Abstract
Classical swine fever virus (CSFV) replicates efficiently in cell lines and monocytic cells, including macrophages (MΦ), without causing a cytopathic effect or inducing interferon (IFN) secretion. In the present study, the capacity of CSFV to interfere with cellular antiviral activity was investigated. When the porcine kidney cell line SK-6 was infected with CSFV, there was a 100-fold increased capacity to resist to apoptosis induced by polyinosinic-polycytidylic acid [poly(IC)], a synthetic double-stranded RNA. In MΦ, the virus infection inhibited poly(IC)-induced alpha/beta IFN (type I IFN) synthesis. This interference with cellular antiviral defense correlated with the presence of the viral Npro gene. Mutants lacking the Npro gene (ΔNpro CSFV) did not protect SK-6 cells from poly(IC)-induced apoptosis, despite growth properties and protein expression levels similar to those of the wild-type virus. Furthermore, ΔNpro CSFV did not prevent poly(IC)-induced type I IFN production in MΦ but rather induced type I IFN in the absence of poly(IC) in both MΦ and the porcine kidney cell line PK-15, but not in SK-6 cells. With MΦ and PK-15, an impaired replication of the ΔNpro CSFV compared with wild-type virus was noted. In addition, ΔNpro CSFV, but not wild-type CSFV, could interfere with vesicular stomatitis virus replication in PK-15 cells. Taken together, these results provide evidence for a novel function associated with CSFV Npro with respect to the inhibition of the cellular innate immune system.
Classical swine fever (CSF) is a highly contagious disease of pigs caused by the classical swine fever virus (CSFV) and leads to important economic losses worldwide. CSFV, together with bovine viral diarrhea virus (BVDV) and border disease virus (BDV), form the genus Pestivirus within the family Flaviviridae. The two other genera of the family are the genus Flavivirus and the genus Hepacivirus (55). Pestiviruses carry an RNA genome which possesses a 5′ untranslated region (5′UTR), a single large open reading frame (ORF), and a 3′UTR. The 5′UTR functions as an internal ribosomal entry site for cap-independent translation initiation. The ORF encodes a polyprotein of approximately 3,900 amino acids which is processed into 12 mature proteins by virus-encoded and host cell proteases (for a review, see reference 35). The first protein encoded is the nonstructural protein Npro. It exhibits autoproteolytical activity and cleaves itself off the downstream nucleocapsid protein C (43, 47, 56). Interestingly, it has no corresponding counterpart in flaviviruses and hepatitis C virus and was found to be dispensable for virus replication in cell culture (52). The Npro gene of the moderately virulent strain vA187-1 and of the highly virulent strain vEy-37 has been deleted or replaced with the murine ubiquitin gene, which substitutes for Npro in the generation of the authentic nucleocapsid protein. Such mutants lacking the Npro gene (ΔNpro CSFV) displayed no major alteration of growth characteristics in the porcine kidney cell line SK-6 but were avirulent in pigs (52; D. Mayer, M. A. Hofmann, and J. D. Tratschin, submitted for publication).
Monocytes and macrophages (MΦ) are among the main targets for CSFV infection (29, 53). They are potential factors in the spread of CSFV to different tissues, are major reservoirs for infectious virus, and are involved in CSFV-induced immunomodulation (29). Although CSFV replication in MΦ is sensitive to interferon (IFN), the virus does not induce IFN in these cells (28). Considering that MΦ possess the potential to produce alpha/beta IFN (type I IFN) in response to virus infection (39), it is plausible that CSFV may be capable of counteracting cellular antiviral activity. In this vein, Schweizer and Peterhans (44) recently demonstrated that noncytopathogenic (ncp) BVDV interfered with polyinosinic-polycytidylic acid [poly(IC)]-induced apoptosis in primary bovine turbinate cells and IFN induction in bovine MΦ. As CSFV is also ncp, the present work sought to determine the capacity of CSFV to interfere with apoptosis in the swine kidney cell line SK-6 and with IFN-α/β production in porcine MΦ. The results demonstrated that resistance to poly(IC)-induced cell death and control of IFN induction were dependent on the presence of the Npro gene, indicating a function of Npro in innate immune evasion of CSFV.
MATERIALS AND METHODS
Cells and viruses.
The porcine kidney cell lines SK-6 (27) (kindly provided by M. Pensaert, Faculty of Veterinary Medicine, Ghent University, Ghent, Belgium) and PK-15 (American Type Culture Collection, Manassas, Va.) were propagated in Earle's minimum essential medium supplemented with 7% horse serum. Monocytes were isolated by culture of peripheral blood mononuclear cells (PBMC) from specific pathogen-free pigs at 4 × 106 cells/ml for 18 h in a mixture of Dulbecco's modified Eagle medium, 10% porcine serum, 2 mM l-glutamine, and 25 mM HEPES, as described earlier (29, 34). Then, the adherent cells, representing monocytic cells partially differentiated to MΦ (6), were detached and reseeded at 0.5 × 106 cells/ml in the above medium for 1 h before infection. The highly virulent CSFV strains Eystrup and Brescia were kindly provided by H.-J. Thiel, Justus-Liebig-Universität, Giessen, Germany. CSFV strain Riems, a vaccine virus derived from a lapinized C strain (2), was kindly provided by G. Schirrmeier, Riemser Arzneimittel Gmbh, Insel Riems, Germany. CSFV vA187-1 and vEy-37 were derived from the full-length cDNA clones pA187-1 (42), vRiems, and pEy-37 (D. Mayer, M. A. Hofmann, and J. D. Tratschin, submitted for publication), respectively. The ΔNpro CSFV vA187-ΔNpro-Ubi (formerly named vA187-Ubi), in which the Npro gene was replaced by the murine ubiquitin gene in the vA187-1 backbone, has been previously described (52). CSFV vA187-ΔNpro, an alternative ΔNpro CSFV for which the Npro gene was replaced by the residues methionine and glycine and initially described as being noninfectious (52), was reconstructed and resulted in replication-competent virus rescued from in vitro transcripts with high specific infectivity. CSFV vEy-ΔNpro was constructed in analogy to vA187-ΔNpro by deleting the Npro gene in the pEy-37 cDNA clone. All cDNA-derived viruses were obtained by electroporation of SK-6 cells with the respective in vitro transcripts as described elsewhere (37). For the IFN bioassays, vesicular stomatitis virus (VSV) strain Indiana (American Type Culture Collection) was used at a multiplicity of infection (MOI) of 5 50% tissue culture infectious doses (TCID50)/cell.
Assay for cell survival after poly(IC) treatment.
