Abstract
Structural differences between poxvirus and human mRNA capping enzymes recommend cap formation as a target for antipoxviral drug discovery. Genetic and pharmacologic analysis of the poxvirus capping enzymes requires in vivo assays in which the readout depends on the capacity of the viral enzyme to catalyze cap synthesis. Here we have used the budding yeast Saccharomyces cerevisiae as a genetic model for the study of poxvirus cap guanine-N7 methyltransferase. The S. cerevisiae capping system consists of separate triphosphatase (Cet1), guanylyltransferase (Ceg1), and methyltransferase (Abd1) components. All three activities are essential for cell growth. We report that the methyltransferase domain of vaccinia virus capping enzyme (composed of catalytic vD1-C and stimulatory vD12 subunits) can function in lieu of yeast Abd1. Coexpression of both vaccinia virus subunits is required for complementation of the growth of abd1Δ cells. Previously described mutations of vD1-C and vD12 that eliminate or reduce methyltransferase activity in vitro either abolish abd1Δ complementation or elicit conditional growth defects. We have used the yeast complementation assay as the primary screen in a new round of alanine scanning of the catalytic subunit. We thereby identified several new amino acids that are critical for cap methylation activity in vivo. Studies of recombinant proteins show that the lethal vD1-C mutations do not preclude heterodimerization with vD12 but either eliminate or reduce cap methyltransferase activity in vitro.
The m7GpppN cap structure of eukaryotic mRNAs is formed cotranscriptionally by the sequential action of three enzymes: an RNA triphosphatase that removes the gamma phosphate of the nascent transcript to yield a 5′ diphosphate end, a GTP:RNA guanylyltransferase that adds a GMP residue to the diphosphate end in a 5′-to-5′ orientation, and an RNA (guanine-N7) methyltransferase that uses S-adenosylmethionine to methylate the N7 position of the cap guanine (34, 35). The capping enzyme of vaccinia virus is a heterodimer of vD1 (844 amino acids [aa]) and vD12 (287 aa) polypeptides that catalyzes all three capping reactions (24, 38). The triphosphatase, guanylyltransferase, and methyltransferase active sites are arrayed in modular fashion within the vD1 subunit. The amino-terminal 60-kDa segment of vD1 (from aa 1 to 545) is an autonomous triphosphatase-guanylyltransferase domain in which the triphosphatase active site is located in the proximal half and the guanylyltransferase active site is in the distal half (8, 9, 12, 26, 27, 28, 37, 41, 42) The methyltransferase domain consists of the carboxyl-terminal portion of the vD1 subunit (vD1-C) heterodimerized with the vD12 subunit (7, 15, 16, 17, 20, 21, 29). The methyltransferase active site is located within vD1-C, which has a weak intrinsic methyltransferase activity that is stimulated 30- to 50-fold by the vD12 subunit, which is itself catalytically inert. Thus, the native vaccinia virus cap methyltransferase consists of a catalytic subunit (vD1-C) and a regulatory subunit (vD12).
The physical organization of the poxvirus capping apparatus is quite distinct from that of cellular capping enzymes (34, 35). For example, the triphosphatase, guanylyltransferase, and methyltransferase components of the yeast capping apparatus reside on three different polypeptides encoded by three genes (CET1, CEG1, and ABD1, respectively). The metazoan capping enzyme consists of two components, a bifunctional triphosphatase-guanylyltransferase polypeptide and a separately encoded methyltransferase polypeptide. The cap methyltransferases of fungi and metazoa are monomeric enzymes. In the budding yeast Saccharomyces cerevisiae, the cap methylating enzyme Abd1 is essential for cell viability (22, 23, 40). Cap methyltransferases from other unicellular eukarya (Ccm1, Pcm1, and Ecm1 of Candida albicans, Schizosaccharomyces pombe, and Encephalitozoon cuniculi, respectively) can function in lieu of Abd1 to sustain yeast cell growth (14, 30). The human cap methyltransferase Hcm1 can also replace Abd1 in vivo, attesting to the conservation of structure and function between cap methylating enzymes from yeast to humans (30). Cellular cap methyltransferases display local amino acid sequence similarity to the vaccinia virus methyltransferase catalytic subunit but not to the stimulatory subunit (40).
The structural differences between the poxvirus and human capping enzymes recommend cap formation as a target for antipoxviral drug discovery. The stimulatory subunit of the viral methyltransferase is an especially attractive molecular target, because there is no discernible homolog of vD12 in the known proteomes of any metazoan species. Although structure-function analyses of vaccinia virus cap methyltransferase have been carried out with purified vD1-C and D12 proteins expressed in bacteria, there has been no genetic analysis of the role of cap N7 methylation in poxvirus replication in vivo. Mutations that elicit temperature-sensitive virus growth phenotypes have been mapped to the two capping enzyme subunits (6, 13), but the picture is complicated because the capping enzyme plays multiple additional roles in viral gene expression. Capping enzyme serves as a transcription termination factor during the synthesis of viral early mRNAs (19, 36) and as an initiation factor during the transcription of intermediate genes (39). Surprisingly, the vD1 and vD12 temperature-sensitive mutant viruses display no gross defect in viral gene expression at the restrictive temperature but instead are defective in resolving concatameric DNA replication intermediates into the hairpin telomeres of the mature viral genome (6, 13). The telomere phenotype has no obvious connection to the known mRNA processing functions of the vD1 and vD12 proteins.
Genetic and pharmacologic studies of poxvirus capping enzymes would be expedited by the development of in vivo assays in which the functional readout is clearly and exclusively dependent on the capacity of the viral gene product to catalyze cap synthesis. Previously, Ho et al. validated the concept of using the budding yeast S. cerevisiae as a genetic model for the study of the RNA triphosphatase and guanylyltransferase components of the poxvirus capping enzyme (18). Ho et al. showed that expression of vD1(1-545) in S. cerevisiae complemented the growth of mutant strains lacking the endogenous RNA triphosphatase or guanylyltransferase enzymes provided that vD1(1-545) was fused to a cellular protein capable of targeting it to the RNA polymerase II (Pol II) transcription complex.
