA cloning. The PCR product of mCAT1-HA was also ligated to pMXpuro retrovirus vector DNA (19).
The expression plasmids encoding R− Env of Mo-MLV and Friend MLV (Fr-MLV) (18) were constructed by PCR using a mutant primer that replaces the codon for valine at amino acid position 617 with a termination codon. The PCR product was ligated into the pTargeT expression vector by TA cloning.
Cells, retrovirus vectors, and N-glucan processing inhibitors.
NIH 3T3, F10, XC, 293T, BOSC23 (22), and TELCeB6 (3) cells were cultured in Dulbecco's modified Eagle's medium (Sigma) at 37°C in 5% CO2. CHO cells were cultured in Ham's F-12 nutrient mixture medium (Gibco BRL). The culture media were all supplemented with 8% fetal bovine serum (HyClone).
BOSC23 ecotropic packaging cells (22) were transfected with mCAT1-HA or mCAT1-M3 mutant-expressing retroviral vector DNA. Culture supernatant of the transfected BOSC23 cells was inoculated into XC and NIH 3T3 cells. Several drug-resistant cell clones were isolated, and mCAT1-HA-expressing cell clones were selected by Western immunoblotting using the anti-HA antibody. mCAT1-M3 mutant-expressing cell clones were selected by Northern hybridization.
The mCAT1-HA-expressing XC and NIH 3T3 cells were treated with tunicamycin (Sigma), 1-deoxynojirimycin (dNM) (Sigma), or 1-deoxymannojirimycin (dMM) (Sigma).
Syncytium formation assay.
The expression plasmids encoding the wild-type Env protein (R+ Env) or Env lacking the R peptide (R− Env) were transfected into 293T cells using the TransIT-LT1 reagent (Mirus). Two days after transfection, aliquots of the target cell culture were inoculated onto the transfected 293T cells. These cells were fixed and stained with 1% methylene blue in methanol 24 h later, and the number of syncytia per microscopic field was determined.
Transduction assay.
The ecotropic retrovirus vector was generated by stable transfection of TELCeB6 cells, which express the Mo-MLV gag and pol genes and the retroviral vector genome containing Escherichia coli lacZ (3), with the wild-type Mo-MLV or Fr-MLV Env expression plasmid. Target cells were exposed to the retrovirus vector for 24 h in the presence of Polybrene (4 μg/ml). After being cultured for an additional 24 h in fresh medium, the cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and the blue cells were counted to estimate the transduction titer.
Western immunoblotting.
Cell lysates were prepared, and aliquots containing 60 μg of protein were subjected to sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis and subsequent Western immunoblotting using the mouse anti-HA epitope monoclonal antibody (Covance) and horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (Bio-Rad).
Immunofluorescence assay.
XC cells expressing mCAT1-HA were fixed with cold methanol. The cells were treated with the mouse anti-HA antibody and then with an anti-mouse IgG conjugated with indocarbocyanine (Cy3) (Sigma). They were observed by immunofluorescence microscopy.
RESULTS
Altered glycosylation in XC cells.
To analyze the ecotropic MLV receptor protein in XC cells, a retroviral vector DNA encoding the mCAT1-HA protein was constructed and transfected to BOSC23 ecotropic packaging cells (22). The retrovirus vector also contains the puromycin resistance gene (19). Culture supernatant of the transfected BOSC23 cells was inoculated into XC and NIH 3T3 cells. Several puromycin-resistant cell clones were isolated, and cell clones expressing mCAT1-HA were selected by Western immunoblotting using the anti-HA antibody. CHO cells expressing the mCAT1-HA were as susceptible as those expressing the untagged mCAT1 to Mo-MLV vector carrying the lacZ reporter gene. The transduction efficiency of Mo-MLV vector in the mCAT1-HA-expressing XC cells was comparable to that in the parental cells. The R+ Env protein induced syncytia in XC cells expressing mCAT1-HA as efficiently as in the normal XC cells. This result indicates that mCAT1-HA does not inhibit XC cell-specific syncytium formation by the R+ Env protein and that XC cell syncytia are induced through interaction between the R+ Env and mCAT1-HA proteins.
