Abstract
Ribosome biogenesis in eukaryotes depends on the coordinated action of ribosomal and nonribosomal proteins that guide the assembly of preribosomal particles. These intermediate particles follow a maturation pathway in which important changes in their protein composition occur. The mechanisms involved in the coordinated assembly of the ribosomal particles are poorly understood. We show here that the association of preribosomal factors with pre-60S complexes depends on the presence of earlier factors, a phenomenon essential for ribosome biogenesis. The analysis of the composition of purified preribosomal complexes blocked in maturation at specific steps allowed us to propose a model of sequential protein association with, and dissociation from, early pre-60S complexes for several preribosomal factors such as Mak11, Ssf1, Rlp24, Nog1, and Nog2. The presence of either Ssf1 or Nog2 in complexes that contain the 27SB pre-rRNA defines novel, distinct pre-60S particles that contain the same pre-rRNA intermediates and that differ only by the presence or absence of specific proteins. Physical and functional interactions between Rlp24 and Nog1 revealed that the assembly steps are, at least in part, mediated by direct protein-protein interactions.
The synthesis of ribosomes is one of the major metabolic pathways of a cell. In Saccharomyces cerevisiae, ribosome assembly begins in the nucleolus after the transcription of two rRNA precursors, the 35S RNA (precursor of the 18S, 5.8S, and 25S rRNAs) and the pre-5S RNA, by RNA polymerases I and III, respectively. The synthesized pre-rRNAs are modified extensively at multiple positions specified by small nucleolar ribonucleoparticles (snoRNPs) or specific enzymes (1, 22, 33). During rRNA maturation, the 5′ and 3′ external transcribed sequences (ETS) and internal transcribed sequence 1 (ITS1) and ITS2 are removed from the 35S precursor RNA by well-ordered cleavages and trimming events, which require the enzymatic activities of helicases and endo- and exonucleases (19, 37).
Cotranscriptional assembly of ribosomal and nonribosomal proteins in the nucleolus gives rise to a large ribonucleoprotein particle corresponding to the 90S preribosomal complexes described more than 20 years ago (35) and recently characterized biochemically (8, 14). These early preribosomal complexes are further converted to smaller pre-40S (43S) and pre-60S (66S) particles, precursors of the mature small and large ribosomal subunits. The pre-40S complexes, each containing a precursor of the 18S rRNA, are exported into the cytoplasm, where they give rise to the mature 40S ribosomal particles (36). Most of the large ribosomal subunit proteins are absent from the 90S preribosomes (8, 14) and associate in the nucleolus with the pre-rRNA, probably concomitantly with the formation of the pre-60S particles. During pre-60S particle maturation, 27S pre-rRNA intermediates are converted into 25S and 5.8S mature rRNAs by successive and well-ordered steps. Several pre-60S particles, which differ in their RNA and protein compositions, are generated successively in the nucleolus and nucleoplasm and then exported to the cytoplasm (for reviews see references 11, 19, and 37).
The existence of ribosome precursors in the form of discrete particles during eukaryotic ribosome biogenesis was described many years ago in experiments using HeLa and yeast cells (for a review on these early results see reference 38). However, a detailed characterization of the preribosomal complexes emerged only recently due to the development of affinity purification methods and large-scale protein identification by mass spectrometry (3, 8, 10, 14, 15, 24, 31). In addition to these results, the high-throughput identification of protein complexes in yeast (13, 17) generated a large amount of data and offered a global view of the factors physically associated with preribosomal particles. To date, more than 120 nonribosomal proteins are predicted to be physically associated in large ribonucleoprotein complexes along the maturation pathway from the nucleolus to the cytoplasm. The physical association with the preribosomes is, for many of these conserved factors, the only hint as to their function.
Previous studies on preribosomal particle composition showed that different tagged proteins were associated with different sets of preribosomal factors in multiple preribosomal complexes (24; for a review, see reference 11). However, especially for early pre-60S particles of highly complex composition, little is known about the identities of the different particles and the association or dissociation of specific factors. A major limitation in the study of ribosome assembly comes from the difficulties in isolating homogenous preribosomal intermediates. By blocking ribosome biogenesis at specific steps and by analyzing the changes in the composition of early pre-60S intermediates, we were able to show that specific protein association and dissociation steps are required for the maturation of 27S-containing pre-60S particles during ribosome biogenesis. This novel approach should be able to elucidate the order of the assembly of preribosomal factors on, and their disassembly from, the particles and the mechanisms governing this coordinated process. We show here that direct protein-protein interactions are likely to be needed for the assembly of multiple complexes during ribosome assembly.