SK-6 cells were either mock infected or infected with CSFV for 48 to 72 h and then seeded in duplicate columns in a 96-well plate. For each infected culture, the first column was treated with serial dilutions of poly(IC) (Sigma-Aldrich, Inc.) in complete medium, and the second column was mock treated. Cell survival was then monitored at 24 and 48 h after poly(IC) treatment by crystal violet staining. Briefly, cells were washed once with phosphate-buffered saline (PBS) and then fixed and stained for 30 min with 1% formaldehyde-0.5% crystal violet in 30% ethanol. After extensive washing with water, the stained cells were lysed with isopropanol containing 0.6% sodium dodecyl sulfate and 40 mM HCl. Optical density was measured at 595 nm (OD595). To normalize the values between the different virus-infected cultures, the mean OD595 of eight untreated wells was set to 100% cell survival for each culture. The percentage of optical density after poly(IC) treatment was then expressed as percent cell survival.
Immunohistochemistry and FCM.
For detection of CSFV-infected cells, the E2-specific monoclonal antibody (MAb) HC/TC26 (20) and the NS3-specific MAb C16 (19) (kindly provided by I. Greiser-Wilke, Hannover Veterinary School, Hannover, Germany) were used for the detection of viral antigen in situ and by flow cytometry (FCM) (48). Briefly, mock- and CSFV-infected cells were fixed and permeabilized with a cell permeabilization kit (Harlan Sera-Lab, Crawley Down, United Kingdom) before labeling with either MAb C16 or HC/TC26 (both at 1 μg/0.5 × 106 cells). Rabbit anti-mouse immunoglobulin G conjugated with phycoerythrin (DAKO A/S, Glostrup, Denmark) was used as secondary reagent. The percentage of infected cells was calculated by subtraction of the background values obtained by C16 staining of mock-infected cells from the values of virus-infected cells. For in situ immunohistochemistry in SK-6 cells, monolayers were dried at 37°C for 30 min and fixed at 80°C for 2 h. PBS-0.01% Tween 20 was used as dilution and wash buffer, and the incubations with primary and secondary antibodies were performed at 37°C for 30 min. Bound primary antibody was labeled with horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin (DAKO A/S) and visualized by adding chromogen solution as described previously (36). Viral antigen detection by FCM was performed as previously described (29).
Analysis of apoptosis by FCM.
AnnexinV binding on the surface of apoptotic cells expressing phosphatidylserine and propidium iodide (PI) incorporation by dead cells were analyzed by using standard protocols, as previously described (49). Briefly, cells were detached by trypsin treatment, resuspended in PBS at a concentration of 105 cells/ml, and labeled with 2 μg of AnnexinV-fluorescein isothiocyanate/ml in 140 mM NaCl-2.5 mM CaCl2-10 mM HEPES (pH 7.4) for 10 min. After addition of 100 ng of PI/ml (Sigma-Aldrich), the samples were analyzed by FCM. For analysis of the sub-G1 DNA content, the cells were fixed with 75% ethanol at 0°C for 2 min. DNA was then stained with 50 μg of PI/ml and treated with 100 μg of RNase cocktail/ml (Promega) for 30 min at 37°C prior to analysis by FCM.
IFN assays.
IFN bioactivity was detected with a bioassay based on the antiviral effect against VSV in the porcine kidney cell line PK-15. A confluent monolayer of the cells, seeded in 96-well plates, was incubated with samples for 18 h before VSV infection with an infectious dose resulting in a complete cytopathic effect (cpe) within 24 h in nontreated controls. The cpe was quantified by crystal violet staining as described above. Serial dilutions of recombinant porcine IFN-α (32) (PBL Biomedical Laboratories, Alexis, Switzerland, and kindly provided by B. Charley, INRA, Jouy-en-Josas, France) were prepared in the respective culture medium to establish standard curves for quantification. For neutralization of IFN-α, 250 μl of sample was incubated for 30 min on ice with 1 μl of rabbit polyclonal antibody against porcine IFN-α (>250 neutralization U/μl as determined by the manufacturer; PBL Biomedical Laboratories).
Type I IFN activity was also assayed using an Mx/CAT reporter gene assay described for the quantification of bovine IFN-α/β and kindly provided by Martin D. Fray (Institute of Animal Health, Compton, Newbury, Berkshire, United Kingdom) (14). Briefly, MDBK-t2 cells maintained under blasticidin selection were seeded in six-well plates at a density of 106 cells/well. After 24 h, samples diluted 1:5 [or 1:40 if poly(IC) was present in the sample] or porcine IFN-α standard were added to the cells in 1 ml of Earle's minimal essential medium supplemented with 7% heat-inactivated fetal calf serum. The cells were then incubated for 24 h prior to lysis and chloramphenicol acetyltransferase (CAT) quantification using a CAT enzyme-linked immunosorbent assay (Roche Biochemicals, Rotkreuz, Switzerland). Rabbit anti-porcine IFN-α antibody completely blocked activity of recombinant porcine IFN-α in both assays (data not shown), validating their use for quantification of bioactive porcine IFN-α. In some experiments, virus was inactivated by dialysis against 100 mM glycine (pH 2) at 4°C for 24 h followed by an overnight dialysis step against PBS. Alternatively, virus infectivity was neutralized by incubation for 1 h on ice with an excess of MAb HC/TC26 (20). Virus inactivation was monitored on SK-6 cells using immunohistochemistry with MAb HC/TC26.
Kinetics of virus replication.
Cells were inoculated for 1 h at an MOI of 1 for SK-6 cells and 5 (based on the titer determined on SK-6 cells) for PK-15 cells and MΦ. The cells were then washed with PBS one and five times, respectively, prior to addition of fresh medium. At various times postinfection (p.i.), replication was stopped by freezing the respective cultures to −70°C, and the cells were lysed by two cycles of freezing and thawing. The culture medium was then clarified by centrifugation at 1,000 × g for 10 min at 4°C and stored in aliquots at −70°C. The titer (TCID50 per milliliter) was determined by end-point dilution on SK-6 cells and immunoperoxidase staining using MAb HC/TC26 (see above).
Statistical analyses.
The statistical analyses used a two-sample Student's t test. Differences were considered significant when P < 0.05.
RESULTS
CSFV protects SK-6 cells from poly(IC)-induced apoptosis.
The capacity of CSFV to interfere with cell death induced in SK-6 cells by the synthetic double-stranded RNA (dsRNA) poly(IC) was analyzed. Survival of uninfected versus CSFV-infected SK-6 cells following poly(IC) treatment was monitored (Fig. 1). Cells infected with CSFV prior to poly(IC) addition showed an elevated viability compared with uninfected cells (Fig. 1A). With the CSFV-infected cells, around 50% cell survival was observed when the poly(IC) concentration was as high as 20 μg/ml; in contrast, uninfected cells did not survive poly(IC) concentrations in excess of 0.1 μg/ml. This higher resistance to poly(IC)-induced cell death was independent of the virus strain used for infection: the highly virulent vEy-37, the moderately virulent vA187-1, and the avirulent Riems strain conferred similar levels of protection. To confirm that all the cells were infected by CSFV in the cultures used for the cell survival assays, cultures parallel to those employed in Fig. 1A were immunostained for viral glycoprotein E2 expression at the time of poly(IC) addition. As shown in Fig. 1B, almost all of the cells stained for this protein, albeit with variable intensity.