In order to determine the molecular sizes of the mCAT1-HA proteins in XC and NIH 3T3 cells, Western immunoblotting was performed using the anti-HA antibody. The mCAT1-HA protein expressed in XC and NIH 3T3 cells showed high heterogeneity (Fig. 1). At the shorter exposure time, several different bands were detected. The molecular size of mCAT1-HA in XC cells was smaller than that in NIH 3T3 cells. Tunicamycin treatment reduced the molecular sizes of mCAT1-HA proteins in XC and NIH 3T3 cells. The molecular sizes of the mCAT1-HA proteins expressed in tunicamycin-treated XC and NIH 3T3 cells were similar, and they had much lower heterogeneity than in untreated cells. The same result was observed in other puromycin-resistant cell clones expressing the mCAT1-HA protein.
FIG. 1.
Altered N glycosylation in XC cells. Normal XC and NIH 3T3 cells and mCAT1-HA-expressing XC and NIH 3T3 cells were treated with 100 ng of tunicamycin/ml for 24 h. Cell lysates were prepared from the cells. Western immunoblotting was performed using the anti-HA antibody.
A 40-kDa molecule which was reacted with the anti-HA antibody was not detected in mCAT1-HA-expressing NIH 3T3 cells and tunicamnycin-treated mCAT1-HA-expressing XC and NIH 3T3 cells, but it was detected in other cells. It is unknown why tunicamycin treatment of mCAT1-HA-expressing XC cells induced the disappearance of the 40-kDa molecule but that of control XC cells did not.
The molecular sizes of the mCAT1-HA proteins in CHO and 293T cells were similar to those in NIH 3T3 cells (data not shown). This result indicates that the mCAT1-HA protein is modified by N glycosylation in XC cells but that N-linked glycosylation of mCAT1-HA in XC cells is different from that in NIH 3T3, CHO, and 293T cells.
N glycosylation is responsible for XC cell-specific syncytium formation by R+ Env protein.
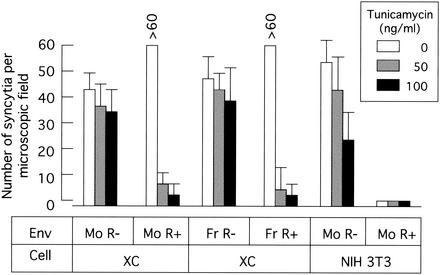
To know whether N glycosylation in XC cells is involved in XC cell-specific syncytium formation by the R+ Env protein, the fusion activity of the R+ Env protein was measured in tunicamycin-treated XC cells. XC cells were pretreated with tunicamycin (50 or 100 ng/ml) for 24 h. The tunicamycin-treated or untreated XC cells were plated onto 293T cells expressing the R+ Env protein of Mo-MLV or Fr-MLV and cultured for 24 h in the absence of tunicamycin to exclude the effect of tunicamycin on expression and N glycosylation of the Env protein in 293T cells. Tunicamycin treatment significantly and reproducibly inhibited the fusion activity of the R+ Env proteins of Mo-MLV and Fr-MLV in XC cells (Fig. 2). However, tunicamycin treatment slightly inhibited the fusion activity of the R− Env protein in XC and NIH 3T3 cells. This result indicates that N glycosylation in XC cells is required for XC cell-specific syncytium formation by the R+ Env protein.
FIG. 2.
Tunicamycin inhibits XC cell-specific syncytium formation by the R+ Env protein. 293T cells were transfected with the plasmid encoding the R− or R+ Env protein of Mo-MLV (Mo) or Fr-MLV (Fr). XC and NIH 3T3 cells were pretreated with tunicamycin (50 or 100 ng/ml) for 24 h. The transfected 293T cells were added to the pretreated XC and NIH 3T3 cells and cultured for 24 h in the absence of tunicamycin. The cells were stained with 1% methylene blue, and the number of syncytia per microscopic field was determined. The error bars indicate standard deviations.
