Abstract
The 11th influenza A virus gene product is an 87-amino-acid protein provisionally named PB1-F2 (because it is encoded by an open reading frame overlapping the PB1 open reading frame). A significant fraction of PB1-F2 localizes to the inner mitochondrial membrane in influenza A virus-infected cells. PB1-F2 appears to enhance virus-induced cell death in a cell type-dependent manner. For the present communication we have identified and characterized a region near the COOH terminus of PB1-F2 that is necessary and sufficient for its inner mitochondrial membrane localization, as determined by transient expression of chimeric proteins consisting of elements of PB1-F2 genetically fused to enhanced green fluorescent protein (EGFP) in HeLa cells. Targeting of EGFP to mitochondria by this sequence resulted in the loss of the inner mitochondrial membrane potential, leading to cell death. The mitochondrial targeting sequence (MTS) is predicted to form a positively charged amphipathic α-helix and, as such, is similar to the MTS of the p13II protein of human T-cell leukemia virus type 1. We formally demonstrate the functional interchangeability of the two sequences for mitochondrial localization of PB1-F2. Mutation analysis of the putative amphipathic helix in the PB1-F2 reveals that replacement of five basic amino acids with Ala abolishes mitochondrial targeting, whereas mutation of two highly conserved Leu to Ala does not. These findings demonstrate that PB1-F2 possesses an MTS similar to other viral proteins and that this MTS, when fused to EGFP, is capable of independently compromising mitochondrial function and cellular viability.
Most influenza A virus (IAV) isolates carry an additional open reading frame overlapping the PB1 gene that encodes a small protein termed PB1-F2 (4). PB1-F2 has a number of unusual features compared with other influenza virus gene products or, for that matter, the gene products of other viruses. The levels of expression of PB1-F2 differ greatly among infected cells in a manner that is independent of the levels of expression of other viral proteins. PB1-F2 localizes to the mitochondria in ∼50% of cells with detectable PB1-F2 expression and in the nucleus and/or cytosol in the remaining cells. Mitochondrial localization is an intrinsic property of the protein that can be observed outside of the context of IAV infection, since it occurs following either microinjection of a full-length synthetic version of the protein or expression from a transgene encoding PB1-F2. A fully synthetic version of the PB1-F2 induces cell death when incubated with cells or following its introduction into cells via microinjection. In the context of viral infection, PB1-F2 is associated with cell type-specific enhancement of cell death, which occurs in monocytes but not epithelial cells that support IAV replication.
PB1-F2 is one of a rapidly growing list of viral proteins from a number of viral families that localize to the mitochondria and influence cell viability (2). Given the central importance of mitochondria in controlling programmed cell death (7, 12, 17), it is easy to understand why it would be in the interest of viruses to modulate mitochondrial function.
There are many questions to be answered regarding the mitochondrial targeting of PB1-F2 and how mitochondrial localization impacts mitochondrial function. In this communication we identify the region of the protein necessary and sufficient for mitochondrial localization and demonstrate the effects of mitochondrial localization on its ability to promote cell death.
MATERIALS AND METHODS
Oligonucleotides.
The oligonucleotides used in this study were all purchased from Bio-Synthesis, Inc. (Lewisville, Tex.). Upstream (5′) oligonucleotides used for PCR amplification of PB1-F2 fragments consisted of a SalI restriction enzyme recognition sequence followed by an ATG translation initiation codon followed by six coding triplets. Downstream (3′) oligonucleotides consisted of a BamHI recognition site and a CG spacer to maintain reading frames with green fluorescent protein (EGFP) of plasmid pEGFP-N1 followed by 18 nucleotides, the reverse complement of which codes for the last six codons of the PB1-F2 fragment. These were as follows: 5′ PB1-F2 SalI (CGTCGACATGGGACAGGAACAGGATACA); 5′ PB1-F2 Sal42 (CGTCGACATGTGCCAGAAGACAATGAAC); 5′ PB1-F2 Sal52 (CGTCGACATGCCCAAACAGATTGTGTAT); 5′ PB1-F2 Sal65 (CGTCGACATGAGGAATCCCATCCTGGTA); 5′ PB1-F2 Sal73 (CGTCGACATGAAAACTCGTGTATTGAAA); 3′ PB1-F2 Bam87 (CGGATCCCGCTCGTGTTTGCTGAACAA); 3′ PB1-F2 Bam72 (CGGATCCCGCAAAAATACCAGGATGGG); 3′ PB1-F2 Bam51 (CGGATCCCGCATAACCACTTGGTTCAT); and 3′ PB1-F2 Bam41 (CGGATCCCGGTGGCCCATCAATCGGGT).
The oligonucleotides used for site-directed mutagenesis were as follows: PB1-F2p13helixAtop (CCCATCCTGGTATTTCTACGTGTTTGGAGACTGTGTGCAAGGCGATTGTTCAGCAAAC); PB1-F2 p13helixAbot (GTTTGCTGAACAATCGCCTTGCACACAGTCTCCAAACACGTAGAAATACCAGGATGGG); PB1-F2 LALAtop (CTGGTATTTGCGAAAACTCGTGTAGCGAAACGATGGAGG); PB1-F2 LALAbot (CCTCCATCGTTTCGCTACACGAGTTTTCGCAAATACCAG); PB1-F2 L72IL-77Itop (CCCATCCTGGTATTTATCAAAACTCGTGTATTGAAACG); PB1-F2 L72IL-77Ibot (CGTTTCAATACACGAGTTTTGATAAATACCAGGATGGG); PB1-F2 L77Itop (GAAAACTCGTGTAATAAAACGATGGAGGTTG); PB1-F2 L77Ibot (CAACCTCCATCGTTTTATTACACGAGTTTTC); PB1-F2 5Atop (GGTATTTTTGGCAACTGCTGTATTGGCAGCATGGGCGTTGTTCAGC); and PB1-F2 5Abot (GCTGAACAACGCCCATGCTGCCAATACAGCAGTTGCCAAAAATACC).
Oligonucleotide pairs which were directly phosphorylated, annealed, and ligated into plasmid vectors were as follows: PB1-F2 69-85top (TCGACATGCTGGTATTTTTGAAAACTCGTGTATTGAAACGATGGAGGTTGTTCAGCAAG); PB1-F2 69-85bot (GATCCTTGCTGAACAACCTCCATCGTTTCAATACACGAGTTTTCAAAAATACCAGTACG); PB1-F2 72-82top (TCGACATGTTGAAAACTCGTGTATTGAAACGATGGAGGTTG); PB1-F2 72-82bot (GATCCAACCTCCATCGTTTCAATACACGAGTTTTCAATACG); PB1-F2 69-82top (TCGACATGCTGGTATTTTTGAAAACTCGTGTATTGAAACGATGGAGGTTG); PB1-F2 69-82bot (GATCCAACCTCCATCGTTTCAATACACGAGTTTTCAAAAATACCAGTACG); PB1-F2 72-85top (TCGACATGTTGAAAACTCGTGTATTGAAACGATGGAGGTTGTTCAGCAAG); and PB1-F2 72-85bot (GATCCTTGCTGAACAACCTCCATCGTTTCAATACACGAGTTTTCAATACG).
