Abstract
Genetic studies have shown that retinoic acid (RA) signaling is required for mouse retina development, controlled in part by an RA-generating aldehyde dehydrogenase encoded by Aldh1a2 (Raldh2) expressed transiently in the optic vesicles. We examined the function of a related gene, Aldh1a1 (Raldh1), expressed throughout development in the dorsal retina. Raldh1−/− mice are viable and exhibit apparently normal retinal morphology despite a complete absence of Raldh1 protein in the dorsal neural retina. RA signaling in the optic cup, detected by using a RARE-lacZ transgene, is not significantly altered in Raldh1−/− embryos at embryonic day 10.5, possibly due to normal expression of Aldh1a3 (Raldh3) in dorsal retinal pigment epithelium and ventral neural retina. However, at E16.5 when Raldh3 is expressed ventrally but not dorsally, Raldh1−/− embryos lack RARE-lacZ expression in the dorsal retina and its retinocollicular axonal projections, whereas normal RARE-lacZ expression is detected in the ventral retina and its axonal projections. Retrograde labeling of adult Raldh1−/− retinal ganglion cells indicated that dorsal retinal axons project to the superior colliculus, and electroretinography revealed no defect of adult visual function, suggesting that dorsal RA signaling is unnecessary for retinal ganglion cell axonal outgrowth. We observed that RA synthesis in liver of Raldh1−/− mice was greatly reduced, thus showing that Raldh1 indeed participates in RA synthesis in vivo. Our findings suggest that RA signaling may be necessary only during early stages of retina development and that if RA synthesis is needed in dorsal retina, it is catalyzed by multiple enzymes, including Raldh1.
Vertebrate eye development requires vitamin A (retinol) as demonstrated in gestational vitamin A deficiency studies, where ocular defects are one of the most common malformations observed (51). The effect of retinol on eye development is mediated by its active metabolite retinoic acid (RA), which is most likely produced locally by several retinoid dehydrogenases expressed in the eye (8). RA administration can rescue ocular defects observed during gestational vitamin A deficiency (7), and withdrawal of RA during deficiency leads to ocular defects (6). RA functions as a ligand for three nuclear RA receptors (RARα, RARβ, and RARγ) that are all expressed in the eye during development (36). Mice carrying single null mutations of RARs appear to be relatively normal during embryogenesis, and most mice survive postnatally (22, 24), although RARβ mutant mice do exhibit a minor eye defect (retrolenticular membrane) (11). When two RARs are knocked out together, many embryonic defects are observed in the eye and other organs, and lethality is observed at birth (23, 29). Consistent defects of the eye in double RAR mutants include microphthalmia, coloboma of the retina, and abnormalities of the cornea, eyelids, and conjunctiva (23). Thus, RARs mediate the functions of vitamin A during eye development, as the defects observed are essentially the same as those seen during gestational vitamin A deficiency.
Production of RA from retinol involves a two-step oxidation pathway (8). First, retinol is oxidized to retinaldehyde by cytosolic retinol dehydrogenases, which are members of the alcohol dehydrogenase family (32, 33), or by microsomal retinol dehydrogenases, which are members of the short-chain dehydrogenase/reductase family (19, 37). The second step, oxidation of retinaldehyde to RA, is performed by several members of the aldehyde dehydrogenase (ALDH) family, which are officially known in the mouse as Aldh1a1 (also called Raldh1 or Aldh1), Aldh1a2 (also called Raldh2), and Aldh1a3 (also called Raldh3) (8, 48). These three enzymes, often referred to as retinaldehyde dehydrogenases (Raldh), share approximately 70% amino acid sequence identity and have distinct tissue-specific expression patterns during mouse eye development: Raldh1 in the dorsal neural retina (16, 25, 27), Raldh2 transiently in the optic vesicle (14, 38, 54), and Raldh3 in the ventral neural retina, dorsal retinal pigment epithelium (RPE), and lens (12, 20, 26, 31, 47).
Through analysis of mouse embryos carrying a RARE-lacZ RA reporter transgene in which β-galactosidase activity marks RA activity (41), it has been observed that a peculiar dorsoventral pattern of RA activity is established in the developing mouse eye that may play a role in retina development (49). At embryonic day 8.75 (E8.75), RA activity is localized in the optic vesicle neuroepithelium (where Raldh2 is expressed) as well as in the surface ectoderm adjacent to the optic vesicle that will later undergo lens induction (where Raldh3 is expressed) (30). The presence of Raldh2 mRNA and protein in the optic vesicle is very transient, lasting only until E9.0 (14, 49), but Raldh3 expression in the surface ectoderm persists, and Raldh1 and Raldh3 begin to be expressed in the retina from E9.5 onwards in distinct dorsal or ventral domains (12, 20, 31, 47). By E13.5, a tripartite organization of RA activity within the eye is established, with both dorsal and ventral retina having distinct RA activity domains separated by a central retinal zone having no RA activity (49). It has been shown that RA in the neural retina initially accumulates dorsally and then builds up ventrally (46), with the central zone likely being the result of Cyp26a1 (encoding an RA-degrading P450 enzyme) expressed in just that location (10, 28). In RARE-lacZ embryos from E13.5 to E18.5, β-galactosidase activity is observed in the axonal projections from the dorsal and ventral retinal ganglion cells to the diencephalon but not in the central axons derived from cells expressing Cyp26a1, leaving a central zone in the diencephalon without β-galactosidase activity.
The role of this extraordinary pattern of RA distribution in eye development remains to be solved. The dorsoventral stripes of RA activity could be involved in regulating eye dorsoventral polarity, eye growth, retinal neuron differentiation, and/or retinal ganglion cell axonal pathfinding to establish connections with the visual center of the brain. This is being approached genetically by analyzing mouse embryos lacking various RA-generating enzymes. Targeted disruption of Raldh2 prevents embryonic growth beyond E8.5 and results in an elimination of RA synthesis in the optic vesicle neuroepithelium, although RA activity and Raldh3 expression still remain in the eye field surface ectoderm, which gives rise to the lens (30). Raldh2−/− embryos develop optic vesicles with normal dorsoventral polarity, suggesting that RA is not needed to establish the dorsoventral axis (30). Raldh2−/− embryos are rescued by maternal administration of RA from E7.25 to E8.25, which evidently replaces the early function of Raldh2 in both the trunk mesoderm and optic vesicle, allowing embryos to undergo relatively normal development to E10.5, including normal expression of Raldh1 and Raldh3 in the optic vesicle plus normal RA activity in the eye as monitored by RARE-lacZ expression (30). Thus, Raldh2 function may be needed early in the optic vesicle to establish expression of Raldh1 and Raldh3 in the retina, which then directs local RA synthesis from E9.5 to E13.5, when the dorsoventral stripes of RA activity are established. In order to further dissect the mechanism of retinoid signaling in the eye, we now report the effect of a loss of Raldh1 on retina development.
MATERIALS AND METHODS
Generation of Raldh1 null mutant mice.