Tunicamycin treatment of NIH 3T3 (Fig. 2) and F10 (data not shown) cells did not make the R+ Env protein fusogenic. This result supports the previous finding that CHO cells expressing a glycosylation-defective variant of the ecotropic MLV receptor isolated from XC cells (xcCAT1) do not respond to syncytium induction by the R+ Env protein (13).
Tunicamycin treatment did not affect cellular distribution of mCAT1-HA protein.
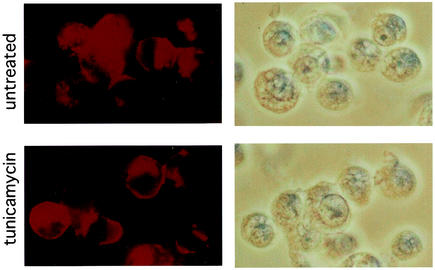
To determine the effect of tunicamycin treatment on cellular distribution of the mCAT1-HA protein, immunofluorescence analysis of mCAT1-HA-expressing XC cells was performed using the anti-HA antibody. XC cells expressing mCAT1-HA were treated with 100 ng of tunicamycin/ml for 24 h. The XC cells were fixed with methanol and blotted with the mouse anti-HA antibody and then with a Cy3-conjugated anti-mouse IgG antibody. Microscopic fields for images were selected randomly. Fluorescence images were obtained by 10-s exposure to compare signal strengths in untreated and treated cells. The mCAT1-HA protein was detected on the cell surfaces and in the cytoplasm of untreated and tunicamycin-treated XC cells (Fig. 3). No effect of tunicamycin treatment on the cellular distribution of the mCAT1-HA protein was detected.
FIG. 3.
Cellular distributions of mCAT1-HA protein in untreated and tunicamycin-treated XC cells. XC cells expressing the mCAT1-HA protein were treated with 100 ng of tunicamycin/ml for 24 h. Untreated (top) and treated (bottom) XC cells were fixed with cold methanol. The cells were blotted with the anti-HA antibody and then with a Cy3-conjugated anti-mouse IgG antibody. Microscopic fields to be imaged were selected randomly. Fluorescence images were obtained by 10-s exposures to compare signal strengths in untreated and treated cells. Fluorescence and transmitted-light images of the same fields are shown on the left and right, respectively.
Effects of dNM and dMM on syncytium formation by the Env protein.
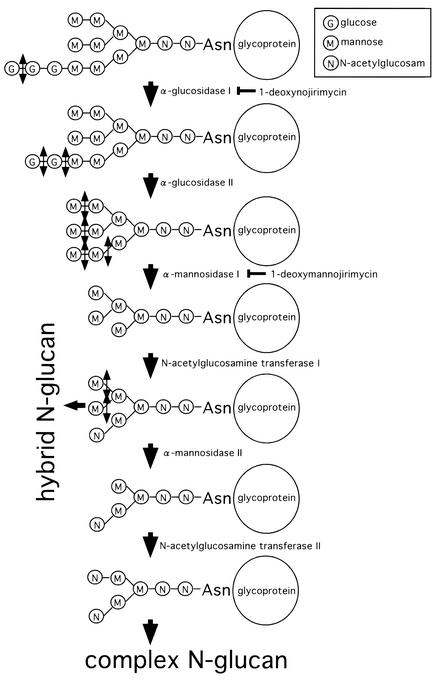
To know the relationship between N-glucan structure and membrane fusion by the Env protein, the effects of N-glucan processing inhibitors, dNM and dMM, on syncytium formation by the Env proteins and on susceptibility to ecotropic MLV vector infection were analyzed. The compounds dNM and dMM inhibit glucosidase I and mannosidase I, respectively (6, 17, 30) (Fig. 4). XC cells were treated with dNM or dMM (2 mM) for 24 h and added to Env protein-expressing 293T cells. As a result, it was found that syncytium formation by the R+ and R− Env proteins in XC cells was not affected by these compounds (data not shown).
FIG. 4.
N-Glucan processing pathway. dNM and dMM inhibit glucosidase I and mannosidase I, respectively.