MATERIALS AND METHODS
Yeast strains, plasmids, and oligonucleotides.
The strains used in this study are listed in Table 1. They were generated by homologous recombination using PCR products to transform either the BMA64 or the MGD353-13D strains (4). PCR fragments used to generate conditional mutants or TAP-tagged proteins were synthesized from pFA6a-kanMX6-PGal1-3HA or pBS1479, respectively, by using oligonucleotides designed previously (23, 29). For recombinant-protein expression, PCR fragments containing the whole NOG1, RPL24, RPL24B, and RPL5 open reading frames (ORFs) were introduced into a vector derived from pGEX4-T (Amersham Biosciences) at NcoI/SalI sites. The genes were subcloned into pET16 (Novagen) at NcoI/XhoI sites. The sequences of oligonucleotides used for Northern hybridization and primer extension analysis were as follows: CS14, CAT GGC TTA ATC TTT GAG AC; MFR422, CTA CTC GGT CAG GCT C (5S); MFR457, GCT TAA AAA GTC TCT TCC CGT CC (snRNA-U2); MFR221, CCA AGT TGG ATT CAG TGG CTC (snRNA-U3). The sequences of the other oligonucleotides were previously described (31).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| MGD353-13D | MATatrp1-289 ura3-52 ade2 leu2-3, 112 Arg4(RV−) | 29 |
| BMA64 | MATa/alpha ura3-1/ura3-1 Δtrp1/Δtrp1 ade2-1/ade2-1 leu2-3,112/leu2-3, 112 his3-11,15/his3-11,15 | 4 |
| LMA148 | MATaura3-1 Δtrp1 ade2-1 leu2-3,112 his3-11,15 GAL1::NOG2/KanMX6 | 31 |
| LMA158 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) Nog1-TAP/TRP1 | 31 |
| LMA160 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) Rlp24-TAP/TRP1 | 31 |
| LMA174 | MATatrp1-289 ura3-52 ade2 leu2-3,112, Arg4(RV−) GAL1::NOG1/KanMX6 | This study |
| LMA176 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::NOG2/KanMX6 Nog1-TAP/TRP1 | This study |
| LMA177 | MATatrp1-289 ura3-52 ade2 leu2-3,112, Arg4(RV−) GAL1::NOG2/KanMX6 Rlp24-TAP/TRP | This study |
| LMA178 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::RLP24/KanMX6 | This study |
| LMA181 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::RLP24/KanMX6 Nog1-TAP/TRP1 | This study |
| LMA182 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::NOG1/KanMX6 Rlp24-TAP/TRP1 | This study |
| LMA205 | MATa/α ura3-1/ura3-1 Δtrp1/Δtrp1 ade2-1/ade2-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 GAL1::GFP-Rlp24/TRP1 | This study |
| LMA226 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::NOG1/KanMX6 Rpl24B-GFP/TRP1 | This study |
| SC1101 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) Ssf1-TAP/URA3 | 13 |
| LMA233 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::RLP24/KanMX6 Ssf1-TAP/URA3 | This study |
| LMA234 | MATatrp1-289 ura3-52 ade2 leu2-3,112 Arg4(RV−) GAL1::NOG1/KanMX6 Ssf1-TAP/URA3 | This study |
Similarity searches for Rlp24 homologues.
The amino acid sequence encoded by yeast ORF YLR009W (RLP24) was used for Blast searches of nonredundant databases. Similar sequences were retrieved and compared with the Rpl24-related sequences, and multiple alignments were obtained with ClustalX (34). For the Guillardia theta nucleomorph genome, the partial sequence of an Rpl24-like protein whose gene had no initiator ATG was also used in the final alignment.
High-copy-number suppressor genetic screen.
The LMA160 (Rlp24-TAP) strain was transformed with a yeast genomic high-copy-number vector library constructed in pFL46S (a gift from F. Lacroute, Gif-sur-Yvette, France). The transformants were grown on solid synthetic minimal medium lacking leucine at 25°C. Colonies having a growth rate superior to that of LMA160 transformed with an empty vector were selected.
RNA extraction, Northern blotting, and primer extension.