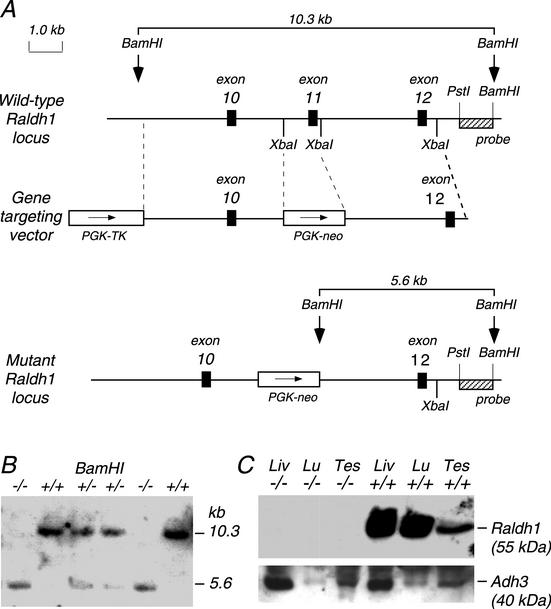
The methodology for production of null mutant mice has been described previously (17, 30). Briefly, a mouse Raldh1 cDNA (16) was used as a probe to isolate an 11-kb Raldh1 genomic clone containing exons 10 to 12 derived from a mouse 129/SvJ genomic library prepared in lambda FIX-II (Stratagene) by using conventional procedures (42). This DNA was used to construct a gene-targeting vector in which exon 11 was deleted, being replaced by a phosphoglycerate kinase-neomycin-positive selectable marker in the same transcriptional orientation as Raldh1; a phosphoglycerate kinase-thymidine kinase gene was also included as a negative selectable marker (Fig. 1). The upstream homology consisted of a 4.1-kb SalI-XbaI fragment including exon 10, and the downstream homology was a 3.6-kb XbaI fragment containing exon 12. The Raldh1 gene targeting vector was introduced by electroporation into mouse embryonic stem (ES) cells (R1 cells from strain 129/Sv) and subjected to positive selection with G418 and negative selection with ganciclovir. We identified two targeted ES cell lines (clones 147 and 168) by Southern blot analysis with a 3′ probe external to the targeting vector (1-kb PstI-BamHI fragment within intron 12) that detected both a wild-type 10.3-kb BamHI fragment and a mutant 5.6-kb BamHI fragment in which exon 11 was deleted. Both clones were microinjected into C57BL/6 blastocysts, and chimeric mice for each clone were produced. Matings of chimeric mice with C57BL/6 mice produced Raldh1+/− heterozygous mutant mice from each clone which were then mated to generate homozygous mice. As each clone resulted in the same phenotype, all further experiments were performed with clone 168. All genotyping was performed by Southern blotting.
FIG. 1.
Raldh1 gene targeting. (A) The wild-type Raldh1 gene contains 13 exons, and gene targeting with the replacement vector shown creates a mutant Raldh1 locus in which exon 11 has been deleted. (B) Southern blot showing the genotypes of mice obtained from heterozygous matings. (C) Western blot showing a complete lack of Raldh1 protein detection in Raldh1−/− adult liver (Liv), lung (Lu), and testis (Tes), whereas Raldh1 protein is detected in all three tissues of wild-type mice; as a control, Adh3 protein is detected in both Raldh1−/− and wild-type tissues.
In situ hybridization and immunohistochemistry.
Mouse embryos were subjected to whole-mount in situ hybridization as described previously, using digoxigenin-labeled antisense RNA probes (30). Antisense RNA probes were prepared from a cDNA carrying mouse Tbx5 (3) and from an expressed sequence tag (EST) carrying a portion of mouse Aldh8a1 (GenBank accession number BF781998) (Invitrogen, Grand Island, N.Y.), an ortholog of human ALDH8A1 (ALDH12) (21).
Whole-mount immunohistochemistry was performed as described previously, using antibodies generated against full-length mouse Raldh1 protein (14) and Adh3 protein (13). Also, an affinity-purified polyclonal antibody against mouse Raldh3 was generated by using protein expressed from a mouse cDNA previously described (31). Briefly, Raldh3 was expressed in Escherichia coli as a glutathione S-transferase fusion protein by using a portion of the mouse Raldh3 cDNA containing codons 85 to 512 inserted into the pGEX-4T-2 plasmid vector (Pharmacia Biotech, Uppsala, Sweden). Rabbit polyclonal antiserum was generated from four monthly injections of 200 μg of Raldh3 protein. The antiserum was affinity purified by using the cognate mouse Raldh3-glutathione S-transferase fusion protein attached to polyvinylidene fluoride membrane as described in our previous studies (5, 14). Western blotting was performed as previously described (14).
Detection of embryonic RA activity.
RA activity was detected in situ in embryos carrying the RAREhsplacZ (RARE-lacZ) transgene, in which lacZ is controlled by an RA response element (RARE) (41). Raldh1−/− mice were mated to RARE-lacZ mice to generate Raldh1−/− male mice carrying one copy of RARE-lacZ identified by Southern blot analysis. Embryos used for RARE-lacZ analysis were derived from matings of Raldh1−/− females and Raldh1−/−:RARE-lacZ−/+ males, to ensure that all embryos with RARE-lacZ expression contained no more than one copy of RARE-lacZ. Embryos were stained for 6 h for lacZ expression (β-galactosidase activity) with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as the substrate (15). Stained E16.5 eyes were embedded in paraffin and sectioned at 20 μm.
Morphological analysis of the retina.
After enucleation, eyes were immersed overnight in fixative composed of 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) at 4°C, followed by cryoprotection by soaking in 20% sucrose overnight at 4°C. The eyes were then frozen in Tissue-Tek O.C.T. compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan) on dry ice, and 8-μm-thick cryostat sections were cut, thaw mounted onto glass slides coated with poly-l-lysine, and air dried overnight at 4°C. Sections were stained with hematoxylin and eosin and subjected to morphological analysis as described previously (18).
Retrograde labeling of retinal ganglion cells.
To specifically label and identify retinal ganglion cells, these cells were retrogradely labeled via injection into their projection sites, the superior colliculi, using a modification of a method that we have previously described (18). Mice were anesthetized with 1% isoflurane and 70% N2O for all experimental manipulations and placed in a small stereotactic instrument. The bregma was identified and marked, and small windows were drilled over each hemisphere, leaving the dura intact. By using a stereotactic measuring device and a Hamilton injector, 5% FluoroGold (aminostilbamidine; Molecular Probes, Eugene, Oreg.) was injected into the superior colliculi on both sides (coordinates for the injection were 2.95 mm caudal from the bregma, 1 mm lateral from the superior sagittal suture, and 2 mm deep from the surface of the brain). Four days after injection of FluoroGold, mice were euthanatized with an overdose of pentobarbital, and the eyes were removed. Eyecups were prepared by removing the anterior segments in PBS solution and fixed in 4% paraformaldehyde for 20 min. The retinas were then carefully dissected from the eyes, prepared as a flat whole mount in PBS solution, mounted on glass slides, and examined by epifluorescence microscopy to visualize the retinal ganglion cells.
ERG.
Electroretinography (ERG) was performed similarly to that previously reported (44). Wild-type and Raldh1−/− mice were dark adapted overnight before each experiment, and ensuing procedures were performed under dim red light. Each mouse was anesthetized with a mixture of 60 mg of ketamine per kg and 20 mg of xylazine per kg administered intraperitoneally. The right pupil was dilated with 2.5% phenylephrine hydrochloride and 0.5% tropicamide eye drops (Wilson Ophthalmic Corp., Mustang, Okla.). In order to maintain body temperature at 37°C, a rectal thermometer (Barnant, Barrington, Ill.) was inserted and the mouse was placed on a variable heating block centered at the edge of a Ganzfeld dome. A ground electrode was inserted subcutaneously in the right leg; a needle placed subcutaneously in the forehead served as the reference electrode. EyeGel (hydroxypropyl methylcellulose; Eye Supply USA, Inc., Tampa, Fla.) was placed on the right eye to improve conduction. A saline-moistened cotton wick contacted the cornea as the recording electrode.
ERG recordings were performed in a full-field Ganzfeld dome with an 11-V light bulb. The light stimulus was obtained from a Grass PS33 stroboscope (Astro-Med, Inc., West Warwick, R.I.) placed 8 cm in front of the eye and subtended 20° of visual angle. The ERG responses were amplified and averaged by a PowerLab data acquisition system (ADInstruments, Grand Junction, Colo.).
Data collection consisted of three consecutive phases: three sensitivity curves in the dark, a 5-min intense photobleach, and recovery of dark adaptation. The sensitivity curve consisted of five flashes of increasing light intensity, which was repeated twice (2-min delay between the brightest and dimmest intensities). The flash duration was 10 μs. The intensity of stimulation (0.8 × 103 μW/cm2) was changed by the interposition of neutral density filters and/or by varying the flash intensity manually. After the sensitivity curves were performed, an adapting light (500 cd/m2) was turned on for 5 min (photobleach period). Recordings were continued at 1-Hz intervals. When the adapting light was turned off, recordings were collected every minute (interstimulus interval of 20 s, mean of three flashes) for at least 25 min after the photobleach.
