Abstract
Human immunodeficiency virus type 1 (HIV-1) subtype C viruses have been found to almost exclusively use the chemokine receptor CCR5 as a coreceptor for entry, even in patients with advanced AIDS. We have characterized subtype C virus isolates from 28 patients from Harare, Zimbabwe, 20 of whom were receiving antiretroviral treatment. Virus from 10 of the treated patients induced syncytium formation (SI virus) when cultured with MT2 cells. Only non-syncytium-inducing (NSI) virus was cultured from the peripheral blood mononuclear cells of the eight patients who had not received treatment. The majority of these subtype C SI viruses were capable of using both CCR5 and CXCR4 as coreceptors for viral entry, and the consensus V3 loop sequences from the SI viruses displayed a high net charge compared to those of NSI viruses. While those on treatment had reverse transcriptase (RT) and protease mutations, there was no clear association between RT and protease drug resistance mutations and coreceptor tropism. These results suggest that CXCR4-tropic viruses are present within the quasispecies of patients infected with subtype C virus and that antiretroviral treatment may create an environment for the emergence of CXCR4 tropism.
Human immunodeficiency virus type 1 (HIV-1) infects target cells through binding of its envelope protein, gp120, to both CD4 and a second cellular coreceptor, leading to fusion of the viral envelope with the plasma membrane of the target cell. While several chemokine receptors including CCR1 and CCR3 (43) may mediate HIV-1 entry, CCR5 (R5) and CXCR4 (X4) are the predominant coreceptors (reviewed in reference 6). In general, X4-tropic viruses are recognized through syncytium induction (SI phenotype) in laboratory T-cell lines such as MT2 cells, while R5-tropic viruses do not induce syncytia (NSI phenotype) and are associated with macrophage infection.
Most studies of HIV-1 transmission have shown that R5 tropism is the commonly transmitted phenotype, irrespective of the transmission route (41, 42, 44). Early studies of subtype B viruses demonstrated that predominant coreceptor use by HIV-1 may switch during the course of infection from R5 to X4 (21, 32, 34). This change in coreceptor use, initially identified as a change from NSI to SI phenotype, occurs in up to 50% of patients infected with HIV-1 subtype B and correlates with a higher rate of disease progression (18, 20). Although this switch was initially identified in untreated patients, X4-tropic virus has been identified from patients receiving highly active antiretroviral (ARV) therapy as well (15, 27, 40). A change from R5 tropism to X4 tropism associated with disease progression has also been shown previously for HIV-1 subtypes A, D, and CRF_01AE (1, 8, 19, 37).
HIV-1 subtype C virus predominates in southern Africa as well as India (9, 25, 36, 38) and is the most prevalent HIV-1 subtype globally (26). In contrast to subtype B, most studies of subtype C viruses have demonstrated an overwhelming predominance of R5-tropic viruses at various stages of disease, including advanced AIDS (2, 4, 30, 37). Although a few X4-tropic viruses have been reported elsewhere (5, 36, 37, 39), X4 tropism and SI phenotype among subtype C viruses are rarer than among subtype B viruses in the later stages of disease. These observations have been made among untreated patients since drug therapy is unusually rare in the areas where subtype C predominates. In the present study, we analyzed virus isolates from a consecutive cohort of subjects, who had enrolled in an ARV treatment access program in Harare, Zimbabwe. A high frequency of SI phenotype and X4 tropism was seen in these HIV-1 subtype C isolates, exclusively among patients with prolonged ARV drug exposure.
(This work was presented in part at the Ninth Conference on Retroviruses and Opportunistic Infections, Seattle, Wash., 24 to 28 February 2002, and the First IAS Conference on HIV Pathogenesis and Treatment, Buenos Aires, Argentina, 8 to 11 July 2001.)
Isolation of SI viruses among individuals receiving HIV drug therapy.