Here we tested the feasibility of using yeast as a genetic system to study the function of the poxvirus cap methyltransferase. We report that the vaccinia virus methyltransferase functions in lieu of the yeast Abd1 when the vD1-C and vD12 proteins are coexpressed in vivo. Genetic complementation by the viral methyltransferase did not require fusion to a cellular targeting protein. We exploit the yeast-based genetic system to identify constituents of the poxvirus catalytic and regulatory subunits that are important for cap methylation in vivo and in vitro.
MATERIALS AND METHODS
Yeast expression plasmids for vaccinia virus cap methyltransferase.
The open reading frame encoding the methyltransferase catalytic subunit vD1-C (aa 498 to 844) was PCR amplified from pET14-His-D1(498 to 844) (20). The PCR product was inserted into yeast expression vector pYN132 (CEN TRP1) such that expression of vD1-C was under the control of the yeast TPI1 promoter. The TPI1-vD1-C cassette was then inserted into a multicopy 2μm TRP1 vector. CEN TRP1 and 2μm TRP1 plasmids carrying mutated versions of vD1-C (H682A-Y683A, D598A, G600A, G602A, Y683S, S684A, and F685A) were constructed similarly using the respective mutant pET14-D1 (aa 498 to 844) plasmids (20) as templates for PCR amplification. Other vD1-C mutants were generated via the two-stage PCR overlap extension method and then cloned into yeast CEN and 2μm expression vectors. All DNA inserts were sequenced completely to ensure that no unwanted coding mutations were introduced during amplification and cloning.
The vD12 gene encoding the stimulatory subunit of the vaccinia virus cap methyltransferase was excised from pET16-D12 (31) by digestion with NdeI and BamHI and then inserted into pYN132. The expression cassette TPI1-vD12 was then transferred into plasmids pRS413 (CEN HIS3) and pRS423 (2μm HIS3). The same strategy was employed to generate the plasmids for expression in yeast of the mutant vD12 proteins N42A-Y43A, L61A-K62A, F176A-K177A, and F245A-L246A.
Plasmid shuffle assay for cap methyltransferase activity in vivo.
S. cerevisiae strain YBS40 lacks the chromosomal ABD1 locus encoding the yeast cap methyltransferase (30). Growth of YBS40 depends on the maintenance of plasmid p360-ABD1(CEN URA3 ABD1). YBS40 was cotransformed with yeast expression plasmids carrying wild-type or mutant vD1-C and vD12. Trp+His+ transformants were selected at 30°C on agar medium lacking tryptophan and histidine. Cells were then streaked at 37, 30, 25, and 19°C on agar medium containing 0.75 mg of 5-FOA/ml. The plates were incubated at 37, 30, 25, and 19°C for up to 8 days. The lethal alleles were those that failed to yield colonies on 5-FOA at any temperature. Individual FOA-resistant colonies arising at permissive temperature were patched to yeast extract-peptone-dextrose (YEPD) agar. Cells were then streaked on YEPD agar, and growth was scored at different temperatures.
Coexpression of vD1-C and vD12 proteins in bacteria and enzyme purification.
The wild-type vD12 gene was inserted into pET3c to yield pET3-D12. The wild-type vD1-C open reading frame was cloned into pET16b. NdeI-XhoI fragments encoding mutated versions of vD1-C were excised from the respective pYN132-vD1-C constructs and transferred into the pET16b vector. To generate coexpression plasmids, the expression cassettes for wild-type vD1-C and the alanine mutants were excised from the pET16b plasmids with BglII and HindIII and then inserted between the BamHI and HindIII sites of pET3-D12. Transcription of the vD1-C and vD12 genes in the coexpression vector was driven by separate tandemly oriented T7 promoters. The plasmids were transformed into Escherichia coli BL21(DE3).
E. coli BL21(DE3)/pET transformants were inoculated into Luria-Bertani medium containing 0.1 mg of ampicillin/ml and grown at 37°C until the A600 reached 0.3. The cultures (500 ml) were placed on ice for 30 min, adjusted to either 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) [for His-vD1-C/D12, His-vD1-C(R682A)/vD12, His-D1-C(F780A)/vD12, and His-D1-C(F814A-F815A)/vD12 expression] or 0.15 mM IPTG (for all others). The cultures were incubated for 18 to 20 h at 17°C with continuous shaking. Cells were harvested by centrifugation, and the pellets were stored at −75°C. All subsequent procedures were performed at 4°C.
Cell pellets were thawed on ice and suspended in 25 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 10% glycerol). Cell lysis was achieved by addition of lysozyme and Triton X-100 to achieve final concentrations of 0.25 mg/ml and 0.1%, respectively. Insoluble material was removed by centrifugation at 17,000 rpm for 30 min in a Sorvall SS34 rotor. The supernatants were mixed for 1 h with 2.5 ml of Ni-nitrilotriacetic acid-agarose resin (Qiagen) that had been equilibrated with lysis buffer. The slurries were poured into columns and then washed with lysis buffer. The columns were eluted stepwise with IMAC buffer (50 mM Tris-HCl [pH 8.0], 0.15 M NaCl, 10% glycerol, 0.1% Triton X-100) containing 50, 200, and 500 mM imidazole. The polypeptide compositions of the fractions were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant vaccinia virus proteins were recovered in the 200 mM imidazole eluate. The concentrations of His-vD1-C and vD12 proteins were determined by SDS-PAGE analysis of protein samples in parallel with 0.5, 1.0, 1.5, and 2.0 μg of bovine serum albumin. The gels were fixed and stained with Coomassie blue dye. The staining intensities of the polypeptides were quantified using a Digital Imaging and Analysis system (Alpha Innotech Corporation). The concentrations of the wild-type and the mutant D1 proteins were calculated by interpolation to the bovine serum albumin standard curve.
Methyltransferase assay.