Plasmid construction.
Using Platinum Taq High-Fidelity polymerase (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions, fragments containing PB1-F2 coding sequence were PCR amplified with upstream and downstream oligonucleotides. Resulting PCR product DNAs were directly ligated into plasmid pCR-Blunt II-TOPO (Invitrogen). After DNA sequence verification, SalI-to-BamHI fragments of these plasmids were ligated into SalI- and BamHI-digested pEGFP-N1 (BD Biosciences Clontech, Palo Alto, Calif.). The resultant plasmids were pPB1-F2 1-87EGFP, pPB1-F2 1-72EGFP, pPB1-F2 1-51EGFP, pPB1-F2 1-41EGFP, pPB1-F2 42-87EGFP, pPB1-F2 52-87EGFP, pPB1-F2 65-87EGFP, pPB1-F2 73-87EGFP, and pPB1-F2 52-72EGFP. In these plasmids, the spans of numbers indicate the amino acids of PB1-F2 fused to EGFP. All EGFP fusion constructs contained a flexible linker sequence between the PB1-F2 sequence and EGFP (RDPPVAT). For finer mapping of the mitochondrial targeting sequence (MTS) of PB1-F2, complementary oligonucleotide pairs having SalI and BamHI cohesive ends and containing an ATG translation initiation codon followed by coding triplets were used. These oligonucleotide pairs were phosphorylated using polynucleotide kinase (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions. Phosphorylated oligonucleotide pairs were directly ligated into SalI- and BamHI-digested pEGFP-N1. The resultant plasmids were pPB1-F2 69-85EGFP, pPB1-F2 72-82EGFP, pPB1-F2 69-82EGFP, and pPB1-F2 72-85EGFP.
Mutagenesis.
Plasmid pPB1-F2 1-87EGFP, consisting of the full-length PB1-F2 open reading frame fused in frame with that of EGFP, was used as a template for all mutagenesis. The QuickChange site-directed mutagenesis kit (Stratagene, Cedar Creek, Tex.) was used according to the manufacturer's instructions. The resultant plasmids were pPB1-F2 1-87L72AL77AEGFP (containing leucine 72-to-alanine and leucine 77-to-alanine mutations), pPB1-F2 1-87L72IL-77IEGFP (containing leucine 72-to-isoleucine and leucine 77-to-isoleucine mutations), pPB1-F2 1-87L77IEGFP (containing a leucine 77-to-isoleucine mutation), and pPB1-F2 1-875AEGFP (containing alanine mutations at lysine 73, arginine 75, lysine 78, arginine 79, and arginine 81). Finally, mutation pPB1-F2 1-71p13helixEGFP consisted of PB1-F2 amino acids 1 to 71, followed by the p13II sequence LRVWRLCARRL, fused in frame to EGFP.
Microscopy.
Confocal microscopy was performed using either a Zeiss Axioplan fluorescence microscope equipped with an MRC-1024 laser scanning confocal system (controlled by Lasersharp 3.2 image acquisition and analysis software from Bio-Rad Laboratories [Hercules, Calif.]) or a Leica SP2 confocal system equipped with UV-Vis lasers from Leica Microsystems (Mannheim, Germany). Images were obtained with a 63×, 1.4-numerical-aperture Plan-Apochromat oil immersion or a 40×, 1.2-numerical-aperture water immersion dipping objective. To assess patterns of subcellular localization of various PB1-F2-EGFP constructs, EGFP fluorescence in transfected HeLa cells was examined in living cells or after fixation with 3.2% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. To assess localization to mitochondria, living cells were labeled with tetramethylrhodamine ethyl ester (TMRE) (50 nM) or MitoTracker Red CMXRos (100 nM) from Molecular Probes (Eugene, Oreg.) according to the manufacturer's instructions. Mitochondrial transmembrane potential (ΔΨm) was monitored using TMRE. For time-lapse analysis, cells were plated on Delta T culture dishes and temperature was controlled using the Delta TC3 system from Bioptechs (Butler, Pa.). Cells were imaged at 37°C in phenol red-free Dulbecco's minimal Eagle's medium supplemented with 10% fetal bovine serum and 20 mM HEPES (pH 7.2). Levels of phosphatidylserine exposed on the outside of cells were determined by binding of Alexa 647-labeled annexin V. Propidium iodide (PI), ethidium homodimer 1, and Hoechst 33342 (all from Molecular Probes) were used to assess cell viability and nuclear morphology. PB1F2EGFP, TMRE, PI, Alexa 647-labeled annexin V, and Hoechst 33342 fluorescence images were acquired using 488-, 568-, 568-, 633-, and 364-nm laser lines, respectively. For illustrations, images were cropped and montaged using Photoshop 7.0 software from Adobe Systems (Mountain View, Calif.). A series of images over time (at 10- to 30-s intervals) were obtained by sequential line scanning to avoid signal crossover (i.e., each line of the 512-line image was scanned sequentially using laser line no. 1 and then laser line no. 2; this required 3.6 s). Laser power was kept at very low intensity to limit cytotoxicity. Using Premiere 6.0 software from Adobe Systems, time-lapse movies were created from the sequential image series.
Fluorescence resonance energy transfer (FRET) experiments were performed to determine the proximity of the 65-87-EGFP fusion protein to the inner mitochondrial membrane labeled by TMRE. Using the Leica SP2 confocal system, FRET levels were measured by the acceptor photobleaching method (1). The SP2 spectral detector enables the recording of spectral information of the specimen with 5-nm-bandwidth resolution. To utilize the full dynamic range of the system, gain and offset values were set to prevent pixel saturation and the minimum values in the images were near zero. Image series were recorded using the advanced time-lapse software module of the system. Five prebleaching images were recorded of the donor (EGFP at 488 nm) and of the acceptor (TMRE at 568 nm) by sequential line scanning. The acceptor (TMRE) was photobleached by parking the 568-nm laser line at full power for 5 to 6 s over defined points of interest; this reduced the fluorescence to background levels. Postbleaching series of images were obtained for both the donor and acceptor. The fluorescence intensities of the two channels were plotted over time. Using the mean fluorescence intensities in the photobleached area, pixel-by-pixel FRET efficiencies were calculated from the following image arithmetic operation: (donor postbleaching − donor prebleaching)/(donor postbleaching).