FIG. 1.
CSFV-mediated cell survival after poly(IC) treatment is strain independent. Cell survival of SK-6 cells infected with three CSFV strains of different virulence was monitored by crystal violet staining 48 h after poly(IC) treatment. (A) The percentage of cell survival was calculated from the ratio of the OD595 of each poly(IC)-treated well to the average OD595 of the corresponding eight untreated wells (see Materials and Methods). The mean values with standard deviation from three independent experiments in SK-6 cells infected with CSFV strains vEy-37 (⋄), vA187-1 (□), and vRiems (○) or mock- infected cells (▵) are shown for decreasing poly(IC) concentrations. (B) Seventy-two hours p.i., at the time of poly(IC) addition, parallel cultures were analyzed for expression of the viral glycoprotein E2 by using immunoperoxidase staining.
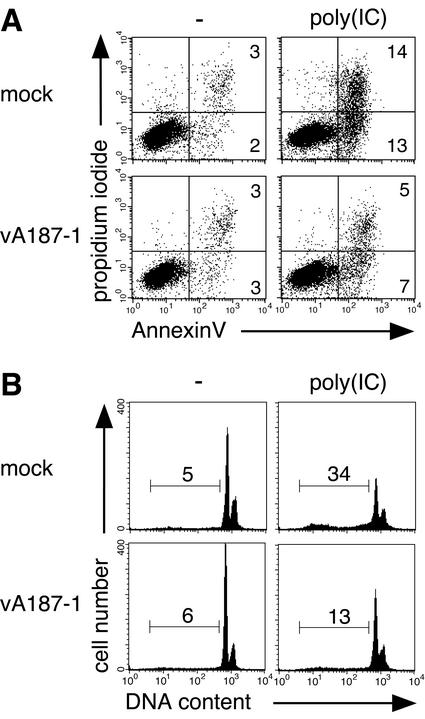
In order to demonstrate that the enhanced cell survival to poly(IC) was indeed due to protection from apoptosis, AnnexinV binding, PI incorporation, and sub-G1 DNA content were analyzed. SK-6 cells, either uninfected or infected with vA187-1, were treated with 100 μg of poly(IC)/ml for 40 h prior to FCM analysis. The results of a representative experiment are shown in Fig. 2. In the uninfected cell culture, a five- to sixfold increase of AnnexinV-positive cells was noted following treatment with poly(IC) (Fig. 2A). When the cells had been previously infected with CSFV, this high concentration of poly(IC) induced only a twofold increase in the AnnexinV-positive population (Fig. 2A). Similar ratios were observed for the sub-G1 DNA content (Fig. 2B).
FIG. 2.
CSFV protects SK-6 cells from poly(IC)-induced apoptosis. SK-6 cells were infected with CSFV vA187-1 or mock infected and treated after 72 h with 100 μg of poly(IC)/ml or mock treated as control. After 40 h, AnnexinV binding and PI incorporation (A) and sub-G1 DNA content (B) were analyzed by FCM. Numbers in the gated areas indicate the percentage of events. This data are representative of two independent experiments.
CSFV interferes with poly(IC)-induced type I IFN production in MΦ.
For these analyses, MΦ were either mock infected or infected with CSFV strains of different virulence prior to IFN induction by poly(IC). Supernatants from mock- or CSFV-infected MΦ cultures, which did not receive poly(IC), displayed no evidence of antiviral activity either in the VSV assay (Fig. 3) or through detectable CAT expression in the Mx/CAT reporter gene assay (data not shown). The same results were obtained in both assays with and without prior inactivation of the virus present in the MΦ supernatants, demonstrating that wild-type CSFV does not induce antiviral activity or IFN-α/β production in MΦ, PK-15, and MDBK-t2 cells (data not shown). After poly(IC) stimulation of mock-treated MΦ, up to 40 U of type I IFN/ml were detected in eight independent experiments. In contrast, with CSFV-infected MΦ, poly(IC) induced only 0.5 to 3 U of type I IFN/ml. Despite this relatively high variability between the experiments, CSFV-infected MΦ always produced only 2 to 10% of the IFN induced in mock-infected cultures. A representative experiment is shown in Fig. 3. These observations were independent of the CSFV strain used for infection. The type I IFN activity measured in the Mx/CAT reporter gene assay confirmed the results obtained in the VSV assay (data not shown). Furthermore, preincubation of the samples with rabbit anti-porcine IFN-α antibodies abolished the antiviral activity and the CAT response, respectively (data not shown), indicating that the antiviral activity was essentially due to IFN-α. These data demonstrated that both virulent and avirulent CSFV strains were capable of inhibiting the secretion of type I IFN by poly(IC)-treated MΦ.
FIG. 3.
CSFV interferes with poly(IC)-induced interferon production in MΦ. MΦ were infected with CSFV strains or cDNA-derived recombinant viruses as indicated on the x axis (MOI, 5 TCID50/cell) or mock infected with SK-6 supernatant (mock) or with supernatant of mock-transfected SK-6 cells (mock tf) and cultured for 24 h at 38°C. Then, some cultures were left without treatment (black bars) or treated with 10 μg of poly(IC)/ml (white bars) for another 24 h. Antiviral activity in virus-inactivated cell culture supernatants was measured in PK-15 cells with the VSV antiviral bioassay and quantified based on recombinant IFN-α, as described in Materials and Methods. The detection limit was 0.5 U of IFN-α/β/ml.
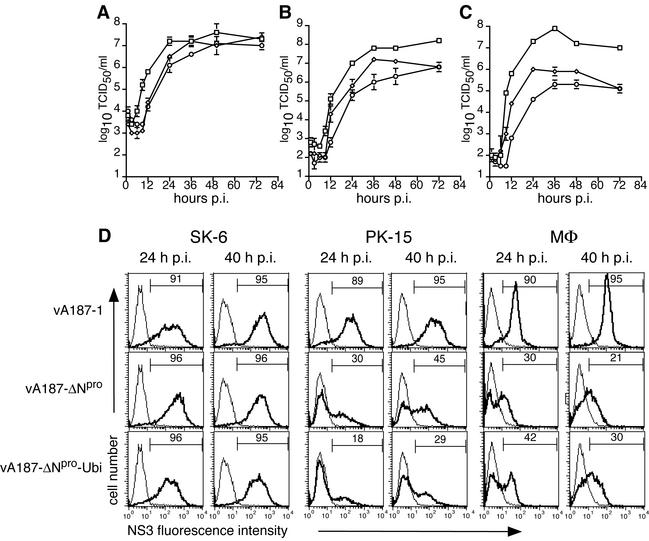
The replication characteristics of ΔNpro CSFV are dependent on the cell type.