High-pressure liquid chromatography (HPLC) quantitation of retinoids in adult mouse liver.
all-trans-RA, all-trans-retinol, and retinyl esters in wild-type and Raldh1−/− liver from untreated mice and mice treated with a dose of all-trans-retinol were quantitated. The mice examined were age- and weight-matched adult males. For retinol treatment, all-trans-retinol (Sigma) was dissolved in acetone-Tween 20-water (0.25:5:4.75, vol/vol), and a dose of 50 mg/kg was injected orally as described previously to measure metabolism of retinol to RA (4). After 2 h, liver was collected and subjected to retinoid analysis. For untreated and retinol-treated liver samples, all extraction and analytical procedures were carried out in a darkened room to protect retinoids from exposure to light.
Liver (0.5 g) was homogenized in 1 ml of PBS (0.01 M) (pH 7.4), and 150 μl (10%) was removed for analysis of retinyl esters (see below). To the remaining homogenate (used for analysis of all-trans-RA and all-trans-retinol), 1.5 ml of methanol was added and mixed by vortexing. This mixture was extracted twice with 5 ml of hexane. The hexane layers were collected, combined, and evaporated under vacuum. The residue was dissolved in 150 μl of HPLC mobile phase (see below), and a portion of this was injected into the HPLC system.
Retinyl esters were quantitated as previously described (43), using liver samples subjected to saponification to convert retinyl esters to retinol. Briefly, 150 μl of the liver homogenate described above (representing 0.05 g of liver) was mixed with 1.5 ml of a freshly prepared solution composed of 5% potassium hydroxide, 94% ethanol, and 1% pyrogallol and then incubated at 65°C for 15 min. The saponified liver sample was then mixed with 0.6 ml of water and extracted twice with 5 ml hexane, the hexane layers were combined and evaporated, and the residue was dissolved in 150 μl of HPLC mobile phase. The liver retinyl ester content was obtained from the difference between the total retinol measurement (from saponified liver) and the unesterified retinol measurement (from nonsaponified liver).
Reversed-phase HPLC analysis was performed on a Waters 2695 HPLC system with a SUPLEX pkB-100 analytical column (250 by 4.6 mm) (Supelco) at a flow rate of 1 ml/min and a column temperature of 35°C. The mobile phase consisted of 2% ammonium acetate, glacial acetic acid, acetonitrile, and methanol (16:3:79:2). Detection of retinoids was performed with a photodiode array detector (Waters model 2996) which collected spectra at between 200 and 450 nm. Standard solutions of retinoids (all-trans-retinol and all-trans-RA; Sigma) were used to obtain the calibration curves. Characteristic peak spectra and retention times were used to identify each retinoid, and quantitation of peak areas was calculated at λmax with Waters Millennium Chromatography Manager software. The detection limit for the all-trans-RA standard was 0.2 ng per sample, thus allowing identification and quantitation of all-trans-RA in untreated liver.
RESULTS
Growth and survival of Raldh1−/− mice.
The gene-targeting strategy for production of Raldh1 null mutant mice is shown in Fig. 1A. Mouse Raldh1 (Aldh1a1) consists of 13 exons spanning about 45 kb and encodes a mature protein of 500 amino acids in length (16). The deletion of exon 11 generated here is predicted to eliminate Raldh1 sequence from exon 11 to 13 (residues 400 to 500). If mRNA is produced by aberrant RNA splicing of exon 10 to 12, this would create a frameshift mutation at residue 400 and a stop codon soon after, eliminating Raldh1 sequence downstream of residue 400. ALDH crystallographic studies indicate that deletion of residues 400 to 500 will lead to a null phenotype, as this region contains a portion of the substrate-binding pocket and the tetramerization domain which is essential for enzyme function and stability (35, 45).
Raldh1+/− (clone 168) mutant mice were mated, and offspring from nine litters resulted in 88 offspring with the following genotypes as determined by Southern blotting: 14 Raldh1−/− (16%), 49 Raldh1+/− (56%), and 25 Raldh1+/+ (28%). An example of the Southern blot genotyping is shown Fig. 1B. Matings of Raldh1+/− (clone 147) mice gave 29 mice in three litters with the following genotypes: 8 Raldh1−/− (28%), 14 Raldh1+/− (48%), and 7 Raldh1+/+ (24%). The presence of a large number of surviving homozygous mice at close to the expected Mendelian ratio of 25% derived from two independent mutant ES cell lines indicates that the Raldh1 mutation does not lead to a lethal phenotype. Mating of male and female homozygous Raldh1 mutant mice resulted in the production of litters of normal size (∼8 to 10 pups) with no apparent gross abnormalities or defects in survival or growth, indicating that Raldh1−/− mice are relatively healthy and fertile.
Western blot analysis demonstrated a total lack of Raldh1 protein in Raldh1−/− adult liver, lung, and testis, whereas the protein was apparent in wild-type mice (Fig. 1C). As a control, another enzyme, Adh3, was detected in both mutant and wild-type tissues (Fig. 1C). We also used immunohistochemistry to demonstrate a lack of Raldh1 detection in Raldh1−/− newborn retina (data not shown) and embryonic retina (see Fig. 3E and F). This confirms that an Raldh1 null mutant was generated. Evidently, a lack of the Raldh1 tetramerization domain may have destabilized any truncated protein produced, leading to very quick turnover.
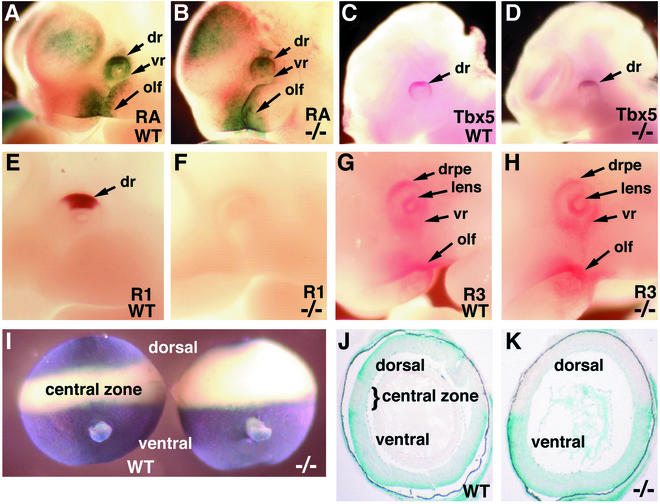
FIG. 3.
Effect of Raldh1−/− genotype on the embryonic dorsal retina. (A and B) RA detection in E10.5 wild-type (WT) and Raldh1−/− embryos carrying the RARE-lacZ transgene. (C and D) Tbx5 mRNA observed at E10.5 by whole-mount in situ hybridization. (E to G) Raldh1 protein (E and F) and Raldh3 protein (G and H), both detected at E10.5 by whole-mount immunohistochemistry. (I) RA detection in E16.5 dissected whole eyes by using the RARE-lacZ transgene (shown from a posterior view with optic nerve observed in the ventral portion). (J and K) Frontal sections through the eyes shown in panel I. dr, dorsal retina; drpe, dorsal RPE; lens, lens vesicle; olf, olfactory region; R1, Raldh1 protein; R3, Raldh3 protein; vr, ventral retina.
Retinal morphology in Raldh1−/− mice.
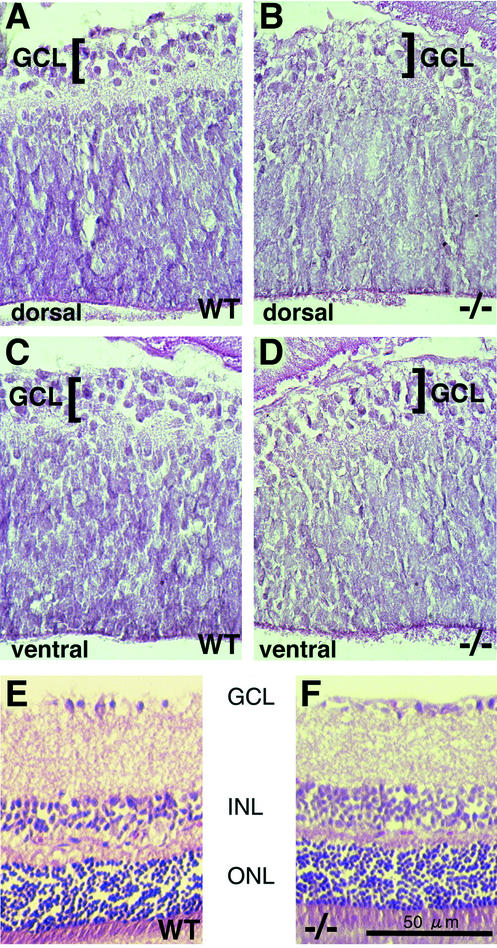
At the newborn stage, the mouse retina has a defined ganglion cell layer, but development of other layers is still under way. There were no significant morphological differences in the retinas of wild-type and Raldh1−/− newborn mice from the dorsal (Fig. 2A and B) or ventral (Fig. 2C and D) regions, either in terms of retinal thickness or with regard to the presence of a ganglion cell layer. Also, no morphological difference was observed in the adult dorsal retina, where staining of sections demonstrated that the Raldh1−/− retina developed normally, displaying the expected lamination (ganglion cell layer, inner nuclear layer, and outer nuclear layer) with the same degree of thickness for each layer as that of wild-type mice (Fig. 2E and F).