Protein samples were adjusted with 50 mM Tris-HCl (pH 7.5)-5 mM dithiothreitol to contain ∼20 fmol of the vD1-C subunit per μl. Aliquots (2 μl) of the protein solutions and serial dilutions thereof were assayed for cap methylation in reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 7.5), 5 mM dithiothreitol, 50 μM AdoMet, and 15 fmol of 32P cap-labeled poly(A) RNA (33). The mixtures were incubated at 37°C for 5 min. The reactions were quenched by heating to 95°C for 5 min. The mixtures were adjusted to 50 mM sodium acetate (pH 5.5) and then digested with 5 μg of nuclease P1 for 1 h at 37°C. The digests were spotted on polyethyleneimine cellulose thin-layer chromatography plates that were developed with 0.35 M (NH4)2SO4. The extent of methylation of the cap [expressed as m7GpppA/(m7GpppA+GpppA)] was determined by scanning the chromatogram with a phosphorimager.
RESULTS
Vaccinia virus cap methyltransferase is active in yeast.
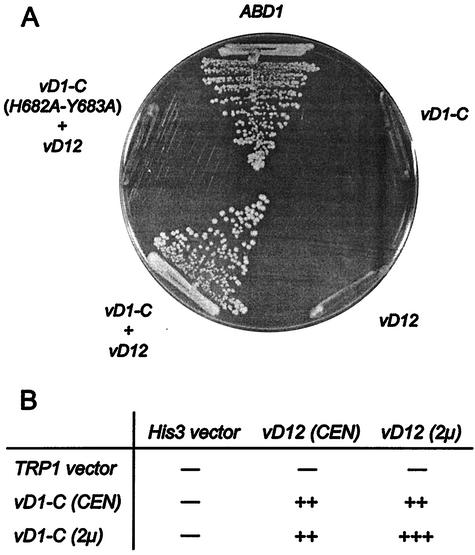
We used the plasmid shuffle technique to test the capacity of the vaccinia virus cap methyltransferase to complement the abd1Δ mutation of S. cerevisiae (Fig. 1). abd1Δ cells containing a copy of ABD1 on a CEN URA3 plasmid were cotransformed with vD1-C (2μm TRP1) and vD12 (2μm TRP1) expression plasmids. ABD1 (CEN TRP1) served as a positive control. Transformants were selected and then tested for growth on medium containing 5-fluoroorotic acid (5-FOA), a drug that selects against the URA3 ABD1 plasmid. Transformants that contained both vD1-C and vD12 on 2μm plasmids readily formed 5-FOA-resistant colonies, but cells transformed singly with either vD1-C or vD12 on a 2μm plasmid did not (Fig. 1). Thus, the heterodimeric vaccinia virus methyltransferase can function in yeast and both subunits are necessary for cap methylation in vivo.
FIG. 1.
Complementation of abd1Δ cells by vaccinia virus cap methyltransferase. (A) Growth on 5-FOA medium. Yeast strain YBS40 (abd1Δ) was transformed with 2μm plasmids containing either vD1-C (TRP1), vD12 (HIS3), vD1-C plus vD12, or vD1-C (H682A-Y683A) plus vD12. A control transformation was performed with a CEN TRP1 plasmid containing the ABD1 gene. Single transformants were streaked to medium containing 5-FOA. The plate was photographed after incubation for 4 days at 30°C. (B) vD1-C and vD12 were introduced into YBS40 cells on CEN and 2μm plasmids in combinations as indicated and were tested by plasmid shuffle. Strains that failed to grow on 5-FOA are indicated by minuses. The growth of 5-FOA survivors was assessed on rich medium and recorded after 2 days of incubation at 30°C. +++ indicates that the colony size was indistinguishable from that of ABD1 cells, and ++ indicates that the strain formed slightly smaller colonies than wild-type cells.
To assess the influence of gene dosage on in vivo methyltransferase activity, abd1Δ cells were transformed with various combinations of vD1-C and vD12 on either multicopy 2μm or single-copy CEN plasmids. All combinations yielded 5-FOA-resistant strains, which were then tested for growth on rich (YEPD) agar medium. Cells expressing vD1-C and vD12 on 2μm plasmids grew as well as wild-type ABD1 cells (Fig. 1B), whereas cells carrying either vD1-C or vD12 on a CEN plasmid formed slightly smaller colonies.
Vaccinia virus methyltransferase activity is essential for abd1Δ complementation.
The vD1-C mutation H682A-Y683A abolishes methyltransferase activity in vitro without affecting subunit heterodimerization (21). Here we found that a vD1-C (H682A-Y683A) mutant on a 2μm plasmid failed to complement abd1Δ when cotransformed with vD12 (Fig. 1A). This result indicates that methyltransferase catalytic activity is essential for complementation of abd1Δ by the poxvirus capping enzyme.
Six other previously characterized vD1-C mutants that retained the capacity to heterodimerize with vD12 were also tested for complementation of abd1Δ. CEN and 2μm plasmids expressing the vD1-C mutants were introduced into abd1Δ cells along with CEN or 2μm plasmids expressing wild-type vD12 (Table 1). Transformants were selected and streaked on 5-FOA agar plates at 19, 30, and 37°C. vD1-C alleles G600A and Y683S were lethal at all temperatures, whether on single-copy or multicopy plasmids. We surmise that lethality is a consequence of the defect in catalyzing cap methylation by the mutant proteins, insofar as the purified recombinant G600A/vD12 and Y683S/vD12 heterodimers exhibited 4% and 0.1% of wild-type methyltransferase activity in vitro, respectively (21).
TABLE 1.
Effects of biochemically characterized vD1-C mutations on cap methyltransferase function in vivoa
| vD1-C allele + vD12 | Growth at temp (°C):b
|
||
|---|---|---|---|
| 19 | 30 | 37 | |
| CEN | |||
| WTc | ++ | ++ | ++ |
| D598A | + | − | − |
| G600A | − | − | − |
| G602A | + | + | + |
| Y683S | − | − | − |
| S684A | ++ | ++ | + |
| F685A | ++ | ++ | + |
| 2μm | |||
| WT | +++ | +++ | +++ |
| D598A | + | + | − |
| G600A | − | − | − |
| G602A | +++ | ++ | ++ |
| Y683S | − | − | − |
| S684A | +++ | ++ | ++ |
| F685A | +++ | ++ | ++ |
The indicated alleles of vD1-C were tested by plasmid shuffle for complementation of abd1Δ as described in Methods and Materials. CEN vD1-C plasmids were cotransformed with CEN vD12 (CEN), and 2μm vD1-C plasmids were cotransformed with 2μm vD12 (2μm).