It has been previously shown that vA187-ΔNpro-Ubi, a mutant CSFV in which Npro was replaced with the murine ubiquitin gene, replicated to titers similar to those of the parent virus vA187-1 in SK-6 cells (52). Experiments in pigs suggested a crucial role of Npro for the expression of virulence (D. Mayer, M. A. Hofmann, and J. D. Tratschin, submitted for publication). Therefore, the growth characteristics of two independent ΔNpro CSFVs, namely, vA187-ΔNpro and vA187-ΔNpro-Ubi, were compared in the SK-6 and PK-15 cell lines and in MΦ. In SK-6 cells, the two ΔNpro CSFVs and the parent vA187-1 replicated to similar titers of approximately 107 TCID50/ml within 48 h (Fig. 4A). However, analysis of the exponential growth phase of these viruses demonstrated that the replication of the ΔNpro CSFV was delayed by 6 h (Fig. 4A). This delay was also observed in PK-15 cells (Fig. 4B) and MΦ (Fig. 4C), but in these cells the maximum titer for ΔNpro CSFV was 1.5 to 2 log10 TCID50/ml lower than the titer of the parent virus. In addition to these differences in virus titer, FCM analysis of infected PK-15 and MΦ demonstrated differences in the expression of the viral protein NS3 (Fig. 4D). For the parent virus vA187-1, 89 to 95% of the cells stained positive for NS3 after 24 h and 40 h. With the vA187-ΔNpro or vA187-ΔNpro-Ubi mutants, at the same MOI as the parent virus, only part of the PK-15 cells and MΦ was positive for NS3 (Fig. 4D).
FIG. 4.
Virus replication in SK-6, PK-15, and Mφ. CSFV vA187-1 (□), vA187-ΔNpro (⋄), and vA187-ΔNpro-Ubi (○) were used to infect SK-6 cells (A), PK-15 cells (B), and PBMC-derived MΦ (C) at an MOI of 1 TCID50/cell for SK-6 and 5 TCID50/cell for PK-15 and MΦ. At various times p.i., virus was harvested and the titer was determined on SK-6 cells by endpoint dilution. Each point represents the mean of results of three parallel experiments for SK-6 and PK-15 cells and two parallel experiments for MΦ, with error bars showing the standard deviation of the mean. (D) NS3 expression in SK-6, PK-15, and MΦ infected with vA187-1 and ΔNpro CSFV (MOI, 5 TCID50/cell) was analyzed 24 h and 40 h p.i. by FCM (bold lines). The light lines represent background fluorescence of NS3 staining in uninfected controls, and the percentage of NS3-positive cells is indicated for each graph. The experiment was repeated twice with similar results.
ΔNpro CSFV does not protect SK-6 cells from poly(IC)-induced cell death.
The replication characteristics in MΦ and PK-15 cells of ΔNpro CSFV compared with the parent virus pointed to a possible role of Npro in CSFV escape from particular cellular antiviral defenses. In addition to producing IFN, eukaryotic cells can also respond to viral infection by entering into apoptotic cell suicide (5), which is also part of the cellular antiviral system. Wild-type CSFV can certainly interfere with the induction of both apoptosis and IFN (see above). Consequently, it was of interest to determine the influence of the presence of the Npro gene on the capacity of CSFV to protect cells against poly(IC)-induced cell death.
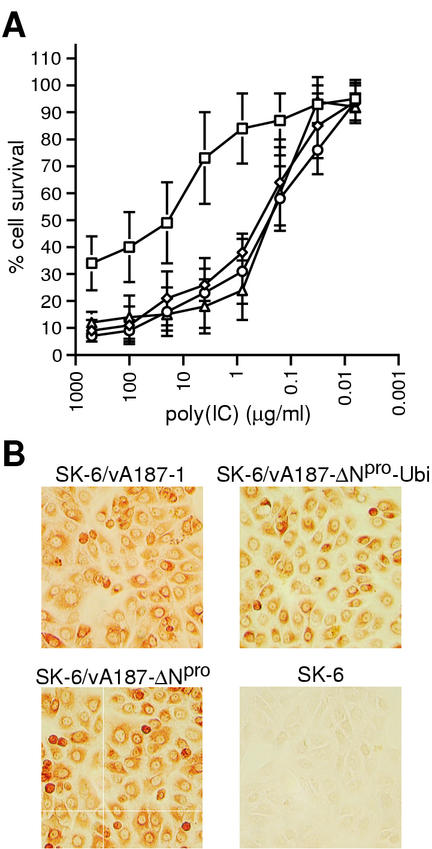
Unlike the wild-type vA187-1, neither vA187-ΔNpro nor vA187-ΔNpro-Ubi could protect SK-6 cells from poly(IC)-induced cell death (Fig. 5A). The rate of poly(IC)-induced cell death of SK-6 cells infected with ΔNpro CSFV was comparable to that of mock-infected cells. At the time of poly(IC) addition, parallel cultures were stained for expression of glycoprotein E2. They displayed a similar immunostaining pattern for all three viruses (Fig. 5B), in accordance with the FCM analysis of the relative NS3 expression (Fig. 4D, SK-6 cells) and with the replication characteristics of all three viruses in SK-6 cells (Fig. 4A). This confirmed that the cell cultures were equivalently infected and expressed comparable levels of viral proteins at the time of poly(IC) stimulation.
FIG. 5.
ΔNpro CSFV does not protect SK-6 cells from poly(IC)-induced cell death. (A) SK-6 cells were infected with CSFV vA187-1 (□), vA187-ΔNpro (⋄), and vA187-ΔNpro-Ubi (○) or were mock infected (▵) 72 h prior to treatment with poly(IC) or mock treatment. Cell survival was monitored 48 h later by crystal violet staining and measurement of OD595. The percentage of cell survival was calculated from the ratio of the OD595 of poly(IC)-treated wells to that of mock-treated wells, in analogy to the data shown in Fig. 1. The mean values with standard deviation from four independent experiments are shown for decreasing poly(IC) concentrations. (B) At the time of poly(IC) addition, parallel cultures were analyzed for expression of the viral glycoprotein E2 using immunoperoxidase staining.
In order to determine if the requirement for Npro in protection from poly(IC)-induced cell death was strain independent, poly(IC)-treated cell survival assays were repeated with a ΔNpro CSFV of the highly virulent strain vEy-37 (vEy-ΔNpro). Similar to the results obtained for the vA187-1-derived viruses, SK-6 cells infected with vEy-ΔNpro were not protected from poly(IC)-induced cell death and behaved like the uninfected cells (data not shown).
ΔNpro CSFV does not prevent poly(IC)-induced type I IFN production in MΦ.