Cells were broken with glass beads, and total RNAs were subjected to phenol-chloroform extraction and finally resolved on 1% agarose-6% formaldehyde gels or 5% acrylamide-urea gels. RNAs were transferred to Hybond-N+ (Amersham Biosciences) membranes probed with various 32P-labeled oligonucleotides complementary of specific regions of the intermediate and mature rRNAs.
Sucrose gradients and protein analysis.
Total-protein extracts were prepared from exponentially growing yeast cells and separated on 10 to 50% sucrose gradients. Fractions of the gradient were collected, total proteins were precipitated, and specific proteins were detected by immunoblotting. Rabbit polyclonal Rlp24- and Nog2-specific antibodies were generated by using recombinant glutathione S-transferase (GST)-Rlp24 and GST-Nog2 purified from Escherichia coli. Nog1-specific antibodies were obtained in a similar manner but with native Nog1 expressed in bacteria and purified as inclusion bodies.
In vitro binding assay.
The proteins were expressed in native form or in fusions with the GST in the BL21 CodonPlus RP (Stratagene) strain. Expression was induced by 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 h at 37°C in exponentially growing cells. Protein extracts were obtained in a French press by using 10 volumes of a buffer containing 50 mM MOPS (morpholinepropanesulfonic acid), pH 7, 150 mM NaCl, 5 mM EDTA, complete protease inhibitors (Roche), and 1 mM dithiothreitol (DTT). The extracts, clarified by centrifugation at 20,000 × g for 45 min, were stored frozen at −70°C. After 45 min of preincubation a mixture of equal (0.5-ml) volumes of extracts from strains expressing GST-tagged and untagged proteins was bound to 50 μl of glutathione-Sepharose beads (Amersham Biosciences). Following 1 h of incubation and eight washes with 0.7 ml of buffer (20 mM MOPS [pH 7], 150 mM NaCl, 1 mM DTT, 0.1% (Sigma Igepal CA-630) the bound proteins were eluted by boiling the beads in 40 μl of denaturing buffer for electrophoresis.
Complex purification.
Strains expressing Rlp24 and Nog1 with C-terminally fused TAP tags were used for complex purification by a slight modification of the original tandem affinity purification protocol (29) starting from 4 liters of yeast culture. For the analysis of RNAs associated with Nog1 and Rlp24 the first step of the tandem affinity purification protocol was performed with a lysis buffer containing 20 mM vanadyl ribonucleoside complex (New England Biolabs) and a TEV protease digestion buffer that contained 0.1 U of recombinant RNasin/μl (Promega). The RNAs contained in the TEV eluate were extracted twice with phenol-chloroform and precipitated.
Mass spectrometry identification of proteins.
Proteins were identified by peptide mass fingerprinting with a Voyager DE-STR matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer. Systematic and band-specific analyses of gels were performed. The Investigator Progest system (Genomic Solutions) was used for digestion with modified porcine trypsin (Promega), peptide purification, and preparation of MALDI plates. A minimum of four matching peptides (at 30-ppm error) were considered necessary for positive protein identifications.
Immunofluorescence methods and electron microscopy.
The proteins in fusion with the TAP tag were detected by immunofluorescence according to methods described previously (28). Hoechst 33258 at 2 μg/ml was used to stain DNA. Pre-25S rRNAs were localized with a Cy3-conjugated oligomeric probe complementary to the 5′ part of ITS2 as described previously (5). Electron microscopy was performed as previously described (31) using riboprobes complementary to the ITS2 sequence. To evaluate the distribution of pre-rRNAs in the nuclei of mutant cells after in situ hybridization for electron microscopy, 18 to 20 sections of nuclei displaying both the nucleolus and the nucleoplasm were randomly photographed per strain. On each picture, the labeling densities of the nucleolus and of the nucleoplasm were calculated on the basis of the ratio of the number of gold particles to the surface area.
RESULTS
Rlp24 shuttles between the nucleus and the cytoplasm.
We recently isolated pre-60S particles containing three novel proteins, Nog1, Nog2, and Rlp24 (31). Sequence similarity searches showed that Rlp24 is highly conserved in Archaea and Eukarya and belongs to the family of eukaryotic Rpl24e proteins (Fig. 1A). All the eukaryotic genomes completely sequenced so far encode at least one pair of Rpl24-like and Rlp24-like proteins. These observations prompted us to study the cellular role of Rlp24 in ribosome biogenesis.