+++ indicates colony sizes indistinguishable from those of strains bearing wild-type ABD1, ++ indicates slightly reduced colony sizes, + indicates that only pinpoint colonies were formed, and −(minus) indicates no growth.
WT, wild type.
D598A supported the emergence of 5-FOA-resistant colonies only at low temperatures. Cells carrying the vD1-C-D598A mutant on a single-copy plasmid formed pinpoint colonies at 19°C but failed to grow above this temperature. Increasing the copy number of the D598A and vD12 genes allowed formation of pinpoint colonies at 19 and 30°C but no growth at 37°C (Table 1). It was reported previously that the purified D598A/vD12 heterodimer was 50% as active as the wild-type vaccinia virus methyltransferase in vitro (21). Thus, the conditional growth phenotype may reflect the decrement in methyltransferase catalytic activity.
5-FOA-selected cells containing G602A on a CEN plasmid formed pinpoint colonies on YEPD medium at 19, 30, and 37°C (Table 1). Increasing the dosage of the G602A and vD12 genes improved growth at all temperatures. The size of the 2μm G602A/vD12 colonies at 19°C was indistinguishable from that of “wild-type” 2μm vD1-C/vD12 cells; the colony size of the mutant strain was slightly smaller than that of the wild-type strain at 30 and 37°C (Table 1). Earlier studies showed that the methyltransferase activity of the purified G602A/vD12 heterodimer was identical to that of wild-type vaccinia virus methyltransferase (21). The present finding that vD1-C-G602A/vD12 cells displayed a severe slow growth phenotype that was suppressed by overexpression of both components of the cap methyltransferase suggests that (i) the G602A mutation can affect protein folding or stability when vD1-C is expressed in yeast or (ii) the G602A mutation can reduce the affinity of vD1-C for the vD12 subunit in yeast.
S684A and F685A strains expressing the methyltransferase subunits from CEN plasmids grew as well as wild-type vD1-C/vD12 cells at 19 and 30°C but formed smaller colonies at 37°C (Table 1). Increasing the dosage of the S684A and F685A genes resulted in improved growth at both 19 and 37°C. These results are consistent with biochemical studies showing that purified recombinant S684A/vD12 and F685A/vD12 heterodimers exhibited 60 to 80% of wild-type methyltransferase activity in vitro (21).
Genetic screen for new mutations affecting cap methyltransferase function in vivo.
Our initial genetic analysis of biochemically defined mutants of vaccinia virus cap methyltransferase indicated that (i) lesions in the catalytic subunit that abolish activity in vitro were consistently lethal in vivo and (ii) the yeast complementation assay was sensitive to mutations resulting in partial loss of cap methyltransferase activity which resulted in conditional phenotypes and/or growth defects that were suppressible by increased gene dosage. We therefore sought to exploit the yeast-based genetic assay as a primary screening method for identification of new structural components of the catalytic subunit that contribute to methyltransferase function.
We used the plasmid shuffle procedure to test the effects of single-alanine substitutions in vD1-C at positions Asp604, Asp620, and Arg632, which are conserved in all poxvirus D1 orthologs (Fig. 2) as well as in cellular cap methyltransferases (4, 32, 40). The vD1-C alleles D604A, D620A, and R632A were lethal at all temperatures, whether on single-copy or multicopy plasmids (Table 2). These findings are concordant with previous studies showing that the equivalent Asp and Arg chains were essential for the in vivo activity of yeast cap methyltransferase Abd1 and human cap methyltransferase Hcm1 (23, 30, 32, 40).
FIG. 2.
Sequence alignment of poxvirus vD1-C orthologs. The amino acid sequence of vaccinia virus D1 (vv) from aa 573 to 843 was aligned with the sequences of orthologous polypeptides of molluscum contagiosum virus (mc), fowlpox virus (fp), lumpy skin disease virus (ls), Amsacta moorei entomopoxvirus (Am), and Melanoplus sanguinipes entomopoxvirus (Ms). Positions of side chains with identity or similarity in all six poxvirus proteins are indicated by a caret below the aligned sequences. Missense mutations at positions (marked by •) and double-alanine mutations (indicated by double dashes) were tested for in vivo function in yeast. The conserved amino acids at which mutations resulted in unconditional lethality in yeast are highlighted by shading.
TABLE 2.
New alanine-scanning mutagenesis of vD1-Ca
| vD1-C allele + vD12 | Growth at temp (°C):b
|
||
|---|---|---|---|
| 19 | 30 | 37 | |
| CEN | |||
| WTc | ++ | ++ | ++ |
| D604A | − | − | − |
| Y608A-F609A | − | − | − |
| D620A | − | − | − |
| R632A | − | − | − |
| F667A | ++ | ++ | ++ |
| F672A | ++ | ++ | ++ |
| D676A-W677A | − | − | − |
| E763A-Y764A | − | − | − |
| F775A | + | + | + |
| Y778A | ++ | ++ | ++ |
| F780A | + | + | − |
| D784A | + | + | − |
| F814A-F815A | − | − | − |
| 2μm | |||
| WT | +++ | +++ | +++ |
| D604A | − | − | − |
| Y608A-F609A | − | − | − |
| D620A | − | − | − |
| R632A | − | − | − |
| D676A-W677A | − | − | − |
| E763A-Y764A | − | − | − |
| F775A | +++ | +++ | +++ |
| F780A | ++ | ++ | ++ |
| D784A | +++ | +++ | + |
| F814A-F815A | + | + | + |
The indicated alleles of vD1-C were tested by plasmid shuffle for complementation of abd1Δ. CEN vD1-C plasmids were contransformed with CEN vD12 (CEN), and 2μm vD1-C plasmids were cotransformed with 2μm vD12 (2μm).