FIG. 2.
Analysis of retinal morphology in newborn and adult Raldh1−/− mice. Hematoxylin-eosin staining is shown for retinal sections of wild-type (WT) newborn dorsal retina (A), Raldh1−/− newborn dorsal retina (B), WT newborn ventral retina (C), and Raldh1−/− newborn ventral retina (D). Also, shown is hematoxylin-eosin staining of adult dorsal retina sections for WT (E) and Raldh1−/− (F) mice. Bar, 50 μm (E and F). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer.
RA activity and specification of dorsal retina.
Raldh1−/− embryos were examined to determine if a defect in retinoid signaling was apparent in the developing dorsal retina. Embryos carrying the RARE-lacZ RA reporter transgene (41) were used to localize RA signaling activity in and around the developing eye. Treatment of RARE-lacZ embryos with RA has previously been shown to upregulate RARE-lacZ expression in all cell types, but normally only a subset of cells express the transgene, including the eye, presumably due to local embryonic RA synthesis (41). β-Galactosidase activity detected in embryos carrying RARE-lacZ is dependent upon a combination of local RA synthesis, degradation, and diffusion, and conclusions obtained are limited by the concentration threshold for transgene activation and the half-life of β-galactosidase, which is several hours longer than that of RA (30). Despite these stipulations, recent studies on Raldh2−/− embryos carrying RARE-lacZ have demonstrated that this transgene is a precise marker for the presence of RA, as it was demonstrated that loss of Raldh2 function resulted in selective elimination of RARE-lacZ expression in cells where Raldh2 is normally expressed, i.e., paraxial, somitic, and lateral plate mesoderm as well as the optic pit neuroepithelium at E8.5 (30).
In E10.5 wild-type embryos, RARE-lacZ expression was detected at high levels in the dorsal retina and at lower levels in the ventral retina (Fig. 3A). E10.5 Raldh1−/− embryos maintained this pattern of RARE-lacZ expression in the eye, with no significant difference from wild-type levels being observed (Fig. 3B). This presumably indicates the existence of another source of RA synthesis for the dorsal eye in addition to Raldh1. We examined the dorsal retina for expression of the dorsal-specific marker Tbx5. Raldh1−/− embryos were found to express Tbx5 in the dorsal retina similarly to wild-type embryos (Fig. 3C and D). Thus, the dorsal retina appears to have been specified in the absence of Raldh1 function.
RA-generating enzymes in the Raldh1−/− eye.
As the dorsal retina of Raldh1−/− embryos did not show a significant reduction in RA activity, we examined E10.5 embryos for the presence of Raldh proteins in the eye by using whole-mount immunohistochemistry. We have previously shown that Raldh2 protein is undetectable in the wild-type eye at E10.5, although it is easily observed at E8.5 (14). We observed that Raldh2 protein was also undetectable in the eyes of E10.5 Raldh1−/− embryos (data not shown), thus indicating that Raldh2 activity was not upregulated to compensate for a lack of Raldh1. Raldh1 protein was abundant in the dorsal neural retina of wild-type E10.5 embryos but was completely absent in Raldh1−/− embryos, thus validating the effectiveness of the gene disruption (Fig. 3E and F). Raldh3 protein was localized at E10.5 in the dorsal RPE and lens vesicle, weakly in the ventral retina, and strongly in the olfactory pit, with no significant difference being observed between wild-type and Raldh1−/− embryos (Fig. 3G and H). This indicates that Raldh3 activity is not significantly upregulated or downregulated in the eye when Raldh1 is missing. We did not detect Raldh3 protein in the dorsal neural retina, consistent with previous studies showing that Raldh3 mRNA is expressed in the dorsal RPE but not the dorsal neural retina of E10.5 mouse embryos (20, 31). Thus, Raldh3 in the dorsal RPE could be responsible for RARE-lacZ expression remaining dorsally in the Raldh1−/− embryonic eye (Fig. 3B).
It cannot be ruled out that another RA-generating enzyme is responsible for RARE-lacZ expression in the E10.5 eyes of Raldh1−/− embryos. ALDH8A1 (also known as ALDH12) encodes another RA-generating enzyme identified in humans and rats (21). We identified an EST that encodes a mouse ortholog (Aldh8a1) sharing 87% nucleotide sequence identity with human ALDH8A1 (GenBank accession number BF781998). However, examination of mouse embryos by in situ hybridization with this EST revealed no expression of Aldh8a1 in the eye or any other tissues at E10.5 (data not shown).
RA activity in the eye at E16.5.
At E10.5, as shown above, RA generation in the eye is just getting under way, and the tripartite dorsoventral pattern of RA activity previously detected in the retina from E13.5 to birth by using RARE-lacZ has not yet been established (49). We examined wild-type retinas in E16.5 embryos carrying RARE-lacZ and observed the characteristic well-defined dorsoventral stripes of RA activity, i.e., a ventral stripe encompassing the entire ventral half of the retina, terminating just dorsal to the optic nerve, and a dorsal stripe encompassing only the upper quarter of the retina, with the intervening central zone devoid of RA activity (Fig. 3I). In Raldh1−/− embryos, the dorsal stripe of RA was missing, but the ventral stripe and its border were unaffected (Fig. 3I). Raldh1−/− eyes at E16.5 did not appear to be significantly different in size from wild-type eyes. Sections through the eyes confirmed that wild-type eyes expressed RARE-lacZ in both the dorsal and ventral retina but not the central zone, whereas Raldh1−/− eyes expressed RARE-lacZ only in the ventral retina (Fig. 3J and K). These findings suggest that the Raldh1−/− mutation does indeed eliminate RA synthesis in the dorsal retina by E16.5. As previous studies have shown that Raldh3 mRNA is expressed in the dorsal RPE only until E12.5 (20), there is no alternate source of RA for the dorsal retina at E16.5 (unlike the situation at E10.5 described above).
Retinal ganglion cell axonal projections.
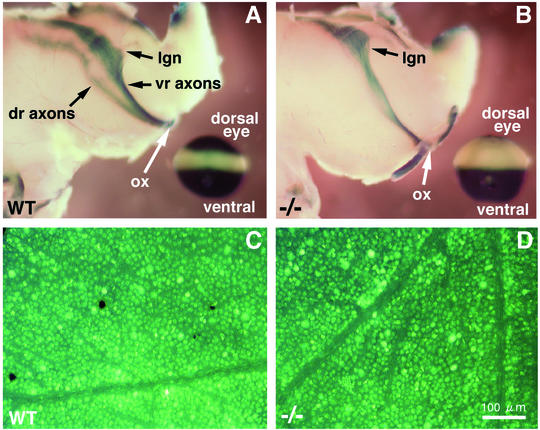
RARE-lacZ expression in the cell bodies of dorsal and ventral retinal ganglion cells gives rise to β-galactosidase, which has previously been detected in embryonic dorsal and ventral axons projecting into the brain (superior colliculus), with axons from the central region of the retina being devoid of β-galactosidase (49). Thus, β-galactosidase is a good marker to monitor growth of dorsal and ventral axons but does not necessarily indicate that RA action is occurring in the axons. In wild-type embryonic brain at E16.5 we also observed β-galactosidase activity in the dorsal and ventral retina axon tracts but not in the central retinal axons (Fig. 4A). Upon analysis of Raldh1−/− embryos at E16.5, ventral retinal axons contained β-galactosidase activity in the brain as normal, but there was a complete absence of β-galactosidase in the region where the dorsal retinal axons should be located (Fig. 4B). This correlates with a lack of β-galactosidase detection in the E16.5 Raldh1−/− dorsal retina cell bodies (Fig. 3I and K). This suggests that Raldh1 expression in dorsal retinal ganglion cell bodies leads to RA generation that induces β-galactosidase, which is then transported down the dorsal axons as they grow into the brain. Thus, the brain itself is not generating RA along the axon growth pathway. These results provide further evidence that Raldh1 indeed functions to produce RA in the dorsal retina.