Transfection and fluorescence-activated cell sorter analysis.
HeLa cells that had been split 24 h previously were trypsinized, and 2 × 106 cells were resuspended in 2 ml of Dulbecco's minimal Eagle's medium with 10% fetal bovine serum. A 200-μl transfection mixture containing 2 μg of plasmid DNA and 4 μl of Lipofectamine 2000 (Invitrogen) was added per the manufacturer's instructions. Transfected cells were rotated overnight at 37°C in suspension. At 24 h posttransfection, cells were centrifuged and washed once in PBS (Invitrogen). Cells were then layered on an equal volume of Ficoll and centrifuged at 913 × g for 20 min. Cells from the buffer-Ficoll interface were again washed in PBS and resuspended in 1 ml of PBS. Cells were stained with 50 nM TMRE (Molecular Probes) in the presence of 40 μM verapamil for 20 min at 37°C, followed by direct analysis on a FACScan cytometer (BD Biosciences, San Jose, Calif.) using a 488-nm laser. A GFP signal was acquired in channel FL1. TMRE signal was acquired in channel FL2. Verapamil was added to negate effects of expressed genes on multidrug resistance protein-mediated export of TMRE from cells. Similar results were obtained in experiments in which verapamil was omitted.
RESULTS
Sequence analysis.
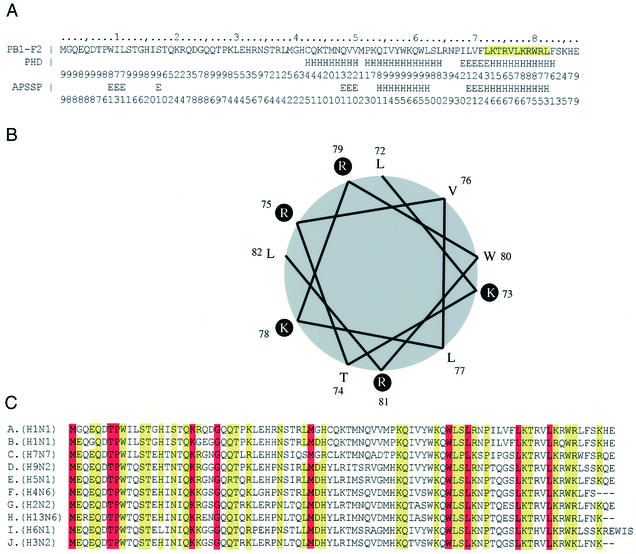
BLAST searches between the sequence of PB1-F2 encoded by PR8 and non-influenza virus sequences deposited in GenBank failed to reveal any significant sequence similarities. Lacking such clues regarding PB1-F2 structure, we turned to the software programs PHD (13) and APSSP (http://imtech.ernet.in/raghava/apssp) to predict regions of secondary structure (Fig. 1A). Both programs strongly predict α-helical regions between amino acids 54 to 62 and 73 to 82. The predicted helix at amino acids 73 to 82 was found to contain an 11-amino-acid sequence with obvious similarity to the amphipathic α-helix of the human T-cell leukemia virus type 1 (HTLV-I) p13II protein, which is responsible for mitochondrial targeting of this protein (5, 7).
FIG. 1.
PB1-PB1-F2 sequence comparison and structural predictions. (A) The sequence of PB1-F2 is shown along with structural predictions (H, helix; E, extended) by PHD and APSSP programs with respect to helix formation. (B) Helical wheel representation of a predicted amphipathic helix formed by residues 72 to 82 of PB1-F2. (C) Sequences of PB1-F2 representing all IAV subtypes for which sequences are available. Residues present in all 90 IAV strains examined are highlighted in red; those conserved in >90% of the strains are highlighted in yellow. Strains are indicated as follows: A, Puerto Rico/8/34 (H1N1); B, FM/1/47 (H1N1); C, Equine/London/1416/73 (H7N7); D, Chicken/Korea/38349-p96323/96 (H9N2); E, Goose/Guangdong/1/96 (H5N1); F, Duck/Nanchang/662/98 (H4N6); G, Leningrad/134/47/57 (H2N2); H, Gull/Maryland/704/77 (H13N6); I, Teal/Hong Kong/W312/97 (H6N1); J, NT/60/68 (H3N2).
Specifically, LKTRVLKRWRL of PB1-F2 and LRVWRLCARRL of HTLV-I p13II both have three Leu residues and are rich in basic amino acids, possessing five and four basic residues, respectively. A helical wheel representation (created using the program GCG Lite [Accelrys, San Diego, Calif.]) demonstrates the potential of PB1-F2 to form amphipathic helices (Fig. 1B). Two other retroviruses encode small proteins with an MTS that are likely to form amphipathic helices: one is the bovine leukemia virus (like HTLV-I, a deltaretrovirus), which encodes G4 protein (10), and the other is human immunodeficiency virus 1, which encodes viral protein R (VPR) (9).
Using the AlignX component of Vector NTI Suite (Informax, Inc., Bethesda, Md.), we aligned the amino acid sequences of predicted PB1-F2 gene products from 74 IAV isolates as a further method of gauging potential functional activities of PB1-F2 domains. Representative sequences encoded by PB1-F2 genes from different IAV subtypes are shown in Fig. 1C. Amino acids that are completely conserved among all 74 isolates are colored red, and those conserved in more than 90% of isolates are colored yellow. Notably, residues within the predicted amphipathic helix are highly conserved, with two Leu residues being among only 10 residues that are completely conserved among the 74 sequences. The basic residues are also highly conserved and, with only a few exceptions, are replaced by other basic residues.
This sequence conservation is consistent with the proposed helical structure for this region, but it is not conclusive, particularly since PB1-F2 overlaps with the PB1 open reading frame, so conserved residues in PB1 may restrict the latitude of acceptable substitutions in PB1-F2. On the other hand, more than 85% of residues in PB1-F2 differ between influenza virus isolates and the protein is much more variable than the overlapping region of PB1.
Plasmid-mediated expression of PB1-F2-EGFP fusion proteins.
