Abstract
Epstein-Barr virus (EBV)-encoded oncogene latent membrane protein (LMP) 1, which is consistently expressed in multiple EBV-associated malignancies, has been proposed as a potential target antigen for any future vaccine designed to control these malignancies. However, the high degree of genetic variation in the LMP1 sequence has been considered a major impediment for its use as a potential immunotherapeutic target for the treatment of EBV-associated malignancies. In the present study, we have employed a highly efficient strategy, based on ex vivo functional assays, to conduct an extensive sequence-wide analysis of LMP1-specific T-cell responses in a large panel of healthy virus carriers of diverse ethnic origin and nasopharyngeal carcinoma patients. By comparing the frequencies of T cells specific for overlapping peptides spanning LMP1, we mapped a number of novel HLA class I- and class II-restricted LMP1 T-cell epitopes, including an epitope with dual HLA class I restriction. More importantly, extensive sequence analysis of LMP1 revealed that the majority of the T-cell epitopes were highly conserved in EBV isolates from Caucasian, Papua New Guinean, African, and Southeast Asian populations, while unique geographically constrained genetic variation was observed within one HLA A2 supertype-restricted epitope. These findings indicate that conserved LMP1 epitopes should be considered in designing epitope-based immunotherapeutic strategies against EBV-associated malignancies in different ethnic populations.
The Epstein-Barr virus (EBV) is not only one of the most widespread human viruses, but, somewhat paradoxically, it is also linked to a range of neoplasms (1). These include various B- and T-cell non-Hodgkin's lymphomas, Hodgkin's disease (HD), and several lymphoepithelioma-like carcinomas, of which nasopharyngeal carcinoma (NPC) is the archetype. The association of EBV with these tumors and the oncogenic potential of EBV in vitro are well documented (6, 17). CD8+ T-cell activity has an important role in controlling EBV infections by recognizing small peptides derived from infected cells presented on the surface by major histocompatibility complex (MHC) class I molecules. EBV-specific cytotoxic T lymphocyte (CTL) preparations can be generated in vitro by stimulating memory T cells from peripheral blood of healthy virus carriers with cells of autologous EBV-transformed lymphoblastoid cell lines (LCLs) (7, 16). Within an LCL, EBV expresses six nuclear antigens (EBNAs 1, 2, 3A, 3B, 3C, and LP) and two latent membrane proteins (LMP1 and 2). Of these, the EBNA3 family (EBNA 3A, 3B, 3C) is immunodominant for CTL responses over a wide range of HLA backgrounds (7, 16).
In both HD and NPC, the tumor cells express viral proteins known to provide target epitopes for CTLs. These two malignancies express EBNA1, BARF0, LMP1, and LMP2. EBNA1 includes a unique glycine-alanine repeat (GAr), which acts as a cis-inhibitory signal for proteasomal degradation and thus blocks endogenous presentation of CTL epitopes within this antigen (12). CTL presentation of BARF0 epitopes is impaired by the differential splicing of viral transcripts, resulting in dominant protein isoforms from which the CTL determinants are deleted (9). As opposed to EBNA1 and BARF0, both LMP1 and LMP2 are strong targets for EBV-specific CTLs, and therefore much attention has been directed at identifying target epitopes within these two antigens (8, 10, 13). Since it is well established that immunization with whole viral proteins does not elicit an efficient CTL response, interest has been directed toward vaccines and immunotherapeutic strategies based on defined epitope sequences. This is particularly the case with oncogenic viruses such as EBV, since individual viral genes such as LMP1, introduced in recombinant vectors, have the potential to initiate oncogenic processes. Previous studies have suggested that although LMP2 epitope sequences are generally well conserved (10), the inclusion of LMP1 epitopes in future vaccine design may be significantly compromised, as the virus isolates recovered from different geographic regions display a high degree of genetic variation within the LMP1 gene (5, 14, 19, 20).
In the present study, we adopted the gamma interferon (IFN-γ)-based ELISPOT assay to conduct an extensive sequence-wide analysis of LMP1-specific T-cell responses in a large panel of ethnically diverse healthy virus carriers and NPC patients. This approach was combined with the extensive sequence analysis of virus isolates from different geographic regions of the world to investigate the degree of genetic variation within the CTL epitopes mapped from LMP1. Based on these assays, we found that LMP1 protein includes a number of potential epitopes, which are targeted by both HLA class I- and class II-restricted T cells. Unexpectedly, sequence analysis of virus isolates from different geographic regions of the world revealed that most of these epitopes are highly conserved and are efficiently recognized by individuals of diverse ethnic origin. These findings provide an opportunity to exploit the highly conserved LMP1 epitopes as immunotherapeutic tools for the treatment of EBV-associated malignancies such as NPC and HD.
MATERIALS AND METHODS
Establishment and maintenance of cell lines.
EBV-transformed LCLs were established from seropositive donors by exogenous virus transformation of peripheral B cells using the B95-8, BL74, and QIMR-WIL virus isolates. These cell lines were routinely maintained in RPMI 1640 supplemented with 2 mM l-glutamine, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml plus 10% fetal calf serum (growth medium). In addition, the peptide transporter (TAP)-negative B × T hybrid cell line 0.174 × CEM.T2 (referred to as T2) (18) was used for peptide stabilization assays.
To generate phytohemagglutinin (PHA) blasts, peripheral blood mononuclear cells (PBMC) were stimulated with PHA (CSL Ltd., Melbourne, Australia), and after 3 days of culture, growth medium containing MLA 144 supernatant and highly purified recombinant human interleukin-2 (rIL-2) was added (7). PHA blasts were propagated by twice-weekly replacement of rIL-2 and MLA supernatant (no further PHA added) for up to 6 weeks.
Virus isolates.
LCLs were established from a panel of unrelated healthy EBV-seropositive African, Caucasian, and Papua New Guinean donors by spontaneous outgrowth from peripheral lymphocytes cultured in the presence of 0.1 μg of cyclosporine A/ml (15). A total of 29 spontaneous LCLs (11 Caucasian, 11 Papua New Guinean, 2 African and 5 Southeast Asian) were used to recover the resident EBV isolate from each individual. In addition, 16 virus isolates from Southeast Asia were directly sequenced from EBV-carrying NPC biopsy samples.
PCR and DNA sequencing of EBV gene fragment.