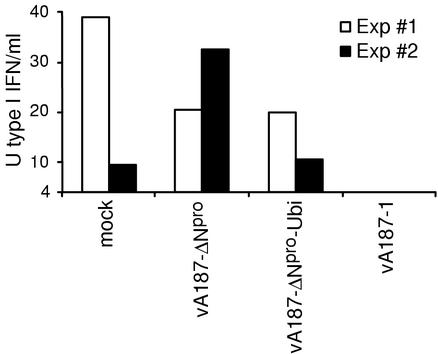
In the light of the lower levels of replication observed for the two ΔNpro CSFVs in MΦ than for the wild-type virus, the capacity of ΔNpro CSFV to prevent poly(IC)-induced type I IFN production was analyzed. Porcine MΦ were mock infected or infected with vA187-1, vA187-ΔNpro, or vA187-ΔNpro-Ubi and cultured for 24 h before poly(IC) stimulation. Similar to the results presented in Fig. 3, poly(IC)-stimulated mock-infected MΦ produced 10 to 40 U of type I IFN, whereas vA187-1-infected MΦ did not respond to poly(IC) stimulation by producing detectable type I IFN (Fig. 6). However, compared to the data obtained with the VSV assay (see Fig. 3), the detection limit was eight times higher (4 U of type I IFN/ml) when the Mx/CAT reporter gene assay was used to measure samples containing poly(IC). This was caused by IFN induction in the MDBK-t2 cells when poly(IC) was present at concentrations above 0.25 μg/ml. Nevertheless, CSFV-infected MΦ produced no or very low levels of type I IFN after poly(IC) stimulation in a total of eight independent experiments. In contrast, in five independent experiments, ΔNpro CSFV-infected MΦ responded to poly(IC) with type I IFN production comparable to that of mock-infected MΦ. Two of these experiments are shown in Fig. 6. Although the level of IFN production was variable between experiments, these differences were statistically significant when a background IFN production corresponding to the level of the detection limit was used for the statistical analyses (v187-ΔNpro, P = 0.01; vA187-ΔNpro-Ubi, P = 0.03). Incubation of the MΦ supernatants with anti-porcine IFN-α antibody prior to addition into the Mx/CAT assay abolished the CAT response (data not shown).
FIG. 6.
ΔNpro CSFV does not inhibit poly(IC)-induced type I IFN synthesis in MΦ. MΦ were mock infected or infected at an MOI of 5 TCID50/cell with either of the two ΔNpro CSFVs or with the parent vA187-1, as indicated on the x axis, and cultured for 24 h. Then, the cultures were treated with 10 μg of poly(IC)/ml for 24 h in the first experiment (white bars) and 40 h in the second, independent experiment shown (black bars). IFN-α/β activity was measured in cell culture supernatants using the Mx/CAT reporter gene assay and recombinant porcine IFN-α for quantitation. The detection limit is given by the lower limit of the y axis (4 U of type I IFN/ml).
ΔNpro CSFV induces type I IFN in MΦ and PK-15 cells but not in SK-6 cells.
In the absence of poly(IC), no detectable IFN was induced in MΦ and PK-15 cells by any of the wild-type CSFV strains tested. This contrasted with either vA187-ΔNpro or vA187-ΔNpro-Ubi, for which, in the absence of any poly(IC) stimulation, supernatants of infected cells contained between 1.5 and 8.5 U of type I IFN/ml for MΦ (Fig. 7A) and between 3 and 7.5 U of type I IFN/ml for PK-15 cells (Fig. 7B). For the MΦ, the results of four independent experiments using MΦ preparations from separate batches of PBMC are shown, with the type I IFN quantified in the Mx/CAT reporter gene assay. The levels of type I IFN were between 5 and 25 times lower than those observed after poly(IC) stimulation of uninfected MΦ (see Fig. 3 and 6). ΔNpro CSFV-infected MΦ induced statistically significantly more IFN compared to wild-type CSFV and mock-infected Mφ, even when the calculations were based on background IFN levels at the detection limit of the assay (P < 0.03). Comparable results were obtained with PK-15 cells, with less variability (Fig. 7B). Interestingly, incubation of the supernatants of ΔNpro CSFV-infected cells with anti-porcine IFN-α antibody prior to analysis in the Mx/CAT assay could completely block the IFN activity in the MΦ supernatants, whereas the activity in the PK-15 supernatants was only slightly reduced (data not shown). This suggests that MΦ produce mainly IFN-α, whereas PK-15 cells mainly secrete IFN-β in response to infection with ΔNpro CSFV. Similar experiments were performed with supernatants from SK-6 cells infected with vA187-1, vA187-ΔNpro, or vA187-ΔNpro-Ubi or stimulated with poly(IC) as control. None of the supernatants harvested at 12-h intervals between 6 and 65 h p.i. revealed any detectable type I IFN activity in the Mx/CAT reporter gene assay, regardless of whether virus was inactivated or not prior to the IFN assay (data not shown). This demonstrates that SK-6 cells, in contrast to MΦ and PK-15 cells, do not produce type I IFN in response to infection with ΔNpro CSFV.
FIG. 7.
ΔNpro CSFV induces type I IFN synthesis in MΦ and PK-15 cells. MΦ (A) and PK-15 cells (B) were mock-infected or infected at an MOI of 5 TCID50/cell with either of the two ΔNpro CSFVs or with the parent vA187-1, as indicated on the x axis. The results of four independent experiments are shown. For MΦ, the samples were harvested and analyzed at 24, 36, 40, and 48 h p.i. (Exp. #1 to 4, respectively). For PK-15 cells, the samples were analyzed at 40 h p.i. Type I IFN activity was measured in cell culture supernatants using the Mx/CAT reporter gene assay and recombinant porcine IFN-α for quantitation. The detection limit is given by the lower limit of the y axis (0.5 U of type I IFN/ml).
ΔNpro CSFV interferes with VSV replication in PK-15 cells.