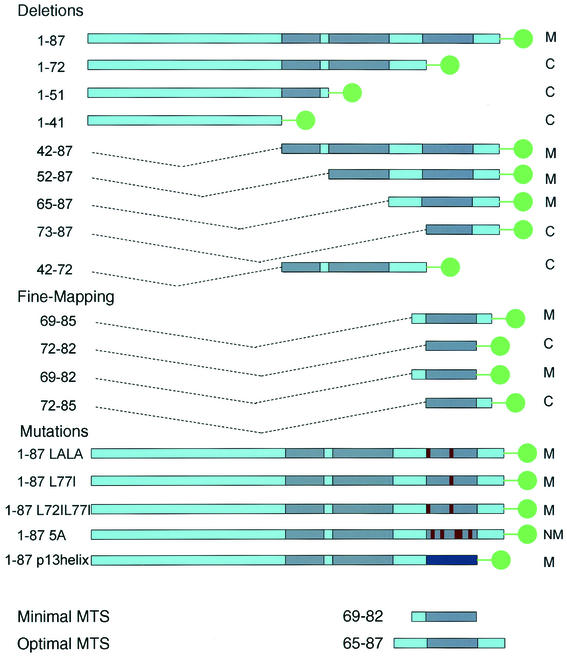
To map the MTS of PB1-F2, we generated a panel of cDNAs encoding amino- and carboxy-terminal deletion mutations appended to EGFP as a reporter protein (Fig. 2). This approach has been used successfully to identify the MTSs present in retroviral proteins (5, 10). These constructs were designed to encompass or omit potential secondary structural elements at amino acids 42 to 50, 52 to 64, 67 to 72, and 73 to 83. Using the cytomegalovirus early promoter to control expression of the fusion proteins, proteins were expressed by plasmid-mediated transfection.
FIG. 2.
Chimeric PB1-F2-EGFP fusion proteins utilized in this study. The residues incorporated in fusion proteins are shown on the left. Green dots signify EGFP. Dark blue segments represent predicted helices. Red segments represent substituted residues. All fusion proteins contained a 7-residue linker (RDPPVAT) between the PB1-F2 sequence and EGFP. The intracellular localization of the fusion proteins is summarized on the right as follows: M, mitochondrial; C, cytoplasmic and/or nuclear; NM, nonmitochondrial.
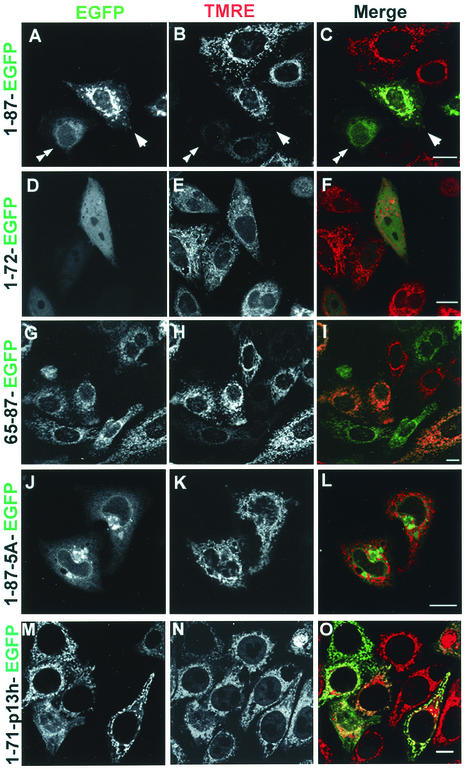
Transfection of the control plasmid pEGFP, expressing EGFP alone, results in a diffuse green fluorescence in the cytoplasm and nucleus of HeLa cells. This reflects the lack of trafficking signals in EGFP in conjunction with its compact structure, which enables it to freely traverse nuclear pores. In cells expressing full-length PB1-F2 fused with EGFP (1-87-EGFP), by contrast, fluorescence was primarily localized to filamentous cytoplasmic structures that in number and appearance are likely to represent mitochondria (Fig. 3A). This was confirmed by staining cells with TMRE, which is an inner mitochondrial membrane potential (ΔΨm)-sensitive dye (Fig. 3B and C). Constructs expressing PB1-F2 amino acids 65 to 87 (Fig. 3G to I), 42 to 87, and 52 to 87 also exhibited mitochondrial localization (data not shown), whereas constructs expressing PB1-F2 amino acids 1 to 72 (Fig. 3D to F), 1 to 51, 1 to 41, 73 to 87, and 52 to 72 exhibited EGFP-like diffuse cytoplasmic and/or nuclear fluorescence, demonstrating the loss of the MTS (data not shown).
FIG. 3.
Intracellular targeting of PB1-F2-EGFP fusion constructs. HeLa cells were labeled with the ΔΨm-sensitive mitochondrial dye TMRE, and images were acquired live at 16 h posttransfection with plasmids expressing PB1-F2-EGFP fusion proteins as indicated. Fluorescence images are presented as unmerged EGFP (left column), TMRE (middle column), and merged fluorescence (right column; EGFP is shown in green, and TMRE is shown in red). The full-length (1 to 87) chimeric protein (A to C) localizes to mitochondria (arrowheads) but also to the nucleus, nuclear membrane, and cytoplasm (double arrowheads). A deletion mutant (1 to 72) lacking the last 15 amino acids (D to F) displays uniform cytoplasmic and nuclear localization and is clearly absent from mitochondria. Among all the chimeric proteins, that consisting of PB1-F2 residues 65 to 87 exhibits the most complete mitochondrial localization (G to I). Replacing the 5 positively charged residues in the MTS of the full-length construct with Ala (1-87-5A) targeted the fusion protein to the nuclear membrane and another cytoplasmic structure (J to L). Replacing the MTS with the MTS of p13II in the full-length construct (1-71-p13 h-EGFP) targeted the fusion protein to mitochondria (M to O). Bars, 10 μm.
Among the fusion proteins that localized to mitochondria, there was considerable variation in the degree of mitochondrial localization. All of the fusion proteins that localized to the mitochondria, including 1-87-EGFP, were also present in the nucleus, nuclear membrane, and cytoplasm of a significant fraction of cells. While this is similar to the intracellular localization of PB1-F2 in IAV-infected cells, it differs from PB1-F2 expressed via transfection or a recombinant vaccinia virus, which is much more uniformly distributed in a mitochondrial pattern (4). It appears, therefore, that fusion with EGFP or interaction with other influenza virus proteins disfavors to a degree the intrinsic MTS in PB1-F2. One thing that is clear, however, is that 65-87-EGFP exhibits an extent of mitochondrial localization superior to 1-87-EGFP or any other construct tested.