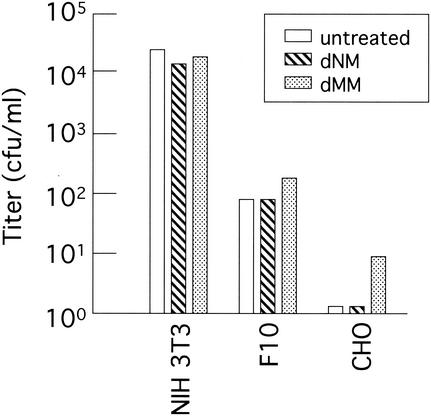
The effects of these inhibitors on the transduction efficiency of the ecotropic MLV vector were analyzed. Target cells were pretreated with dNM or dMM (2 mM) for 24 h. The cells were washed with phosphate-buffered saline to remove the compound and inoculated with the culture supernatant of TELCeL6 cells transfected with the Mo-MLV Env expression plasmid. Treatment of NIH 3T3, F10, and CHO cells with dNM did not affect the transduction titer of the Mo-MLV vector (Fig. 5). The treatment of CHO cells, but not of NIH 3T3 and F10 cells, with dMM, however, increased the transduction titer. This experiment was performed three times, and similar results were observed.
FIG. 5.
Effects of dNM and dMM on Mo-MLV vector transduction. NIH 3T3, F10, and CHO cells were pretreated with dNM or dMM (2 mM) for 24 h. The Mo-MLV vector carrying the lacZ gene was inoculated into the pretreated cells. The inoculated cells were cultured for 2 days in the absence of the inhibitors. The transduction titer was determined by X-Gal staining of the cells.
The molecular sizes of mCAT1-HA proteins in the dNM- and dMM-treated NIH 3T3 cells were smaller than those in untreated NIH 3T3 cells, as expected (Fig. 6). Treatment of XC cells with dNM did not change the molecular size of mCAT1-HA compared to that in control XC cells. The molecular size of the mCAT1-HA protein in XC cells was similar to that in dNM-treated NIH 3T3 cells. These results suggest a defect of glucosidase I in XC cells. To confirm this, XC cells were transfected with a glucosidase I expression plasmid (10), and the molecular size of mCAT1-HA and the fusogenic activity of the R+ Env protein were analyzed. Unfortunately, the molecular size of mCAT1-HA was not affected in the glucosidase I-transfected XC cells, and syncytium formation by the R+ Env protein was not suppressed (data not shown). This result indicates that the altered N glycosylation observed in XC cells is not induced by a defect of glucosidase I function.
FIG. 6.
Molecular size of the mCAT1-HA protein in dNM- or dMM-treated cells. mCAT1-HA-expressing XC and NIH 3T3 cells were treated with dNM or dMM (2 mM) for 24 h. Cell lysates were prepared from the cells and subjected to 7.5% polyacrylamide gel electrophoresis. Western immunoblotting was performed using the anti-HA antibody.
Syncytia were induced by R+ Env in XC cells expressing an mCAT1 mutant lacking both of two N glycosylation sites.
XC cells originally express xcCAT1 that has an amino acid substitution at one of two N-linked glycosylation sites (13). To know whether N-glucan linked to the ecotropic MLV receptor is required for XC cell-specific syncytium formation by the R+ Env protein, XC cells were inoculated with a retrovirus vector encoding an mCAT1 mutant, M3, which has amino acid substitutions at both of two N glycosylation sites (31). The R+ Env protein, however, induced syncytia in XC cells expressing the mutant mCAT1, and tunicamycin treatment of the cells largely suppressed XC cell-specific syncytium formation by R+ Env (data not shown), as in the control XC cells. This result suggests that N-glucan in a molecule(s) other than the receptor is required for XC cell-specific syncytium formation by the R+ Env protein.
DISCUSSION
Altered N glycosylation in XC cells.