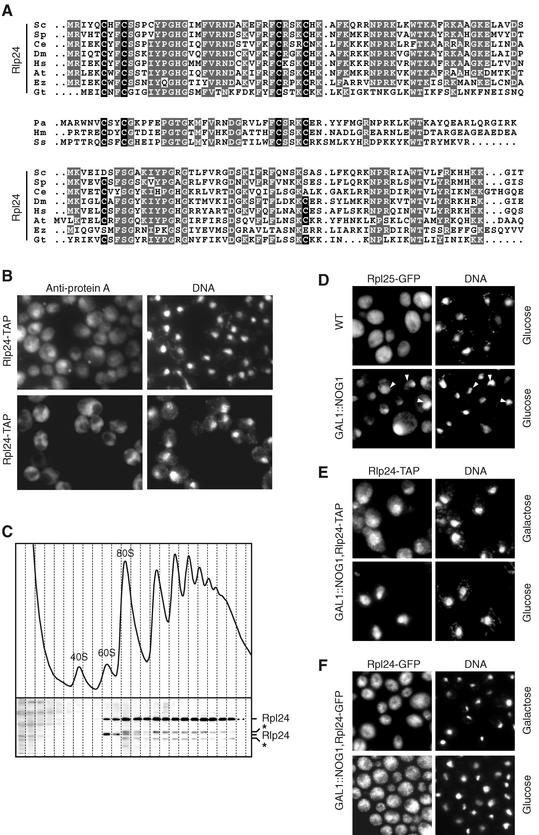
FIG. 1.
Rlp24 and Rpl24 are homologous proteins of different cellular localizations. (A) N-terminal sequences of Rlp24-like and Rpl24-like proteins encoded by genes present in eukaryotes or Archaea were aligned with ClustalX. Gray, amino acid residues identical in several sequences; black, conserved cysteines of the putative zinc finger present in Rlp24 sequences. Accession numbers (TrEMBL, SwissProt, or Genpept) for theRlp24-like sequences are as follows: Saccharomyces cerevisiae (Sc), Q07915; Schizosaccharomyces pombe (Sp), Q10353; Caenorhabditis elegans (Ce), Q17606; Drosophila melanogaster (Dm), Q9VGN9; Homo sapiens (Hs), Q9UHA3; Arabidopsis thaliana (At), O22165; Encephalitozoon cuniculi (Ez), gi-19074000; G. theta nucleomorph (Gt), gi-13812425; Pyrococcus abyssi (Pa), Q9V0W3; H. marismortui (Hm), P14116; Sulfolobus sulfataricus (Ss), Q980Q6. Accession numbers for the Rpl24-like sequences are as follows: S. cerevisiae, P04449; S. pombe, O74884; C. elegans, O01868; D. melanogaster, Q9VJY6; H. sapiens, P38663; A. thaliana, Q9LF73; E. cuniculi, gi-19074515 (starting residue 14). For the G. theta nucleomorph pseudo-Rpl24, see Materials and Methods. (B) Unusual localization of Rlp24 from the nucleolus to the cytoplasm. Cells expressing Rlp24-TAP and Rpl24-TAP under the control of their own promoters were fixed with formaldehyde, washed, and incubated with anti-protein A antibodies (Sigma) followed by Cy3-conjugated secondary antibodies (Jackson Immunoresearch). (C) Rlp24 is neither free nor associated with polysomes. The sedimentation behavior of Rpl24-TAP was assessed by sucrose gradient ultracentrifugation. Twenty-four fractions were collected, and tagged proteins were detected by immunoblotting using peroxidase-antiperoxidase conjugates (Sigma) and Rlp24-specific antibodies. Asterisks, cross-reactive bands. The same profile of sedimentation was obtained with an Rlp24-TAP strain. (D) Cytoplasmic Rlp24 localization depends on active ribosome biogenesis. Preribosomes are blocked in the nucleus in absence of Nog1. Wild-type (WT) and GAL1::NOG1 cells expressing Rpl25-GFP were grown for 16 h in glucose-containing medium. The fluorescence signals of GFP and Hoechst 33342-stained DNA were observed by microscopy. Arrowheads, position of the nucleoplasm. The localizations of Rlp24-TAP (E) and Rpl24-GFP (F) expressed under the control of their own promoters were observed in GAL1::NOG1 cells either grown exclusively on galactose or shifted to glucose-containing medium for 16 h. Rlp24-TAP was detected as described for panel B. Similar results (data not shown) were obtained when we blocked pre-60S export by overexpressing the Nmd3Δ100 dominant-negative allele (16).