Taken together, these results imply that the predicted amphipathic α-helix identified as potentially similar to that of HTLV-I p13II functions as the PB1-F2 MTS. Specifically, amino acids 65 to 87 are sufficient for mitochondrial localization, whereas residues 1 to 72 and 73 to 87 are insufficient for mitochondrial localization. The failure of residues 73 to 87 (which contain the predicted amphipathic helix) to function as an MTS suggested that the short hydrophobic sequence upstream is also important for proper targeting. We decided to explore in detail the contribution of the preceding hydrophobic residues to the targeting.
Fine mapping of the MTS.
Working on the basis of these results, we generated plasmids that express PB1-F2 amino acids 69 to 85, 72 to 82, 69 to 82, or 72 to 85 of PB1-F2 fused at the N terminus of EGFP. Of these partial PB1-F2 gene products, sequences spanning amino acids 69 to 85 (LVFLKTRVLKRWRLFSK) and 69 to 82 (LVFLKTRVLKRWRL) alone targeted EGFP to mitochondria, albeit less efficiently than the sequence spanning amino acids 65 to 87, which demonstrated the most complete mitochondrial targeting of all of the sequences tested (all of the localization data are summarized in Fig. 2). Deletion of the three amino terminal residues from 69 to 85 or 69 to 82 completely abolished mitochondrial targeting. Notably, despite the apparently important role of these residues in mitochondrial targeting, various IAV isolates demonstrate nonconservative substitutions at residues 69 to 71, suggesting that a variety of residues can support mitochondrial targeting, though the mitochondrial localization of PB1-F2 from these isolates remains to be established.
We next examined the effect of replacement of basic residues with neutral residues in the predicted amphipathic helix. Replacement of the two Lys residues with Ala reduced MTS function. MTS function was completely abolished by substituting Ala for Lys or Arg at positions 73, 75, 78, 79, and 81 in 1-87-EGFP (construct 1-87 5A-EGFP in Fig. 2 and Fig. 3J to L). Instead, the fusion protein localized to the nuclear membrane and condensed cytoplasmic structures of an uncertain nature. Based on the two predictive programs, these substitutions should not interfere with helix formation, which suggests that mitochondrial targeting requires an amphipathic α-helix and not simply an α-helix. These results imply that the five positively charged residues are required for mitochondrial targeting. Replacement of highly conserved Leu residues at positions 72 and 77 with Ile or Ala did not abolish mitochondrial localization.
Finally, we replaced amino acids 72 through 87 in 1-87-EGFP with the 11-residue MTS from HTLV-I p13II, LRVWRLCARRL (Fig. 3 M to O). The fusion protein localized to mitochondria, demonstrating the functional interchangeability of the two sequences for mitochondrial targeting. Since there is structural evidence that the p13II forms an amphipathic helix (6), this provides additional evidence that the analogous sequence in PB1-F2 behaves similarly.
Localization of PB1-F2 65-87-EGFP within mitochondria.
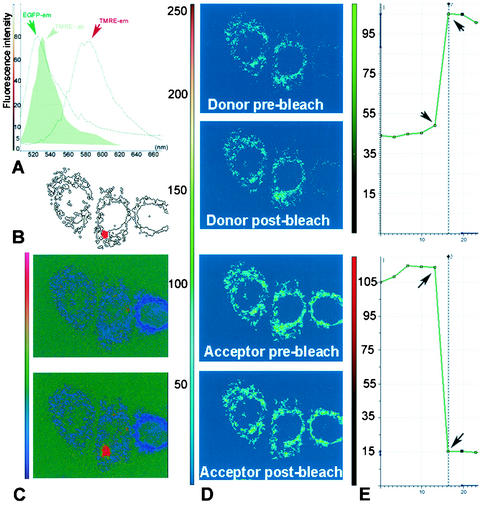
Having established that residues 65 to 87 serve as an MTS, it was of interest to further characterize the location of 65-87-EGFP within mitochondria. Taking advantage of the autofluorescence of EGFP and the known location of TMRE in the mitochondrial matrix, we used FRET to determine whether EGFP is in close proximity to TMRE. When fluorescent molecules are intimately associated (less than 10 nm apart), the fluorescent emissions of donor fluors can be captured by acceptor fluors with relatively high efficiency. When this occurs, the intensity of emission detected from the donor fluor is decreased, a process known as quenching. This occurrence can be detected by photodestruction (photobleaching) of the acceptor fluor, which results in increased emission detected from the donor fluor. EGFP and TMRE constitute a well-matched FRET pair, since there is considerable overlap between the EGFP emission and TMRE excitation spectra (Fig. 4A).
FIG. 4.
FRET analysis of 65-87-EGFP-TMRE localization. (A) Using a Leica AOBS-SP2 confocal system, EGFP and TMRE emission spectra were recorded by wavelength scanning (lambda scan) between 500 and 660 nm with a 5-nm detection window. Fluorescence intensity plots show significant overlap between EGFP emission spectra and TMRE excitation spectra (green filled histogram) and sufficient separation between the TMRE excitation and emission spectra. (B) Spot bleaching (area marked in red) of the acceptor was performed as described in Material and Methods. (C) The ratio images of donor-acceptor fluorescence before (upper panel) and after (lower panel) photobleaching are indicated together with the look-up table on the left side of the figure. Notice that the only change is in the region of photobleaching, which demonstrates a large increase in donor signal. (D) Confocal images of the donor acquired using a 488-nm excitation line and emission between 500 and 550 nm obtained pre- and postbleaching reveal dequenching of the donor. Using a 568-nm excitation line and emission between 580 and 620 nm, sequential scan images of the acceptor were also acquired pre- and postbleaching; these reveal effective bleaching of the acceptor. (E) These panels depict the average pixel intensity profiles over time of the bleached region, with the images pre- and postbleaching marked by arrows. Notice the increase in intensity of EGFP (donor; upper graph) signal that occurs concomitantly with the bleaching-induced decrease in the TMRE (acceptor; bottom graph) signal.
To establish whether TMRE quenches 65-87-EGFP, we examined the effect of photobleaching TMRE on the intensity of EGFP fluorescence. Photobleaching of TMRE results in an immediate increase in the EGFP fluorescence of 65-87-EGFP (Fig. 4C to D), which is indicative of close spatial proximity of the two fluors. The measured FRET efficiency of 40% is consistent with a distance of less than 50 Å between EGFP and TMRE. Importantly, we observed no FRET between the TMRE and 1-72-EGFP which was present in the nucleus and/or cytoplasm or enhanced yellow fluorescent protein (EYFP) targeted to the mitochondrial matrix by addition of the MTS of subunit VIII of human cytochrome c oxidase (note that EYFP is an equally suitable FRET partner for TMRE) (data not shown).