Specific oligonucleotide primers flanking different epitopes within the LMP1 gene were selected for PCR amplification. The resulting PCR products were purified using QIAquick spin columns (Qiagen Inc., Chatsworth, Calif.) and sequenced in both directions using a PRISM ready reaction dye deoxy terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, Calif.) in accordance with the manufacturer's protocol.
Synthetic peptides.
The amino acid sequences of the peptides were derived from the published LMP1 sequences from the Caucasian prototype 1 EBV strain B95-8. A panel of 45 peptides of 17 amino acids in length, overlapping by 8 residues and spanning the entire LMP1 sequence, was synthesized on an automated peptide synthesizer by the Merrifield solid phase method (Mimotopes Pty Ltd., Melbourne, Australia). Peptide aliquots were dissolved in 20% dimethyl sulfoxide at 2 mg/ml.
EBV-seropositive donors.
A panel of 42 healthy virus carriers and NPC patients of diverse ethnic origin were recruited for this study. These donors included 25 Caucasian, 14 Thai, 6 Thai-Chinese, 3 Indian, and 1 Vietnamese person. Each of the donors was HLA typed using both serological and/or PCR-based DNA typing methods.
IFN-γ ELISPOT assay.
The ELISPOT assay was used to assess whether stimulation of PBMC from a large panel of seropositive donors with LMP1 peptides could induce IFN-γ expression in T cells (2). Briefly, 96-well mixed cellulose ester membrane plates (Millipore, Bedford, Mass.) were precoated with 100 μl/well of 10 μg/ml of anti-IFN-γ monoclonal antibody (MAb) 1-D1K (Mabtech, Stockholm, Sweden) overnight at 4°C. The plate was washed six times with phosphate-buffered saline (PBS), and unreactive sites were blocked with 5% fetal bovine serum. PBMC from healthy EBV carriers of known HLA types were separated from whole blood by Ficoll-Hypaque (Sigma) density gradient centrifugation. PBMC were added in triplicate wells at 2.5 × 105 cells in a volume of 100 μl/well, and then peptides were added onto the PBMC at a final concentration of 5 μg/ml. PBMCs were also tested at different concentrations in triplicate wells at 2.5 × 105, 1.5 × 105, and 1 × 105 cells/well. PBMC with various peptide concentrations, namely, 10, 5, and 1 μg/ml, were assayed. For negative controls, PBMC were incubated in growth medium alone without addition of peptides. The plate was incubated at 37°C with 5% CO2 overnight (about 16 to 20 h) and thereafter washed three times with PBST (0.05% Tween-20 in PBS) and three times with PBS before 100 μl/well of 1-μg/ml anti-IFN-γ mAb 7-B6-1 (Mabtech, Stockholm, Sweden) was added; the plate was then incubated at room temperature for 3 to 4 h. After incubation, the plate was washed again thrice each with PBST and PBS, and 100 μl/well of 1-μg/ml streptavidin-alkaline phosphatase conjugate (Sigma) was added. The plate was incubated at room temperature for 2 h. Wells were washed again three times each with PBST and PBS, and individual IFN-γ-producing cells were detected as dark spots after about 30 min to 1 hour of color reaction with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium in an alkaline phosphatase-conjugated substrate (BCIP/NBT; Sigma). Spots were counted automatically using image analysis software (ImagePro) (2) and were expressed as spot-forming cells (SFC) per 106 PBMCs. The number of IFN-γ-secreting T cells was calculated by subtracting the negative control value from the SFC count.
Depletion of CD4+ and CD8+ cells.
CD4+ cells or CD8+ cells were depleted from fresh PBMC, using anti-human CD4 or CD8 immunomagnetic beads (Dynabeads M450-CD4 and M450-CD8), respectively (Dynal, Oslo, Norway), in accordance with the manufacturer's instructions. Efficient depletion was confirmed by staining the depleted PBMC by dual staining with fluorescein isothiocyanate-conjugated anti-CD8 and phycoerythrin-conjugated anti-CD4 antibody and flow cytometry on a Coulter EPICS XL cytometer. The protocol reliably depleted >95% of the CD4+ or CD8+ cells, as confirmed by FACScan analysis. Cells were recounted, resuspended in growth medium, and set up in triplicate ELISPOT assays.
Establishment of polyclonal CTL lines and LMP1-specific CTL clones.
Polyclonal CTL lines and LMP1-specific CTL clones were established according to previously published methods (8, 15). Briefly, 2 × 106 PBMC were stimulated with 1 × 106 autologous lymphocytes (responder-to-stimulator ratio of 2:1) pulsed with 10 μM peptide for 1 h in 2 ml in a 24-well plate. After 3 days, culture medium with IL-2 (10 U/ml) was added, and the cells were further expanded. These lymphocytes were restimulated on day 7 with γ-irradiated (8,000 rad) autologous LCLs. After 10 days in culture medium, the cells were used as polyclonal effectors in a standard 51Cr-release assay against peptide-sensitized autologous PHA blasts.
To generate peripheral blood CTL clones specific for the LMP1-derived peptides, PBMC of healthy donors were reactivated by combining 2 × 106 PBMC with peptide-sensitized (10 μg of peptide/ml) autologous PBMC (1 × 106) in 2-ml wells of a 24-well plate in culture medium. After 3 days, the cells were seeded onto 0.35% agarose and maintained in T-cell growth medium containing rIL2 (50 IU/ml). After another 3 days, growing clones were transferred to 96-well round-bottom tissue culture plates (Life Technologies) and cultured in T-cell growth medium containing rIL2 (50 to 100 IU/ml).
T cell-T cell killing.
This technique (4) is based on the ability of cells in CTL culture to present peptide antigens to each other which, given the appropriate peptide, results in CTL-CTL killing. CTL clones were incubated (about 300 cells/well) in 25 μl of T-cell medium containing 10 μM peptide. Lysis of cells was then assessed with an inverted microscope after overnight incubation at 37°C.
Cytotoxicity assays.
Target cells were presensitized with synthetic peptide epitopes and then incubated with 51Cr for 90 min. Following incubation, these cells were washed in growth medium and used as targets in a standard 5-h 51Cr-release assay (3, 15).
MHC stabilization assays.
