Abstract
The adenovirus (Ad) fiber protein mediates Ad binding to the coxsackievirus and Ad receptor (CAR) and is thus a major determinant of viral tropism. The fiber contains three domains: an N-terminal tail that anchors the fiber to the viral capsid, a central shaft region of variable length and flexibility, and a C-terminal knob domain that binds to cell receptors. Ad type 37 (Ad37), a subgroup D virus associated with severe ocular infections, is unable to use CAR efficiently to infect host cells, despite containing a CAR binding site in its fiber knob. We hypothesized that the relatively short, inflexible Ad37 fiber protein restricts interactions with CAR at the cell surface. To test this hypothesis, we analyzed the infectivity and binding of recombinant Ad particles containing modified Ad37 or Ad5 fiber proteins. Ad5 particles equipped with a truncated Ad5 fiber or with a chimeric fiber protein comprised of the Ad5 knob fused to the short, rigid Ad37 shaft domain had significantly reduced infectivity and attachment. In contrast, placing the Ad37 knob onto the long, flexible Ad5 shaft allowed CAR-dependent virus infection and cell attachment, demonstrating the importance of the shaft domain in receptor usage. Increasing fiber rigidity by substituting the predicted flexibility modules in the Ad5 shaft with the corresponding regions of the rigid Ad37 fiber dramatically reduced both virus infection and cell attachment. Cryoelectron microscopy (cryo-EM) single-particle analysis demonstrated the increased rigidity of this chimeric fiber. These studies demonstrate that both length and flexibility of the fiber shaft regulate CAR interaction and provide a molecular explanation for the use of alternative receptors by subgroup D Ad with ocular tropism. We present a molecular model for Ad-CAR interactions at the cell surface that explains the significance of fiber flexibility in cell attachment.
The primary receptor for most human adenoviruses (Ad) is CAR, a 46-kDa member of the immunoglobulin (Ig) superfamily (5, 39). This receptor is distributed on many cell types in vivo, and its location in tight junctions on polarized epithelial cells (12) plays a role in intercellular dissemination of the virus (45). Apart from Ad types 2 and 5 (subgroup C), there is relatively little information on the identity of receptors used by the 51 different Ad serotypes belonging to six major subgroups (A to F). Previous studies suggested that the majority of Ad serotypes, with the exception of those belonging to subgroup B (e.g., Ad3 and Ad7), recognize CAR, but these studies measured receptor interactions under nonphysiological conditions (6, 29). A comparison of AB loop sequences in the fiber knob domain of different Ad types (28) revealed that only subgroup B Ad have a divergent sequence and therefore would not be expected to recognize CAR. A crystal structure of the Ad12 fiber knob domain in complex with the first (D1) Ig-like domain of CAR (6) and mutagenesis studies (28) showed that receptor binding occurs on the lateral surface of the fiber knob rather than on the top of this molecule, as had originally been predicted (50).
Ad belonging to subgroup D, including Ad37, are frequently associated with epidemic keratoconjunctivitis (10). This disease is characterized by a high rate of transmission and, although rarely, can result in the ulceration of the cornea and loss of visual function. Despite the ability of Ad37 particles and recombinant Ad37 knob to bind CAR in an overlay blot assay (49), these virus types cannot use this receptor efficiently (2, 49). Instead, sialic acid on one or more membrane glycoproteins has been reported to mediate Ad37 binding to various cell types (2). We demonstrated that Ad37 binds to 50- and 60-kDa proteins on conjunctival cells (49). A lysine residue at position 240 located in the CD loop at the top of the fiber knob is important for Ad37 binding to cells (19). This site is distinct from a region located in the AB loop, which provides the major interactions with CAR on the lateral surface of the fiber (6).
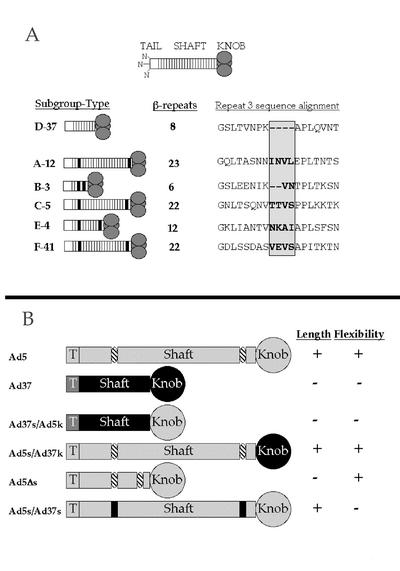
These previous studies, however, do not explain why Ad37 fails to use CAR effectively on cells. One possibility is that other regions of the Ad37 fiber protein, including the shaft domain, influence CAR interaction. The fiber shaft domain is comprised of a variable number of ∼15 amino acid repeats (11) that forms a triple β-spiral fold (42). A single repeat is characterized by a consensus sequence that codes for two β-strands connected by a half-buried type I or II β-turn and a surface-exposed loop of variable length. Different Ad types have different shaft lengths dependent upon the number of β-repeats (Fig. 1A). For example, the long and highly homologous Ad2 and Ad5 fiber shaft domains have 22 repeats while the short Ad37 shaft has only 8 repeats.
FIG. 1.
Ad fiber protein architecture from the six Ad subgroups and chimeric fiber proteins. (A) Ad fibers of different subgroups contain a variable number of β-repeats in the shaft, resulting in the variable length of the fibers, shown on the left. Putative flexibility repeats are denoted in black. Sequences of the third repeat of the fiber shaft from each representative serotype is aligned and shown on the right. The 2- or 4-amino-acid insertion in the middle of the repeat is in bold and is shaded. (B) Chimeric fibers were designed to test the importance of fiber length and flexibility based on alignments of Ad5 and Ad37 fiber proteins. Ad5 regions are shown in light gray, and Ad37 regions are shown in black. The putative flexibility repeats are denoted in striped ovals. Pluses and minuses on the right indicate the relative fiber length or flexibility.
Previous studies have shown that shortening the long (i.e., Ad5) fiber shaft reduces virus infectivity (32). The potential role of flexibility in fiber-receptor interactions, however, has been somewhat overlooked. Negative-stain images of the Ad2 fiber revealed that this protein is not perfectly straight, and a kink has frequently been observed in the N-terminal region of the molecule (11). Consistent with these earlier findings, cryo-EM image reconstructions of Ad2 particles (35, 37) show strong density only in the N-terminal section of the fiber protein. A sequence comparison of different fiber proteins may provide a molecular explanation for fiber flexibility (11). The third repeat of most Ad types has a nonconsensus sequence motif that contains two to four additional amino acids (Fig. 1A). These additional residues are thought to allow bending of the fiber shaft. The third repeat is predicted to lie in approximately the same location in the shaft, ∼60 Å from the surface of the virus capsid, as the kink observed in cryo-EM image reconstructions of Ad2 particles (9, 37). Interestingly, the nonconsensus third repeat is not present in subgroup D Ad serotypes, such as Ad37 (11). Moreover, most Ad types, with the exception of subgroup D viruses, have a KLGXGLXFD/N consensus motif in the last complete shaft repeat (i.e., repeat 21 in Ad2). This sequence may allow a small degree of bending between the knob and shaft domains, as has been observed in the fiber crystal structure (42). Given the distinct molecular composition of subgroup D Ad, we predicted that these viruses would have a short, rigid fiber protein. This was supported by cryo-EM image reconstructions (9) that revealed the complete structure for Ad37 fiber, including the distal knob domain. Chroboczek et al. (11) originally proposed that the bending of the fiber would facilitate the interaction of the RGD loop of the penton with cell surface integrins. In addition, we hypothesized that, in the context of the entire virus capsid, the length and/or flexibility of the fiber shaft can also modulate knob-CAR interaction at the cell surface. As a consequence, the short, rigid Ad37 shaft would restrict knob binding to CAR at the cell surface.
In the studies reported here, we analyzed the role of fiber shaft in CAR interactions by using Ad5 particles pseudotyped with truncated or chimeric Ad37/Ad5 fiber proteins. Complementary cryo-EM and gene delivery experiments correlated fiber structure with viral infectivity. These studies provide evidence for the presence of flexibility modules in subgroup C Ad fibers and demonstrate that flexibility as well as length regulates receptor interactions at the cell surface.