The molecular size of the mCAT1-HA protein expressed in XC cells was smaller than those in NIH 3T3, CHO, and 293T cells due to altered N glycosylation in the XC cells (Fig. 1). It has been reported that the molecular size of the SU subunit of Fr-MLV Env protein expressed in XC cells is smaller than that in NIH 3T3 cells due to altered N glycosylation (9). The molecular size of mCAT1-HA in XC cells was similar to that in dNM-treated NIH 3T3 cells and was not changed by treatment of the XC cells with dNM (Fig. 6). This result suggested the possibility that glucosidase I does not function in XC cells. Transfection of XC cells with a glucosidase I expression plasmid (10), however, did not affect the molecular size of the mCAT1-HA protein and syncytium formation by the R+ Env protein. This result indicates that the altered N glycosylation observed in XC cells is not induced by the defect of glucosidase I enzyme. This is confirmed by the fact that the treatment of XC cells with dMM, which inhibits mannosidase I, a downstream enzyme of glucosidase I in the N-glucan processing pathway (Fig. 4), makes the molecular size of the mCAT1-HA protein smaller (Fig. 6). A more downstream enzyme in N-glucan processing may be defective, or unknown N glycosylation may occur in XC cells.
Alteration of carbohydrate structure linked to glycoproteins is frequently observed in transformed cells (20). XC cells were established from a rat tumor induced by Rous sarcoma virus (RSV) (12). The altered glycosylation detected in XC cells might have resulted from cellular transformation by RSV, although increased branching of N-linked glucans is a common feature of transformed cells (20).
In addition to unglycosylated mCAT1-HA protein, molecules with higher molecular weight were detected in tunicamycin-treated XC cells, although only the unglycosylated form was detected in tunicamycin-treated NIH 3T3 cells (Fig. 1). It is not clear whether the larger molecules detected in tunicamycin-treated XC cells were modified by other posttranslational events or were still N glycosylated.
N glycosylation is required for XC cell-specific syncytium formation by R+ Env.
The treatment of XC cells with tunicamycin significantly suppressed XC cell-specific syncytium formation by the R+ Env protein but did not affect R− Env-induced syncytium formation (Fig. 2) and the cellular distribution of the mCAT1-HA protein (Fig. 3). This result indicates that N-linked glycosylation in XC cells is required for XC cell-specific syncytium formation by the R+ Env protein. Because the treatment of XC cells with dMM, which made the molecular size of the mCAT1-HA protein smaller, had no effect on syncytium formation by the R+ Env protein, we cannot conclude that the altered N glycosylation observed in XC cells is required for syncytium formation of XC cells by the R+ Env protein.
The treatment of XC cells with tunicamycin suppressed XC cell-specific syncytium formation by the R+ Env protein but did not affect syncytium formation by R− Env and the transduction titer of the ecotropic MLV vector (Fig. 2) (13). This result suggests that the mechanism by which R+ Env induces XC cell syncytia is different from that of membrane fusion by the R− Env protein required for viral entry into host cells. Tunicamycin treatment slightly inhibited syncytium formation by the R− Env protein in NIH 3T3 cells (Fig. 2). N glycosylation of the receptor or of another molecule(s) might be involved in syncytium formation by the R− Env protein in NIH 3T3 cells.
N-Glucan processing inhibitors increase Mo-MLV vector transduction in CHO cells.
The N-glucan processing inhibitor dMM increased Mo-MLV vector transduction in CHO cells but not in NIH 3T3 cells (Fig. 5). N glycosylation of the CAT1 molecule completely blocks Mo-MLV infection in hamster cells but only partially blocks it in rat cells (13). N-Glucan linked to CAT1 protein would physically mask the Env binding motif of CAT1 protein in hamster cells but not in mouse cells. dMM treatment is thought to make the N-glucan mask of hamster CAT1 protein smaller, resulting in higher transduction efficiency of the Mo-MLV vector. The treatment of CHO cells with dNM, however, did not increase the transduction titer of the Mo-MLV vector. dMM treatment induced smaller molecular size of the mCAT1-HA protein than dNM treatment (Fig. 6). Therefore, dMM has higher activity to increase the transduction titer of the Mo-MLV vector than dNM.
R+ Env protein induces syncytium formation in XC cells expressing an mCAT1 mutant lacking both of two glycosylation sites.