To assess MHC binding by the HLA A2-restricted LMP1 epitope variants from EBV isolates, T2 cells (2 × 105) were incubated with 200 μl of each of the peptides (100 μg/ml) at 26°C for 14 to 16 h, followed by incubation at 37°C for 2 to 3 h. After the incubations, HLA A2 expression was measured by fluorescence-activated cell sorting using a specific monoclonal antibody (HB82; ATCC).
RESULTS
LMP1-specific T-cell responses in ethnically diverse healthy virus carriers.
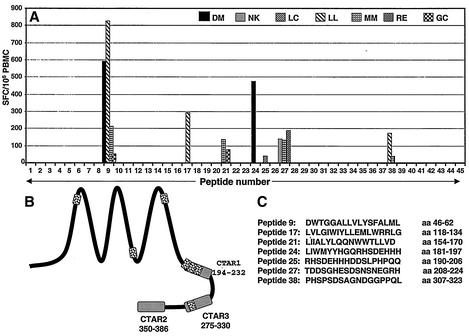
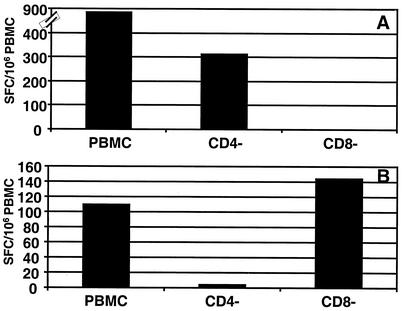
To comprehensively profile the memory T-cell responses against the LMP1 in seropositive virus carriers, we used the ELISPOT assay, which allowed rapid ex vivo profiling of LMP1-specific immune response without prolonged in vitro culture. PBMC isolated from a panel of seropositive healthy virus carriers were stimulated with the complete set of overlapping LMP1 peptides, and the cells that produced IFN-γ were detected. Representative data from the ELISPOT assays using a complete set of overlapping peptides (17 amino acids in length, overlapping by 8 residues) are presented in Fig. 1. Of the 21 healthy donors assessed, 7 donors showed a strong ex vivo response to 7 of the 45 peptides (SFC range, 39 to 824 SFC/106 PBMC) (Fig. 1A). These potential epitopes are distributed throughout the sequence including the transmembrane domains and C terminus (Fig. 1B). A list of all the 17-mer peptides recognized by the different donors is shown in Fig. 1. To identify the T-cell subsets responding to these peptides, PBMC from the seven donors who recognized 17-mer peptides were fractionated into CD4-depleted and CD8-depleted populations and then tested again in ELISPOT assays. Five of these peptides included reactivity by both CD4+ and CD8+ T cells, which suggested that these sequences include both MHC class I- and class II-restricted epitopes. On the other hand, peptide DWTGGALLVLYSFALML clearly showed only CD8+ T-cell-dependent reactivity, while the T-cell response to the peptide TDDSGHESDSNSNEGRH was predominantly mediated by CD4+ cells (Fig. 2A and B). The CD4-dependent reactivity of the peptide TDDSGHESDSNSNEGRH was consistent with the previous observations by Leen and colleagues, who mapped an HLA DQ2-restricted epitope within this sequence (11).
FIG. 1.
Ex vivo profiling of LMP1-specific T-cell responses in a panel of healthy seropositive individuals. (A) PBMC from healthy seropositive individuals were stimulated with overlapping synthetic peptides (10 μg/ml) from LMP1, and IFN-γ production was measured in ELISPOT assays as described in Material and Methods. The results are expressed as SFC/106 PBMC. (B) Schematic distribution of T-cell reactivity within the LMP1 protein. Each of the shaded regions refers to T-cell reactivity toward a 17-mer sequence which includes multiple class I- and/or class II-restricted minimal epitopes (see Table 1). (C) Amino acid sequences of the peptide epitopes recognized by healthy virus carriers in ELISPOT assays.
FIG. 2.
Ex vivo detection of IFN-γ secretion by CD4+ and CD8+ T-cell populations following stimulation with LMP1 peptides. CD4+ cells or CD8+ cells were depleted from fresh PBMCs using anti-human CD4 or CD8 immunomagnetic beads, as described in Materials and Methods. These cells were resuspended in growth medium and stimulated with DWTGGALLVLYSFALML (A) and TDDSGHESDSNSNEGRH (B) peptides (10 μg/ml) and then tested for IFN-γ secretion using ELISPOT assays.
Fine mapping of LMP1-specific T-cell responses.
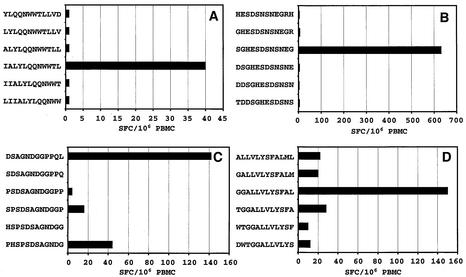
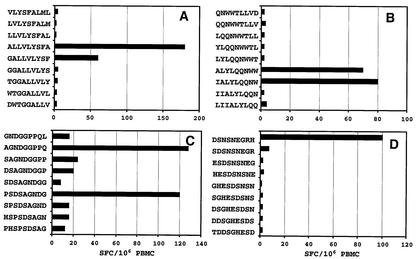
Fine mapping of the LMP1-specific CD8+ T-cell responses was achieved using truncations of the 17-mer peptides in ELISPOT assays. In the initial set of experiments, 12-mer overlapping peptides were used to map the immunogenic region within the 17-mer peptides. Representative data from these overlapping peptides for four individual donors are shown in Fig. 3A to D. A total of twelve 12-mer sequences were identified as potential determinants (see Table 1). Based on these 12-mer peptides, overlapping 9-mer peptides spanning the 12-mer peptides were used to define the minimal epitope sequences. Representative data from four different donors are presented in Fig. 4A to D. A total of 18 9-mer sequences were recognized as potential minimal epitope sequences (Table 1). Two of these minimal sequences (YLLEMLWRL and YLQQNWWTL) have previously been identified as CTL epitopes (8), while other sequences have been identified for the first time in this study. Of these 18 minimal sequences, we were able to define HLA class I restriction for six sequences restricted through HLA A2 and/or B57/B58. Although we were unable to define the HLA restriction for the remaining 12 sequences, the analysis carried out precludes their restriction through HLA A2 and HLA B57/B58.
FIG. 3.