Low-passage-number viral isolates were obtained from 28 blood samples collected from patients at The Centre in Harare, Zimbabwe. Between 1 × 106 and 10 × 106 isolated peripheral blood mononuclear cells (PBMCs) were cocultured with 5 × 106 phytohemagglutinin-stimulated donor PBMCs for 7 to 14 days. Virus supernatant was harvested when the culture contained more than 300 pg of p24gag (p24 enzyme-linked immunosorbent assay; Perkin-Elmer Life Science, Boston, Mass.)/ml. HIV-1 plasma virus was quantified with the Roche Amplicor monitor, version 1.5 (Roche Diagnostics Corporation, Indianapolis, Ind.). The median virus load for these 28 samples was 4.51 log10 copies/ml. CD4+ and CD8+ cells were enumerated with a three-color flow cytometer (Becton Dickinson, Mountain View, Calif.) in the Public Health Hematology Laboratory, University of Zimbabwe Medical School. The median numbers of CD4+ and CD8+ cells were 74.5 (range, 20 to 179) and 457 (range, 342 to 949), respectively (Table 1). Twenty of these 28 infected individuals had received ARV treatment with at least two drugs, usually two reverse transcriptase (RT) inhibitors, for an average of 14.4 months. The eight individuals who had not received treatment had similar viral loads as those of individuals on treatment (P = 0.9, Mann-Whitney test) but had higher numbers of CD4+ cells (P = 0.014, Mann-Whitney test) and CD8+ cells (P = 0.053, Mann-Whitney test). Virus supernatant was cultured with MT2 cells for up to 14 days, and from 2 and 8 days in culture, 10 of the 28 samples induced syncytium formation. There was no difference in median virus load or CD4+ cell counts (P = 0.97, Mann-Whitney test) between patients with SI viruses and those with NSI viruses. However, SI viruses were observed only among 10 of 20 isolates from patients who had received ARV therapy compared to none of eight untreated subjects (P = 0.01, Fisher's exact test). Since the patients receiving therapy had lower numbers of CD4 cells, we examined the relationship between treatment and SI virus for 24 patients with fewer than 300 CD4+ cells. The association remained significant (P = 0.047, Fisher's exact test). Further studies of larger numbers of treated and untreated patients will be necessary to determine whether treatment may select SI virus in subtype C-infected individuals.
TABLE 1.
Characteristics of individuals with SI and NSI viruses in Harare, Zimbabwe
| Characteristic and type of value | All (n = 28) | SI virus (n = 10) | NSI virus with ARV treatmenta (n = 10) | NSI virus without ARV treatment (n = 8) |
|---|---|---|---|---|
| Age (yr) | ||||
| Median | 40 | 41.5 | 39.5 | 35 |
| Range | 30-61 | 31-61 | 33-65 | 30-51 |
| Time on treatment (mo), mean | 14.4 | 13.2 | 18.2 | NAb |
| RNA viral load (log10 copies/ml) | ||||
| Median | 4.51 | 3.84 | 4.00 | 4.64 |
| Interquartile range | 3.26-5.02 | 3.00-4.65 | 3.01-4.78 | 4.44-5.00 |
| No. of CD4+ cells | ||||
| Median | 74.5 | 56.5 | 41 | 186 |
| Interquartile range | 20-179 | 11-101 | 13-158 | 84-372 |
| No. of CD8+ cells | ||||
| Median | 457 | 442 | 393 | 738 |
| Interquartile range | 342-949 | 352-751 | 256-949 | 573-1,208 |
Includes one patient who had received treatment for 2 weeks.
Envelope sequences correspond to subtype C.