XC cells originally express a variant of the ecotropic MLV receptor that lacks one of two N glycosylation sites (13). The R+ Env protein induced syncytia in XC cells expressing a mutant mCAT1 (M3) (31) lacking both of the N glycosylation sites. Tunicamycin treatment suppressed syncytium formation by the R+ Env protein in those cells, as in the control XC cells.
It has been reported that the M3 mutant functions as an ecotropic receptor as efficiently as the wild-type mCAT1 (31) and that mCAT1 functions as an ecotropic receptor better that rCAT1 (13, 28). Though XC cells originally express both xcCAT1 and rCAT1, tunicamycin treatment of XC cells did not increase susceptibility to Mo-MLV vector transduction. This result suggests that xcCAT1 saturates the receptor activity of the cell (13). Therefore, in XC cells overexpressing the M3 mutant, the ecotropic Env protein should preferentially bind to M3 and xcCAT1 rather than rCAT1. If N glycosylation of the ecotropic receptor is required for XC cell-specific syncytium formation by the R+ Env protein, syncytium formation should not be induced by the interaction between the R+ Env and M3 proteins and the level of syncytium formation by R+ Env in M3-expressing XC cells should decrease. However, the R+ Env protein induced syncytium formation in the M3-expressing XC cells as efficiently as in normal XC cells. Although it is possible that the N-glycosylated receptor is dominant for induction of XC cell syncytia by the R+ Env protein, if so, tunicamycin treatment of XC cells should not significantly inhibit syncytium formation by R+ Env. It is possible that glycosylated receptor remains in tunicamycin-treated XC cells, and R+ Env-expressing 293T and pretreated XC cells were cocultured for 24 h in the absence of tunicamycin. It is most likely that N glycosylation of a molecule(s) other than the ecotropic receptor is required for XC cell-specific syncytium formation by the R+ Env protein. A molecule(s) concerned with cell adhesion may be involved in syncytium formation by the Env protein but not in viral entry into cells. There is some evidence that cell adhesion molecules are involved in syncytium formation by retrovirus envelope proteins (2, 4, 8, 21).
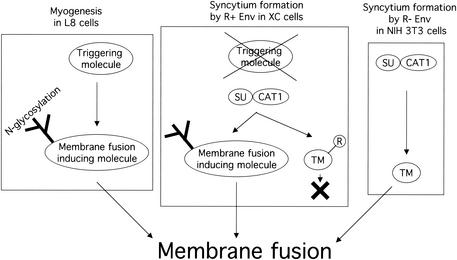
Conclusion.
Finally, XC cells have a specific mechanism by which the R+ Env protein can induce syncytium formation (9), although the R peptide inhibits syncytium formation in other susceptible cells (23, 24). It is unlikely that XC cells are defective in inhibiting syncytium formation by the R peptide. XC cells could have a positive mechanism to overcome the inhibitory effect of the R peptide on syncytium formation (Fig. 7). The mechanism to induce syncytium formation by the R+ Env protein in XC cells could require N-linked glycosylation of unknown molecules other than the ecotropic MLV receptor, although N glycosylation in rat and hamster cells negatively controls Mo-MLV infection. The specific mechanism in XC cells to induce syncytium formation by the R+ Env protein should not be associated with membrane fusion by the R− Env protein required for retroviral entry into host cells. It has been reported that LFA-1 is required for human immunodeficiency virus-mediated cell fusion but not for viral transmission (21).
FIG. 7.
Hypothesis of syncytium formation by R+ Env protein in XC cells. L8 myoblast cells express membrane fusion-triggering and -inducing molecules involved in myogenesis. N glycosylation of the inducing molecule is required for the induction of membrane fusion. The membrane fusion-inducing molecule is activated through the triggering molecule. The triggering molecule corresponds to the surface subunit (SU) in syncytium formation by the Env protein. The membrane fusion-inducing molecule is active in XC cells, but the triggering molecule is not. The SU protein activates the membrane fusion-inducing molecule interacting with the receptor CAT1 instead of the triggering molecule. The R peptide-containing transmembrane subunit (TM) is inactive to induce membrane fusion.