Mapping of a minimal epitope sequence using ELISPOT assays. PBMC from donors MM (A and B), RE (C) and LL (D) were stimulated with overlapping peptides (5 μg/ml), and IFN-γ production was measured in ELISPOT assays, as described in Materials and Methods. The results are expressed as SFC/106 PBMC.
TABLE 1.
Comprehensive list of LMP1 sequences recognized by EBV-specific T cells from healthy virus carriers
| Peptide no. | Epitope sequence | 12-mer sequence(s) recognized | Minimal sequence(s) recognized | CTL reactivitya | HLA restrictionb | Reference or source |
|---|---|---|---|---|---|---|
| 9 | DWTGGALLVLYSFALML | ALLVLYSFALML | ALLVLYSFAL | Y | HLA A2 | This study |
| GALLVLYSFALM | LLVLYSFAL | Y | ND | This study | ||
| DWTGGALLVLYS | ALLVLYSFA | Y | HLA A2 | This study | ||
| GGALLVLYSFAL | VLYSFALML | Y | HLA A2 | |||
| 17 | LVLGIWIYLLEMLWRRLG | IYLLEMLWRLG | YLLEMLWRL | Y | HLA A2 | 8 |
| 21 | LIIALYLQQNWWTLLVD | IALYLQQNWWTL | IALYLQQNW | Y | HLA B57/B58 | This study |
| YLQQNWWTLLVD | ALYLQQNWW | Y | ND | This study | ||
| YLQQNWWTL | Y | HLA A2 | 8 | |||
| QNWWTLLVD | NT | ND | This study | |||
| LYLQQNWWT | NT | ND | This study | |||
| 24 | LIWMYYHGQRHSDEHHH | YYHGQRHSDEHH | QRHSDEHHH | ND | ND | This study |
| IWMYYHGQRHSD | GQRHSDEHH | ND | ND | This study | ||
| YYHGQRHSD | NT | ND | This study | |||
| WMYYHGQRH | NT | ND | This study | |||
| 27 | TDDSGHESDSNSNEGRH | SGHESDSNSNEG | ESDSNSNEG | Y | HLA DQ2 | 11 |
| DSNSNEGRH | ND | ND | This study | |||
| 38 | PHSPSDSAGNDGGPPQL | SDSAGNDGGPPQ | AGNDGGPPQ | ND | ND | This study |
| DSAGNDGGPPQL | PSDSAGNDG | NT | ND | This study |
Y, yes; NT, not tested; ND, not detected.
ND, not defined.
FIG. 4.
Mapping of a minimal epitope sequence using ELISPOT assays. PBMC from donors LL (A), MM (B), and RE (C and D) were stimulated with overlapping peptides (1 μg/ml), and IFN-γ production was measured in ELISPOT assays as described in Materials and Methods. The results are expressed as SFC/106 PBMC.
Characterization of novel LMP1 CTL epitopes.
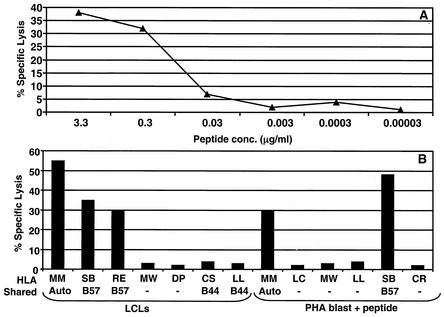
To further characterize the minimal T-cell epitopes defined by ELISPOT assays, we generated polyclonal and clonal CTL lines specific for these epitopes. PBMC from the healthy seropositive donors were stimulated with synthetic peptide epitopes, and the CTL clones or polyclonal lines were established and tested in standard 51Cr-release assays. A series of representative data for two of the novel epitopes (IALYLQQNW and ALLVLYSFA) is presented in Fig. 5 and 6. The IALYLQQNW peptide-specific CTL clones were generated using initial stimulation with peptide-sensitized autologous PBMC followed by continuous restimulation with irradiated autologous LCL. These CTL clones were screened for IALYLQQNW-specific CTL activity by 51Cr-release assays against serially diluted peptide-sensitized autologous PHA blast. Data presented in Fig. 5A clearly showed that CTL recognition of these target cells was dependent on the amount of prepulsed peptide. In the next set of experiments, these CTL clones were tested against a panel of LCLs and peptide-sensitized PHA blasts. Target cells sharing only HLA B57 with the CTLs were recognized by the IALYLQQNW-specific CTL clone, indicating that this epitope is restricted through the HLA B57 allele (Fig. 5B).
FIG. 5.
(A) CTL recognition of the IALYLQQNW epitope by CTL clone MM23. PHA blasts were sensitized with serial dilutions of the peptides and then exposed to an IALYLQQNW-specific CTL clone, MM22. (B) HLA class I restriction analysis for IALYLQQNW-specific CTL clone MM22. Autologous and allogeneic EBV-transformed LCLs and PHA blasts, some sharing HLA class I alleles with donor MM, were exposed to the CTL clone MM22. PHA blasts were presensitized with IALYLQQNW peptide. An effector-to-target ratio of 2:1 was used in the assay.
FIG. 6.
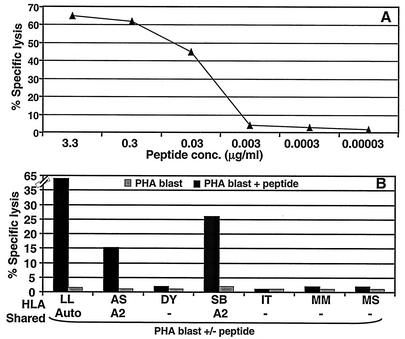
(A) CTL recognition of the ALLVLYSFA epitope by a CTL line from donor LL. PHA blasts were sensitized with serial dilutions of the peptides and then exposed to an ALLVLYSFA-specific CTL line. (B) HLA class I restriction analysis for an ALLVLYSFA-specific CTL line from donor LL. Autologous and allogeneic PHA blasts, some sharing HLA class I alleles with donor LL, were exposed to the epitope-specific CTL line in the absence or presence of the peptide. An effector-to-target ratio of 10:1 was used in the assay.