+++ indicates colony sizes indistinguishable from those of strains bearing wild-type ABD1, ++ indicates slightly reduced colony sizes, + indicates that only pinpoint colonies were formed, and − indicates no growth.
WT, wild type.
We then tested the effects of single-alanine substitutions at five aromatic residues (Phe667, Phe672, Phe775, Tyr778, and Phe780) and one acidic residue (Asp784) that are conserved in other poxvirus capping enzymes (Fig. 2). We focused on aromatic residues in light of crystallographic findings for several proteins involved in cap formation or cap recognition, which indicate that the protein-bound cap guanine is stabilized by π-stacking interactions with aromatic amino acids (5). We found that FOA-selected cells containing either the F667A, F672A, or Y778A alleles on CEN plasmids grew as well as wild-type vD1-C/vD12 cells on rich medium at all temperatures tested (Table 2). We therefore concluded that the Phe667, Phe672, and Tyr778 side chains were not important for methyltransferase activity in vivo.
In contrast, the F775A mutation elicited a growth defect at all temperatures when present in single copy. However, the growth phenotype was suppressed completely at all temperatures by increased gene dosage (Table 2). Similarly, the F780A and D784A mutations in single copies resulted in slow growth at 19 and 30°C and no growth at 37°C and these phenotypes were suppressed by increases in gene copy number (Table 2). For example, 2μm F780A cells displayed improved growth at permissive temperatures and a gain of viability at 37°C. 2μm D784A cells grew as well as wild-type cells at 19 and 30°C, although they remained temperature sensitive, forming pinpoint colonies at 37°C (Table 2). It is noteworthy that three conserved amino acids at which single-alanine mutations conferred dosage-suppressible growth defects (Phe775, Phe780, and Asp784) are clustered together in the vD1-C polypeptide.
Finally, we tested the effects of four new double-alanine mutations in adjacent conserved residues of vD1-C (Fig. 2). The vD1-C alleles Y608A-F609A, D676A-W677A, and E763A-Y764A were lethal at all temperatures, whether on single-copy or multicopy plasmids (Table 2). F814A-F815A was lethal in single copy but supported the formation of pinpoint colonies when present in high copy together with 2μm vD12 (Table 2).
Mutational effects on methyltransferase activity in vitro.
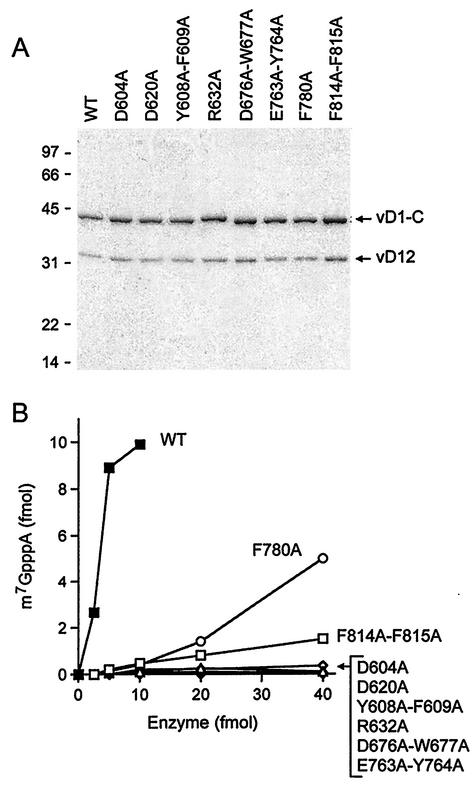
Eight of the new vD1-C mutants were produced as recombinant proteins in bacteria in parallel with the wild-type protein and subjected to biochemical analysis of subunit heterodimerization and cap methylation activity. The mutants selected for in vitro analysis were either lethal in yeast at high gene dosage (D604A, Y608A-F609A, D620A, R632A, D676A-W677A, and E763A-Y764A) or displayed growth defects of various severity at high gene dosage (F780A and F814A-F815A). The His-tagged mutant vD1-C polypeptides were coexpressed in bacteria with the untagged wild-type vD12 subunit. Purification from soluble bacterial extracts was achieved by Ni-agarose affinity chromatography, which selects for the His-tagged vD1-C subunit. Polypeptides corresponding to vD1-C and vD12 were the predominant species recovered after the affinity chromatography of the wild-type enzyme (Fig. 3A, lane WT). The preparations of His-tagged mutants D604A, Y608A-F609A, D620A, R632A, D676A-W677A, E763A-Y764A, F780A, and F814A-F815A also contained the vD12 subunit. The extents of purification were comparable for the wild-type and mutant methyltransferases, and the ratios of the vD1-C and vD12 subunits were similar for the wild-type and mutant enzymes, as gauged by the results of SDS-PAGE and staining with Coomassie blue dye (Fig. 3A). We conclude that the D1-C-Ala mutations selected for their severe in vivo phenotypes in yeast had no significant effect on vD1-C interaction with vD12 when the vaccinia virus subunits were coexpressed in E. coli.
FIG. 3.
Mutational effects on cap methyltransferase activity in vitro. (A) Enzyme purification. Aliquots of the Ni-agarose fractions of recombinant His-vD1-C/vD12 (containing 1 μg of the vD1-C subunit) were analyzed by SDS-PAGE. The polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kilodaltons) of marker proteins are indicated at the left. (B) Cap methyltransferase activity. Activity was assayed and quantitated as described in Methods and Materials. The extent of cap methylation is plotted as a function of the amount of recombinant vD1-C included in the reaction mixture.