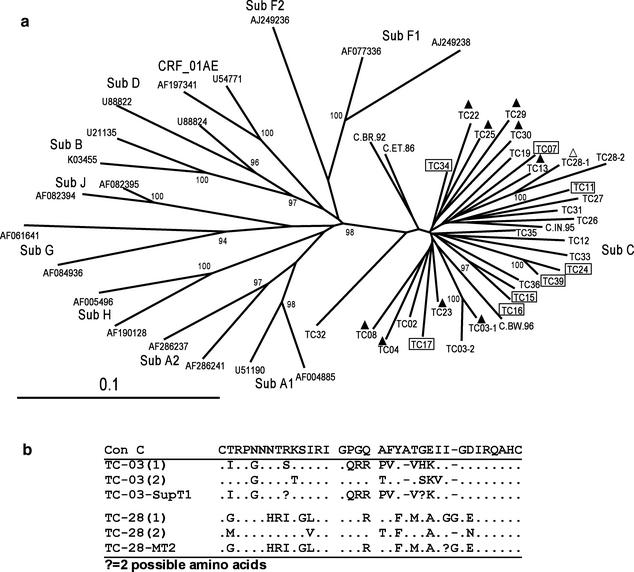
The C2-V5 region of the env gene (650 nucleotides [nt]) was amplified and sequenced from viral RNA extracted from the low-passage-number virus stock by using primers ED5-ED12 and ES7-ES8 (7). env PCR products were sequenced by cycle sequencing and dye terminator methods on an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, Calif.). Sequences were aligned with CLUSTAL W (35), adjusted manually with BioEdit (13) to ensure a correct reading frame, and then stripped of gaps so that 529 nt were compared for all sequences. Distances between sequences were generated by the Kimura two-parameter and maximum-likelihood methods by using DNADIST within PHYLIP, version 3.752 (10). Phylogenetic trees were constructed by neighbor joining with NEIGHBOR. Bootstrap analysis was done with SEQBOOT (100 data sets), DNADIST, NEIGHBOR, and CONSENSE within PHYLIP. All 28 env sequences cluster with subtype C reference sequences (Los Alamos HIV sequence database) with a bootstrap value of 98% (Fig. 1a). Percent pairwise distances were calculated with the SynScan program (12), a program found within the Resources section of the Stanford HIV RT and Protease Sequence database (http://hivdb.stanford.edu). RT and protease sequences corresponded to subtype C virus as well (data not shown), and no evidence of recombination with other HIV-1 subtypes within env or pol was found after analysis with the Los Alamos Recombination Identification Program (http://hiv-web.lanl.gov). In comparison with 82 other subtype C env sequences from GenBank, there was no significant clustering of Zimbabwean samples with high bootstrap values. The sequences were dispersed among the southern African sequences and were distinct from the Indian subtype C cluster (data not shown). The env genes from two pairs of viruses, TC-15-TC-16 and TC-24-TC-39 (pairwise distances of 8.37 and 9.34%, respectively) were more closely related, compared to an average distance of 13.4% between each of the other pairs, and were obtained from sexual partners.
FIG. 1.
SI and NSI virus envelope sequences cluster with subtype C HIV. (a) env (C2-V5) RT-PCR product derived from low-passage-number virus stock was sequenced and aligned with CLUSTAL W. The phylogenetic tree was made from gap-stripped sequences of 529 nt each. Bootstrap values from 100 data sets that have a greater than 90% frequency are indicated on the tree. Boxed samples indicate patients who were not receiving treatment. Filled triangles indicate samples with R5-X4 coreceptor usage, while the open triangle indicates X4 usage only. GenBank accession numbers for subtype C reference sequences are as follows: C.BR.92, U52953; C.BW.96, AF110967; C.IN.95, AF067155; C.ET.86, U46016. The accession numbers for the other reference sequences are given in the figure. (b) TC-03 sequence 1 and sequence 2 and TC-28 sequence 1 and sequence 2 correspond to two distinct yet closely related groups of viruses. env V3 loop sequences from PBMC virus stock and MT2 or SupT1 virus stock were compared to consensus C sequence (Con C).
We were unable to clearly determine a consensus env sequence by direct sequencing of PCR product for 10 viruses, and therefore consensus sequences for these 10 viruses were determined from four to eight clones each. Within each group of clones, the pairwise distance ranges from 1.67 to 6.71%. Among these cloned env sequences, TC-03 and TC-28 each exhibited two distinct virus populations. The consensus sequences for these distinct groups are indicated on the phylogenetic tree (Fig. 1a). TC-03 consensus sequences 1 and 2 differed by 4.71%, while TC-28 consensus sequences 1 and 2 differed by 7.88%. Figure 1b shows the V3 loop sequences for the consensus sequence of each group from low-passage-number virus isolate and virus grown from MT2 or SupT1 cells. Only TC-03 consensus sequence 1 corresponds to virus that grew in SupT1 cells, indicating that this virus population uses X4 while population 2 likely uses R5 for virus entry. Likewise, TC-28 from PBMC culture also contains two distinct but closely related virus populations, with TC-28 consensus sequence 1 aligning closely with virus cultured from MT2 cells. env from virus grown in MT2 cells was sequenced for all other SI viruses, and these sequences aligned closely with sequences from PBMC-cultured virus. Envelope sequence obtained directly from plasma RNA for six cases closely corresponded with the PBMC virus sequences (data not shown).