XC cells were established from a muscle sarcoma induced by RSV (27). It has been reported that the R+ Env protein induces syncytium formation in a spontaneously transformed cell line of L8 myoblast cells, fu-1 (11, 33, 34). The L8 cell line is widely used as an in vitro model system of myogenesis. This suggests that the mechanism to induce myotube differentiation is involved in XC cell-specific syncytium formation by the R+ Env protein. It has been reported that protein N glycosylation participates in myotube differentiation (16, 26). XC cells, but not other susceptible cells, may express a cellular molecule that induces membrane fusion in myogenesis (Fig. 7). N glycosylation of the molecule is required for its membrane fusion activity. However, XC cells lack a triggering molecule to initiate membrane fusion. The Env protein may function as the triggering molecule by binding to the receptor.
Acknowledgments
We thank J. M. Cunningham for the mCAT1 cDNA clone, F.-L. Cosset for the TELCeB6 packaging cells, E. Bause for the glucosidase I expression plasmid, T. Kitamura for the pMXpuro retroviral vector DNA, D. Kabat for the N glycosylation-defective mutant (M3) of mCAT1, R. Watanabe for the rCAT1 cDNA clone, T. Yanagi for technical advice on the immunofluorescence assay, and H. Sato and N. Yamamoto for help. We also thank A. Koshiyama for secretarial work and R. Fujita, M. Katane, E. Takao, and N. Sasaki for experimental support.
Y. Kubo was a special research fellow of RIKEN. This work was supported by a Gene Science Research grant from RIKEN to H. Amanuma and by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science to Y. Kubo.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scaden, and J. M. Cunningham. 1989. A putative murine ecotropic receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Barbeau, B., J. Fortin, N. Genois, and M. J. Tremblay. 1998. Modulation of human immunodeficiency virus type 1-induced syncytium formation by the conformational state of LFA-1 determined by a new luciferase-based syncytium quantitative assay. J. Virol. 72:7125-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosset, F.-L., Y. Takeuchi, J. L. Battini, R. A. Reiss, and M. K. Collins. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daenke, S., S. A. McCracken, and S. Booth. 1999. Human T-cell leukaemia/lymphoma virus type 1 syncytium formation is regulated in a cell-specific manner by ICAM-1, ICAM-3 and VCAM-1 and can be inhibited by antibodies to integrin β2 or β7. J. Gen. Virol. 80:1429-1436. [DOI] [PubMed] [Google Scholar]
- 5.Green, N., T. M. Shinnick, O. Witte, A. Ponticelli, J. G. Sutcliffe, and R. A. Lerner. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. USA 78:6023-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruters, R. A., J. J. Neefjes, M. Tersmette, R. E. Y. de Goede, A. Tulp, H. G. Huisman, F. Miedema, and H. L. Ploegh. 1987. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature 330:74-77. [DOI] [PubMed] [Google Scholar]
- 7.Henderson, L. E., R. Sowder, T. D. Copeland, G. Smythers, and S. Oroszlan. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 52:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildreth, J. K., A. Subramanium, and R. A. Hampton. 1997. Human T-cell lymphotropic virus type 1 (HTLV-1)-induced syncytium formation mediated by vascular cell adhesion molecule-1: evidence for involvement of cell adhesion molecules in HTLV-1 biology. J. Virol. 71:1173-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, J. S., and R. Risser. 1993. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J. Virol. 67:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalz-Fuller, B., E. Bieberich, and E. Bause. 1995. Cloning and expression of glucosidase I from human hippocampus. Eur. J. Biochem. 231:344-351. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman, S. J., and C. M. Parks. 1977. Loss of growth control and differentiation in the fu-1 variant of the L8 line of rat myoblasts. Proc. Natl. Acad. Sci. USA 74:3888-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klement, V., W. P. Rowe, J. W. Hartley, and W. E. Pugh. 1969. Mixed culture cytopathogenicity: a new test for growth of murine leukemia viruses in tissue culture. Proc. Natl. Acad. Sci. USA 63:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubo, Y., T. Ono, M. Ogura, A. Ishimoto, and H. Amanuma. 2002. A glycosylation-defective variant of the ecotropic murine retrovirus receptor is expressed in rat XC cells. Virology 303:338-344. [DOI] [PubMed] [Google Scholar]