MATERIALS AND METHODS
Cell lines and virus strains.
HEK-293T cells (13) and A549 lung carcinoma cells (American Type Culture Collection, Manassas, Va.) were cultured in Dulbecco's modified Eagle medium (Gibco BRL, Rockville, Md.) containing 10% fetal bovine serum (FBS; Mediatech, Herndon, Va.). AE1-2a cells (15) were obtained from Michael Kadan (Genetic Therapy, Inc./Novartis, Summit, N.J.) and were maintained in improved modified Eagle medium (IMEM; Mediatech) containing 10% FBS, 200 μg of hygromycin B/ml (Calbiochem, San Diego, Calif.), and 200 μg of neomycin sulfate/ml (Calbiochem). AE1-2a-derived cell lines 633 and 761, expressing the Ad5 (44) and Ad37 (49) fiber proteins, respectively, were maintained in a solution containing IMEM, 10% FBS, 200 μg of hygromycin B/ml, 200 μg of neomycin sulfate/ml, and 300 μg of zeocin/ml (Invitrogen, Carlsbad, Calif.).
Ad5.GFP.ΔF/5F- and Ad5.GFP.ΔF/37F-pseudotyped Ad5 vectors were produced as previously described (49) in fiber-complementing 633 and 761 cells, respectively. Fiberless Ad5.GFP.ΔF vectors were produced in 293 cells, as previously described (43). Briefly, 293 or fiber-expressing cells were infected at 75 to 80% confluency with Ad5.GFP.ΔF/5F. Fiberless or fiber-pseudotyped particles were purified 2 to 3 days postinfection by ultracentrifugation on 16 to 40% cesium chloride (CsCl) gradients and were dialyzed into a solution of 10 mM Tris (pH 8.1), 150 mM sodium chloride (TBS), and 10% glycerol.
Construction of chimeric fiber proteins.
Chimeric Ad fiber proteins were constructed by using gene splicing by overlap extension PCR (18) from Ad5 and Ad37 fiber gene fragments. PCR and mutagenic primers are listed in Table 1.
TABLE 1.
PCR, overlap extension PCR, and mutagenic primersa
| Construct | Primer | Sequence |
|---|---|---|
| Ad37s/Ad5k | L37 | TGT CTT GAA TCC AAG ATG∗ AAG CGC GCC CGC CCC AGC GAA GAT GAC TTC |
| 37s5k-3 | TGG AGC TGG TGT GGT CCA CAA AGT GCG CGT GTC ATA TTC TGG GTT CCA | |
| 5k-5 | ACT TTG TGG ACC ACA CCA GCT CCA | |
| fiber3 | CAT AAC GCG GCC GCT TCT TTA TTC TTG GGC | |
| Ad5s/Ad37k | fiber5 | ATG GGA TCC AAG ATG∗ AAG CGC GCA AGA CCG |
| 5s-3 | TGG TGT GGT CCA CAA AGT TAG CTT ATC ATT | |
| 5s37k-5 | AAG CTA ACT TTG TGG ACC ACA CCA GAC ACA TCT CCA AAC TGC ACA ATT | |
| 37fr | AAA CAC GGC GGC CGC TCT TTC ATT CTT G | |
| Ad5Δs | short3 | GCT TAG GTT AAC CTC AAG CTT TTT CTT GGT TTT TTT GAG AGG TGG GCT |
| short5 | AGC CCA CCT CTC AAA AAA ACC AGG AAA AAG CTT GAG GTT AAC CTA AGC | |
| Ad5s/Ad37s | rep3-3 | ATC AGT ATT AAC TTG CAG TGG AGC CTT AGG GTT TAC AGT TAG GCT TCC GGC CTC GTC CAG AGA GAG GCC GTT |
| rep3-5 | GGA AGC CTA ACT GTA AAC CCT AAG GCT CCA CTG CAA GTT AAT ACT GAT TCA AAC ATA AAC CTG GAA ATA TCT | |
| rep7-3 | ATC ATT GTC AAA TGT CAA CCC TTC TCT TGC TCT TAC ATT TAT ACC AAT GTT GTA ATC AAA TTC TAG GCC ATG | |
| rep7-5 | ATT GGT ATA AAT GTA AGA GCA AGA GAA GGG TTG ACA TTT GAC AAT GAT GGT GCC ATT ACA GTA GGA AAC AAA |
Introduced restriction sites are in bold. The start codons are denoted by an asterisk. Mutagenic bases are underlined.
Ad37s/Ad5k.
Approximately 108 particles of wild-type Ad37 (American Type Culture Collection) were mixed with a PCR master mix (1× ThermoPol Buffer, 300 μM each dATP, dTTP, dGTP, and dCTP, and 2 U of vent DNA polymerase; New England Biolabs, Beverly, Mass.) and 200 nM primers L37 and 37s5k-3 to amplify amino acids 1 to 184 of the Ad37 fiber. Mutations were incorporated into the Ad37 fiber tail to make the sequence more closely match the Ad5 tail (49). These mutations change the first seven amino acids of the tail from MSKRLRV of Ad37 to MKRARPS of Ad5 to facilitate fiber incorporation into the Ad5 vector capsid. This PCR mixture was heated to 94°C for 5 min and was subjected to 1 cycle of 94°C for 1 min, 45°C for 1.5 min, and 72°C for 2 min, then 30 cycles of 94°C for 1 min, 50°C for 1.5 min, and 72°C for 2 min, and a final extension step of 72°C for 5 min (program 1). In a second reaction, instead of Ad37 virus, pDV67, an expression plasmid for the Ad5 fiber (44), was mixed with PCR master mix. Primers 5k-5 and fiber3 were added to this reaction mixture to amplify amino acids 400 to 581 of the Ad5 fiber. This PCR mixture was heated to 94°C for 5 min and was subjected to 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and a final extension step of 72°C for 5 min (program 2).
The first-step PCR products were gel purified from 10 μl of the 100-μl reaction mixtures in a 1% low-melting-point agarose gel. The gel-purified PCR products were melted, and 10 μl of each was mixed together with a solution containing 1× PCR Buffer, an additional 3 mM MgCl2, 300 μM each deoxynucleoside triphosphate, 0.8 μM each L37 5′ primer and fiber3 3′ primer, and 5 U of Taq DNA polymerase (Gibco BRL). Program 1 was used for the overlap extension PCR.
The PCR product was cloned into the pCR2.1 cloning vector by using the TOPO TA Cloning kit (Invitrogen). The plasmid was transformed into TOP10 Escherichia coli cells (Invitrogen) and was purified from cultured cells by using the Qiagen Plasmid Mini Spin kit (Valencia, Calif.). The chimeric fiber gene was excised from pCR2.1 and was ligated into the BamHI and NotI sites of pCDNA3.1zeo(+) (Invitrogen). The Ad5 tripartite leader (TPL) was excised from pDV55 by using BamHI and BglII and was inserted into the BamHI site in front of the chimeric fiber gene in the expression vector.
Vectors containing Ad5s/Ad37k, Ad5Δs, and Ad5s/Ad37s genes preceded by the Ad5 TPL, designated pLP23, pLP32, and pLP43, respectively, were constructed in the same fashion as Ad37s/Ad5k by using the primers, templates, and PCR programs listed in Table 2.
TABLE 2.