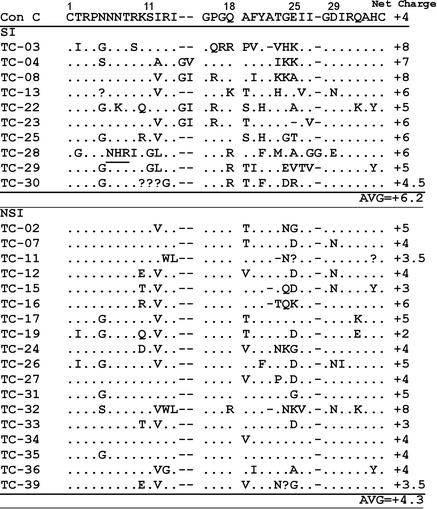
In HIV-1 subtype B, the SI phenotype is associated with basic amino acids at positions 11, 25, and 29 within the 35-amino-acid V3 loop region (14), while CRF01_AE SI viruses are characterized by basic amino acids at positions 8, 11, and 18 within the same region (19). Therefore, we were interested in characterizing the V3 loop region of the SI subtype C viruses in our study. Half (5 of 10) of the SI viruses in our cohort had a basic amino acid residue at position 18, and 7 of the 10 SI viruses had an increase of two or more basic amino acids in addition to those found in the consensus sequence within the V3 region. One of the 10 subtype C SI viruses had a basic residue at position 8 but none at position 11 or 29. Three of the 10 had a positive charge at position 25, and one virus had a positive charge at position 32 (Fig. 2). Among the NSI viruses, 3 of 18 had a basic amino acid at position 25 and two had a basic amino acid at position 32. The increase in basic amino acid residues also affects the net charge of the V3 region; the consensus V3 loops of the SI viruses had a higher average net charge (+6.2) than did those of the NSI viruses (+4.3; P < 0.01, Student's t test) (Fig. 2). Among subtype B viruses, loss of a potential glycosylation site within the V3 loop is associated with SI viruses (22, 31). In our cohort, only one SI virus, TC-28, did not have the potential glycosylation site sequence (NXS/T) within the V3 loop. Overall, these subtype C viruses were characterized by basic amino acids within the crown region (amino acids 15 to 18) and/or between amino acids 21 and 25. Other regions may contribute to X4 tropism independently of the V3 loop. In studies of dualtropic viruses, residues within the V4 and V5 loops of gp120, as well as residues within gp41, contributed to X4 tropism (16, 33). One NSI virus, TC-32, displayed V3 loop characteristics similar to those of the SI viruses, such as a high positive net charge of +8, another indication that other regions of env contribute to tropism selection.
FIG. 2.
Subtype C SI viruses have an increase in the number of basic amino acids compared to that in the env V3 loop consensus sequence (Con C) as well as a higher average net charge. The net charge of the sequence is derived from the summation of charges from positive and negative amino acids. A potential glycosylation site is underlined. Sequences for TC-03 and TC-28 correspond to virus that grew from MT2 cells; the remaining sequences are from low-passage-number PBMC stock.
Coreceptor usage of SI viruses.
We determined coreceptor usage for each of the SI viruses by using GHOST-X4 and GHOST-R5 cells infected with virus derived from MT2 culture or PBMC virus stock (3). A day prior to infection, cells from each GHOST cell line were seeded in 24-well plates at 2 × 104 cells/well in 0.5 ml. On the following morning, the cells were infected with diluted virus isolates. Plates were centrifuged at 930 × g for 30 min (29), and cells harvested after 48 h postinfection were analyzed with a FACScan flow cytometer (Becton Dickinson, San Jose, Calif.) for green fluorescent protein expression in the presence of 2% (wt/vol) formaldehyde. Because GHOST cell derivatives express a low but detectable level of X4, 5% fluorescent cells was considered the cutoff value for positive coreceptor-dependent infectivity. Of the 10 SI viruses from MT2 culture, 9 appeared to use both R5 and X4 as coreceptors (Table 2). The SI virus that did not have a V3 loop glycosylation site, TC-28, exclusively utilized X4. Similar results for coreceptor tropism were obtained with PBMC culture virus stock with the exception of TC-28, which used both R5 and X4, indicating that the X4 virus was preferentially selected by the MT2 cells.