Many of the proteins involved in the maturation of the preribosomes are located in the nucleolus and nucleoplasm. Surprisingly, the localization of Rlp24, detected as a C-terminal fusion with the TAP tag (29), was not limited to the nucleus. A significant amount of Rlp24-TAP fusion protein was also observed in the cytoplasm (Fig. 1B, top). In some of the cells, Rlp24-TAP, although visible in the nucleoplasm and the cytoplasm, showed a preferential nucleolar staining. When we analyzed the localization of Rpl24, the ribosomal homologue of Rlp24, under similar experimental conditions, we observed a specific cytoplasmic localization with no nuclear staining, as expected for a ribosomal protein (Fig. 1B, bottom). Since the C-terminal TAP tag had a slight negative effect on the growth rate of the Rlp24-tagged strain, we verified that the observed localization, unusual for a preribosomal factor, was not the result of the tag. A similar localization was observed for an N-terminal GFP fusion when overexpressed in a diploid strain (data not shown).
We wondered whether the cytoplasmic fraction of Rlp24 is in a free form, associated with active ribosomes, or associated with late pre-60S particles. Total-protein extracts from a strain expressing C-terminally TAP-tagged Rpl24B were fractionated by ultracentrifugation, and the positions of both tagged Rpl24 and native Rlp24 were assessed by immunoblotting (Fig. 1C). Rpl24-TAP was found to sediment at positions corresponding to polysomes and free 60S ribosomal particles, whereas endogenous Rlp24 sedimented only in the 60S fractions. Like the native form of Rlp24, Rlp24-TAP was exclusively found in the 60S fractions and not in the upper part of the gradient, showing that all the Rlp24-TAP protein, including its cytoplasmic fraction, is associated with a large complex.
To analyze the origin of the cytoplasmic fraction of Rlp24, we tested the effects of blocking pre-60S export on Rlp24 intracellular distribution. A ribosomal protein (Rpl25) fused with the green fluorescent protein (GFP) has been previously used to detect pre-60S export defects. The corresponding signal shows nuclear or nucleolar accumulation if the export or maturation of the ribosomes is impaired (18). We used a strain depleted of Nog1 in which preribosomes are blocked in the nucleus, as demonstrated by the retention of the Rpl25-GFP marker (Fig. 1D). Under these conditions, Rlp24-TAP was entirely relocated to the nucleus, with some of the signal more concentrated in the nucleolus (Fig. 1E). Similar results were obtained with a strain depleted of Nog2 (data not shown). In contrast, when we looked at the localization of the ribosomal Rpl24 in fusion with GFP, we observed a decrease in only the cytoplasmic staining, with no nuclear accumulation when the export of the ribosomes was blocked (Fig. 1F). These results indicate that, unlike ribosomal Rpl24, Rlp24 shuttles between the nucleus and the cytoplasm. It further suggests that the cytoplasmic localization of Rlp24 is dependent on active ribosome biogenesis and export. In contrast to most of the other factors involved in 60S maturation, which are confined to the nucleus, Rlp24 seems to remain associated with pre-60S particles even after export to the cytoplasm.
Rlp24 and Nog1 are present in similar preribosomal complexes.
We have previously shown that Rlp24 and Nog1 are associated with pre-60S particles with similar protein profiles (31). Nog1 is a nuclear protein found in association with the nuclear pore complexes (27, 30) and identified in many preribosomal complexes (see reference 11 and references therein). We decided to characterize in detail the protein and RNA compositions of both Nog1- and Rlp24-associated particles. Complexes associated with each of these proteins were isolated by tandem affinity purification (29) using strains expressing the C-terminally TAP-tagged fusion proteins. The proteins present in these purified complexes were separated by electrophoresis (Fig. 2A) and identified by MALDI-TOF mass spectrometry. The results of these identifications and the corresponding positions of proteins on a typical gel are listed in Table 2. In addition to ribosomal proteins of the large subunit and common contaminants (see supplemental data at http://www.pasteur.fr/recherche/unites/Gim/index.html), 45 nonribosomal proteins were identified. Most of them could be found in both Nog1- and Rlp24-associated complexes, consistent with the similar electrophoretic profiles. An obvious exception was Mak11 (protein band 21); this protein, previously found to be essential for 60S formation (25), was present in larger amounts in the Rlp24-associated complex than in the Nog1-associated complex, as determined by the intensity of protein staining (Fig. 2A). We also identified Mak11 as a component of Ssf1-associated particles (not shown), and it was recently reported to be a component of Nsa3/Cic1-associated complexes (24). With the exception of four novel proteins, all the identified proteins have been previously found associated with preribosomal particles (3, 10, 13, 15, 17, 24). Of the four new proteins, Dbp7 was previously characterized as a trans-acting factor involved in 60S biogenesis (7). Vps75, identified in a genetic screen for factors affecting carboxypeptidase Y secretion (6), was not previously identified in association with multiprotein complexes and might be a new component of preribosomes. Finally, the functions of Yil096c and Yor252w are unknown. In a large-scale protein localization study, Yil096c was found to be nuclear (21). The expression profiles of both YIL096C and YOR252W under stress conditions (12) are similar to the expression profiles of known preribosomal genes, strongly suggesting the involvement of these new factors in ribosome biogenesis.