Using a similar approach described above, a novel HLA-A2-restricted CTL epitope was identified. The fine specificity of the ALLVLYSFA epitope-specific CTL line was confirmed by dose-response analysis of the minimal peptide (Fig. 6A). The HLA A2 restriction of the ALLVLYSFA epitope was determined using autologous and allogeneic PHA blasts. Target cells expressing HLA A2 and prepulsed with peptide ALLVLYSFA were recognized by specific CTL line (Fig. 6B).
Another 17-mer peptide, TDDSGHESDSNSNEGRH, showeda CD4+ T-cell response in ELISPOT assays in 3 of 21 healthy donors, with the magnitude of responses within the range of 131 to 190 SFC/106 PBMC (Fig. 1A). Polyclonal CTL lines established from two different donors showed recognition of peptide-coated autologous PHA blasts (data not shown). Two donors (MM and RE) recognized minimal peptides (ESDSNSNEG and DSNSNEGRH, respectively), but these minimized epitopes did not generate CTL lines. It is interesting that Leen and colleagues have previously mapped an HLA DQ2-restricted T-cell epitope within this 17-mer sequence (Table 1).
Recognition of minimal HLA class I- and class II-restricted LMP1 T-cell epitopes by ethnically diverse healthy individuals and NPC patients.
In order to determine the frequency of recognition of the LMP1 T-cell epitopes, PBMC from a panel of healthy virus carriers and NPC patients of diverse ethnic origin were screened by ELISPOT assays. A detailed summary of this analysis is presented in Table 2. Four different HLA class I-restricted and one HLA class II-restricted T-cell epitopes were included in this analysis. Strong responses were observed against the HLA-A2-restricted YLLEMLWRL, YLQQNWWTL, and ALLVLYSFA epitopes in 35 to 45% of the donors tested. These responses were more consistently seen in Caucasian donors, while these epitopes were less frequently recognized by Southeast Asian donors (both healthy individuals and NPC patients). The HLA-B57-restricted IALYLQQNW epitope was recognized by six of eight HLA B57-positive individuals. Previous studies with HIV CTL epitopes have shown that similar HLA class I molecules may present the identical peptides as CTL epitopes (5). Since the common subtypes of HLA B57 and HLA B58 differ by only a few amino acids and share similar peptide-binding motifs, we explored the possibility that IALYLQQNW may also be restricted through the HLA B58 allele. Indeed, five of nine HLA B58 healthy seropositive donors analyzed in our study showed an equally strong response to the minimal IALYLQQNW epitope (Table 2). These observations suggested that this epitope displayed dual HLA restriction for both HLA B57 and HLA B58. The HLA class II-restricted LMP1 epitope, TDDSGHESDSNSNEGRH, was recognized by all three healthy virus carriers tested (Table 2).
TABLE 2.
Frequency of T-cell responses to HLA class I- and HLA class II-restricted LMP1 epitopes in ethnically diverse healthy individuals and NPC patients
| Epitope | HLA type of subject | Subject code | Ethnic origin | T-cell response (SFC/106 PBMC) |
|---|---|---|---|---|
| ALLVLYSFA | A2 | SB | Caucasian | 480 |
| A2 | LL | Caucasian | 192 | |
| A2 | GC | Caucasian | 96 | |
| A2 | NK | Caucasian | 88 | |
| A2 | EL | Caucasian | 80 | |
| A2 | SE | Caucasian | 10 | |
| A2 | TL | Vietnamese | 128 | |
| A2 | WS | Caucasian | 592 | |
| A2 | PR | Thai-Chinese | 16 | |
| A2 | PA | Thai-Chinese | 0 | |
| A2 | WE | Thai | 0 | |
| A2 | SUr | Thai | 0 | |
| A2 | CHu | Thai | 0 | |
| A2 | DS | Caucasian | 1184 | |
| A2 | SOm | Thai-Chinese | 24 | |
| A2 | CI (NPC) | Thai-Chinese | 0 | |
| A2 | PAi (NPC) | Thai | 0 | |
| A2 | MAn (NPC) | Thai | 0 | |
| IALYLQQNW | B57 | KG | Caucasian | 1952 |
| B57 | RE | Indian | 897 | |
| B57 | SB | Caucasian | 455 | |
| B57 | MM | Caucasian | 80 | |
| B57 | SUm | Thai-Chinese | 0 | |
| B57 | SO | Thai | 48 | |
| B57 | AN | Thai | 100 | |
| B57 | CHa | Thai | 0 | |
| B58 | MB | Indian | 240 | |
| B58 | JD | Indian | 955 | |
| B58 | TL | Vietnamese | 504 | |
| B58 | ORa | Thai-Chinese | 186 | |
| B58 | UBo | Thai | 78 | |
| B58 | MAn (NPC) | Thai | 0 | |
| B58 | SUc (NPC) | Thai | 0 | |
| B58 | CI (NPC) | Thai-Chinese | 0 | |
| B58 | PAc (NPC) | Thai | 0 | |
| TDDSGHESDSNSNEGRH | NDa | NK | Caucasian | 141 |
| ND | MM | Caucasian | 131 | |
| ND | RE | Indian | 131 | |
| YLLEMLWRL | A2 | NK | Caucasian | 160 |
| A2 | AS | Caucasian | 100 | |
| A2 | JF | Caucasian | 0 | |
| A2 | WS | Caucasian | 66 | |
| A2 | CV | Caucasian | 20 | |
| A2 | LL | Caucasian | 42 | |
| A2 | EL | Caucasian | 300 | |
| A2 | TL | Vietnamese | 136 | |
| A2 | PR | Thai-Chinese | 0 | |
| A2 | PA | Thai-Chinese | 0 | |
| A2 | WE | Thai | 0 | |
| A2 | SUr | Thai | 0 | |
| A2 | CHu | Thai | 0 | |
| A2 | SUm | Thai-Chinese | 0 | |
| A2 | CI (NPC) | Thai-Chinese | 0 | |
| YLQQNWWTL | A2 | NK | Caucasian | 244 |
| A2 | AS | Caucasian | 110 | |
| A2 | JF | Caucasian | 44 | |
| A2 | WS | Caucasian | 38 | |
| A2 | CV | Caucasian | 26 | |
| A2 | LL | Caucasian | 28 | |
| A2 | EL | Caucasian | 100 | |
| A2 | TL | Vietnamese | 196 | |
| A2 | PR | Thai-Chinese | 0 | |
| A2 | PA | Thai-Chinese | 0 | |
| A2 | WE | Thai | 0 | |
| A2 | SUr | Thai | 0 | |
| A2 | CHu | Thai | 0 | |
| A2 | SUm | Thai-Chinese | 0 | |
| A2 | CI (NPC) | Thai-Chinese | 0 |
ND, not determined.