TABLE 2.
Coreceptor tropism for SI viruses
| Sample | % Positive cells
|
Tropism | |
|---|---|---|---|
| R5 | X4 | ||
| TC-03 | 30 | 22 | X4/R5 |
| TC-04 | 63 | 19 | X4/R5 |
| TC-08 | 28 | 43 | X4/R5 |
| TC-13 | 52 | 37 | X4/R5 |
| TC-22 | 10 | 34 | X4/R5 |
| TC-23 | 39 | 46 | X4/R5 |
| TC-25 | 49 | 15 | X4/R5 |
| TC-28 | 4 | 53 | X4 |
| TC-29 | 14 | 11 | X4/R5 |
| TC-30 | 23 | 13 | X4/R5 |
RT and protease drug resistance mutations from patients with SI and NSI viruses.
RT and protease sequencing was available for the 19 patients who had received drug therapy for at least 3 months; these sequences have been previously described (17). One treated patient had received drug therapy for only 2 weeks and was not included in this analysis. Tables 3 and 4 show the ARV treatment histories and RT and protease drug resistance mutations according to the SI and NSI phenotypes. All 19 patients had received two drugs or more, including one nucleoside RT inhibitor; all but two had also received a protease inhibitor. No significant differences between the SI and NSI groups were seen in the number of drugs used, in the number of RT and protease mutations per patient, and in the specific mutation prevalences. A trend toward a higher mutation rate at position 219 in the SI group was seen (4 of 10 versus 0 of 9; P = 0.054, Fisher exact test). All occurrences of the major protease mutation L90M were seen in the SI group (three of nine patients).
TABLE 3.
RT resistance mutations from viruses with SI and NSI phenotypes
| Virus | RT inhibitor drug(s)a
|
RT mutationb
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Present | Past | 41 M | 44 E | 67 D | 69 T | 70 K | 74 L | 77 F | 98 A | 103 K | 118 V | 151 Q | 179 V | 181 Y | 184 M | 215 T | 219 K | |
| SI viruses | ||||||||||||||||||
| TC-03 | HYD + DDI + AZT | DDI + HYD | N | N | R | L | M | I | E/Q | |||||||||
| TC-04 | ABC + EFV | D4T + NVP | ||||||||||||||||
| TC-08 | 3TC + D4T | AZT | I | V | ||||||||||||||
| TC-13 | 3TC + AZT | DDI + HYD | ||||||||||||||||
| TC-22 | NVP | HYD + DDI + AZT | L | N | V | Y | ||||||||||||
| TC-23 | AZT + DDC | G | R | Q | ||||||||||||||
| TC-25 | EFV | D4T + NVP | ||||||||||||||||
| TC-28 | 3TC + AZT | N | R | V | I | Q | ||||||||||||
| TC-29 | 3TC + AZT | P | V | N | R | I | I | V | Q | |||||||||
| TC-30 | 3TC + AZT | V | ||||||||||||||||
| NSI viruses | ||||||||||||||||||
| TC-02 | 3TC + AZT | |||||||||||||||||
| TC-12 | DDI + HYD | |||||||||||||||||
| TC-19 | 3TC + AZT | L | N | R | V | |||||||||||||
| TC-26 | 3TC + AZT | 3TC + AZT, 3TC | ||||||||||||||||
| TC-27 | DDI + AZT | AZT | L | N | R | G | V | Y | ||||||||||
| TC-31 | D4T + 3TC | L | N | V | I | Y | ||||||||||||
| TC-32 | 3TC + AZT | DDI + AZT + EFV | V | |||||||||||||||
| TC-33 | 3TC + AZT | D4T + DDI | ||||||||||||||||
| TC-35 | 3TC + AZT | DDC + AZT | L | R | V | Y | ||||||||||||
Abbreviations: HYD, hydroxyurea; DDI, didanosine; AZT, zidovudine; ABC, abacavir; EFV, efavirenz; D4T, stavudine; NVP, nevirapine; 3TC, lamivudine; DDC, zalcitabine.