Supernatants from MΦ infected with ΔNpro CSFV, as opposed to supernatants from wild-type CSFV-infected MΦ, induced a clear antiviral activity against VSV in PK-15 cells. This activity could be neutralized with anti-IFN-α antibody only if the infectious CSFV in the MΦ supernatants was inactivated (data not shown). This indicated that in contrast to wild-type virus, ΔNpro CSFV could interfere directly with VSV replication upon coinfection of PK-15 cells. In order to confirm this hypothesis, PK-15 cells were infected with 10-fold dilutions of either vA187-1, vA187-ΔNpro, or vA187-ΔNpro-Ubi produced in SK-6 cells, starting with an MOI of 10 TCID50/cell. As mentioned above, supernatants of SK-6 cells infected with either wild-type or ΔNpro CSFV do not contain type I IFN activity, as determined in the Mx/CAT reporter gene assay. After 24 h, the CSFV-infected PK-15 cell cultures were superinfected with VSV at an MOI of 5 TCID50/cell, a dose resulting in complete cpe in uninfected control cells within 24 h. As opposed to vA187-1, both ΔNpro CSFVs protected PK-15 cells from the VSV-induced cpe in a dose-dependent manner (Fig. 8A). This protection was dependent on the presence of infectious virus, since preincubation of the CSFV inocula with a virus-neutralizing dose of anti-E2 MAb abolished the interference with VSV-induced cpe (Fig. 8B). Preincubation of the CSFV inocula with a neutralizing dose of anti-IFN-α antibody, however, did not reverse the interference effect (Fig. 8C). At the time of VSV challenge, a duplicate control culture treated as above was immunostained for CSFV E2 expression. The percentage of infected cells was dependent on the MOI, and while it was reduced by the anti-E2 MAb, there was no effect of the anti-IFN-α antibodies (data not shown). Taken together, these results suggest that CSFV lacking the Npro gene, as opposed to wild-type virus, is capable of interfering with VSV replication in PK-15 cells. The presence of the Npro gene in wild-type CSFV is therefore required for preventing the induction of an antiviral response in CSFV-infected cells.
FIG. 8.
ΔNpro CSFV interferes with VSV replication in PK-15 cells. PK-15 cells were infected at an MOI of 10, 1, 0.1 and 0.01 TCID50/cell with ΔNpro CSFV and with the corresponding parent vA187-1 as indicated. Cells were cultured for 24 h prior to superinfection with VSV at an MOI of 5 TCID50/cell, resulting in complete cpe within 24 h in uninfected controls. Twenty-four hours after VSV infection, cell survival was monitored by crystal violet staining and measurement of OD595. The experiments were performed in the absence (A) or presence (B) of a neutralizing titer of MAb HC/TC26 or in the presence of polyclonal anti-IFN-α serum (C). Results from a representative experiment out of three are shown.
DISCUSSION
The autoproteinase Npro is the first protein expressed from the single large ORF of all pestiviruses (43, 47, 56) and has no counterpart within the other genera of the family Flaviviridae (55). When CSFV, BVDV, and BDV are compared, the amino acid sequence identity of Npro is higher than 70% (40) and the residues Glu22, His49, and Cys69, essential for the proteolytic activity of Npro (43), as well as the residues Cys168 and Ser169 surrounding the cleavage site (47), are conserved. Tratschin et al. demonstrated earlier that Npro of CSFV was dispensable for virus replication in cell culture (52). Nevertheless, it has been retained during evolution. Therefore it was tempting to postulate that Npro exerted additional functions in vivo besides simply cleaving itself off the nascent polyprotein. The first piece of evidence was obtained with the observation that the lack of Npro completely abolished the virulence of the moderately virulent strain vA187-1 (52) and of the highly virulent strain vEy-37 (D. Mayer, M. A. Hofmann, and J. D. Tratschin, submitted for publication).
In the present study, we analyzed the interaction of CSFV with cellular antiviral defense in vitro. For this purpose, we used cell stimulation with the synthetic dsRNA macromolecule poly(IC), a well-known biological response modifier, as a model for activation of nonspecific antiviral activities, i.e., apoptosis in the porcine SK-6 cell line and type I IFN induction in porcine monocytic cells. It was shown earlier that preinfection of cells with BVDV suppressed the poly(IC)-induced interference with VSV replication and cpe, a characteristic exploited in a diagnostic assay for the detection of BVDV (33). Rossi and Kiesel, however, suggested that the poly(IC)-induced interference with VSV was a non-IFN-based mechanism (41). More recent data published by Schweizer and Peterhans clearly demonstrated that BVDV blocked poly(IC)-induced type I IFN transcription and secretion (44). In addition, BVDV completely protected bovine turbinate cells from poly(IC)-induced apoptosis. A very recent study reported similar observations for hepatitis A virus that was also shown to inhibit poly(IC)-induced IFN-β synthesis and apoptosis (8). For both BVDV and CSFV, the interference with antiviral activities was recognized a long time ago with the observation that these viruses enhanced the multiplication of Newcastle disease (ND) virus in cell culture (24, 31). This phenomenon was designated END (for “exaltation of ND virus”) and applied to a quantitative assay for BVDV (24) and CSFV (30). More recently, BVDV was also shown to enhance replication of orbiviruses (38). It was suggested that the mechanism of END was a result of suppression of IFN production (13, 51). The results presented herein demonstrate that CSFV, by analogy to BVDV, is capable of preventing poly(IC)-mediated induction of cellular antiviral activities. Together with the reports mentioned above, our data indicate that the interference with cellular antiviral defense might be a general feature of most pestivirus strains. It is therefore not surprising that for both BVDV (44) and CSFV (this study), the inhibition of IFN induction was independent of the origin and virulence of the virus strain analyzed. Interestingly, Shimizu and coworkers reported an attenuated CSFV strain that did not suppress IFN production and was itself an effective IFN inducer (46, 54). This strain had been attenuated by serial passage in pigs followed by multiple passages in swine and bovine testicle cells. Finally, after 30 passages in guinea pig cells, a clone inducing interference with ND virus was isolated and designated GPE− (END-negative). The CSFV-mediated inhibition effect on the heterologous interference caused by the CSFV strain GPE− against VSV replication in cell culture was exploited in a diagnostic assay named reverse interference method (54).
The phenotype of the strain GPE− in cell culture is reminiscent of the characteristics we observed with the CSFV mutants lacking the Npro gene. In contrast to the wild-type parent virus, ΔNpro CSFV did not suppress poly(IC)-induced type I IFN production in MΦ and apoptosis in SK-6 cells, indicating a crucial role for Npro in the interference of the virus with antiviral pathways.
It is important to note that after infection with either vA187-1 or ΔNpro CSFV, all SK-6 cells were infected and expressed comparable amounts of glycoprotein E2 and of nonstructural protein NS3 at the time of poly(IC) addition. This indicates that the loss of the capacity of ΔNpro CSFV to interfere with poly(IC)-induced apoptosis is not associated with reduced viral replication or protein synthesis. Nevertheless, the fact that only part of the MΦ population expressed detectable virus antigen when infected with ΔNpro CSFV has to be taken into account when interpreting the regained poly(IC) responsiveness of MΦ cultures infected with ΔNpro CSFV compared with wild-type virus. Nevertheless, the role of Npro in the inhibition of antiviral processes was substantiated by the observation that CSFV lacking Npro actively induced type I IFN in MΦ and PK-15 cells as well as resistance to VSV-induced cpe in PK-15 cells. The latter could be mediated in part through the activation of intrinsic antiviral activity. As a consequence of the ΔNpro CSFV-mediated antiviral activity, part of the MΦ and PK-15 population probably becomes resistant to CSFV replication and might still respond to poly(IC). Thus, the observation that ΔNpro CSFV does not prevent poly(IC)-induced type I IFN production in MΦ could be the result of two overlapping mechanisms, namely, the loss of an inhibitory activity of Npro on type I IFN production and the reduced viral replication due to the induction of an antiviral state in the MΦ culture.