Sequence analysis of HLA class I-restricted LMP1 CTL epitopes in EBV isolates from diverse geographic regions.
Both in the present work and in previous studies from our laboratory (8), target epitopes in LMP1 were identified using CTLs reactivated with the reference type1 EBV strain, B95-8. However, if such epitopes are to form the basis of an effective CTL therapy for EBV-associated malignancies, we must first determine the extent to which these epitope sequences are conserved among other EBV strains from different world populations. Therefore, a large panel of EBV isolates from healthy virus carriers and NPC biopsies were sequenced across the DNA regions encoding the LMP1 epitopes. A detailed summary of the sequence analysis of four LMP1 epitopes (IALYLQQNW, ALLVLYSFA, YLLEMLWRL, and YLQQNWWTL) is presented in Table 3. The HLA A2-restricted (ALLVLYSFA and YLQQNWWTL) and HLA B57- and B58-restricted (IALYLQQNW) epitopes were highly conserved in virus isolates from Caucasian, African, and Southeast Asian donors (Table 3). Only a small number of virus isolates showed minor variation within the epitope sequences. Although the YLQQNWWTL and IALYLQQNW epitopes were generally conserved in Papua New Guinean isolates, alterations in the ALLVLYSFA were more common. Every single isolate showed an identical change from serine to alanine at position 7 (Table 3).
TABLE 3.
Sequences of HLA class I-restricted LMP1 epitopes in EBV isolates from Caucasian, African, Chinese, Papua New Guinean, and Indonesian individualsa
| Virus origin | Epitope sequence | HLA restriction | No. of isolates |
|---|---|---|---|
| B95.8 | GCC CIC CTT GTC CTC TAT TCC TTT GCT | ||
| ALLVLYSFA | HLA A2 | ||
| Caucasian | --------------------------- | 4 | |
| - - - - - - - - - | |||
| --G------------------------ | 1 | ||
| - - - - - - - - - | |||
| African | --------------------------- | 2 | |
| - - - - - - - - - | |||
| S.E. Asian (Spon) | --G------------------------ | 4 | |
| - - - - - - - - - | |||
| S.E. Asian (NPC) | --G------------------------ | 14 | |
| - - - - - - - - - | |||
| Papua New Guinean | --A--- T-G--------- G-------- | 7 | |
| - - - - - - A - - | |||
| --G--------------- G-------- | 1 | ||
| - - - - - - A - - | |||
| B95.8 | ATT GCT CTC TAT CTA CAA CAA AAC TGG | ||
| IALYLQQNW | HLA B57 and B58 | ||
| Caucasian | --------------------------- | 10 | |
| - - - - - - - - - | |||
| -----------------C--------- | 1 | ||
| - - - - - H - - - | |||
| African | --------------------------- | 2 | |
| - - - - - - - - - | |||
| S. E. Asian (Spon) | --------------------------- | 4 | |
| - - - - - - - - - | |||
| S. E. Asian (NPC) | --------------------------- | 14 | |
| - - - - - - - - - | |||
| C-------------------------- | 1 | ||
| L - - - - - - - - | |||
| Papua New Guinean | --------------------------- | 10 | |
| - - - - - - - - - | |||
| --------------------C------ | 1 | ||
| - - - - - - H - - | |||
| B95.8 | TAC TTA TTG GAG ATG CTC TGG CGA CTT | ||
| YLLEMLWRL | HLA A2 | ||
| Caucasian | -----C--------T------------ | 1 | |
| - F - - I - - - - | |||
| --------------------------- | 5 | ||
| - - - - - - - - - | |||
| -----C--------T------ G-G--- | 4 | ||
| - F - - I - - G - | |||
| --------------T------------ | 1 | ||
| - - - - I - - - - | |||
| African | --------------T------------ | 2 | |
| - - - - I - - - - | |||
| S.E. Asian (Spon) | -----C--------T--------G--- | 3 | |
| - F - - I - - - - | |||
| -----C-----C--T--------G--- | 1 | ||
| - F - I - - - - | |||
| S.E. Asian (NPC) | -----C--------T--------G--- | 15 | |
| - F - - I - - - - | |||
| Papua New Guinean | -----C--------T--------G--- | 4 | |
| - F - - I - - - - | |||
| -----C-----A--T--------G--- | 6 | ||
| - F - - I - - - - | |||
| --- C-- A-------T------------ | 1 | ||
| - - M - I - - - - | |||
| B95.8 | TAT CTA CAA CAA AAC TGG TGG ACT CTA | ||
| YLQQNWWTL | HLA A2 | ||
| Caucasian | --------------------------- | 10 | |
| - - - - - - - - - | |||
| --------C------------------ | 1 | ||
| - - H - - - - - - | |||
| African | --------------------------- | 2 | |
| - - - - - - - - - | |||
| S.E. Asian (Spon) | --------------------------- | 4 | |
| - - - - - - - - - | |||
| S.E. Asian (NPC) | --------------------------- | 15 | |
| - - - - - - - - - | |||
| Papua New Guinean | --------------------------- | 10 | |
| - - - - - - - - - | |||
| -----------C--------------- | 1 | ||
| - - - H - - - - - |
Boldface indicates CTL epitope sequence from prototype B95.8 isolate.
Sequence analysis of the HLA A2-restricted epitope YLLEMLWRL in EBV isolates from diverse ethnic origins revealed an interesting pattern of genetic variation. Five out of 11 Caucasian isolates showed the prototype B95-8 sequence for the YLLEMLWRL epitope, whereas the remaining isolates displayed changes affecting one or more residues (Table 3). In four isolates, leucine at position 2, methionine at position 5, and arginine at position 8 were substituted for phenylalanine, isoleucine, and glycine, respectively. The other two isolates showed alterations at positions 2 and 5 or only at position 5. Sequence analysis of virus isolates from Southeast Asian individuals revealed an identical set of substitutions in almost all isolates which included alterations at position 2 (L→F) and position 5 (M→I). One isolate showed mutation at position 4 (E→D). It is important to stress here that virus isolates from both spontaneous LCLs and NPC biopsy samples displayed generally identical patterns of mutations (Table 3). No B95-8-like sequence was detected in any of the isolates from Southeast Asia. Interestingly, the dominant genetic variant form of the YLLEMLWRL epitope seen in the Southeast Asian population (YFLEILWRL) was also frequently detected in virus isolates from the Papua New Guinean region.