FIG. 4.
Investigation of retinal ganglion cell axonal projections to the brain in Raldh1−/− mice. (A and B) RARE-lacZ expression in retinal ganglion cell axonal projections into the embryonic brain at E16.5 in wild-type (WT) (A) and Raldh1−/− (B) embryos; in the lower left corner a RARE-lacZ-stained eye from the same embryo is shown for orientation. It should be stressed that when the dorsal and ventral retina axon tracts pass beyond the optic chiasm, they innervate the spatially reversed portions of the brain (ventral-lower and dorsal-upper regions, respectively). (C and D) For adult mice, retrograde labeling of retinal ganglion cells is shown for the WT (C) and Raldh1−/− (D) genotypes. Bar, 100 μm (C and D). dr, dorsal retina; lgn, lateral geniculate nucleus; ox, optic chiasm; vr, ventral retina.
As Raldh1−/− dorsal retinal axons were not labeled with β-galactosidase in embryonic brain at E16.5, we tested adult Raldh1−/− mice to determine if dorsal axons had indeed innervated the brain. Retinal ganglion cells were labeled in a retrograde fashion with FluoroGold administered to the adult brain. Both wild-type and Raldh1−/− mice displayed a characteristically fine-dotted pattern of fluorescence in the perikarya of the dorsal retina, where Raldh1 is normally expressed, indicating that dorsal retinal axons had indeed reached the brain in the absence of Raldh1 function (Fig. 4C and D). Labeled retinal ganglion cells were observed throughout the dorsoventral axis of the adult retina indicating that a lack of RA activity in the dorsal retina did not block growth of dorsal, central, or ventral axons to targets in the brain (data not shown).
Examination of visual function by electroretinography.
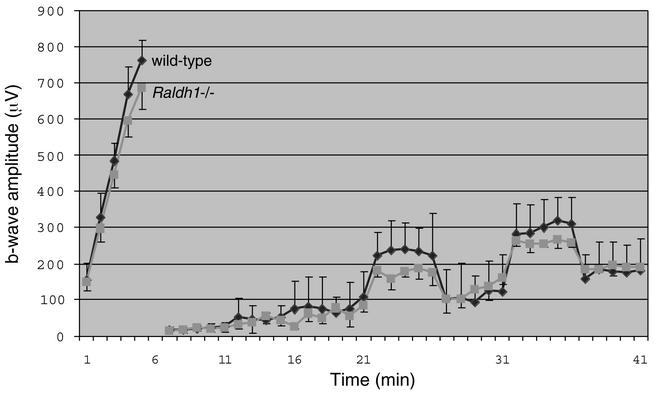
In the adult retina, RA has previously been proposed to have light-adaptive effects that may modulate synaptic transmission of visual information (50, 53). As Raldh1 protein normally persists in the adult dorsal retina (27), ERG was employed to examine the effect of a loss of Raldh1 on adult visual function. There was no evidence of retinal degeneration in Raldh1−/− mice compared to wild-type mice, as shown by data which indicated similar maximum ERG b-wave amplitudes during the initial dark-adapted period (Fig. 5, data on left). With regard to recovery of dark adaptation after a photobleach, the ERG results demonstrated that Raldh1−/− mice displayed a rate of recovery of dark adaptation similar to that of wild-type mice (Fig. 5, data on right). These findings indicate that a loss of Raldh1 does not significantly affect the rate of visual signal transduction.
FIG. 5.
Visual function assessed by ERG. Female wild-type mice (n = 5) and Raldh1−/− mice (n = 4) at approximately 210 days of age were examined. No statistically significant difference was observed between the two genotypes. Error bars indicate standard deviations.
Defective metabolism of retinol to RA in Raldh1−/− mice.
The lack of RARE-lacZ expression observed in the dorsal eye of E16.5 Raldh1−/− embryos suggests that Raldh1 is normally catalyzing RA synthesis in that location. Therefore, we examined adult Raldh1−/− mice to determine if they have reduced metabolism of retinol to RA in liver, a tissue which contains high levels of Raldh1 protein in wild-type mice but in which no Raldh1 protein is detected in Raldh1−/− mice (Fig. 1C). all-trans-RA, all-trans-retinol, and retinyl esters were quantitated by HPLC in wild-type and Raldh1−/− adult liver, either in untreated mice (steady-state levels) or 2 h after treatment of mice with a dose of all-trans-retinol (Fig. 6; Table 1).
FIG. 6.
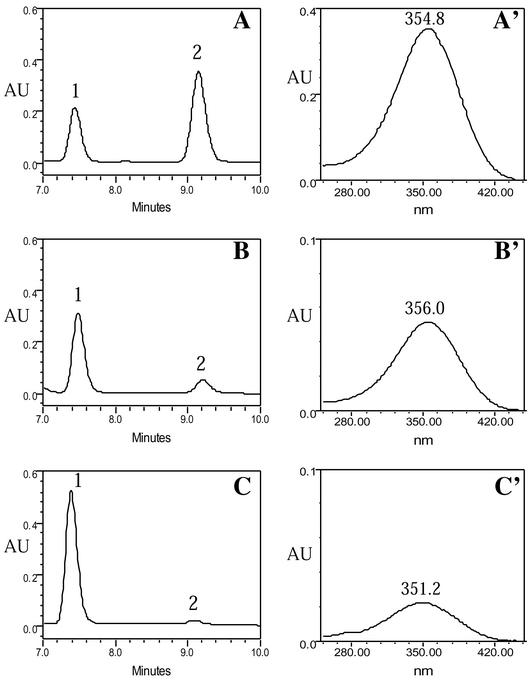
Effect of Raldh1 genotype on retinoid levels following a dose of retinol. all-trans-RA and all-trans-retinol were examined by HPLC in liver 2 h after a 50-mg/kg oral dose of all-trans-retinol. Shown are HPLC chromatograms at 355 nm (A to C) and, for each chromatogram, the corresponding spectra of peak 2 showing λmax (A′ to C′). (A) Characteristic retention times for 2-μg standard samples of all-trans-retinol (peak 1) and all-trans-RA (peak 2). (B and C) Wild-type liver (B) and Raldh1−/− liver (C) following retinol treatment (in both cases, 25-μl samples representing 75 mg of liver were loaded). The spectrum of all-trans-RA (peak 2) in the standards (A′) is very similar to the spectra of peak 2 in wild-type liver (B′) and Raldh1−/− liver (C′), thus verifying peak 2 as all-trans-RA in the liver samples. Also, the spectrum for all-trans-retinol (peak 1) in the standards matched the spectra for peak 1 in the liver samples (not shown). AU, absorbance units.
TABLE 1.
Quantitation of retinoids in livera
| Group | Genotype | all-trans-RA (ng/g) | all-trans-Retinol (μg/g) | Retinyl esters (μg/g) |
|---|---|---|---|---|
| Untreated | Wild type | 1.5 ± 0.2 | 8.5 ± 1.0 | 571 ± 59 |
| Raldh1−/− | 1.6 ± 0.3 | 14.4 ± 1.22b | 1,278 ± 68c | |
| Retinol treated | Wild type | 3,100 ± 480 | 19.5 ± 0.2 | 570 ± 10 |
| Raldh1−/− | 990 ± 60b | 36.2 ± 3.2c | 1,244 ± 57c |
Liver from mice treated with retinol (50 mg/kg) was examined 2 h after oral dosing. All values are means ± standard errors. (n = 4).
P < 0.05 (Student's t test).
P < 0.01 (Student's t test).