The recombinant proteins were assayed for their ability to catalyze AdoMet-dependent conversion of 32P cap-labeled poly(A) [GpppA(pA)n] to methylated cap-labeled poly(A) [m7GpppA(pA)n] (boldface characters indicate the labeled phosphorus). The specific activities of the D604A, D620A, Y608A-F609A, D676A-W677A, R632A, and E763A-Y764A enzymes were <1% of the wild-type value (Fig. 3B). Note that these six mutations were lethal in vivo. The F780A and F814A-F815A enzymes displayed low residual methyltransferase activity (approximately 7.1% and 2.2% of the wild-type specific activity, respectively) (Fig. 3B). The growth phenotypes of these two mutants in vivo at high gene dosage (++ for F780A and + for F814A-F815A) correlated with their relative cap methyltransferase activities in vitro. We conclude from these experiments that abd1Δ complementation is an effective primary screen for pinpointing essential structural features of the catalytic subunit of the poxvirus cap methyltransferase.
Effects of mutations in the D12 subunit of vaccinia virus cap methyltransferase.
Complementation of abd1Δ cells by vaccinia virus cap methyltransferase is dependent on the stimulatory vD12 subunit (Fig. 1). Thus, the genetic assay should be sensitive to mutations of the vD12 subunit that either diminish its affinity for the catalytic subunit or affect its allosteric activation. Saha and Shuman previously identified and characterized double-alanine mutations of adjacent amino acids of vD12 that compromised its interaction in vitro with vD1-C (31). Here we tested the effects of such mutations on the in vivo activity of the vaccinia virus methyltransferase in yeast. vD12 mutants L61A-K62A, F176A-K177A, F245A-L246A, and N42A-Y43A were cloned into CEN and 2μm yeast expression vectors, which were then introduced into yeast abd1Δ cells together with the wild-type vD1-C gene on a CEN or 2μm plasmid. Complementation was tested by plasmid shuffle, and 5-FOA survivors were streaked on YEPD agar at 19, 30, and 37°C (Table 3).
TABLE 3.
Effects of vD12 mutations on abd1 complementationa
| vD12 allele + vD1-C | Growth at temp (°C):b
|
||
|---|---|---|---|
| 19 | 30 | 37 | |
| CEN | |||
| WTc | ++ | ++ | ++ |
| N42A-Y43A | ++ | + | − |
| L61A-K62A | ++ | ++ | + |
| F176A-K177A | − | − | − |
| F245A-L246A | ++ | ++ | − |
| 2μm | |||
| WT | +++ | +++ | +++ |
| N42A-Y43A | +++ | +++ | +++ |
| L61A-K62A | +++ | +++ | +++ |
| F176A-K177A | + | + | − |
| F245A-L246A | +++ | +++ | +++ |
The indicated alleles of vD12 were tested by plasmid shuffle for complementation of abd1Δ. CEN vD12 plasmids were cotransformed with CEN vD1-C (CEN) and 2μm vD12 plasmids were cotransformed with 2μm vD1-C (2μm).
+++ indicates colony sizes indistinguishable from those of strains bearing wild-type ABD1, ++ indicates slightly reduced colony sizes, + indicates that only pinpoint colonies were formed, and − indicates no growth.
WT, wild type.
F176A-K177A was lethal at all temperatures in single copy and viable but severely defective in high copy, i.e., 2μm F176A-K177A cells formed pinpoint colonies at 19 and 30°C and failed to grow at 37°C. The three other vD12 mutants displayed less drastic phenotypes in single copy that were suppressed completely by increased gene dosage (Table 3). Cells bearing L61A-K62A or F245A-L246 in single copy grew as well as wild-type vD1-C cells at 19 and 30°C but were inviable at 37°C. CEN N42A-Y43A cells grew as well as wild-type vD1-C cells at 19°C, grew more slowly at 30°C, and failed to form colonies at 37°C. Yet 2μm L61A-K62A, F245A-L246, and N42A-Y43A cells displayed +++ growth at all temperatures.
The findings that vD12 mutations that affect subunit heterodimerization in vitro display temperature sensitivity in vivo and dosage suppression suggest that (i) the effects of the vD12 mutations on its affinity for the catalytic subunit are exacerbated at 37°C and (ii) weakened subunit affinity can be compensated by simple mass action, whereby increasing the concentration of both subunits by raising gene copy number allows formation of a threshold level of active heterodimer that suffices for yeast viability.
DISCUSSION
Virus-encoded cap methylating enzymes are potential targets for antiviral drugs, because the pharmacological sensitivities of the viral cap methylation reactions can differ from those of the host cell. This notion is supported by the broad-spectrum efficacy of S-adenosyl homocysteine (SAH) hydrolase inhibitors against poxviruses and many RNA viruses that encode their own cap-methylating enzymes (2, 10). SAH is a direct product and a competitive inhibitor of cap methylation reactions. Pharmacological inhibition of SAH hydrolase elevates intracellular levels of SAH by preventing its cleavage to homocysteine and adenosine. The inference from the antiviral action of SAH hydrolase inhibitors is that viral cap methyltransferases are more sensitive to SAH than are the numerous cellular enzymes that use AdoMet as a substrate. Blocking a cellular enzyme to interdict viral capping is an indirect approach that can be supplanted by targeting the viral cap methylating enzymes directly. The poxvirus cap guanine-N7 methyltransferase is an attractive drug target, because it is uniquely dependent on a virus-encoded regulatory subunit of which there is no homolog in the host cell. The discovery of new antipoxviral drugs is an immediate concern, given the threat of smallpox as a biological weapon and the inevitability of complications as prophylaxis with live vaccinia virus is resumed.
The yeast-based genetic system we describe here provides a valuable tool for basic and applied studies of the poxvirus cap methyltransferase. We show that vaccinia virus cap methyltransferase can function in lieu of the essential yeast enzyme Abd1 and that its in vivo activity requires coexpression of the catalytic vD1-C and stimulatory vD12 subunits. Hence the methylation function of the viral enzyme can be studied in vivo independent of the other roles played by the capping enzyme during poxvirus replication (transcription initiation, transcription termination, and telomere resolution). The availability of isogenic yeast strains containing mammalian versus poxvirus cap methyltransferases provides a means of drug discovery aimed at blocking viral cap formation (30). For example, any compound that is selectively cytotoxic to a vD1-C/vD12 strain but not to an HCM1 strain is a strong candidate for a specific inhibitor of poxvirus cap methylation.