- 14.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and E. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, D. G., and A. D. Miller. 1992. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J. Virol. 66:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsumoto, Y., and A. Klip. 1992. Developmental regulation of the subcellular distribution and glycosylation of GLUT1 and GLUT4 glucose transporters during myogenesis of L6 muscle cells. J. Biol. Chem. 267:4957-4962. [PubMed] [Google Scholar]
- 17.Montefiori, D. C., W. E. Robinson, Jr., and W. M. Mitchell. 1988. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 85:9248-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obata, M., H. Amanuma, Y. Harada, N. Sagata, and Y. Ikawa. 1984. Env-related leukemogenic genes (gp55 genes) to two closely related polycythemic strains of Friend spleen focus-forming virus possess different recombination points with an endogenous mink cell focus-forming virus env gene. Virology 136:435-438. [DOI] [PubMed] [Google Scholar]
- 19.Onishi, M., S. Kinoshita, Y. Morikawa, A. Shibuya, J. Phillips, L. L. Lanier, D. M. Gorman, G. P. Nolan, A. Miyajima, and T. Kitamura. 1996. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 24:324-329. [PubMed] [Google Scholar]
- 20.Orntoft, T. F., and E. M. Vestergaard. 1999. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis 20:362-371. [DOI] [PubMed] [Google Scholar]
- 21.Pantaleo, G., L. Butini, C. Graziosi, G. Poli, S. M. Schnittman, J. J. Greenhouse, J. I. Gallin, and A. S. Fausi. 1991. Human immunodeficiency virus (HIV) infection in CD4+ T lymphocytes genetically deficient in LFA-1: LFA-1 is required for HIV-mediated cell fusion but not for viral transmission. J. Exp. Med. 173:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragheb, J. A., and W. F. Anderson. 1994. PH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rein, A., A. J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p12E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schult, A., and A. Rein. 1985. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology 145:335-339. [DOI] [PubMed] [Google Scholar]
- 26.Spearman, M. A., J. C. Jamieson, and J. A. Wright. 1987. Studies on the effect of glycoprotein processing inhibitors on fusion of L6 myoblast cell lines. Exp. Cell Res. 168:116-126. [DOI] [PubMed] [Google Scholar]
- 27.Svoboda, J. 1960. Presence of chicken tumour virus in the sarcoma of the adult rat inoculated after birth with Rous sarcoma tissue. Nature 186:980-981. [DOI] [PubMed] [Google Scholar]
- 28.Takase-Yoden, S., and R. Watanabe. 1999. Contribution of virus-receptor interaction to distinct viral proliferation of neuropathogenic and nonneuropathogenic murine leukemia viruses in rat glial cells. J. Virol. 73:4461-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavoloni, N., and A. Rudenholz. 1997. Variable transduction efficiency of murine leukemia retroviral vector on mammalian cells: role of cellular glycosylation. Virology 229:49-56. [DOI] [PubMed] [Google Scholar]
- 30.Walker, B. D., M. Kowalski, W. C. Goh, K. Kozarsky, M. Krieger, C. Rosen, L. Rohrschneider, W. A. Haseltine, and J. Sodroski. 1987. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc. Natl. Acad. Sci. USA 84:8120-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, H., E. Klamo, S. E. Kuhmann, S. L. Kozak, M. P. Kavanaugh, and D. Kabat. 1996. Modulation of ecotropic murine retroviruses by N-linked glycosylation of the cell surface receptor/amino acid transporter. J. Virol. 70:6884-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, C. A., and M. V. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong, P. K., P. H. Yuen, and S. J. Kaufman. 1977. Induction of syncytia by Moloney murine leukemia virus in myoblasts defective in differentiation. J. Virol. 21:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, P. K., P. H. Yuen, and S. J. Kaufman. 1977. Mechanism of murine leukemia virus-induced fusion in rat myoblasts defective in differentiation. J. Virol. 23:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]