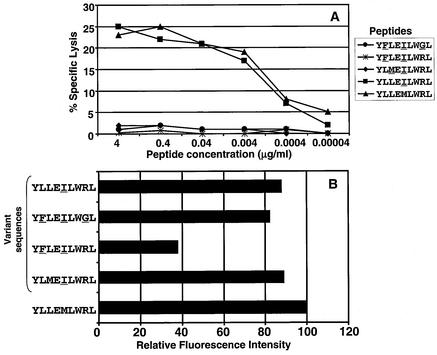
To further characterize the genetic variants of the YLLEMLWRL epitope, synthetic peptides for each of the variant sequences were synthesized and assessed for HLA binding and immunological recognition by EBV-specific CTLs. Data presented in Fig. 7A show that the synthetic peptides for most of the variant sequences were significantly less efficient than the prototypic (B95-8) YLLEMLWRL peptide at sensitizing autologous PHA blast to lysis by a specific CTL clone. Only one variant sequence, YLLEILWRL, was recognized as efficiently as the prototype epitope (Fig. 7A). Furthermore, incubation of T2 cells with the variant peptide sequence (YFLEILWRL) frequently seen in Southeast Asian isolates was unable to rescue HLA A2 expression (Fig. 7B). Other variant peptides (YFLEILWGL, YLMΕILWRL, and YLLEILWRL), however, significantly increased MHC expression in T2 cells, suggesting that the loss of antigenicity of these variants is due to inappropriate T-cell receptor interaction with the MHC-peptide complex rather than the loss of MHC binding.
FIG. 7.
(A) CTL recognition of variant and prototypic HLA A2-restricted LMP1 epitope YLLEMLWRL. PHA blasts were sensitized with serial dilutions of each of the peptides and then exposed to the YLLEMLWRL-specific CTL clone, SB7. (B) MHC stabilization analysis on T2 cells using variant and prototypic HLA A2-restricted LMP1 epitope. T2 cells were initially incubated with 200 μl of each of the peptides (10 μg/ml) for 14 to 16 h at 26°C followed by incubation at 37°C for 2 to 3 h. HLA A2 expression on these cells was analyzed by fluorescence-activated cell sorting using HLA A2-specific MAb.
DISCUSSION
An important feature common to HD and NPC is the fact that the expression of EBV antigens is often limited to EBNA1, LMP1, and LMP2. Of these, EBNA1 is protected from processing by the classical HLA class I pathway (12), so LMP1 and LMP2 are the only target antigens available to develop novel strategies to expand antigen-specific T-cell immunity for the treatment of HD and NPC. Therapies aimed at boosting T-cell responses to LMP antigens may be useful in the management of EBV-associated relapsed NPC and HD. We adopted the ELISPOT assay to comprehensively profile the LMP1-specific T-cell responses in a large panel of virus carriers. Of all the donors tested in this study, LMP1-specific T-cell responses were detected in 55 to 60% of the healthy Caucasians. On the other hand, these responses were less frequently detected in both healthy virus carriers and NPC patients from Southeast Asia. One of the interesting features of this analysis was that five of the six CD8+ T-cell epitopes mapped in this study were restricted through HLA A2. One of the possible explanations for this observation is that the LMP1 sequence is rich in leucine residues, which is a dominant anchor residue at positions 2 and 9 for a majority of the HLA A2-binding peptides (8). Thus it is likely that HLA A2-binding epitopes may be more frequently processed and presented from LMP1 protein. On the other hand, it is important to stress here that most of the T-cell reactivity to LMP1 in healthy virus carriers was not targeted only through the HLA A2 and HLA B57 or B58 alleles. In total, we identified 18 minimal 9-mer sequences. Of these, we were able to define HLA class I restriction for only six sequences (five HLA A2-restricted and one HLA B57- or B58-restricted), while HLA restriction for the remaining 12 sequences remained undefined. However, the analysis carried out precludes their restriction through HLA A2 and either HLA B57 or B58. Our repeated attempts to generate CTL lines and clones for mapping HLA restriction for these 12 potential epitope sequences were unsuccessful. In addition, we also identified potential reactivities within the LMP1 sequence which were targeted by EBV-specific CD4+ T cells.
Previous studies have suggested that LMP1 sequences in different geographic regions of the world display a very high degree of variation. This genetic variation has been considered a major impediment to the use of LMP1 as a potential immunotherapeutic target for the treatment of relapsed HD and NPC. Hence it was important to determine the extent to which the B95-8-derived LMP1 epitope sequences are conserved in EBV isolates from different geographic regions of the world. Studying a large panel of peripheral blood-derived EBV isolates from different regions, as well as NPC biopsies, we found that T-cell epitope sequences from LMP1 were generally well conserved. Three (ALLVLYSFA, IALYLQQNW, and YLQQNWWTL) of the four epitopes sequenced here showed only minor sequence variation. The only epitope with major sequence divergence was the HLA A2 supertype-restricted YLLEMLWRL epitope, which displayed an interesting pattern of sequence diversity in EBV isolates from different geographic regions. This was particularly highlighted by the isolates from Southeast Asia, which showed an identical pattern of genetic variation within the YLLEMLWRL epitope region. Moreover, no difference in the pattern of genetic variation was observed between isolates from NPC and spontaneous LCLs. Although common sequence variants of the YLLEMLWRL epitope were also seen in Caucasian and Papua New Guinean isolates, these isolates also displayed additional genetic variation that was quite unique to each specific ethnic group. Antigenic analysis of the genetic variants of the YLLEMLWRL epitope indicated that the variant sequence (YFLEILWRL), prevalent in Southeast Asia, was not only poorly recognized by epitope-specific CTLs but also showed significant loss in HLA A2 binding compared to the B95-8 sequence. In contrast, although some of the other variant sequences (YFLEILWGL and YLMEILWRL) were poorly recognized, their HLA A2 binding was not significantly affected. Although the precise reason and mechanism for such a high degree of genetic variation within this epitope are not known, it is possible that mutation within this epitope in the isolates from Southeast Asia, where NPC is endemic, may provide an advantage in protecting these isolates from the EBV-specific CTL response.