In conjunction with previous evidence from immunoelectron microscopy that PB1-F2 localizes to the inner mitochondrial membrane (4), these data are consistent with the idea that the PB1-F2 MTS targets a substantial fraction of the EGFP fusion protein to the inner mitochondrial membrane in a manner such that the GFP domain is present in the matrix. The enhanced interaction of 65-87-EGFP with TMRE relative to matrix-targeted EYFP might be explained by a higher effective TMRE concentration at the surface of the inner mitochondrial membrane than in the matrix (15) (L. M. Loew, personal communication).
Effect of the PB1-F2 MTS on mitochondrial function.
We previously reported that the presence of PB1-F2 in mitochondria of IAV-infected or PB1-F2-transfected or -microinjected cells is associated with mitochondrial rounding, swelling, and fragmentation (4). This was usually accompanied by a loss of mitochondrial potential, as indicated by a decreased accumulation of ΔΨm-sensitive dyes such as MitoTracker Red. We examined the effect of expression of the chimeric fusion proteins on mitochondrial potential in transfected HeLa cells, as determined by TMRE fluorescence. In the course of examining the patterns of localization of various EGFP-tagged fusion proteins we observed a decrease of the mitochondrial potential of some cells, as indicated by low levels of TMRE staining (Fig. 3). We explored this finding in a quantitative manner by both flow cytometry and confocal microscopy.
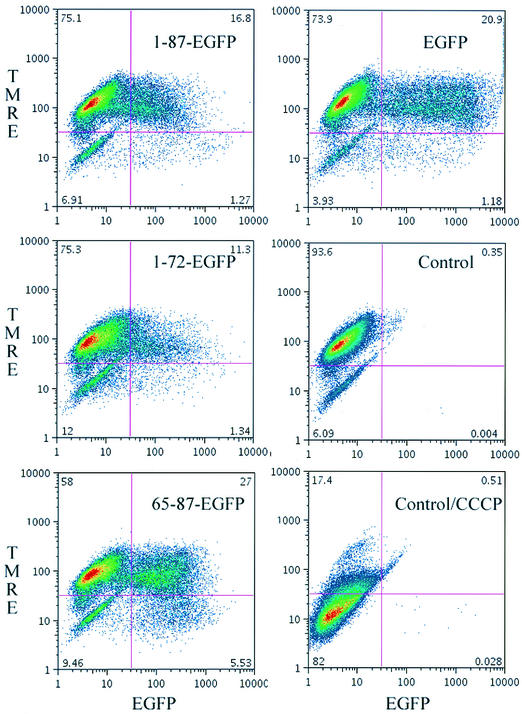
Via flow cytometry, we found that TMRE accumulation in HeLa cells depends on functional mitochondria, as shown by loss of staining following the collapsing of ΔΨm with the protonophore CCCP (Fig. 5, bottom right panel). Transient expression of 65-87 EGFP was associated with a decrease in TMRE staining that was related to the level of GFP expression; i.e., decreased TMRE staining correlated with high levels of GFP fluorescence (Fig. 5). By contrast, cells expressing 1-87-EGFP (top left panel), 1-72-EGFP (center left panel), or EGFP (top right panel) failed to demonstrate such a relationship.
FIG. 5.
Effect of PB1-F2 MTS-targeted protein on ΔΨm cells as detected by flow cytometry. The results of analysis of transiently transfected are shown. At 16 h posttransfection, HeLa cells were incubated with 50 nM TMRE for 30 min and then analyzed for EGFP expression (x axes) and TMRE accumulation (y axes). Control, mock-transfected cells. CCCP was added to cells to demonstrate that TMRE accumulation was based on maintenance of ΔΨm.
The effects of fusion protein expression on mitochondrial function were confirmed by acquisition of laser-scanning confocal microscopy images of randomly chosen fields of transfected cells grown on coverslips and by determination of the fraction of cells exhibiting low levels of TMRE staining (at least 100 cells were counted for each construct examined). Of the untransfected HeLa cells, 23% spontaneously exhibited low levels of TMRE under these conditions. Expression of 1-87-EGFP resulted in a marginal increase in the number of TMRE cells staining at low levels (33%) over the values obtained with EGFP or 1-72-EGFP (27% of cells were low staining). By contrast, fully 60% of 65-87-EGFP-expressing cells demonstrated low levels of TMRE staining.
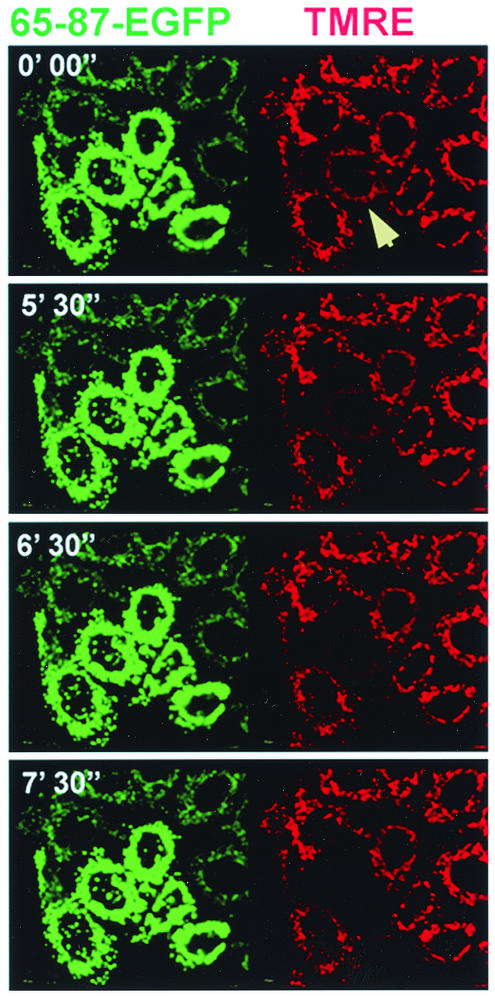
These data indicate that in the context of a fusion protein, the PB1-F2 MTS compromises mitochondrial function. We further investigated the effect of 65-87-EGFP on mitochondrial function by time-lapse microscopy. As seen in Fig. 6, total loss of ΔΨm in a 65-87-EGFP-transfected cell occurred within a span of 2 to 3 min. This seems to be typical, since we observed similar kinetics of loss of TMRE staining in five additional time-lapse movies. Loss of mitochondrial potential eventually leads to cell death which exhibits many of the hallmarks of apoptosis, such as phosphatidylserine exposure on the outer leaflet of the plasma membrane (as determined by annexin V staining), while maintaining membrane integrity (as determined by excluding PI) (Fig. 7).