Although wild-type CSFV does not have the capacity to induce IFN in MΦ, we had previously shown that replication of CSFV was inhibited by pretreatment of the cells with recombinant porcine IFN-α, resulting in a virus titer reduction of 2 log10 TCID50/ml (28). The IFN-mediated titer reduction correlates with the impaired replication observed in MΦ and PK-15 cells with ΔNpro CSFV compared to vA187-1. It has been observed repeatedly that viral genes that were dispensable for virus replication in cell cultures or in mice deficient in a defined antiviral activity were essential to counteract this antiviral activity in the wild-type parent animal or cell. Some examples are NS1 of influenza A virus (16); NSs of Bunyamwera virus (9) and Rift Valley fever virus (7); NS1, NS2, SH, and M2-2 of respiratory syncytial virus (10, 26); the V protein of simian virus 5 (22); and the l-proteinase of foot-and-mouth disease virus (11).
The active induction of antiviral processes in MΦ and PK-15 cells by mutant CSFV lacking the Npro gene argues against the possibility that ΔNpro CSFVs simply have lost their capability to interfere with poly(IC)-induced antiviral activities because of a possible reduced replication level as a result of altered translation initiation due to the modifications of the RNA structures at the boundary between the internal ribosomal entry site and the ORF. Furthermore, the hypothesis that Npro is required for direct interference of CSFV with endogenous antiviral pathways is supported by the data of Npro-dependent resistance to poly(IC)-induced apoptosis in SK-6 cells.
A central starting point in the complex cascade of signal transduction events involved in the endogenous cellular antiviral defense is the appearance of intracellular dsRNA, which is provided by the viral genome itself or formed during replication and transcription of viral genomes (25). The best-characterized pathway induced by viral dsRNA involves activation of the dsRNA-dependent protein kinase (PKR) and of 2′-5′ oligoadenylate synthetase which is followed by activation of eIF-2α and RNaseL, respectively, both leading to apoptosis (for selected reviews, see references 12, 17, and 18). A second important dsRNA-dependent pathway induces type I IFN and other cytokines and chemokines through activation of PKR and NF-κB (4). Type I IFN induction by viruses and dsRNA is also dependent on activation and nuclear translocation of the interferon regulatory factor (IRF) family of transcription factors (reviewed in reference 50). Interestingly, it was shown recently that ncp BVDV induced translocation of IRF-3 into the nucleus without subsequent binding to DNA. Furthermore, ncp BVDV was able to block Semliki Forest virus-induced IFN-β production through a block in the formation of IRF-3-DNA complexes (3). Whether this is also true for CSFV and whether Npro is involved in this process remain to be investigated. A third antiviral pathway involves the IFN-inducible Mx proteins, which are large GTPases with homology to dynamin (21, 23). Nevertheless, the remaining innate resistance of triple-deficient RNaseL, PKR, and Mx1 mice to virus infections indicates the presence of additional pathways (57).
During evolution, viruses have learned to interfere with most if not all of these complex antiviral systems (for selected reviews, see references 1, 5, 15, 18, and 45). Although the cellular pathway with which Npro might interact remains to be characterized, our data suggest a novel function of Npro in counteraction of cellular antiviral defense. The importance of this function of Npro becomes lucent when considering the pathogenesis of CSF. Only the presence of Npro permits efficient infection of monocytic cells, including monocytes, MΦ, and even dendritic cells (data not shown). These cells are among the main targets for CSFV, allow high-level replication, and permit cell-associated spreading and colonization of immunological tissue by CSFV. Furthermore, they appear to play a central role in virus-induced immunomodulation (29). Interestingly, ΔNpro CSFV of both the moderately virulent strain Alfort/187 and the highly virulent strain Eystrup were attenuated in pigs (D. Mayer, M. A. Hofmann, and J. D. Tratschin, submitted for publication). The relationship of this finding with the in vitro data presented here, as well as the role of Npro in the pathogenesis of CSF, remains to be established.
Acknowledgments
This work was in part funded by the Swiss National Science Foundation (grant # 31-56719-99).
We thank Markus Gerber, Luzia Liu, Viviane Neuhaus, René Schaffner, and Valérie TÂche for excellent technical assistance and Christian Griot for continuous support. We also thank Martin D. Fray for providing the Mx/CAT reporter gene assay and Bernard Charley for the recombinant porcine IFN-α.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Immunol. Today 21:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aynaud, J. M. 1988. Principles in vaccination, p. 165-180. In B. Liess (ed.), Classical swine fever and related viral infections. Martinus Nijhoff Publishing, Hannover, Germany.