The yeast genetic system also provides an effective route for identification of amino acids in either subunit that are important for cap methylation. Four individual side chains in the catalytic subunit have been shown to be critical for activity in vivo and in vitro: Asp604, Asp620, Arg632, and Tyr683. All four of these side chains are conserved and essential in cellular cap methyltransferases. Molecular modeling based on the crystal structures of other methyltransferases suggests that the two essential aspartates can contact AdoMet and the essential arginine can interact with the triphosphate bridge of the GpppN cap (4). In the absence of a real atomic structure for any cap guanine-N7 methyltransferase, further genetic and biochemical dissection of structure-activity relationships in vD1-C should continue to reveal similarities and distinctions between the catalytic components of the cellular and poxvirus enzymes.
The utility of the yeast-based genetic system extends to the stimulatory vD12 subunit of the poxvirus capping enzyme. The architecture of the vD12 side of the subunit interface and the mechanism by which vD12 binding activates the catalytic subunit are largely uncharted. Substrate-protein cross-linking studies indicate that activation does not entail D12 stimulation of the binding of AdoMet or the RNA acceptor to the active site on the vD1-C subunit (17, 29), suggesting an allosteric effect at the active site perhaps contingent on simultaneous binding of AdoMet and the GpppRNA methyl acceptor (21). Screening in yeast for mutations of vD12 that display a gene dosage-dependent growth phenotype allows one to identify candidate components of the subunit interface. Moreover, the yeast system offers a potentially powerful means to delineate specific contacts at the subunit interface or the putative allosteric activation site by performing forward genetic screenings for allele-specific suppressors, i.e., selection for single-copy vD1-C mutations that suppress single-copy vD12 mutant phenotypes and vice versa.
Finally, it is noteworthy that the poxvirus cap methyltransferase functions in yeast without attachment to a cellular targeting protein. Capping of pre-mRNA in eukaryotic cells is intimately linked to transcription by RNA Pol II. Cellular capping enzymes are targeted to nascent Pol II transcripts via physical interactions with the phosphorylated carboxyl-terminal domain (CTD) of the largest subunit of Pol II (11, 25). The CTD serves as a scaffold for a multitude of nuclear proteins involved in mRNA maturation (1). Poxviruses replicate entirely in the cytoplasm. Thus, it is sensible that the poxvirus-encoded DNA-dependent RNA polymerase has no counterpart of the CTD (3) and that the poxvirus capping enzyme does not bind to the phosphorylated CTD of nuclear Pol II (25). The earlier finding that the in vivo activity of the vaccinia virus triphosphatase-guanylyltransferase domain in yeast was contingent on its fusion to a cellular CTD-binding protein was taken as evidence in support of the need for a targeting vehicle to attain genetic complementation of cellular cap-forming enzymes by their viral analogs (18). The present study shows that this does not apply to cap methyltransferase.
An intriguing inference from the complementation of abd1Δ by vaccinia virus cap methyltransferase is that there are alternative means to access the 5′ ends of pre-mRNAs for guanine N7 methylation that are independent of physical recruitment to the Pol II CTD, to which the vaccinia virus capping enzyme does not bind. The simplest scenario is that the poxvirus capping enzyme is expressed at sufficiently high level under the control of the yeast TPI1 promoter to encounter its RNA substrate in the yeast nucleus without any need for protein-protein interactions with the transcription elongation complex. An alternative scenario, which runs counter to orthodox views of mRNA processing, is that the poxvirus methyltransferase acts on GpppN-capped yeast mRNAs after they have been transported from nucleus to the cytoplasm. Preliminary experiments in which vaccinia virus cap methyltransferase was expressed in yeast as a fusion to green fluorescent protein suggest that the viral enzyme is distributed diffusely throughout the cell. Finer analysis is needed to determine whether the viral cap methyltransferase is acting in the yeast nucleus, the cytoplasm, or both.
Acknowledgments
This work was supported by NIH grants GM42498, GM52470, and AI053471.
REFERENCES
- 1.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-343. [DOI] [PubMed] [Google Scholar]
- 2.Borchardt, R. T., B. T. Keller, and U. Patel-Thombre. 1984. Neplanocin A: a potent inhibitor of S-adenosylhomocysteine hydrolase and of vaccinia virus multiplication in mouse L929 cells. J. Biol. Chem. 259:4353-4358. [PubMed] [Google Scholar]
- 3.Broyles, S. S., and B. Moss. 1986. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequences and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc. Natl. Acad. Sci. USA 83:3141-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bujnicki, J. M., M. Feder, M. Radlinska, and L. Rychlewski. 2001. mRNA guanine-N7 cap methyltransferases: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure relationships. BMC Bioinformatics 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calero, G., K. F. Wilson, T. Ly, J. L. Rios-Steiner, J. C. Clardy, and R. A. Cerione. 2002. Structural basis of m7GpppG binding to the nuclear cap-binding complex. Nat. Struct. Biol. 9:912-917. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, M. S., and A. M. DeLange. 1991. A temperature-sensitive lesion in the small subunit of vaccinia virus-encoded mRNA capping enzyme causes a defect in viral telomere resolution. J. Virol. 65:4042-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong, P., and S. Shuman. 1992. Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J. Biol. Chem. 267:16424-16429. [PubMed] [Google Scholar]
- 8.Cong, P., and S. Shuman. 1993. Covalent catalysis in nucleotidyl transfer: a KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J. Biol. Chem. 268:7256-7260. [PubMed] [Google Scholar]