FIG. 6.
Effect of PB1-F2 MTS-targeted protein on ΔΨm-microscopy. HeLa cells expressing PB1-F2 65-87-EGFP at 16 h posttransfection were labeled with TMRE, and images were acquired. Images were collected at one frame per 10 s. Selected frames are shown side by side (green, EGFP; red, TMRE) with the cumulative time elapsed (in minutes and seconds) indicated in the upper left corner of each panel. Note the rapid decrease in the mitochondrial TMRE fluorescence (red) of one transfected cell (indicated with an arrow). The entire time-lapse video is provided as supplemental material at http://www.niaid.nih.gov/dir/labs/lvd/bennink.htm.
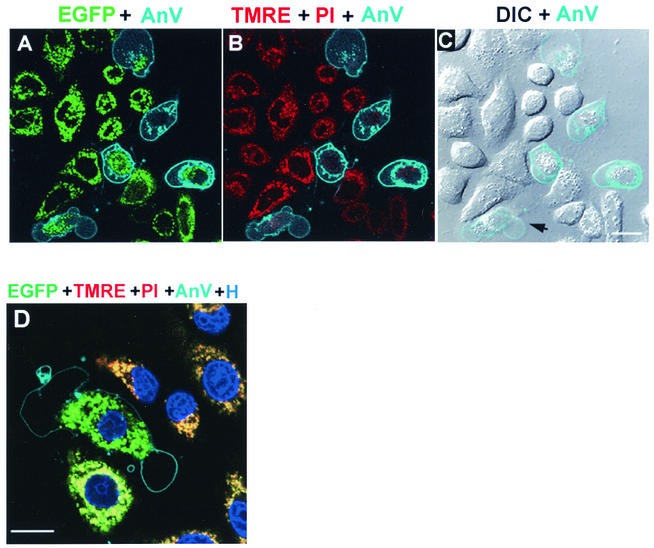
FIG. 7.
Visualization of cell death of PB1-F2 65-87-EGFP-expressing cell. (Top panels) Live HeLa cells expressing PB1-F2 65-87 EGFP at 16 h posttransfection were labeled with TMRE, and images were acquired at 37°C in the presence of annexin V (AnV)-Alexa 647 and PI. (A) A merged image of EGFP (green) and annexin V (cyan) fluorescence shows annexin labeling outlining the periphery of five transfected cells. The vesicular staining probably represents endosomes with internalized annexin V. EGFP fluorescence remained associated with mitochondria in most of the annexin-positive cells. (B) A merged image of TMRE (red) and PI (red) and annexin V (cyan) fluorescence shows a marked decrease of TMRE signal (decreased ΔΨm) in the annexin V-positive cells, while these cells excluded PI from the nucleus, demonstrating the integrity of the plasma membrane (with the same imaging settings, dead cells in other fields exhibited bright nuclear staining). (C) A merged image of differential interference contrast (DIC) in grey and annexin V (cyan) fluorescence shows clear plasma membrane blebbing (arrow) of one of the annexin V-positive cells. Bar, 10 μm. (Bottom panel) HeLa cells at 16 h posttransfection were labeled with TMRE (red) and the cell permeant DNA stain Hoechst 33342 (blue) and imaged in the presence of annexin V-Alexa (cyan) and PI (red) at 15-min intervals up to 2 h. (D) A merged image of a representative field image acquired at 15 min shows the decrease in size of the nuclei due to chromatin condensation in annexin-positive cells. Bar, 10 μm.
The behavior of stably transfected cells expressing PB1-F2-EGFP fusion proteins is consistent with this conclusion. Although it is possible to create cell lines expressing 65-87-EGFP, cells exhibit progressive loss of gene expression. In addition, cells expressing 65-87-EGFP at high levels demonstrate a marked tendency toward loss of TMRE staining and cell death compared to surrounding cells expressing lower levels of protein (data not shown).
Taken together, these findings demonstrate that the PB1-F2 MTS can induce mitochondrial dysfunction and cell death in the context of a chimeric protein. This is not strictly related to the amount of the chimeric protein present in mitochondria, as cells expressing smaller amounts of protein are often observed to die surrounded by cells which remain viable and express larger amounts of protein.
DISCUSSION
These findings demonstrate that similar to p13II and VPR from human immunodeficiency virus 1, PB1-F2 possesses a short MTS that is predicted to form a positively charged amphipathic α-helix. Confirmation of the predicted structure awaits the physical characterization of a synthetic peptide corresponding to this region. This approach has been used to provide strong evidence that the MTS of p13II forms an amphipathic helix. Inasmuch as we show that this sequence is able to replace the MTS of PB1-F2, it is highly likely that PB1-F2 forms a similar structure.
Positively charged amphipathic helices are commonly used to target cellular proteins to mitochondria (8). In the case of proteins destined for the mitochondrial matrix, such sequences are located at the NH2 terminus and are generally cleaved by matrix proteases. For proteins located in the inner mitochondrial membrane, targeting sequences can be located throughout the protein and are not always cleaved upon arrival at their destination. The PB1-F2 MTS appears to fall into the latter category. Interestingly, the presence of the predicted α-helical sequence alone is insufficient for targeting EGFP to mitochondria, which requires a short hydrophobic stretch (of at least 3 residues) preceding the α-helix. It has been suggested that the binding of α-helical targeting sequences to the hydrophobic groove present in the TOM (translocase of the outer mitochondrial membrane) receptor requires an initial interaction of neighboring hydrophobic residues (3). After proper binding of the presequence, the presence of the positively charged amino acid residues becomes important for the actual translocation through the outer and inner membrane of the mitochondria. Such a signal has been characterized for the rat cytochrome P450 2E1 (CYP2E1) protein (11). It is possible in the case of 65-87-EGFP that the MTS functions as a typical NH2-terminal mitochondrial signal in targeting EGFP to mitochondria (inserting the protein with via the NH2 terminus) whereas in the context of the full-length PB-1F2 the MTS acts by inserting the COOH terminus of the protein in a hairpin-like manner. Mitochondrial PB1-F2 expressed by IAV or from a transfected gene remains reactive with antibodies raised to peptides present at the NH2 or COOH termini, suggesting that cleavage of the MTS does not occur. Moreover, we failed to detect biochemical evidence for cleavage of PB1-F2 at the MTS. These findings, in conjunction with evidence that both PB1-F2 and 65-87-EGFP reside in the inner mitochondrial membrane, suggest that the MTS acts in a similar manner in the context of 65-87-EGFP and PB1-F2.