- 3.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 5.Barber, G. N. 2001. Host defense, viruses and apoptosis. Cell Death Differ. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 6.Basta, S., S. M. Knoetig, M. Spagnuolo-Weaver, G. Allan, and K. C. McCullough. 1999. Modulation of monocytic cell activity and virus susceptibility during differentiation into macrophages. J. Immunol. 162:3961-3969. [PubMed] [Google Scholar]
- 7.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brack, K., I. Berk, T. Magulski, J. Lederer, A. Dotzauer, and A. Vallbracht. 1930. 2002. Hepatitis A virus inhibits cellular antiviral defense mechanisms induced by double-stranded RNA. J. Virol. 76:11920-11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinsangaram, J., M. E. Piccone, and M. J. Grubman. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 73:9891-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemens, M. J. 1997. PKR—a protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol. 29:945-949. [DOI] [PubMed] [Google Scholar]
- 13.Diderholm, H., and Z. Dinter. 1966. Interference between strains of bovine virus diarrhea virus and their capacity to suppress interferon of a heterologous virus. Proc. Soc. Exp. Biol. Med. 121:976-980. [DOI] [PubMed] [Google Scholar]
- 14.Fray, M. D., G. E. Mann, and B. Charleston. 2001. Validation of an Mx/CAT reporter gene assay for the quantification of bovine type-I interferon. J. Immunol. Methods 249:235-244. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 17.Gil, J., and M. Esteban. 2000. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5:107-114. [DOI] [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 19.Greiser-Wilke, I., K. E. Dittmar, B. Liess, and V. Moennig. 1992. Heterogeneous expression of the non-structural protein p80/p125 in cells infected with different pestiviruses. J. Gen. Virol. 73:47-52. [DOI] [PubMed] [Google Scholar]
- 20.Greiser-Wilke, I., V. Moennig, C. O. Coulibaly, J. Dahle, L. Leder, and B. Liess. 1990. Identification of conserved epitopes on a hog cholera virus protein. Arch. Virol. 111:213-225. [DOI] [PubMed] [Google Scholar]
- 21.Haller, O., and G. Kochs. 2002. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 22.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 23.Horisberger, M. A. 1992. Interferon-induced human protein MxA is a GTPase which binds transiently to cellular proteins. J. Virol. 66:4705-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inaba, Y., T. Omori, and T. Kumagai. 1963. Detection and measurement of non-cytopathogenic strains of virus diarrhea of cattle by the END method. Arch. Gesamte Virusforsch. 13:425-429. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 26.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210-218. [DOI] [PubMed] [Google Scholar]
- 27.Kasza, L., J. A. Shadduck, and G. J. Christofinis. 1972. Establishment, viral susceptibility and biological characteristics of a swine kidney cell line SK-6. Res. Vet. Sci. 13:46-51. [PubMed] [Google Scholar]
- 28.Knoetig, S. M., K. C. McCullough, and A. Summerfield. 2002. Lipopolysaccharide-induced impairment of classical swine fever virus infection in monocytic cells is sensitive to 2-aminopurine. Antiviral Res. 53:75-81. [DOI] [PubMed] [Google Scholar]
- 29.Knoetig, S. M., A. Summerfield, M. Spagnuolo-Weaver, and K. C. McCullough. 1999. Immunopathogenesis of classical swine fever: role of monocytic cells. Immunology 97:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumagai, T., T. Shimizu, S. Ikeda, and M. Matumoto. 1961. A new in vitro method (END) for detection and measurement of hog cholera virus and its antibody by means of effect of HC virus on Newcastle disease virus in swine tissue culture. I. Establishment of standard procedure. J. Immunol. 87:245-256. [PubMed] [Google Scholar]
- 31.Kumagai, T., T. Shimizu, and M. Matumoto. 1958. Detection of hog cholera virus by its effect on Newcastle disease virus in swine tissue culture. Science 128:366. [DOI] [PubMed] [Google Scholar]
- 32.Lefevre, F., R. L'Haridon, F. Borras-Cuesta, and C. La Bonnardiere. 1990. Production, purification and biological properties of an Escherichia coli-derived recombinant porcine alpha interferon. J. Gen. Virol. 71:1057-1063. [DOI] [PubMed] [Google Scholar]
- 33.Maisonnave, J., and C. R. Rossi. 1982. A microtiter test for detecting and titrating noncytopathogenic bovine viral diarrhea virus. Arch. Virol. 72:279-287. [DOI] [PubMed] [Google Scholar]
- 34.McCullough, K. C., S. Basta, S. Knoetig, H. Gerber, R. Schaffner, Y. B. Kim, A. Saalmuller, and A. Summerfield. 1999. Intermediate stages in monocyte-macrophage differentiation modulate phenotype and susceptibility to virus infection. Immunology 98:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers, G., and H. J. Thiel. 1996. Molecular characterization of pestiviruses. Adv. Virus Res. 47:53-118. [DOI] [PubMed] [Google Scholar]
- 36.Mittelholzer, C., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. Generation of cytopathogenic subgenomic RNA of classical swine fever virus in persistently infected porcine cell lines. Virus Res. 51:125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser, C., P. Stettler, J. D. Tratschin, and M. A. Hofmann. 1999. Cytopathogenic and noncytopathogenic RNA replicons of classical swine fever virus. J. Virol. 73:7787-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura, S., T. Shimazaki, K. Sakamoto, A. Fukusho, Y. Inoue, and N. Ogawa. 1995. Enhanced replication of orbiviruses in bovine testicle cells infected with bovine viral diarrhoea virus. J. Vet. Med. Sci. 57:677-681. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, N. J., Jr., R. G. Douglas, Jr., R. M. Simons, and M. E. Diamond. 1979. Virus-induced interferon production by human macrophages. J. Immunol. 123:365-369. [PubMed] [Google Scholar]
- 40.Roehe, P. M., M. J. Woodward, and S. Edwards. 1992. Characterisation of p20 gene sequences from a border disease-like pestivirus isolated from pigs. Vet. Microbiol. 33:231-238. [DOI] [PubMed] [Google Scholar]
- 41.Rossi, C. R., and G. K. Kiesel. 1980. Factors affecting the production of bovine type I interferon on bovine embryonic lung cells by polyriboinosinic-polyribocytidylic acid. Am. J. Vet. Res. 41:557-560. [PubMed] [Google Scholar]
- 42.Ruggli, N., J. D. Tratschin, C. Mittelholzer, and M. A. Hofmann. 1996. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J. Virol. 70:3478-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rümenapf, T., R. Stark, M. Heimann, and H. J. Thiel. 1998. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J. Virol. 72:2544-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer, M., and E. Peterhans. 2001. Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J. Virol. 75:4692-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu, Y., S. Furuuchi, T. Kumagai, and J. Sasahara. 1970. A mutant of hog cholera virus inducing interference in swine testicle cell cultures. Am. J. Vet. Res. 31:1787-1794. [PubMed] [Google Scholar]
- 47.Stark, R., G. Meyers, T. Rümenapf, and H. J. Thiel. 1993. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J. Virol. 67:7088-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Summerfield, A., S. M. Knoetig, and K. C. McCullough. 1998. Lymphocyte apoptosis during classical swine fever: implication of activation-induced cell death. J. Virol. 72:1853-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Summerfield, A., K. Zingle, S. Inumaru, and K. C. McCullough. 2001. Induction of apoptosis in bone marrow neutrophil-lineage cells by classical swine fever virus. J. Gen. Virol. 82:1309-1318. [DOI] [PubMed] [Google Scholar]
- 50.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 51.Toba, M., and M. Matumoto. 1969. Role of interferon in enhanced replication of Newcastle disease virus in swine cells infected with hog cholera virus. Jpn. J. Microbiol. 13:303-305. [DOI] [PubMed] [Google Scholar]
- 52.Tratschin, J. D., C. Moser, N. Ruggli, and M. A. Hofmann. 1998. Classical swine fever virus leader proteinase Npro is not required for viral replication in cell culture. J. Virol. 72:7681-7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trautwein, G. 1988. Pathology and pathogenesis of the disease, p. 27-54. In B. Liess (ed.), Classical swine fever and related infections. Martinus Nijhoff Publishing, Boston, Mass.
- 54.Tsuchiya, Y., A. Uchimura, H. Tajika, K. Sakamoto, T. Furuya, K. Sato, K. Nanba, and Y. Miura. 1993. Reverse interference method for measurement of hog cholera virus (HCV) and anti-HCV antibody. J. Vet. Med. Sci. 55:233-236. [DOI] [PubMed] [Google Scholar]
- 55.vanRegenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the classification and nomenclature of viruses. The seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 56.Wiskerchen, M., S. K. Belzer, and M. S. Collett. 1991. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J. Virol. 65:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]