Overlap extension PCRs for construction of chimeric fiber proteins
| Reaction | Construct | Templatea | Polymeraseb | 5′ Primer | 3′ Primer | Program | Amplified fiber protein fragment (residues) |
|---|---|---|---|---|---|---|---|
| 1 | Ad37s/Ad5k | wt Ad37 virus | Vent | L37 | 37s5k-3 | 1 | Ad37 (1-184) |
| 2 | Ad37s/Ad5k | pDV67 | Vent | 5k-5 | fiber3 | 2 | Ad5 (400-581) |
| 3 | Ad37s/Ad5k | Rxns 1 and 2 | Taq | L37 | fiber3 | 1 | Ad37 (1-184); Ad5 (400-581) |
| 4 | Ad5s/Ad37k | pDV67 | Vent | fiber5 | 5s-3 | 2 | Ad5 (1-405) |
| 5 | Ad5s/Ad37k | wt Ad37 virus | Vent | 5s37k-5 | 37fr | 2 | Ad37 (188-365) |
| 6 | Ad5s/Ad37k | Rxns 4 and 5 | Taq | fiber5 | 37fr | 1 | Ad5 (1-405); Ad37 (188-365) |
| 7 | Ad5Δs | pDV67 | Vent | fiber5 | short3 | 2 | Ad5 (1-94) |
| 8 | Ad5Δs | pDV67 | Vent | short5 | fiber3 | 2 | Ad5 (317-581) |
| 9 | Ad5Δs | Rxns 7 and 8 | Taq | fiber5 | fiber3 | 1 | Ad5 (1-94 and 317-581) |
| 10 | Ad5s/Ad37s | pDV67 | Vent | fiber5 | rep3-3 | 2 | Ad5 (1-75); Ad37 (74-89) |
| 11 | Ad5s/Ad37s | pDV67 | Vent | rep3-5 | rep7-3 | 2 | Ad5 (95-370); Ad37 (74-89 and 166-171) |
| 12 | Ad5s/Ad37s | pDV67 | Vent | rep7-5 | fiber3 | 2 | Ad5 (387-581); Ad37 (166-171) |
| 13 | Ad5s/Ad37s | Rxns 11 and 12 | Taq | rep3-5 | fiber3 | 2 | Ad5 (95-370 and 387-581); Ad37 (74-89 and 166-171) |
| 14 | Ad5s/Ad37s | Rxns 10 and 13 | Taq | fiber5 | fiber3 | 1 | Ad5 (1-75, 95-370, and 387-581); Ad37 (74-89 and 166-171) |
wt, Wild type; Rxns, reactions.
Vent, vent DNA polymerase (New England Biolabs) Taq, Taq DNA polymerase (Gibco BRL).
All four plasmids, pLP13, pLP23, pLP32, and pLP43, were purified from 500-ml cultures by using the Qiagen Plasmid Maxi kit.
Generation of fiber-pseudotyped Ad.
Ad5-based vectors containing the Ad5 fiber or Ad37 fiber gene replacing the Ad5 fiber directly in the viral genome were generated as previously described (20). As an alternative approach, pseudotyping of green fluorescent protein (GFP)-expressing Ad5 vectors with the Ad5 fiber (Ad5.GFP.ΔF/5F) or the Ad37 fiber (Ad5.GFP.ΔF/37F) has been previously described (49). Ad5 vectors were pseudotyped with new chimeric fibers in a similar fashion. Briefly, 7 μg of the fiber expression vectors pLP13, pLP23, and pLP32 were stably transfected into 5 × 106 AE1-2a cells suspended in IMEM-0.1 mM dithiothreitol by using a Gene Pulser II (Bio-Rad, Richmond, Calif.) at 0.3 kV and 500 μF. Cells were plated overnight in growth medium and were selected by using 600 μg of zeocin/ml, 400 μg of hygromycin B/ml, and 400 μg of neomycin sulfate/ml. Selected colonies were analyzed by immunofluorescence with anti-fiber monoclonal 4D2 antibody (NeoMarkers, Fremont, Calif.) and AlexaFluor 488 goat anti-mouse IgG conjugate (Molecular Probes, Eugene, Oreg.). pLP43 was transiently transfected into 293T cells by using SuperFect transfection reagent (Qiagen) as described by the manufacturer's instructions, and particles producing these fibers were produced as previously described (20).
Fiber-expressing cells were infected at 75 to 80% confluency with Ad5.GFP.ΔF/5F at approximately 2,000 particles per cell. Cells were detached around 72 h postinfection and were lysed by repeated freezing and thawing. Cell debris was removed by centrifugation, and the liberated virus particles were purified in a 16 to 40% CsCl gradient at 111,000 × g for 3 h. Purified virus was dialyzed into TBS-10% glycerol. Protein concentration was determined by a Bradford protein assay (Bio-Rad). Virus concentration was calculated from the protein concentration by using the known molecular weight of Ad2 particles (1 μg = 4 × 109 particles).
Immunoblot analyses of Ad particles.
Five hundred nanograms of virus was denatured by boiling in a 2% sodium dodecyl sulfate (SDS) and 0.2 M 2-mercaptoethanol buffer for 5 min. Viral proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in an 8 to 16% Tris-glycine gel (Novex/Invitrogen) and was transferred to a polyvinyl difluoride membrane. The membrane was blocked in a solution containing 5% (wt/vol) milk in phosphate-buffered saline and 0.02% (vol/vol) Tween-20 (PBS-T) overnight at 4°C. After being blocked the membrane was incubated with 4D2 anti-fiber monoclonal antibody diluted 1:1,000 in milk in PBS-T for 1 h at room temperature. The membrane was washed and incubated with 1:10,000 goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody (Sigma, St. Louis, Mo.) for 30 min at room temperature. After washing the membrane again, the blot was probed with enhanced chemiluminescence reagents (Supersignal West Pico reagents; Pierce, Rockford, Ill.) and was developed on film.
To ensure equal loading of virus samples, the membrane was stripped and reprobed for penton base. The membrane was incubated with a solution of 100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris (pH 6.7) for 1 h at 42°C. After being washed the membrane was probed with 1:500 dilution of a rabbit anti-penton polyclonal antibody (46) for 1 h, washed, probed with 1:5,000 goat anti-rabbit HRP-conjugated antibody (Sigma) for 30 min, and washed again. The penton blot was developed as described above.
Cryo-EM three-dimensional image reconstruction.
Small droplets (3 μl) of a purified viral preparation of fiber-pseudotyped Ad (Ad5s/Ad37s) (∼200 μg/ml) were applied to glow-discharged holey carbon grids. The grids were blotted and vitrified in ethane slush chilled by liquid nitrogen (1). The frozen grids were transferred one at a time to a Gatan 626 cryotransfer holder prechilled with liquid nitrogen or stored under liquid nitrogen. Electron micrographs were recorded by using low-dose conditions on a FEI/Philips CM120 transmission electron microscope equipped with a LaB6 filament and a Gatan slow-scan CCD camera (YAG scintillator; 1024 by 1024 pixels). A nominal magnification of 35,000× was used, yielding a pixel size of 5.2 Å on the molecular scale. Images were collected with defocus values of −1.5, −1.0, and −0.7 μm to generate phase contrast.
Particles were digitally selected from cryo-electron micrographs in 360-by-360-pixel image files by using the QVIEW software package (31), and most of the further image processing was done by using the IMAGIC-5 software package (40). Initial particle orientations were obtained by using a previous reconstruction of Ad5 as the search model (43). Once the initial orientations were obtained, each particle was reconstructed separately to determine if its fibers were straight enough to generate significant reconstructed fiber density. If the reconstruction based on a single Ad particle showed fiber density above the background noise level and along more than 50% of the predicted fiber length, it was selected for inclusion in the data set of Ad particles with the straightest fibers. In this manner, 85 Ad particle images were selected from a total set of 1,236 particle images of the fiber-pseudotyped Ad (Ad5s/Ad37s). Seven out of 403 wild-type Ad5 particle images were selected by using the same criteria. Computational correction for the contrast transfer function (CTF) of the electron microscope was done prior to merging particle images collected with different defocus values as has been previously described (8). We note that without the high coherence of a field emission gun electron microscope, correction of the CTF can only be approximate at best. Even after CTF correction, a weak density ring is still visible around the Ad capsid in slices through the reconstruction. Incomplete correction of the CTF most likely explains why the fiber density is apparently disconnected from the penton base. Four rounds of anchor set refinement were performed with the selected set of particle images by using a 1° angular search step size. This improved the resolution of the reconstructed capsid from 37 to 29 Å. Although the icosahedral capsid density improved, the reconstructed fiber density did not improve with refinement. This observation implies that even the straightest fibers must not all be perfectly aligned along the icosahedral fivefold axes. Thus, we present the unrefined reconstruction based on the selected 85 particle images, which shows the longest fiber density. Presumably small misalignments in the orientations of these particle images serve to bring the density of the slightly bent fibers into better alignment.