In untreated wild-type liver, all-trans-RA was observed at 1.5 ± 0.2 ng/g, whereas in untreated Raldh1−/− liver, all-trans-RA was essentially unchanged at 1.6 ± 0.3 ng/g. However, all-trans-retinol was increased 1.7-fold in untreated Raldh1−/− liver, and the retinyl ester content was increased 2.2-fold (Table 1). The clearly observed increases in liver all-trans-retinol and retinyl esters suggest the possibility that conversion of retinol to RA may be reduced in Raldh1−/− liver, resulting in more retinol being available to be converted to retinyl esters for storage. Thus, even though no change in the steady-state level of RA was observed in Raldh1−/− liver, steady-state RA levels are nearly 10,000-fold lower than steady-state retinol levels, and there could still be a function for Raldh1 in the dynamic control of liver RA production following consumption of a diet containing retinol.
To examine more directly the potential participation of Raldh1 in metabolism of retinol to RA, Raldh1−/− and wild-type mice were given a 50-mg/kg oral dose of all-trans-retinol, and retinoids in liver were quantitated 2 h after treatment. In treated wild-type liver, all-trans-RA was observed at 3,100 ± 480 ng/g, but in treated Raldh1−/− liver, the level of all-trans-RA was only 990 ± 60 ng/g (a 3.1-fold reduction) (Fig. 6; Table 1). Thus, for both Raldh1−/− and wild-type liver, retinol treatment results in a large increase in liver all-trans-RA compared to what is observed in untreated liver, but the increase observed in Raldh1−/− liver is significantly smaller. Liver all-trans-retinol levels following retinol treatment were increased in both Raldh1−/− and wild-type mice, but Raldh1−/− liver contained 1.9-fold-higher levels of all-trans-retinol than wild-type liver, suggesting reduced metabolism of the retinol dose (Table 1). Liver retinyl ester levels were not significantly changed following retinol treatment, with Raldh1−/− mice still having 2.2-fold higher levels than wild-type mice, similar to the results observed for untreated mice (Table 1).
These findings indicate that Raldh1 plays a major role in dynamic metabolism of a dose of all-trans-retinol to all-trans-RA, accounting for nearly 70% of the all-trans-RA produced in the liver following an oral dose of retinol. This provides evidence of an in vivo function for Raldh1 in metabolism of retinaldehyde to RA. As some RA is still generated in the absence of Raldh1, multiple enzymes for metabolism of retinaldehyde to RA exist in the adult mouse.
DISCUSSION
A function for RA in retina development is inferred from studies on rats subjected to gestational vitamin A deficiency (6, 51) as well as mice harboring null mutations of two of the three RARs (23, 29). In these studies, microphthalmia is a consistent observation, with greatly reduced growth of the retina both dorsally and ventrally. Thus, metabolism of retinol to RA appears to be required for growth of the retina. The RA-generating enzymes Raldh1, Raldh2, and Raldh3 are all present in the developing optic neuroepithelium at some point (20, 31), suggesting that all three may play a role in RA signaling needed for retina development. Thus, it is surprising to see here that even though Raldh1 is expressed at high levels in the dorsal retina, Raldh1−/− mice do not have an obvious defect in retina development. As our findings in adult liver have demonstrated that Raldh1 indeed catalyzes RA synthesis in vivo, our observation of a lack of RARE-lacZ expression in the dorsal eyes of E16.5 Raldh1−/− embryos can be attributed to a lack of local RA synthesis normally catalyzed by Raldh1. The apparently normal dorsal retinal ganglion cell axon outgrowth observed in Raldh1−/− embryos at E16.5 when the dorsal retina is devoid of RA, plus normal dorsal retina lamination observed postnatally, indicates that RA signaling must not be needed dorsally at these late stages for retina development. This suggests that dorsal RA signaling in the retina, if necessary, must occur at early stages during formation of the optic cup. Raldh3 expressed during early stages of retina development in the dorsal RPE (adjacent to the dorsal neural retina) as well as the ventral neural retina may provide sufficient RA for dorsal and ventral retina development at the optic cup stage. Alternatively, another RA-generating enzyme may exist for dorsal retina at early stages.
Development and function of the retina in the absence of Raldh1.
We did not observe any defects in optic cup morphology in E10.5 Raldh1−/− embryos, and the dorsal retina-specific marker Tbx5 was expressed as normal, indicating no defect in dorsoventral specification. At the E16.5 and newborn stages, as well as in the adult, no defects in retinal morphology were observed either dorsally, where Raldh1 is expressed, or ventrally We found that Raldh1 is solely responsible for production of RA during the late stages of dorsal eye development (E16.5), when no expression of Raldh3 is observed dorsally. However, loss of dorsal RA at E16.5 did not prevent normal retina development, suggesting that RA is no longer needed by this stage. Raldh1 protein normally persists in the adult dorsal retina (27), suggesting that it may play a role in the adult visual system. As it had been previously proposed that RA may have light-adaptive effects on adult retinal horizontal cells that modulate synaptic transmission of visual information (50, 53), we tested adult dark-adapted mice for visual function by ERG. These results indicate that Raldh1−/− mice do not have a defect in recovery after a photobleach. However, we cannot rule out other, more subtle defects in adult retina function due to loss of Raldh1 function.
Generation of RA in the embryonic retina.
The studies reported here indicate that Raldh1 protein is absent from the developing dorsal retina as well as adult liver, lung, and testis of Raldh1−/− mice. Consistent with that observation, we demonstrated that Raldh1−/− embryos lack expression of the RARE-lacZ RA reporter gene in the dorsal retina at E16.5. Also consistent was our demonstration that Raldh1−/− embryos lack β-galactosidase activity in the dorsal retinal axons as they innervate the brain at E16.5. Thus, our findings support the previous hypothesis that Raldh1 produces RA in the developing dorsal retina (49). However, at early stages of mouse eye development, Raldh1 is not the only RA-generating enzyme dorsally. At E10.5, the folding of the optic vesicle to produce the optic cup has generated two retinal layers lying in close contact, the prospective RPE and neural retina layers. Our studies of Raldh3 protein localization in the E10.5 embryo, as well as previous studies of Raldh3 mRNA localization (20), reveal that Raldh3 is present transiently in the dorsal portion of the RPE (from E9.5 to E12.5). As Raldh3 has 10-fold higher activity than Raldh1 for oxidation of retinaldehyde to RA (12), it can be proposed that RA produced by Raldh3 in the dorsal RPE may be sufficient to stimulate the observed RARE-lacZ expression observed in the dorsal eye of Raldh1−/− embryos.
The fact that at E16.5 we no longer observe RARE-lacZ expression in the Raldh1−/− dorsal retina is consistent with the observation that Raldh3 expression in the dorsal RPE terminates at E12.5 (20). Thus, at E16.5 there is no enzymatic source of RA for the Raldh1−/− dorsal retina, and the retina fails to maintain the dorsal domain of RA activity previously described (49). In E16.5 Raldh1−/− embryos, the ventral retina maintains its domain of RA activity and its sharp horizontal boundary just dorsal to the optic nerve, suggesting that Raldh3 expression ventrally is still producing RA and Cyp26a1 expression in the central domain is still degrading RA as normal. Thus, a loss of Raldh1 does not impair RA generation and degradation in the ventral and central retina.
Function of RA during eye development.
Our findings make it clear that E16.5 Raldh1−/− embryos have no RA detectable in the dorsal retina but that the retina appears to develop normally to the adult stage, thus arguing that RA is not needed after E16.5 for development of the various retinal cell layers. Also, our findings do not support the previous hypothesis that RA produced in the retinal axons during anterograde transport of Raldh1 may be necessary for retinal ganglion cell axonal outgrowth (49). Raldh1−/− mice carrying RARE-lacZ have no β-galactosidase detectable in the dorsal retinal ganglion cell axons as they innervate the brain, but innervation does occur as shown by retrograde labeling, indicating that RA signaling is not needed for dorsal axon growth. As central retinal axons never do have RA signaling (even in wild-type embryos) due to the RA-degrading enzyme Cyp26a1 and yet still reach their targets in the brain, RA signaling may not be needed in general for targeting of any retinal axons, unless ventral axons have a special RA signaling requirement. Although our retrograde labeling studies indicate that dorsal retinal axons do grow and reach the superior colliculus in the absence of Raldh1, our experiments do not rule out the possibility that a lack of Raldh1 changes the retinocollicular topography of dorsal axons in a subtle way.