TOM complexes are believed to have an effective diameter of ∼20 Å, which means that proteins must cross in an unfolded conformation. The problem of maintaining such a conformation is minimized when the MTS is located at the NH2 terminus, since translocation can proceed cotranslationally once the NH2 terminus emerges from the ribosome translation tunnel. The further downstream the MTS, the greater the likelihood that folding will impede translocation. Cells can surmount this difficulty by maintaining the nascent protein in an unfolded conformation or by unfolding the protein as necessary. Folding, therefore, can be used by cells to control the destination of proteins with multiple functions in distinct organelles. This mechanism is apparently used to control the distribution of adenylate kinase, an abundant enzyme that functions in both the cytosol and mitochondria (16).
The placement of the PB1-F2 MTS near the COOH terminus of PB1-F2 may explain why 65-87-EGFP is more efficiently targeted to mitochondria than 1-87-EGFP. It may also contribute to the leaky mitochondrial targeting of PB1-F2 in IAV infections where PB1-F2 localizes to the nucleus and/or cytosol in a substantial number of cells. This does not negate the possibility, however, that the interaction of PB1-F2 with influenza virus proteins, or alterations in the cell induced by IAV infection, modulates the ability of the PB1-F2 MTS to interact with TOM.
The presence of 65-87-EGFP in the inner mitochondrial membrane indicates that residues 65 to 87 are sufficient not only to target the protein to the membrane but also to maintain its presence there. This may reflect the interaction of the MTS with membrane lipids or perhaps with an inner mitochondrial membrane protein. One candidate is the adenine nucleotide translocator (ANT), which is believed to interact with several other viral proteins, including Vpr (2). Intriguingly, Vpr, like PB1-F2, has its MTS located near the COOH terminus, and the COOH terminal domain is responsible for interacting with ANT and for mitotoxicity (14). Inasmuch as the 65-87-EGFP fusion protein induces a loss of ΔΨm resulting in cell death, this is consistent with the idea that this domain interacts with ANT.
Notably, in contrast to fusion proteins expressing the COOH portions of PB1-F2, 1-87-EGFP exhibited only borderline mitotoxicity and cytotoxicity in transiently transfected HeLa cells. In additional experiments, we have found that transient expression of 1-87-EGFP from the same plasmid clearly induces apoptosis in lymphoid cells (F. Hornung, unpublished data). This is consistent with the idea, admittedly speculative, that the amino-terminal portion of PB1-F2 confers cell type specificity on the proapoptotic function of the COOH terminal MTS.
Acknowledgments
We are grateful to Gabriele Burger (Leica Microsystems) for advice on FRET methodology.
REFERENCES
- 1.Bastiaens, P. I., I. V. Majoul, P. J. Verveer, H. D. Soling, and T. M. Jovin. 1996. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15:4246-4253. [PMC free article] [PubMed] [Google Scholar]
- 2.Boya, P., B. Roques, and G. Kroemer. 2001. New EMBO members' review: viral and bacterial proteins regulating apoptosis at the mitochondrial level. EMBO J. 20:4325-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brix, J., K. Dietmeier, and N. Pfanner. 1997. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272:20730-20735. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 5.Ciminale, V., L. Zotti, D. M. D'Agostino, T. Ferro, L. Casareto, G. Franchini, P. Bernardi, and L. Chieco-Bianchi. 1999. Mitochondrial targeting of the p13II protein coded by the x-II ORF of human T-cell leukemia/lymphotropic virus type 1 (HTLV-I). Oncogene 18:4505-4514. [DOI] [PubMed] [Google Scholar]
- 6.D'Agostino, D. M., L. Ranzato, G. Arrigoni, I. Cavallari, F. Belleudi, M. R. Torrisi, M. Silic-Benussi, T. Ferro, V. Petronilli, O. Marin, L. Chieco-Bianchi, P. Bernardi, and V. Ciminale. 2002. Mitochondrial alterations induced by the p13II protein of human T-cell leukemia virus type 1. Critical role of arginine residues. J. Biol. Chem. 277:34424-34433. [DOI] [PubMed] [Google Scholar]
- 7.D'Agostino, D. M., L. Zotti, T. Ferro, G. Franchini, L. Chieco-Bianchi, and V. Ciminale. 2000. The p13II protein of HTLV type 1: comparison with mitochondrial proteins coded by other human viruses. AIDS Res. Hum. Retrovir. 16:1765-1770. [DOI] [PubMed] [Google Scholar]
- 8.Endo, T., and D. Kohda. 2002. Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta 1592:3-14. [DOI] [PubMed] [Google Scholar]
- 9.Jacotot, E., L. Ravagnan, M. Loeffler, K. F. Ferri, H. L. Vieira, N. Zamzami, P. Costantini, S. Druillennec, J. Hoebeke, J. P. Briand, T. Irinopoulou, E. Daugas, S. A. Susin, D. Cointe, Z. H. Xie, J. C. Reed, B. P. Roques, and G. Kroemer. 2000. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 191:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefebvre, L., V. Ciminale, A. Vanderplasschen, D. D'Agostino, A. Burny, L. Willems, and R. Kettmann. 2002. Subcellular localization of the bovine leukemia virus R3 and G4 accessory proteins. J. Virol. 76:7843-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neve, E. P., and M. Ingelman-Sundberg. 2001. Identification and characterization of a mitochondrial targeting signal in rat cytochrome P450 2E1 (CYP2E1). J. Biol. Chem. 276:11317-11322. [DOI] [PubMed] [Google Scholar]
- 12.Ravagnan, L., T. Roumier, and G. Kroemer. 2002. Mitochondria, the killer organelles and their weapons. J. Cell. Physiol. 192:131-137. [DOI] [PubMed] [Google Scholar]
- 13.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 14.Roumier, T., H. L. Vieira, M. Castedo, K. F. Ferri, P. Boya, K. Andreau, S. Druillennec, N. Joza, J. M. Penninger, B. Roques, and G. Kroemer. 2002. The C-terminal moiety of HIV-1 Vpr induces cell death via a caspase-independent mitochondrial pathway. Cell Death Differ. 9:1212-1219. [DOI] [PubMed] [Google Scholar]
- 15.Scaduto, R. C., Jr., and L. W. Grotyohann. 1999. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys. J. 76:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strobel, G., A. Zollner, M. Angermayr, and W. Bandlow. 2002. Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol. Biol. Cell 13:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterhouse, N. J., J. E. Ricci, and D. R. Green. 2002. And all of a sudden it's over: mitochondrial outer-membrane permeabilization in apoptosis. Biochimie 84:113-121. [DOI] [PubMed] [Google Scholar]