The resolution of each reconstruction was assessed by the Fourier shell correlation method with the 0.5 correlation threshold criterion. Soft masks were applied to the two half-reconstructions in order to consider just the icosahedral capsid in the resolution assessment (36). All of the image processing and graphics were performed on Compaq/DEC alpha workstations. The graphics representations were generated with the AVS-5 software package (Advanced Visualization Systems, Inc.).
Infection assay.
Infection assays that used pseudotyped Ad vectors carrying a GFP transgene were performed as previously described (49). Briefly, 50,000 adherent A549 cells were incubated with 20,000 particles per cell with vector for 3 h at 37°C in Dulbecco's modified Eagle medium-10% FBS. Cells were washed three times with saline and were cultured overnight in growth medium. Cells were detached and analyzed by fluorescence-assisted cell sorting in a FACScan cytometer (Becton Dickenson, Franklin Lakes, N.J.). A threshold established by the fluorescence of uninfected cells was used to distinguish infected cells expressing GFP.
Quantitative PCR analyses of virus attachment.
Cultured A549 cells were detached by using 5 mM EDTA for 5 min. Cells were resuspended in PBS and were aliquoted to a density of 1.0 × 106 cells per tube. Particles of virus (1.0 × 109) were added to tubes, and the tubes were rocked for 1 h at 4°C to prevent internalization. Nonspecific Ad binding was determined by the addition of an excess of recombinant Ad5 knob (100 μg/ml). Cells were pelleted by centrifugation and were resuspended in PBS three times. Total sample DNA was extracted from cells and bound virus by using the QIAamp DNA Mini kit (Qiagen) as directed by the manufacturer's instructions.
Five microliters of each 200-μl DNA extract was added to a 45-μl reaction mixture containing 1× TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, Calif.), 300 nM each 5′ primer EGFP553f and 3′ primer EGFP810r, 200 nM probe EGFP734p, and VIC-labeled RNase P Control Reagents (Applied Biosystems). Enhanced green fluorescent protein (EGFP) primers and probes were designed to detect a 258-bp region in the EGFP transgene (21) in the Ad5 vector genome, while RNase P control reagents were designed to amplify a segment of the host cell genomic RNase P gene. After initial denaturation and activation of the AmpliTaq Gold DNA polymerase by heating to 50°C for 2 min and then 95°C for 10 min, the amplicons were amplified with 40 cycles of 15 s at 95°C followed by 1 min at 60°C. Fluorescence of reporter dyes FAM and VIC were measured during each cycle in an ABI Prism 7900 Sequence Detection System (Applied Biosystems). Known amounts of pEGFP-N1 (Clontech, Palo Alto, Calif.) plasmid, an EGFP expression plasmid, and purified cellular DNA were used as standards to measure the number of copies of Ad genomes and cell number in each sample.
Integrin binding ELISA.
Integrin binding to intact virus particles was determined as previously described (27). Briefly, purified pseudotyped Ad were coated to wells of a 96-well plate (Immulon 4 HBX; Dynex Techonologies, Chantilly, Va.) overnight at room temperature. The wells were blocked with Superblock in PBS (Pierce). Virus-coated wells were first incubated for 2 h with various amounts of soluble αvβ5 integrin (27). After being washed the virus-coated wells were incubated for 1 h with a 10-μg/ml concentration of a non-function-blocking anti-αv subunit monoclonal antibody (LM142) (kindly provided by D. Cheresh, The Scripps Research Institute). After additional wash steps the wells were incubated with 1:10,000 goat anti-mouse HRP-conjugated antibody for 1 h. The enzyme-linked immunosorbent assay (ELISA) was developed with 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) substrate and was analyzed by measuring absorbance at 405 nm.
RESULTS
Generation and characterization of fiber-pseudotyped Ad particles.
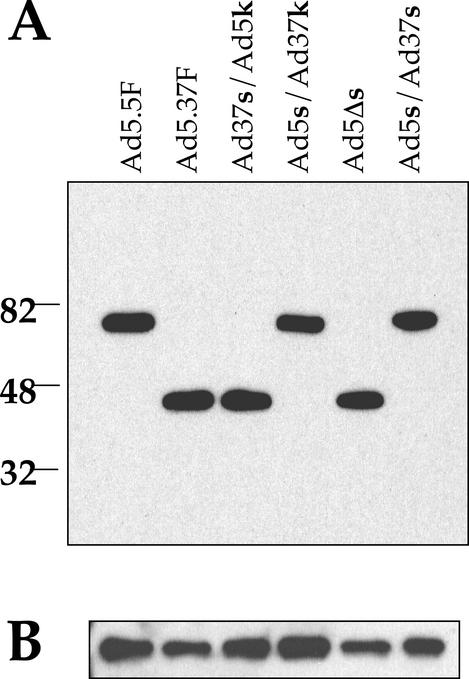
To investigate the role of the fiber shaft in receptor interactions, we generated a panel of Ad5 particles pseudotyped with different fiber proteins (Fig. 1B) by using packaging cell lines (44). The major goal of these studies was to create fiber proteins that vary in length and/or flexibility. Chimeric fiber genes were spliced together by using overlap extension PCR. To confirm that the shaft can modulate CAR binding, the short, rigid shaft of Ad37 was fused to the CAR-binding fiber knob of Ad5 (Ad37s/Ad5k). The reverse fiber protein, with the long, flexible Ad5 shaft fused to the Ad37 knob (Ad5s/Ad37k), was also created. We reasoned that if Ad37 cannot use CAR as a receptor due to its short, rigid shaft, fusing the long, flexible Ad5 shaft to the Ad37 knob should restore CAR binding to the Ad37 knob. To determine the relative importance of the length of the fiber shaft in CAR binding, repeats 4 to 17 of the Ad5 fiber were deleted, leaving a shaft with only eight β-repeats (Ad5Δs). This fiber has the same length as the Ad37 fiber but retains the putative flexibility repeats (3rd and 21st repeats). To determine the importance of flexibility of the shaft, the Ad5 fiber was straightened by replacing the flexibility repeats in the Ad5 shaft with the corresponding third and seventh repeats of the rigid Ad37 fiber (Ad5s/Ad37s). Expression plasmids containing the chimeric fiber genes were stably transfected into AE1-2a cells to generate packaging lines. Pseudotyped Ad particles lacking an endogenous fiber gene (ΔF) were initially propagated in 633 cells (44) and then were passaged through the appropriate fiber-packaging cell line. To verify the content and expected size of different fiber proteins on Ad particles, we analyzed purified particles by immunoblotting with an anti-fiber monoclonal antibody that recognizes the conserved N-terminal tail domain. Each of the chimeric or truncated fiber-pseudotyped Ad particles contained similar amounts of fiber protein of the expected size (Fig. 2A). A comparison of the fiber immunoblots with penton base blots (Fig. 2B) showed that similar amounts of each virus were analyzed.
FIG. 2.
Western blot analysis of pseudotyped Ad vectors. Gradient-purified Ad particles were subjected to SDS-PAGE, transferred to a polyvinyl difluoride membrane, and probed with a monoclonal anti-fiber antibody (A) or a polyclonal anti-penton base antibody (B). An Ad5-pseudotyped fiberless vector (Ad5.5F) or an Ad37-pseudotyped virus (Ad5.37F) with the relevant fiber genes incorporated into the viral genome were included as molecular size references.
The Ad5s/Ad37s fiber protein is less flexible than wild-type Ad fibers.
We hypothesized that replacing the nonconsensus repeats of the Ad5 fiber with that of Ad37 would create a long, rigid fiber protein. To test this hypothesis, we attempted to visualize the Ad5s/Ad37s fiber protein incorporated into an Ad5 pseudotyped virus particle by using cryo-EM and single-particle image reconstruction methods. Over 1,200 cryo-EM particle images of the Ad5s/Ad37s fiber-pseudotyped Ad were collected, along with ∼400 particle images of wild-type Ad5. During the image processing of the fiber-pseudotyped data set it became apparent that some, but not all, of the particle images were contributing to strong fiber density in the reconstructed density map. Thus, each particle image was analyzed separately by calculating a three-dimensional reconstruction based on each single particle image and evaluating the resulting reconstructed fiber density. We selected particles that showed reconstructed fiber density, which was stronger than the background noise level and which extended along more than half of the predicted fiber length. The same selection criteria were applied to the wild-type Ad5 particle images. Subsets of 85 fiber-pseudotyped images and 7 wild-type images, corresponding to 6.8 and 1.7% of the respective data sets, were selected with strong reconstructed fiber density. In order to determine the significance of these findings, a statistical chi square analysis was performed. This test indicated a very small probability (0.0001) that the null hypothesis is true, i.e., that the fiber-pseudotyped Ad particles and wild-type Ad5 particles have equally flexible fibers. We note that the crystal structure of a portion of the fiber shaft (42) indicates that the average diameter of the fiber shaft is 15 Å. For such a thin fiber, a bend of just 2° would be enough to prevent the superposition of fiber density at the distal end of the shaft in a cryo-EM reconstruction. Thus, while our cryo-EM results indicate that the fibers of the pseudotyped particles are less flexible than the wild-type fibers, they probably bend somewhat more than 2° on average.