Amino acids and their positions correspond to consensus wild-type subtype B sequence.
TABLE 4.
Protease resistance mutations from viruses with SI and NSI phenotypes
| Virus | Protease inhibitor drug(s)a
|
Protease mutationb
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Present | Past | 10 L | 20 K | 36 M | 54 I | 63 L | 71 A | 73 G | 74 T | 82 V | 90 L | 93 I | |
| SI viruses | |||||||||||||
| TC-04 | APV | NFV | I | P | L | ||||||||
| TC-08 | SQV | I | T | S | L | ||||||||
| TC-13 | SQV | I | P | I | L | ||||||||
| TC-22 | IDV + SQV | I | P | L | |||||||||
| TC-23 | SQV | I | P | I | M | L | |||||||
| TC-25 | RTV + SQV | IDV | I | P | L | ||||||||
| TC-28 | SQV | I | P | V | S | M | L | ||||||
| TC-29 | SQV | R | I | P | V | M | L | ||||||
| TC-30 | NFV | I | I | L | |||||||||
| NSI viruses | |||||||||||||
| TC-02 | NFV | I | L | ||||||||||
| TC-26 | SQV | I | P | I | L | ||||||||
| TC-27 | SQV | R | I | L | |||||||||
| TC-32 | RTV | I | V | S | L | ||||||||
| TC-33 | IDV | R | I | L | |||||||||
| TC-35 | NFV | IDV or NFV | I | P | S | L | |||||||
| TC-31 | APV | I | I | F | P | L | |||||||
Abbreviations: APV, amprenavir; NFV, nelfinavir; SQV, saquinavir; IDV, indinavir; RTV, ritonavir.
Amino acids and their positions correspond to consensus wild-type subtype B sequence.
Based on isolates from patients with advanced disease, progression to AIDS in subtype C HIV-1 infection does not characteristically include the switch in coreceptor usage that has been observed with high frequency among patients infected with subtype B virus. However, in the Zimbabwean isolates examined here, there is evidence that treatment with ARV drugs may produce the conditions necessary for a change in phenotype. The 20 patients receiving drug therapy had lower numbers of CD4+ cells than did those who had not received ARV therapy, and most had developed resistance to one or more of their prescribed drugs, suggesting inconsistent drug access or use.
It is possible that X4-using SI viruses emerge among treated individuals with low numbers of CD4 cells due to increased patient survival associated with ARV therapy (23). Additionally, recent studies of subtype B-infected patients show that up-regulation of R5 expression on CD4 cells is associated with HIV infection and may be reversed by drug treatment (11, 28). Among patients receiving highly active ARV therapy, reduced expression of R5 on PBMC has been documented early in the course of ARV therapy (24). This may provide one explanation for the unexpected frequency of SI X4-tropic viruses in these drug-treated patients. Whether ARV drug treatment can elicit a similar response in patients with subtype C virus where X4-tropic virus variants are likely to be present at low levels in the pretreatment quasispecies will require longitudinal studies.
Alternatively, we may be witnessing a gradual emergence of X4-tropic viruses within the subtype C population of viruses that is unrelated to drug treatment. Two recent studies of subtype C isolates in South Africa have indicated a higher proportion of X4-tropic viruses than had previously been observed among drug-naïve patients with advanced disease (5, 39). Continued study of subtype C cohorts, both drug treated and drug naïve, may indicate whether X4-tropic subtype C viruses are becoming more common overall or because of specific factors such as drug treatment or specific opportunistic infections.
The transition from R5-tropic to X4-tropic virus is not well understood for any of the HIV-1 subtypes. Although many of these virus isolates appear to use both coreceptors, these may represent a mixture of virus populations, each using a single coreceptor. In two cases, sequencing of multiple clones did demonstrate two distinct populations of env sequences with characteristics suggesting different coreceptor usage. Among the SI isolates which appear to use both types of receptors in the GHOST assay, extensive biological cloning will be needed to clarify the frequency of X4-, R5-, and potentially dualtropic viruses among patients with subtype C virus infection.
Nucleotide sequence accession number.
Envelope nucleotide and amino acid sequences from the 28 study group patients were submitted to GenBank and can be found under the accession numbers AY265929 to AY265958. RT and protease sequences for the treated patients can be found under the accession numbers AY090839 to AY090859.