Overall, these studies provide, for the first time, a comprehensive analysis of LMP1-specific T-cell responses in a large cohort of ethnically diverse human subjects. The data presented here provide an important platform for the development of potential immunotherapeutic strategies for the treatment of EBV-associated NPC and HD. Considering the strong oncogenic potential of LMP1, it is highly unlikely that a vaccine or immunotherapeutic strategy based on full-length LMP1 would ever be a preferred choice for the treatment of NPC and HD. Moreover, consistent with our previous observations, the present study demonstrates that in a majority of the healthy virus carriers, LMP1 protein generally generates very low levels of T-cell responses. This is presumably due to its transmembrane localization, which limits its accessibility to the classic MHC class I pathway. The use of highly conserved LMP1 and LMP2 epitopes as a polyepitope vaccine clearly overcomes both these potential limitations. Indeed, preliminary studies with a polyepitope vaccine based on the LMP1 and LMP2 epitopes have indicated that this approach is highly efficient in generating LMP1- and LMP2-specific T-cell responses and provides therapeutic benefit against tumor challenge (5a).
Acknowledgments
This study was supported by the Cooperative Research Centre for Vaccine Technology and the National Health and Medical Research Council (NH&MRC) of Australia. R.K. is supported by a Senior Research Fellowship (NH&MRC).
REFERENCES
- 1.Anagnostopoulos, I., and M. Hummel. 1996. Epstein-Barr virus in tumours. Histopathology 29:297-315. [DOI] [PubMed] [Google Scholar]
- 2.Bharadwaj, M., P. G. Parsons, and D. J. Moss. 2001. Cost-efficient quantification of enzyme-linked immunospot. BioTechniques 30:36-38. [DOI] [PubMed] [Google Scholar]
- 3.Burrows, S. R., S. J. Rodda, A. Suhrbier, H. M. Geysen, and D. J. Moss. 1992. The specificity of recognition of a cytotoxic T lymphocyte epitope. Eur. J. Immunol. 22:191-195. [DOI] [PubMed] [Google Scholar]
- 4.Burrows, S. R., A. Suhrbier, R. Khanna, and D. J. Moss. 1992. Rapid visual assay of cytotoxic T-cell specificity utilizing synthetic peptide induced T-cell-T-cell killing. Immunology 76:174-175. [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, S. T., D. P. Huang, A. B. Hui, K. W. Lo, C. W. Ko, Y. S. Tsang, N. Wong, B. M. Whitney, and J. C. Lee. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83:121-126. [DOI] [PubMed] [Google Scholar]
- 5a.Duraiswamy, J., M. Sherritt, S. Thomson, J. Tellam, L. Cooper, G. Connolly, M. Bharadway, and R. Khanna. 2003. Therapeutic LMP1 polyepitope vaccine for EBV-associated Hodgkin’s disease and nasopharyngeal carcinoma. Blood 101:3150-3156. [DOI] [PubMed] [Google Scholar]
- 6.Khanna, R., and S. R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19-48. [DOI] [PubMed] [Google Scholar]
- 7.Khanna, R., S. R. Burrows, M. G. Kurilla, C. A. Jacob, I. S. Misko, T. B. Sculley, E. Kieff, and D. J. Moss. 1992. Localization of Epstein-Barr virus cytotoxic T-cell epitopes using recombinant vaccinia: implications for vaccine development. J. Exp. Med. 176:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khanna, R., S. R. Burrows, J. Nicholls, and L. M. Poulsen. 1998. Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur. J. Immunol. 28:451-458. [DOI] [PubMed] [Google Scholar]
- 9.Kienzle, N., M. Buck, S. L. Silins, S. R. Burrows, D. J. Moss, A. Winterhalter, A. Brooks, and R. Khanna. 2000. Differential splicing of antigen-encoding RNA reduces endogenous epitope presentation that regulates the expansion and cytotoxicity of T cells. J. Immunol. 165:1840-1846. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. P., R. J. Tierney, W. A. Thomas, J. M. Brooks, and A. B. Rickinson. 1997. Conserved CTL epitopes within EBV latent membrane protein 2: a potential target for CTL-based tumor therapy. J. Immunol. 158:3325-3334. [PubMed] [Google Scholar]
- 11.Leen, A., P. Meij, I. Redchenko, J. Middeldorp, E. Bloemena, A. Rickinson, and N. Blake. 2001. Differential immunogenicity of Epstein-Barr virus latent-cycle proteins for human CD4+ T-helper 1 responses. J. Virol. 75:8649-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685-688. [DOI] [PubMed] [Google Scholar]
- 13.Meij, P., A. Leen, A. B. Rickinson, S. Verkoeijen, M. B. Vervoort, E. Bloemena, and J. M. Middeldorp. 2002. Identification and prevalence of CD8(+) T-cell responses directed against Epstein-Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. Int. J. Cancer 99:93-99. [DOI] [PubMed] [Google Scholar]
- 14.Miller, W. E., R. H. Edwards, D. M. Walling, N. Raab-Traub. 1994. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J. Gen. Virol. 75:2729-2740. [DOI] [PubMed] [Google Scholar]
- 15.Moss, D. J., I. S. Misko, S. R. Burrows, K. Burman, R. McCarthy, and T. B. Sculley. 1988. Cytotoxic T-cell clones discriminate between A- and B-type Epstein-Barr virus transformants. Nature 331:719-721. [DOI] [PubMed] [Google Scholar]
- 16.Murray, R. J., M. G. Kurilla, J. M. Brooks, W. A. Thomas, M. Rowe, E. Kieff, and A. B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T-cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr Virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 18.Salter, R. D., and P. Cresswell. 1986. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 5:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandvej, K., J. W. Gratama, M. Munch, X. G. Zhou, R. L. Bolhuis, B. S. Andresen, N. Gregersen, and S. Hamilton-Dutoit. 1997. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild-type EBV isolates. Blood 90:323-330. [PubMed] [Google Scholar]
- 20.Sung, N. S., R. H. Edwards, F. Seillier-Moiseiwitsch, A. G. Perkins, Y. Zeng, and N. Raab-Traub. 1998. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and nonendemic regions of China. Int. J. Cancer 76:207-215. [DOI] [PubMed] [Google Scholar]