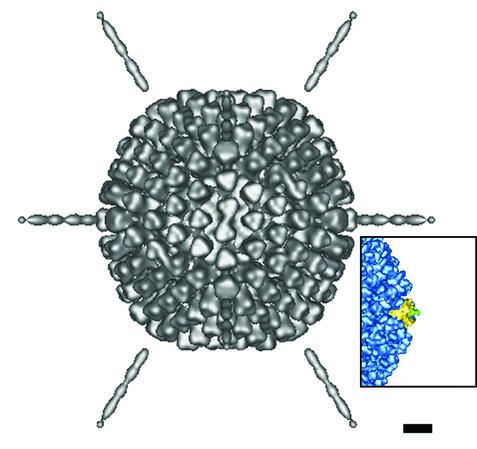
The subset of 85 selected particle images of the fiber-pseudotyped Ad were combined to produce the reconstruction shown in Fig. 3. In this combined reconstruction, fiber density is apparent out to 359 ± 18 Å from the penton base, which is close to the 331 ± 5 Å reported for the Ad2 fiber in intact penton (30). The 5% error range in the length measurement is due to the error range of the microscope magnification value. The discontinuity between the fiber shaft and the penton base may be explained by residual phase-contrast effects (see Materials and Methods). The measured fiber length is roughly equivalent to that expected for the full-length Ad5 fiber. Van Raaij et al. (42) have generated a model of 21 repeats of the Ad2 shaft based on the crystal structure of a portion of the shaft. By their calculations the shaft length of 21 repeats is 265 Å. Together with the fiber knob, which has a length along the fiber axis of ∼50 Å, this implies that the length of the Ad2 fiber shaft plus knob is ∼315 Å. All that is missing is the structure of the N-terminal 44 residues. If we assume the same average axial rise per residue for these 44 residues as found for the shaft, this implies that the N-terminal region is approximately 36 Å long. This yields a total predicted length of 351 Å for the Ad2 fiber (582 residues) and the highly homologous Ad5 fiber (581 residues). We note that even though the predicted flexibility repeat near the fiber knob was replaced in the Ad5s/Ad37s chimeric fiber, we have not reconstructed the full Ad fiber knob. This is presumably because there is still some inherent flexibility between the fiber shaft and the fiber knob.
FIG. 3.
A cryo-EM reconstruction of the pseudotyped (Ad5s/Ad37s) particle viewed along an icosahedral twofold axis. This reconstruction is based on 85 selected particle images with the straightest fibers. Note that the reconstructed fiber density extends 359 Å from the penton base protein of the capsid. The inset shows a cryo-EM reconstruction of Ad5 (43) for comparison. Note the absence of Ad5 fiber (green) density beyond 60 Å from the penton (yellow). Scale bar, 100 Å.
We have demonstrated reconstruction of fiber density out to the full length for the selected particle images of the pseudotyped Ad with the chimeric Ad5s/Ad37s fibers, which are missing the putative flexibility repeats. In addition, we have found a statistically significant number of pseudotyped Ad particles versus Ad5 wild-type particles with reasonably straight fibers. We note that even these selected particles must not have perfectly straight fibers, because during refinement the capsid density gets sharper but the fiber density does not improve. We conclude from the cryo-EM analysis that the Ad5s/Ad37s fibers are less flexible than the wild-type Ad5 fibers and that the chimeric fibers probably cannot bend with as large an angle as the wild type.
The length and flexibility of Ad fibers influence virus infection and cell binding.
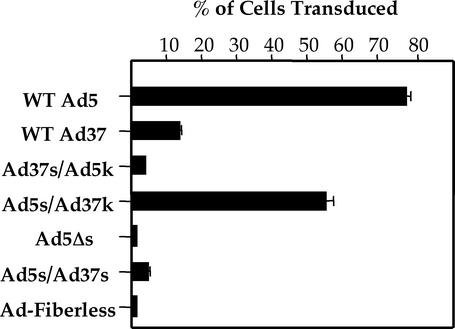
Further studies were undertaken to determine the role of fiber length and flexibility on Ad infection as measured by gene delivery. A549 epithelial cells were infected with pseudotyped Ad5 particles, and then expression of the GFP transgene was measured 24 h postinfection (Fig. 4). Ad5 infects A549 cells by using CAR as its primary receptor (49). Ad particles equipped with the Ad5 wild-type fiber showed approximately eightfold higher infection than viruses equipped with the Ad37 fiber, consistent with previous reports (19, 49). Particles displaying the Ad37 shaft fused to the Ad5 knob (Ad37s/Ad5k), and an Ad5 fiber lacking 14 repeats in the central shaft domain (Ad5Δs) had significantly reduced infectivity, indicating that the fiber shaft domain plays a crucial role in cell infection. Further evidence for this concept was observed by placing the Ad37 knob on the Ad5 fiber shaft (Ad5s/Ad37k). This construct increased virus infectivity nearly to the level of wild-type Ad5 fibers. The enhanced infectivity can be abolished by the addition of an excess of anti-CAR antibody (data not shown). Thus, these findings further support the concept that the length of the fiber shaft regulates cell infection by Ad particles. However, these fiber constructs did not allow us to determine whether fiber flexibility might also play a role in infection. Therefore, we performed further studies to address this issue. As noted above, cryo-EM image analysis indicated that the third repeat in the wild-type Ad2 long shaft, which is very similar to that of Ad5, confers fiber flexibility. Sequence analysis suggested that the 21st repeat adds additional flexibility at the opposite end of the fiber shaft. We next asked whether long Ad5 fibers lacking these repeat elements (Ad5s/Ad37s) were still capable of supporting Ad infection. As shown in Fig. 4, replacing the 3rd and 21st β-repeats of Ad5 with the corresponding regions in the more rigid Ad37 shaft abolished cell infection. Since the truncated Ad5 fiber (Ad5Δs) also had exhibited reduced infectivity, this indicates that flexibility alone is not sufficient to confer efficient virus-host cell interactions. These findings indicate that length and flexibility of the fiber shaft, in concert, regulate cell infection.
FIG. 4.
Ad-mediated gene delivery by fiber-pseudotyped Ad. A549 lung epithelial cells were infected for 3 h at 37°C with Ad particles encoding GFP. Forty-eight hours postinfection cells were analyzed for GFP expression by using flow cytometry. Data represent the averages and standard deviations of triplicate samples. WT, wild type.
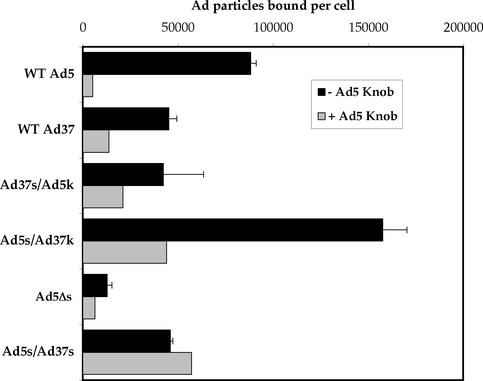
Since Ad infection is primarily regulated by the interaction of the fiber with CAR, we next sought to determine whether modifications of the fiber shaft also altered virus attachment to cells (Fig. 5). Ad5 exhibited higher binding to A549 cells than did Ad37 fiber-pseudotyped virus. Particles containing the truncated Ad5 knob (Ad5Δs) or the Ad37 shaft fused to the Ad5 knob (Ad37s/Ad5k) had reduced binding activity, whereas particles equipped with the Ad5 shaft fused to the Ad37 knob (Ad5s/Ad37k) had substantially increased cell attachment. The increased cell attachment was also reduced by the Ad5 knob, suggesting restoration of CAR binding since Ad5 infection of A549 cells is CAR dependent (49). Importantly, particles containing a straightened Ad5 fiber shaft (Ad5s/Ad37s) exhibited minimal specific cell attachment, indicating that fiber flexibility is needed for cell attachment as well as for cell infection.