- 9.Cong. P., and S. Shuman. 1995. Mutational analysis of mRNA capping enzyme identifies amino acids involved in GTP binding, enzyme-guanylate complex formation, and GMP transfer to RNA. Mol. Cell. Biol. 15:6222-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Clercq, E. 2001. Vaccinia virus inhibitors as a paradigm for the chemotherapy of poxvirus infections. Clin. Microbiol. Rev. 14:382-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabrega, C., V. Shen, S. Shuman, and C. D. Lima. Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II. Mol. Cell, in press. [DOI] [PubMed]
- 12.Gong, C., and S. Shuman. Mapping the active site of vaccinia virus RNA triphosphatase. Virology, in press. [DOI] [PubMed]
- 13.Hassett, D. E., J. I. Lewis, X. Xing, L. DeLange, and R. C. Condit. 1997. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered change-to-alanine mutagenesis and transient dominant selection. Virology 238:391-409. [DOI] [PubMed] [Google Scholar]
- 14.Hausmann, S., C. P. Vivarès, and S. Shuman. 2002. Characterization of the mRNA capping apparatus of the microsporidian parasite Encephalitozoon cuniculi. J. Biol. Chem. 277:96-103. [DOI] [PubMed] [Google Scholar]
- 15.Higman, M. A., N. Bourgeois, and E. G. Niles. 1992. The vaccinia virus mRNA (guanine-N7-) methyltransferase requires both subunits of the mRNA capping enzyme for activity. J. Biol. Chem. 267:16430-16437. [PubMed] [Google Scholar]
- 16.Higman, M. A., L. A. Christen, and E. G. Niles. 1994. The mRNA (guanine-7-) methyltransferase domain of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J. Biol. Chem. 269:14974-14981. [PubMed] [Google Scholar]
- 17.Higman, M. A., and E. G. Niles. 1994. Location of the S-adenosyl-l-methionine binding region of the vaccinia virus mRNA (guanine-7-) methyltransferase. J. Biol. Chem. 269:14982-14987. [PubMed] [Google Scholar]
- 18.Ho, C. K., A. Martins, and S. Shuman. 2000. A yeast-based genetic system for functional analysis of viral mRNA capping enzymes. J. Virol. 74:5486-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, Y., X. Mao, L. Deng, P. Cong, and S. Shuman. 1995. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J. Virol. 69:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao, X., and S. Shuman. 1994. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit: identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J. Biol. Chem. 269:24472-24479. [PubMed] [Google Scholar]
- 21.Mao, X., and S. Shuman. 1996. Vaccinia virus mRNA (guanine-7-) methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photocrosslinking of the cap to the methyl acceptor site. Biochemistry 35:6900-6910. [DOI] [PubMed] [Google Scholar]
- 22.Mao, X., B. Schwer, and S. Shuman. 1995. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 15:4167-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao, X., B. Schwer, and S. Shuman. 1996. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol. Cell. Biol. 16:475-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, S. A., and B. Moss. 1975. Modification of RNA by mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J. Biol. Chem. 250:9330-9335. [PubMed] [Google Scholar]
- 25.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′ Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated C-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myette, J., and E. G. Niles. 1996. Domain structure of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli of a subdomain possessing the RNA 5′-triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J. Biol. Chem. 271:11936-11944. [DOI] [PubMed] [Google Scholar]
- 27.Myette, J., and E. G. Niles. 1996. Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleoside triphosphate phosphohydrolase activities: demonstration that both activities are carried out at the same active site. J. Biol. Chem. 271:11945-11952. [DOI] [PubMed] [Google Scholar]
- 28.Niles, E. G., and L. Christen. 1993. Identification of the vaccinia virus mRNA guanylyltransferase active site lysine. J. Biol. Chem. 268:24986-24989. [PubMed] [Google Scholar]
- 29.Niles, E. G., L. Christen, and M. A. Higman. 1994. Direct photolinkage of GTP to the vaccinia virus mRNA (guanine-7-) methyltransferase GTP methyl acceptor site. Biochemistry 33:9898-9903. [DOI] [PubMed] [Google Scholar]
- 30.Saha, N., B. Schwer, and S. Shuman. 1999. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274:16553-16562. [DOI] [PubMed] [Google Scholar]
- 31.Saha, N., and S. Shuman. 2001. Effects of alanine cluster mutations in the D12 subunit of vaccinia virus mRNA (guanine-N7) methyltransferase. Virology 287:40-48. [DOI] [PubMed] [Google Scholar]
- 32.Schwer, B., N. Saha, X. Mao, H. W. Chen, and S. Shuman. 2000. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics 155:1561-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuman, S. 1989. Functional domains of vaccinia virus mRNA capping enzyme: analysis by limited tryptic digestion. J. Biol. Chem. 264:9690-9695. [PubMed] [Google Scholar]
- 34.Shuman, S. 2000. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1-40. [DOI] [PubMed] [Google Scholar]
- 35.Shuman, S. 2001. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harbor Symp. Quant. Biol. 66:301-312. [DOI] [PubMed] [Google Scholar]
- 36.Shuman, S., S. Broyles, and B. Moss. 1987. Purification and characterization of a transcription termination factor from vaccinia virions. J. Biol. Chem. 262:12372-12380. [PubMed] [Google Scholar]
- 37.Shuman, S., and S. G. Morham. 1990. Domain structure of vaccinia virus mRNA capping enzyme: activity of the Mr 95,000 subunit expressed in Escherichia coli. J. Biol. Chem. 265:11967-11972. [PubMed] [Google Scholar]
- 38.Venkatesan, S., A. Gershowitz, and B. Moss. 1980. Modification of the 5′ end of mRNA: association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7) methyltransferase complex from vaccinia virus. J. Biol. Chem. 255:903-908. [PubMed] [Google Scholar]
- 39.Vos, J. C., M. Sasker, and H. G. Stunnenberg. 1991. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 10:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S. P., and S. Shuman. 1997. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J. Biol. Chem. 272:14683-14689. [DOI] [PubMed] [Google Scholar]
- 41.Yu, L., and S. Shuman. 1996. Mutational analysis of the triphosphatase domain of vaccinia virus mRNA capping enzyme. J. Virol. 70:6162-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, L., A. Martins, L. Deng, and S. Shuman. 1997. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J. Virol. 71:9837-9843. [DOI] [PMC free article] [PubMed] [Google Scholar]