As our studies suggest that RA signaling may not be needed for targeting of any retinal axons to the brain, the tripartite dorsoventral pattern of RA localization observed at E16.5 may simply be a by-product of the RA produced at the optic cup stage that was needed for proper development of the retina. Perhaps expression of Cyp26a1 centrally to remove some of the RA generated by either Raldh1 or Raldh3 is designed to allow the dorsal and ventral regions of the retina to grow at different rates than the central region at the optic cup stage. In Raldh1−/− embryos, RA signaling at the optic cup stage is relatively normal due to Raldh3 or some other enzyme. Thus, it is possible that the function of RA during retina development occurs during the early stages before the full tripartite dorsoventral pattern of RA localization is observed and that this function can be carried out in the absence of Raldh1. If Raldh3 is indeed sufficient for RA generation at the optic cup stage in mouse embryos, this may be specific for mammalian eye development, as it has been demonstrated that chick Raldh3 is not expressed in the dorsal RPE but is limited to the ventral neural retina (12, 47). Thus, it is possible that Raldh1 may be needed in chicken embryos for early development of the dorsal retina.
Zebrafish carrying an RA-responsive reporter gene also possess a tripartite organization of RA detection in the retina with separated dorsal and ventral stripes of RA activity (40). Thus, the dorsoventral pattern of RA activity in the retina is evolutionarily conserved, suggesting an important function. Although Raldh1 has not yet been described for zebrafish, Raldh1 has been found in the amphibian Xenopus, where it functions as an RA-generating enzyme (2) and where it is expressed specifically in the dorsal neural retina just like in chick and mouse (1). As fish and amphibian embryos develop externally, their visual systems must function at a relatively early age compared to avian or mammalian embryos. Thus, Raldh1 may have a function crucial to the early establishment of a functioning visual system in fish or amphibians, and this may not be important in avian or mammalian lines.
Extraocular functions of Raldh1.
It has been reported that Xenopus Raldh1 acts as a binding protein for thyroid hormone (52) and that human Raldh1 binds androgens (39), properties which may exist independent of its enzymatic activity. As we have demonstrated that our null mutant totally lacks Raldh1 protein in E10.5 embryos as well as adult liver, lung, and testis, there may be no residual protein remaining in any tissues that could provide such binding functions. Therefore, if mouse Raldh1 binds either thyroid hormone or androgens, our lack of a significant phenotype in null mutants (including normal reproductive function) suggests that these properties may not be physiologically significant for mice.
However, an alternative retinoid-related function for Raldh1 in the adult liver has been revealed by our studies showing defective metabolism of a dose of retinol to RA in Raldh1−/− mice, accompanied by abnormally high levels of retinol and retinyl esters in the liver. In order to avoid retinol toxicity when the diet is rich in vitamin A, mice metabolize excess retinol to RA primarily in the liver. This has been demonstrated in Adh1−/− mice (lacking alcohol dehydrogenase 1), which have a large reduction in metabolism of a dose of retinol to RA and much higher retinol-induced toxicity than wild-type mice (34). Thus, less toxicity is experienced during efficient conversion of excess retinol to RA as opposed to accumulation of retinol and disposition in any other fashion. Excess RA produced during retinol overdose is efficiently cleared by further oxidative metabolism or glucuronidation followed by excretion in the kidneys or bile (9). Adh1 is expressed highly in the adult liver (13), as is Raldh1 (14). Thus, it is reasonable to propose that one retinoid-related function of Raldh1 in the mouse is to participate with Adh1 in the two-step clearance of excess retinol in adult liver, i.e., oxidation of retinol to retinaldehyde by Adh1 followed by oxidation of retinaldehyde to RA by Raldh1.
Acknowledgments
We thank F. Mic for performing in situ hybridization analysis of Aldh8a1, J. Rossant for the RARE-lacZ RA reporter mouse, and V. Papaioannou for the mouse Tbx5 cDNA probe.
This work was supported by funds from NIH grants R01 EY13969 (to G.D), R01 EY05477 and R01 EY09024 (to S.A.L.), and R01 EY12858 (to W.S.B).
REFERENCES
- 1.Ang, H. L., and G. Duester. 1999. Retinoic acid biosynthetic enzyme ALDH1 localizes in a subset of retinoid-dependent tissues during Xenopus development. Dev. Dyn. 215:264-272. [DOI] [PubMed] [Google Scholar]
- 2.Ang, H. L., and G. Duester. 1999. Stimulation of premature retinoic acid synthesis in Xenopus embryos following premature expression of aldehyde dehydrogenase ALDH1. Eur. J. Biochem. 260:227-234. [DOI] [PubMed] [Google Scholar]
- 3.Chapman, D. L., N. Garvey, S. Hancock, M. Alexiou, S. I. Agulnik, J. J. Gibson-Brown, J. Cebra-Thomas, R. J. Bollag, L. M. Silver, and V. E. Papaioannou. 1996. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev. Dyn. 206:379-390. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D., C. Eckhoff, I. Chahoud, G. Bochert, and H. Nau. 1992. 4-Methylpyrazole partially ameliorated the teratogenicity of retinol and reduced the metabolic formation of all-trans-retinoic acid in the mouse. Arch. Toxicol. 66:652-659. [DOI] [PubMed] [Google Scholar]
- 5.Deltour, L., R. J. Haselbeck, H. L. Ang, and G. Duester. 1997. Localization of class I and class IV alcohol dehydrogenases in mouse testis and epididymis: potential retinol dehydrogenases for endogenous retinoic acid synthesis. Biol. Reprod. 56:102-109. [DOI] [PubMed] [Google Scholar]
- 6.Dickman, E. D., C. Thaller, and S. M. Smith. 1997. Temporally-regulated retinoic acid depletion produces specific neural crest, ocular and nervous system defects. Development 124:3111-3121. [DOI] [PubMed] [Google Scholar]
- 7.Dowling, J. E., and G. Wald. 1960. The biological function of vitamin A acid. Proc. Natl. Acad. Sci. USA 46:587-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duester, G. 2000. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur. J. Biochem. 267:4315-4324. [DOI] [PubMed] [Google Scholar]
- 9.Frolik, C. A. 1984. Metabolism of retinoids, p. 177-208. In M. B. Sporn, A. B. Roberts, and D. S. Goodman (ed.), The retinoids, vol. 2. Academic Press, Orlando, Fla.
- 10.Fujii, H., T. Sato, S. Kaneko, O. Gotoh, Y. Fujii-Kuriyama, K. Osawa, S. Kato, and H. Hamada. 1997. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 16:4163-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghyselinck, N. B., V. Dupé, A. Dierich, N. Messaddeq, J.-M. Garnier, C. Rochette-Egly, P. Chambon, and M. Mark. 1997. Role of the retinoic acid receptor beta (RARβ) during mouse development. Int. J. Dev. Biol. 41:425-447. [PubMed] [Google Scholar]
- 12.Grün, F., Y. Hirose, S. Kawauchi, T. Ogura, and K. Umesono. 2000. Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J. Biol. Chem. 275:41210-41218. [DOI] [PubMed] [Google Scholar]
- 13.Haselbeck, R. J., and G. Duester. 1997. Regional restriction of alcohol/retinol dehydrogenases along the mouse gastrointestinal epithelium. Alcohol. Clin. Exp. Res. 21:1484-1490. [PubMed] [Google Scholar]
- 14.Haselbeck, R. J., I. Hoffmann, and G. Duester. 1999. Distinct functions for Aldh1 and Raldh2 in the control of ligand production for embryonic retinoid signaling pathways. Dev. Genet. 25:353-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Hsu, L. C., W. C. Chang, I. Hoffmann, and G. Duester. 1999. Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem. J. 339:387-395. [PMC free article] [PubMed] [Google Scholar]
- 17.Joyner, A. L. 1993. Gene targeting: a practical approach. IRL Press, Oxford, United Kingdom.