Acknowledgments
We thank Lynde Francis, Director of The Centre, Harare, Zimbabwe, for allowing us access to the patients' unlinked specimens.
This work was supported by the Doris Duke Charitable Foundation.
REFERENCES
- 1.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjorndal, A., A. Sonnerborg, C. Tscherning, J. Albert, and E. M. Fenyo. 1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retrovir. 15:647-653. [DOI] [PubMed] [Google Scholar]
- 3.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S. Paranjape, and D. A. Gadkari. 2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 271:253-258. [DOI] [PubMed] [Google Scholar]
- 5.Cilliers, T., J. Nhlapo, M. Coetzer, D. Orlovic, T. Ketas, W. C. Olson, J. P. Moore, A. Trkola, and L. Morris. 2003. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 77:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 7.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 8.De Wolf, F., E. Hogervorst, J. Goudsmit, E. M. Fenyo, H. Rubsamen-Waigmann, H. Holmes, B. Galvao-Castro, E. Karita, C. Wasi, S. D. Sempala, et al. 1994. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res. Hum. Retrovir. 10:1387-1400. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, U., M. Grez, H. von Briesen, B. Panhans, M. Geissendorfer, H. Kuhnel, J. Maniar, G. Mahambre, W. B. Becker, and M. L. Becker. 1993. HIV-1 strains from India are highly divergent from prototypic African and US/European strains, but are linked to a South African isolate. AIDS 7:23-27. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 11.Gervaix, A., J. Nicolas, P. Portales, K. Posfay-Barbe, C. A. Wyler, M. Segondy, O. Avinens, C. A. Siegrist, J. Clot, J. F. Eliaou, J. Astruc, and P. Corbeau. 2002. Response to treatment and disease progression linked to CD4+ T cell surface CC chemokine receptor 5 density in human immunodeficiency virus type 1 vertical infection. J. Infect. Dis. 185:1055-1061. [DOI] [PubMed] [Google Scholar]
- 12.Gonzales, M. J., J. M. Dugan, and R. W. Shafer. 2002. Synonymous-non-synonymous mutation rates between sequences containing ambiguous nucleotides (Syn-SCAN). Bioinformatics 18:886-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Hoffman, N. G., F. Seillier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtkamp, N., A. Otteken, S. Findhammer, V. Miller, R. Kurth, and A. Werner. 2000. Unexpected coreceptor usage of primary human immunodeficiency virus type 1 isolates from viremic patients under highly active antiretroviral therapy. J. Infect. Dis. 181:513-521. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Q., A. P. Barry, Z. Wang, S. M. Connolly, S. C. Peiper, and M. L. Greenberg. 2000. Evolution of the human immunodeficiency virus type I envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS progression. J. Virol. 74:11858-11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantor, R., L. S. Zijenah, R. W. Shafer, S. Mutetwa, E. Johnston, R. Lloyd, C. Israelski, and D. A. Katzenstein. 2002. HIV-1 subtype C reverse transcriptase and protease genotypes in patients from Zimbabwe failing antiretroviral therapy. AIDS Res. Hum. Retrovir. 18:1407-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson, A., K. Parsmyr, E. Sandstrom, E. M. Fenyo, and J. Albert. 1994. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 32:364-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, K., H. Sato, and Y. Takebe. 1999. Role of naturally occurring basic amino acid substitutions in the human immunodeficiency virus type 1 subtype E envelope V3 loop on viral coreceptor usage and cell tropism. J. Virol. 73:5520-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koot, M., A. B. van't Wout, N. A. Kootstra, R. E. de Goede, M. Tersmette, and H. Schuitemaker. 1996. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 173:349-354. [DOI] [PubMed] [Google Scholar]
- 21.Kuiken, C. L., J. J. de Jong, E. Baan, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J. Virol. 66:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., M. A. Rey-Cuille, and S. L. Hu. 2001. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res. Hum. Retrovir. 17:1473-1479. [DOI] [PubMed] [Google Scholar]