FIG. 5.
Quantitation of fiber-pseudotyped Ad particles binding to A549 cells. A549 cells were incubated with Ad particles at 4°C for 1 h in the presence or absence of soluble Ad5 knob. After washing the cells, total DNA was extracted from each sample and the number of Ad and cellular DNA was determined by quantitative PCR. The knob data are represented by the averages and standard deviations of triplicates, and the + Ad5 knob data are represented by the averages of duplicates. WT, wild type.
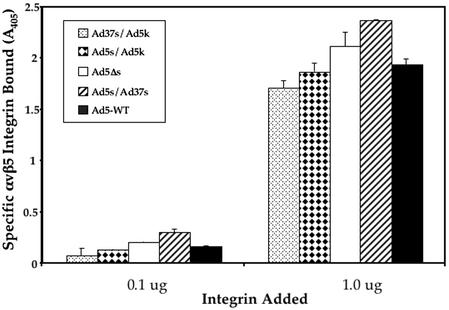
Following Ad binding to CAR, integrin interactions with the penton base protein facilitate virus internalization into cells (3, 47). In previous cryo-EM studies it was observed that the fiber kink, corresponding to the 3rd repeat, is in close proximity to the sites for integrin binding on the penton base protein (8). We therefore considered the possibility that altering fiber flexibility by introducing the 3rd repeat of Ad37 into the Ad5 shaft might also interfere with integrin binding. To investigate this possibility we measured binding of soluble αvβ5 integrin binding to different fiber-pseudotyped Ad (Fig. 6). Soluble αvβ5 integrin has been shown to specifically interact with Ad2 and penton base but not to bovine serum albumin (27). Each of the different Ad particles exhibited similar levels of αvβ5 integrin binding, indicating that fiber shaft modification did not interfere with the association of these secondary receptors.
FIG. 6.
ELISA assay of soluble αvβ5 integrin binding to immobilized Ad particles. Integrin binding was detected by using a non-function-blocking integrin monoclonal antibody as described in Materials and Methods. WT, wild type.
Model of Ad interactions with membrane-associated CAR and αv integrins.
Recent structural analyses have revealed a striking similarity in the structure of the Ad fiber protein and the sigma 1 protein of reovirus (7), the attachment protein for the reovirus receptor, JAM-1 (4). CAR and JAM-1 are both members of the Ig superfamily having two Ig-like ectodomains and are both located in tight junctions on host cells (12, 26, 48). Interestingly, the membrane-distal IgV domains of both CAR and murine JAM (mJAM) homodimerize in similar orientations (22, 41). Also, the dimerization interfaces of both CAR and mJAM contain a central pair of salt bridges (K123-D54 in CAR and R58-E60 in mJAM) that surround the twofold symmetry axes. Given their similar structure, identical locations on polarized epithelial cell membranes, and related host cell function, it is reasonable to propose that the two proteins share a common structure at the cell surface.
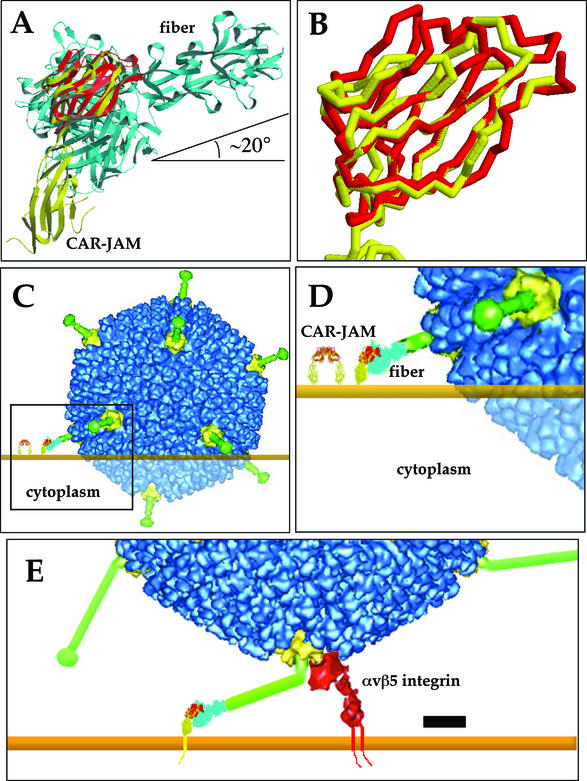
By using TOP, a protein topological comparison program (25), the crystal structures of mJAM, CAR D1 dimer, the Ad12 knob-CAR D1 complex (6), and Ad2 knob plus four β-repeats of the shaft (42) were successively aligned together (Fig. 7A). Because membrane-distal domains of CAR and mJAM are IgV domains and are 23% identical, their β-strands and secondary structures could be closely aligned (Fig. 7B). The resultant chimeric CAR-JAM molecule closely resembles the bent cryo-EM structure of human CAR bound to coxsackievirus B3 (17). Lastly, the 41% identical subgroup C Ad2 knob and shaft, with a nearly identical β-barrel tertiary structure, was easily aligned to the Ad12 knob.
FIG. 7.
Model of Ad interactions with cell surface receptors. By using a protein topology alignment program, TOP (25), the crystal structures of mJAM (yellow), CAR D1 homodimer (monomer shown in red), the CAR D1-Ad12 knob complex, and the Ad2 knob plus four β-repeats (cyan) were successively aligned (A). The entire complex was manually aligned to the cell surface, assuming that the two molecules of the CAR homodimer are equidistant from the cell surface. This panel was created by using Molscript (23). An enlarged view of the aligned membrane distal domains of mJAM and CAR (B) shows that most of the β-strands of the IgV domains overlap. The addition of the density from the cryo-EM reconstruction of the pseudotyped Ad37 vector (9) and the host cell membrane (C), in orange, shows that a large overlap between the Ad capsid and the host cell would occur if the Ad37 knob bound CAR. The steric clash is indicated by transparent density. The CAR-mJAM homodimer model is shown on the left. The region enclosed in the box is enlarged in panel D, showing the details of the model. Long, bent fiber proteins, CAR-mJAM, and the integrin density from the cryo-EM reconstruction of Ad12 complexed with αvβ5 (8) were added to a cryo-EM reconstruction of Ad2 (8) to demonstrate the relative orientations of the Ad2 or Ad5 capsid proteins and virus receptors at the cell surface (E). The lower domain of the cryo-EM integrin density was rotated to reflect the open and active conformation of the integrin. Scale bar, 100 Å. Backbone traces and space filling models in panels B to E were created by using Rasmol 2.7.
The entire fiber-CAR-mJAM complex was manually oriented to the cell surface, assuming that the two molecules from the CAR D1 dimer are equidistant to the planar cell surface. Electron density from a cryo-EM image reconstruction of an Ad5 vector pseudotyped with the Ad37 fiber (9) was added over the crystal structure of the Ad2 knob and shaft and was incorporated in the complex model (Fig. 7C and D). Note that in the resulting model of the Ad37-CAR-host cell complex, the angle between the threefold symmetry axis of the fiber shaft and the cell surface is approximately 20° (Fig. 7A). Also note that in order to position the CAR binding surface of the Ad37 knob in contact with CAR, the model indicates a significant steric collision between Ad37 and the host cell membrane with an overlap of roughly 300 Å. Even if there is a moderately large deviation (±30°) in the bend angles between the two CAR domains versus the two mJAM domains, this would not completely alleviate the steric collision predicted between Ad37 and the host cell membrane. In order to avoid steric interference, the rigid Ad37 fiber would have to be oriented such that the angle between the threefold symmetry axes of the fiber shaft and the cell surface was 60° or greater. Given the high degree of homology between mJAM and CAR and our knowledge of the relative orientations of both JAM domains with respect to the cell surface, it does not seem likely that a productive binding event (geometry) could be achieved for Ad37 and CAR. This molecular model of the Ad37-CAR-host cell complex based on homology modeling as well as on X-ray crystallographic and cryo-EM structures explains why Ad37 fiber cannot support virus binding via CAR at the cell surface despite containing a CAR binding sequence in its fiber knob. The steric constraints imposed by the cell surface and receptor orientation, therefore, play a significant role in alignment of fiber and CAR molecules.