- 18.Kikuchi, M., L. Tenneti, and S. A. Lipton. 2000. Role of p38 mitogen-activated protein kinase in axotomy-induced apoptosis of rat retinal ganglion cells. J. Neurosci. 20:5037-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapshina, E. A., O. V. Belyaeva, O. V. Chumakova, and N. Y. Kedishvili. 2003. Differential recognition of the free versus bound retinol by human microsomal retinol/sterol dehydrogenases: characterization of the Holo-CRBP dehydrogenase activity of RoDH-4. Biochemistry 42:776-784. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., E. Wagner, P. McCaffery, D. Smith, A. Andreadis, and U. C. Dräger. 2000. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech. Dev. 95:283-289. [DOI] [PubMed] [Google Scholar]
- 21.Lin, M., and J. L. Napoli. 2000. cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and identification of a rat ortholog, ALDH12. J. Biol. Chem. 275:40106-40112. [DOI] [PubMed] [Google Scholar]
- 22.Lohnes, D., P. Kastner, A. Dierich, M. Mark, M. LeMeur, and P. Chambon. 1993. Function of retinoic acid receptor gamma in the mouse. Cell 73:643-658. [DOI] [PubMed] [Google Scholar]
- 23.Lohnes, D., M. Mark, C. Mendelsohn, P. Dollé, A. Dierich, P. Gorry, A. Gansmuller, and P. Chambon. 1994. Function of the retinoic acid receptors (RARs) during development. I. Craniofacial and skeletal abnormalities in RAR double mutants. Development 120:2723-2748. [DOI] [PubMed] [Google Scholar]
- 24.Lufkin, T., D. Lohnes, M. Mark, A. Dierich, P. Gorry, M.-P. Gaub, M. LeMeur, and P. Chambon. 1993. High postnatal lethality and testis degeneration in retinoic acid receptor α mutant mice. Proc. Natl. Acad. Sci. USA 90:7225-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCaffery, P., M.-O. Lee, M. A. Wagner, N. E. Sladek, and U. C. Dräger. 1992. Asymmetrical retinoic acid synthesis in the dorsoventral axis of the retina. Development 115:371-382. [DOI] [PubMed] [Google Scholar]
- 26.McCaffery, P., K. C. Posch, J. L. Napoli, L. Gudas, and U. C. Dräger. 1993. Changing patterns of the retinoic acid system in the developing retina. Dev. Biol. 158:390-399. [DOI] [PubMed] [Google Scholar]
- 27.McCaffery, P., P. Tempst, G. Lara, and U. C. Dräger. 1991. Aldehyde dehydrogenase is a positional marker in the retina. Development 112:693-702. [DOI] [PubMed] [Google Scholar]
- 28.McCaffery, P., E. Wagner, J. O'Neil, M. Petkovich, and U. C. Dräger. 1999. Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech. Dev. 82:119-130. [DOI] [PubMed] [Google Scholar]
- 29.Mendelsohn, C., D. Lohnes, D. Décimo, T. Lufkin, M. LeMeur, P. Chambon, and M. Mark. 1994. Function of the retinoic acid receptors (RARs) during development. II. Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 120:2749-2771. [DOI] [PubMed] [Google Scholar]
- 30.Mic, F. A., R. J. Haselbeck, A. E. Cuenca, and G. Duester. 2002. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development 129:2271-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mic, F. A., A. Molotkov, X. Fan, A. E. Cuenca, and G. Duester. 2000. RALDH3, a retinaldehyde dehydrogenase that generates retinoic acid, is expressed in the ventral retina, otic vesicle and olfactory pit during mouse development. Mech. Dev. 97:227-230. [DOI] [PubMed] [Google Scholar]
- 32.Molotkov, A., L. Deltour, M. H. Foglio, A. E. Cuenca, and G. Duester. 2002. Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J. Biol. Chem. 277:13804-13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molotkov, A., X. Fan, L. Deltour, M. H. Foglio, S. Martras, J. Farrés, X. Parés, and G. Duester. 2002. Stimulation of retinoic acid production and growth by ubiquitously-expressed alcohol dehydrogenase Adh3. Proc. Natl. Acad. Sci. USA 99:5337-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molotkov, A., X. Fan, and G. Duester. 2002. Excessive vitamin A toxicity in mice genetically deficient in either alcohol dehydrogenase Adh1 or Adh3. Eur. J. Biochem. 269:2607-2612. [DOI] [PubMed] [Google Scholar]
- 35.Moore, S. A., H. M. Baker, T. J. Blythe, K. E. Kitson, T. M. Kitson, and E. N. Baker. 1998. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure 6:1541-1551. [DOI] [PubMed] [Google Scholar]
- 36.Mori, M., N. B. Ghyselinck, P. Chambon, and M. Mark. 2001. Systematic immunolocalization of retinoid receptors in developing and adult mouse eyes. Investig. Ophthalmol. Vis. Sci. 42:1312-1318. [PubMed] [Google Scholar]
- 37.Napoli, J. L. 1999. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1440:139-162. [DOI] [PubMed] [Google Scholar]
- 38.Niederreither, K., P. McCaffery, U. C. Dräger, P. Chambon, and P. Dollé. 1997. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 62:67-78. [DOI] [PubMed] [Google Scholar]
- 39.Pereira, F., E. Rosenmann, E. Nylen, M. Kaufman, L. Pinsky, and K. Wrogemann. 1991. The 56 kDa androgen binding protein is an aldehyde dehydrogenase. Biochem. Biophys. Res. Commun. 175:831-838. [DOI] [PubMed] [Google Scholar]
- 40.Perz-Edwards, A., N. L. Hardison, and E. Linney. 2001. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev. Biol. 229:89-101. [DOI] [PubMed] [Google Scholar]
- 41.Rossant, J., R. Zirngibl, D. Cado, M. Shago, and V. Giguère. 1991. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5:1333-1344. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Satre, M. A., K. E. Ugen, and D. M. Kochhar. 1992. Developmental changes in endogenous retinoids during pregnancy and embryogenesis in the mouse. Biol. Reprod. 46:802-810. [DOI] [PubMed] [Google Scholar]
- 44.Shang, E. Y., K. Lai, A. I. Packer, J. Paik, W. S. Blaner, M. D. Vieira, P. Gouras, and D. J. Wolgemuth. 2002. Targeted disruption of the mouse cis-retinol dehydrogenase gene: visual and nonvisual functions. J. Lipid Res. 43:590-597. [PubMed] [Google Scholar]
- 45.Steinmetz, C. G., P. G. Xie, H. Weiner, and T. D. Hurley. 1997. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 5:701-711. [DOI] [PubMed] [Google Scholar]
- 46.Stull, D. L., and K. C. Wikler. 2000. Retinoid-dependent gene expression regulates early morphological events in the development of the murine retina. J. Comp. Neurol. 417:289-298. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, R., T. Shintani, H. Sakuta, K. Kato, T. Ohkawara, N. Osumi, and M. Noda. 2000. Identification of RALDH-3, a novel retinaldehyde dehydrogenase, expressed in the ventral region of the retina. Mech. Dev. 98:37-50. [DOI] [PubMed] [Google Scholar]
- 48.Vasiliou, V., A. Bairoch, K. F. Tipton, and D. W. Nebert. 1999. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based upon divergent evolution and chromosomal mapping. Pharmacogenetics 9:421-434. [PubMed] [Google Scholar]
- 49.Wagner, E., P. McCaffery, and U. C. Dräger. 2000. Retinoic acid in the formation of the dorsoventral retina and its central projections. Dev. Biol. 222:460-470. [DOI] [PubMed] [Google Scholar]
- 50.Weiler, R., K. Schultz, M. Pottek, S. Tieding, and U. Janssen-Bienhold. 1998. Retinoic acid has light-adaptive effects on horizontal cells in the retina. Proc. Natl. Acad. Sci. USA 95:7139-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, J. G., C. B. Roth, and J. Warkany. 1953. An analysis of the syndrome of malformations induced by maternal vitamin A deficiency. Effects of restoration of vitamin A at various times during gestation. Am. J. Anat. 92:189-217. [DOI] [PubMed] [Google Scholar]
- 52.Yamauchi, K., J. Nakajima, H. Hayashi, R. Horiuchi, and J. R. Tata. 1999. Xenopus cytosolic thyroid hormone-binding protein (xCTBP) is aldehyde dehydrogenase catalyzing the formation of retinoic acid. J. Biol. Chem. 274:8460-8469. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, D. Q., and D. C. McMahon. 2000. Direct gating by retinoic acid of retinal electrical synapses. Proc. Natl. Acad. Sci. USA 97:14754-14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, D., P. McCaffery, K. J. Ivins, R. L. Neve, P. Hogan, W. W. Chin, and U. C. Dräger. 1996. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur. J. Biochem. 240:15-22. [DOI] [PubMed] [Google Scholar]