- 23.Miller, V., A. N. Phillips, B. Clotet, A. Mocroft, B. Ledergerber, O. Kirk, V. Ormaasen, P. Gargalianos-Kakolyris, S. Vella, and J. D. Lundgren. 2002. Association of virus load, CD4 cell count, and treatment with clinical progression in human immunodeficiency virus-infected patients with very low CD4 cell counts. J. Infect. Dis. 186:189-197. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson, J. K., S. W. Browning, R. L. Hengel, E. Lew, L. E. Gallagher, D. Rimland, and J. S. McDougal. 2001. CCR5 and CXCR4 expression on memory and naive T cells in HIV-1 infection and response to highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 27:105-115. [DOI] [PubMed] [Google Scholar]
- 25.Orloff, G. M., M. L. Kalish, J. Chiphangwi, K. E. Potts, C. Y. Ou, G. Schochetman, G. Dallabetta, A. I. Saah, and P. G. Miotti. 1993. V3 loops of HIV-1 specimens from pregnant women in Malawi uniformly lack a potential N-linked glycosylation site. AIDS Res. Hum. Retrovir. 9:705-706. [DOI] [PubMed] [Google Scholar]
- 26.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 27.Philpott, S., B. Weiser, K. Anastos, C. M. Kitchen, E. Robison, W. A. Meyer III, H. S. Sacks, U. Mathur-Wagh, C. Brunner, and H. Burger. 2001. Preferential suppression of CXCR4-specific strains of HIV-1 by antiviral therapy. J. Clin. Investig. 107:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierdominici, M., A. Giovannetti, F. Ensoli, F. Mazzetta, M. Marziali, M. R. De Cristofaro, D. Santini-Muratori, W. Leti, and F. Aiuti. 2002. Changes in CCR5 and CXCR4 expression and beta-chemokine production in HIV-1-infected patients treated with highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 29:122-131. [DOI] [PubMed] [Google Scholar]
- 29.Pietroboni, G., G. Harnett, and M. Bucens. 1989. Centrifugal enhancement of human immunodeficiency virus (HIV) and human herpesvirus type 6 (HHV-6) infection in vitro. J. Virol. Methods 24:85-90. [DOI] [PubMed] [Google Scholar]
- 30.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, S. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollakis, G., S. Kang, A. Kliphuis, M. I. Chalaby, J. Goudsmit, and W. A. Paxton. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 276:13433-13441. [DOI] [PubMed] [Google Scholar]
- 32.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tersmette, M., R. A. Gruters, F. De Wolf, R. E. de Goede, J. M. Lange, P. T. Schellekens, J. Goudsmit, H. G. Huisman, and F. Miedema. 1989. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 63:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tien, P. C., T. Chiu, A. Latif, S. Ray, M. Batra, C. H. Contag, L. Zejena, M. Mbizvo, E. L. Delwart, J. I. Mullins, and D. A. Katzenstein. 1999. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:147-153. [DOI] [PubMed] [Google Scholar]
- 37.Tscherning, C., A. Alaeus, R. Fredriksson, A. Bjorndal, H. Deng, D. R. Littman, E. M. Fenyo, and J. Albert. 1998. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241:181-188. [DOI] [PubMed] [Google Scholar]
- 38.van Harmelen, J., R. Wood, M. Lambrick, E. P. Rybicki, A. L. Williamson, and C. Williamson. 1997. An association between HIV-1 subtypes and mode of transmission in Cape Town, South Africa. AIDS 11:81-87. [DOI] [PubMed] [Google Scholar]
- 39.Van Rensburg, E. J., T. L. Smith, M. Zeier, B. Robson, C. Sampson, F. Treurnicht, and S. Engelbrecht. 2002. Change in co-receptor usage of current South African HIV-1 subtype C primary isolates. AIDS 16:2479-2480. [DOI] [PubMed] [Google Scholar]
- 40.van Rij, R. P., J. A. Visser, R. M. van Praag, R. Rientsma, J. M. Prins, J. M. Lange, and H. Schuitemaker. 2002. Both R5 and X4 human immunodeficiency virus type 1 variants persist during prolonged therapy with five antiretroviral drugs. J. Virol. 76:3054-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 43.Xiao, L., D. L. Rudolph, S. M. Owen, T. J. Spira, and R. B. Lal. 1998. Adaptation to promiscuous usage of CC and CXC-chemokine coreceptors in vivo correlates with HIV-1 disease progression. AIDS 12:F137-F143. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]