DISCUSSION
The identification of cell receptors for Ad attachment (CAR) and entry (αv integrins) has provided a better understanding of virus-host cell interactions in vitro. However, emerging information suggests that Ad-receptor interactions may be even more complex than the two-receptor model originally proposed. For example, mutation of the binding sites for CAR and integrins on the fiber and penton base protein, respectively, reduces Ad infection in vitro but fails to completely ablate infection of murine hepatocytes (14, 34), suggesting that alternative receptors may promote Ad infection in vivo. Ad belonging to subgroup D, including Ad37, are associated with severe ocular infections. The basis of ocular tropism of these viruses is presently unknown. Interestingly, Ad37 contains a CAR-binding motif in the AB loop of its fiber knob and therefore should allow association with CAR. However, while Ad37 binding to CAR has been demonstrated in virus and knob overlay blot assays, this virus fails to associate with CAR on host cells.
In the studies presented here, we sought to determine whether other regions of the Ad fiber protein impact receptor usage. In particular, we asked whether the length and flexibility of the fiber might be crucial in determining receptor interactions. While previous studies have suggested that fiber length is important for CAR interaction, the role of fiber flexibility has not been explored. In this regard, only the fiber protein of subgroup D Ad lack the third nonconsensus β-repeat as well as a conserved KLGXGLXFD/N motif proximal to the knob domain, suggesting that fibers derived from this subgroup have increased rigidity. In support of this, a cryo-EM image reconstruction of pseudotyped Ad5 particles equipped with the Ad37 fiber showed this fiber to be short and rigid compared to the long and flexible Ad5 fiber. To gain a better understanding of the molecular basis of fiber-CAR interactions, we constructed Ad5 particles with modified fiber proteins that contained either a shortened Ad5 fiber lacking 14 repeats in the central shaft domain or a chimeric fiber comprised of the Ad5 knob fused to the short Ad37 shaft. Functional analyses showed that both of these fiber constructs decreased virus attachment and cell infection. In contrast, a fiber chimera consisting of the Ad37 knob fused to the long Ad5 shaft reconstituted cell infection and virus binding. These findings are consistent with previous reports (32) that found that fiber length is important for cell infection. To determine the role of fiber flexibility, we replaced the 3rd and 21st repeats of the Ad5 fiber with the 3rd and 7th repeats, respectively, of the Ad37 fiber. A cryo-EM image reconstruction showed that this chimeric fiber is considerably less flexible than wild-type Ad5 fiber. Virus particles equipped with this fiber exhibited substantially reduced cell infection as well as virus attachment, indicating that the ability of the protein to bend is important for CAR interactions at the cell surface.
The observation that the fiber is more rigid after replacing the 3rd and 21st β-repeats of the Ad5 fiber demonstrates that one or both of these wild-type repeats is responsible for fiber flexibility. It is likely that the insertion of amino acids TTVS (residues 84 to 87) in the β-turn between the two β-strands of the third repeat would disrupt the triple β-spiral structure in the shaft. The β-turn is somewhat buried inside the shaft, and large insertions of polar residues would drastically affect hydrophobic packing. The structurally related σ1 protein of reovirus also contains a 4- or 6-amino-acid insertion between β-strands in its tail, which is analogous to the Ad fiber shaft. This insertion results in a partially unstructured β-repeat that introduces a 23° bend in the σ1 tail and elongates the tail (7). It is likely that the 3rd repeat of the Ad2 fiber, like reovirus σ1, is also elongated and partially unstructured, allowing the fiber to bend at a large angle.
There is evidence from cryo-EM for a >30o bend or kink in the wild-type Ad2 shaft within 60 to 100 Å from the penton base (9). Previous cryo-EM studies of Ad2, Ad5, and Ad12 indicated fiber density out to only ∼100 Å (Fig. 3, inset), with a kink at ∼60 Å from the penton base (8, 35, 37, 43). Negative-stain EM also showed a hinge in the shaft approximately 70 Å from the tail end of the fiber (30). Displacement of 36 Å by the N-terminal tail and 26 Å by the first two straight fiber shaft repeats places the third β-repeat roughly at the same distance from the penton as the kink seen in the EM studies. When the reconstruction of Ad2 is displayed at a low isosurface to reveal the weakest reconstructed density, a cone-like distribution is observed, indicating a bend in the shaft. The high degree of sequence homology between Ad2 and Ad5 fibers supports the idea that both fibers are probably equally flexible at the third repeat.
There is also evidence from the crystal structure of the Ad2 fiber knob plus four β-repeats (42) that a hinge exists at the interface between the knob and the shaft. In the crystal structure, the threefold axes of the fiber knob and shaft deviate by 2°, starting from the 21st repeat. We have replaced this repeat in the Ad5 fiber, which contains the conserved KLGXGLXFD/N sequence, with that of the Ad37 fiber to form the rigid, long Ad5s/Ad37s fiber. We did not, however, replace residues 394 to 396, where the electron density was weak in the crystal structure. This linker region may form another hinge in the Ad fiber, allowing alteration of the knob's orientation. This would explain the absence of globular electron density corresponding to the knob in the Ad5s/Ad37s particle reconstruction, as the knob would be bent away from the fivefold axes. A large bend at the third repeat plus a ≥2° bend between the knob and the shaft would enable wild-type Ad2 and Ad5 fiber knobs to bind to CAR and avoid a collision at the cell surface (Fig. 7E).
Following CAR interaction with the fiber, αv integrin interactions with the penton base occur in close proximity to the region of the fiber bend. This interaction promotes rapid internalization of virus particles into clathrin-coated vesicles. In these studies we found that virus particles containing either more rigid and/or shortened fiber did not dramatically impede or increase association of soluble αvβ5 integrin. However, these studies do not exclude the possibility that integrin interaction might well be hindered or enhanced in the context of a host cell membrane. In this situation, a more rigid fiber shaft may prevent the close approach of the penton base to the integrin. In Fig. 7E, we present a model of the interaction of wild-type Ad5 with its two host cell receptors, CAR and αvβ5 integrin, based on the cryo-EM αvβ5 integrin density (8). The RGD binding site of an open and active αv integrin extends ∼180 to 200 Å from the cell surface (38). A bend of ∼70° in the third repeat of the fiber would place the ring of five RGD containing protrusions of the penton base within ∼180 Å from the cell surface for integrin interaction. Thus, in the context of the host membrane, flexibility in the fiber shaft may benefit both knob binding to CAR and penton association with αv integrins. Previous studies have reported that the fiber protein dissociates from the virus capsid following receptor interaction at the cell surface (16). Conformational changes occurring in the fiber shaft during receptor binding could also facilitate fiber dissociation after integrin engagement.
As the environment of the host membrane influences the orientations of cell surface receptors, it is important for at least one of the molecules involved in multireceptor viral systems to be flexible. Here we demonstrate that the flexibility of the Ad fiber allows the virus capsid to simultaneously bind CAR and αv integrins. Bending of the αv integrin has also been observed in its crystal structure (51) and may play an important role in its activation (24, 33). In perhaps an analogous situation, the 4- to 6-amino-acid insertion in the third to last beta repeat of the reovirus σ1 tail imparts σ1 with substantial flexibility and likely plays a similar role in orienting the σ1 head for interaction with JAM-1 at the cell surface. For Ad and reovirus, the proper orientation of all capsid and receptor proteins for viral entry requires flexibility of the viral attachment protein.
Virus entry into host cells is a complex process that often involves multiple interactions between the viral capsid and cell receptors. While most studies have focused on the identification and function of viral ligands and their receptors, an emerging concept is that the precise three-dimensional orientation of these molecules is also a contributing factor in virus tropism. This knowledge may provide an opportunity to develop antivirals that prevent cell entry by stabilizing the conformation of virus proteins or their cognate receptors. Increased understanding of the role of Ad fiber flexibility may also aid in the design of CAR-detargeted Ad gene delivery vectors that can be redirected to specific cell types.
Acknowledgments
This work was supported by NIH grants EY11431 and HL54352 to G. R. Nemerow and AI42929 to P. L. Stewart. E. Wu was supported by the LJIS Interdisciplinary Training Program and The Burroughs Wellcome Fund.
We thank Kelly White and Joan Gausepohl for preparation of the manuscript. We also thank Vijay Reddy (The Scripps Research Institute) for his assistance with molecular modeling.
Footnotes
This is manuscript 15323-IMM from The Scripps Research Institute.
REFERENCES
- 1.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. [DOI] [PubMed] [Google Scholar]
- 2.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, M., B. Harfe, and P. Freimuth. 1993. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 67:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 6.Bewley, M. C., K. Springer, and Y.-B. Zhang. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 7.Chappell, J. D., A. E. Prota, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, C. Y., P. Mathias, N. R. Nemerow, and P. L. Stewart. 1999. Structure of adenovirus complexed with its internalization receptor. J. Virol. 73:6759-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, C. Y., E. Wu, S. L. Brown, D. J. Von Seggern, G. R. Nemerow, and P. L. Stewart. 2001. Structural analysis of a fiber-pseudotyped adenovirus with ocular tropism suggests differential modes of cell receptor interactions. J. Virol. 75:5375-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chodosh, J., D. Miller, W. G. Stroop, and S. C. Pflugfelder. 1995. Adenovirus epithelial keratitis. Cornea 14:167-174. [PubMed] [Google Scholar]
- 11.Chroboczek, J., R. W. H. Ruigrok, and S. Cusack. 1995. Adenovirus fiber. Curr. Top. Microbiol. Immunol. 199:163-200. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DuBridge, R. B., P. Tang, H. C. Hsia, P.-M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Einfeld, D. A., R. Schroeder, P. W. Roelvink, A. Lizonova, C. R. King, I. Kovesdi, and T. J. Wickham. 2001. Reducing the native tropism of adenovirus vectors requires removal of both CAR and integrin interactions. J. Virol. 75:11284-11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorziglia, M. I., M. J. Kadan, S. Yei, J. Lim, G. M. Lee, R. Luthra, and B. C. Trapnell. 1996. Elimination of both E1 and E2a from adenovirus vectors further improves prospects for in vivo human gene therapy. J. Virol. 70:4173-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 17.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nature Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528-535. [PubMed] [Google Scholar]
- 19.Huang, S., V. Reddy, N. Dasgupta, and G. R. Nemerow. 1999. A single amino acid in the adenovirus type 37 fiber confers binding to human conjunctival cells. J. Virol. 73:2798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakubczak, J. L., M. L. Rollence, D. A. Stewart, J. D. Jafari, D. J. Von Seggern, G. R. Nemerow, S. C. Stevenson, and P. L. Hallenbeck. 2001. Adenovirus type 5 viral particles pseudotyped with mutagenized fiber proteins show diminished infectivity of coxsackie B-adenovirus receptor-bearing cells. J. Virol. 75:2972-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein, D., B. Bugl, W. H. Gunzburg, and B. Salmons. 2000. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gene Ther. 7:458-463. [DOI] [PubMed] [Google Scholar]
- 22.Kostrewa, D., M. Brockhaus, A. D'Arcy, G. E. Dale, P. Nelboeck, G. Schmid, F. Mueller, G. Bazzoni, E. Dejana, T. Bartfai, F. K. Winkler, and M. Hennig. 2001. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 20:4391-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraulis, P. J. 1991. A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24:946-950. [Google Scholar]
- 24.Liddington, R. C., and M. H. Ginsberg. 2002. Integrin activation takes shape. J. Cell Biol. 158:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, G. 2000. TOP: a new method for protein structure comparisons and similarity searches. J. Appl. Cryst. 33:176-183. [Google Scholar]
- 26.Martin-Padura, I., S. Lostaglio, M. Schneemann, L. Williams, M. Romano, P. Fruscella, C. Panzeri, A. Stoppacciaro, L. Ruco, A. Villa, D. Simmons, and E. Dejana. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathias, P., M. Galleno, and G. R. Nemerow. 1998. Interactions of soluble recombinant integrin αvβ5 with human adenoviruses. J. Virol. 72:8669-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roelvink, P. W., G. M. Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 29.Roelvink, P. W., A. Lizonova, J. G. M. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruigrok, R. W. H., A. Barge, C. Albiges-Rizo, and S. Dayan. 1990. Structure of adenovirus fibre: II morphology of single fibres. J. Mol. Biol. 215:589-596. [DOI] [PubMed] [Google Scholar]
- 31.Shah, A. K., and P. L. Stewart. 1998. Qview: software for rapid selection of particles from digital electron micrographs. J. Struct. Biol. 123:17-21. [DOI] [PubMed] [Google Scholar]
- 32.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimaoka, M., J. Takagi, and T. A. Springer. 2002. Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31:485-516. [DOI] [PubMed] [Google Scholar]
- 34.Smith, T., N. Idamakanti, H. Kylefjord, M. Rollence, L. King, M. Kaloss, M. Kaleko, and S. C. Stevenson. 2002. In vivo hepatic adenoviral gene delivery occurs independently of the coxsackievirus-adenovirus receptor. Mol. Ther. 5:770-775. [DOI] [PubMed] [Google Scholar]
- 35.Stewart, P. L., R. M. Burnett, M. Cyrklaff, and S. D. Fuller. 1991. Image reconstruction reveals the complex molecular organization of adenovirus. Cell 67:145-154. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, P. L., R. B. Cary, S. R. Peterson, and C. Y. Chiu. 2000. Digitally collected cryo-electron micrographs for single particle reconstruction. Microsc. Res. Tech. 49:224-232. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, P. L., C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 16:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takagi, J., B. Petre, T. Walz, and T. Springer. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110:599-611. [DOI] [PubMed] [Google Scholar]
- 39.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 41.van Raaij, M. J., E. Chouin, H. van der Zandt, J. M. Bergelson, and S. Cusack. 2000. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 Å resolution. Structure 8:1147-1155. [DOI] [PubMed] [Google Scholar]
- 42.van Raaij, M. J., A. Mitraki, G. Lavigne, and S. Cusack. 1999. A triple β-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401:935-938. [DOI] [PubMed] [Google Scholar]
- 43.Von Seggern, D. J., C. Y. Chiu, S. K. Fleck, P. L. Stewart, and G. R. Nemerow. 1999. A helper-independent adenovirus vector with E1, E3, and fiber deleted: structure and infectivity of fiberless particles. J. Virol. 73:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Von Seggern, D. J., S. Huang, S. K. Fleck, S. C. Stevenson, and G. R. Nemerow. 2000. Adenovirus vector pseudotyping in fiber-expressing cell lines: improved transduction of Epstein-Barr virus-transformed B cells. J. Virol. 74:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walters, R. W., P. Freimuth, T. O. Moninger, I. Ganske, J. Zabner, and M. J. Welsh. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110:789-799. [DOI] [PubMed] [Google Scholar]
- 46.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin αvβ5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 48.Williams, L. A., I. Martin-Padura, E. Dejana, N. Hogg, and D. L. Simmons. 1999. Identification and characterisation of human junctional adhesion molecule (JAM). Mol. Immunol. 36:1175-1188. [DOI] [PubMed] [Google Scholar]
- 49.Wu, E., J. Fernandez, S. K. Fleck, D. J. Von Seggern, S. Huang, and G. R. Nemerow. 2001. A 50 kDa membrane protein mediates sialic acid-independent binding and infection of conjunctival cells by adenovirus type 37. Virology 279:78-89. [DOI] [PubMed] [Google Scholar]
- 50.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure 2:1259-1270. [DOI] [PubMed] [Google Scholar]
- 51.Xiong, J. P., T. Stehle, T. Diefenbach, B. Zhang, R. Zhang, R. Dunker, D. L. Scott, A. Joachimiak, S. L. Goodman, and M. A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin αvβ3. Science 294:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]