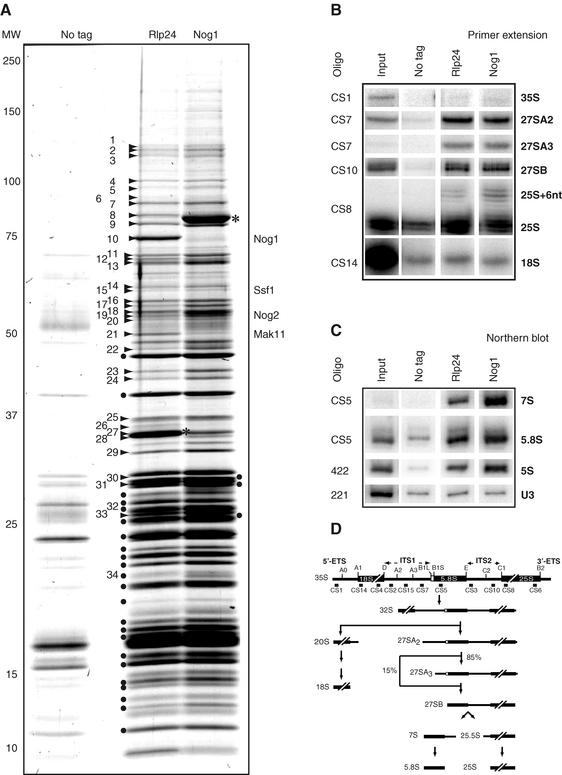
FIG. 2.
Similar compositions of Rlp24- and Nog1-associated preribosomal complexes. Tandem affinity purification of Nog1- and Rlp24-associated complexes was followed by separation of proteins by denaturing electrophoresis on a 5 to 20% polyacrylamide gel and colloidal Coomassie blue staining. Asterisks, positions of the tagged proteins used as baits. Numbers correspond to excised gel bands in which the preribosomal factors listed in Table 2 were identified, with the exception of the band marked 34, identified as ribosomal protein Rpl24A/B. Dots correspond to bands in which ribosomal proteins of the large subunit were identified. The result for an identically treated extract from wild-type cells is shown in the left lane. (B and C) Rlp24 and Nog1 are associated with pre-60S RNAs. RNAs were isolated by one-step purification of extracts of Nog1-TAP, Rlp24-TAP, or wild-type cells and detected by primer extension (B) (35S, 27SA2, 27SA3, 27SB, 25S, and 18S) or Northern blotting (C) (7S, 5.8S, 5S, U3 and U2 snRNA) with radiolabeled oligonucleotides. (D) Schematic representation of the steps involved in pre-rRNA processing in S. cerevisiae. The largest 35S intermediate pre-rRNA undergoes extensive modifications and processing to generate the mature 18S, 5.8S, 25S rRNAs (19). The relative positions of the oligonucleotides used in this study are indicated.
TABLE 2.
Nonribosomal proteins identified in association with Rlp24-TAP and Nog1-TAP
| Spot(s) | Protein | Rlp24 | Nog1 | ORF |
|---|---|---|---|---|
| 1, 2, 3 | Erb1 | + | + | YMR049C |
| Dbp7 | + | YKR024C | ||
| Sda1 | + | + | YGR245C | |
| 5 | Mak5 | + | + | YBR142W |
| 6 | Noc2 | + | + | YOR206W |
| 7 | Nop2 | + | + | YNL061W |
| Rix1 | + | YHR197W | ||
| 9 | Nop7 | + | + | YGR103W |
| 8 | Puf6 | + | + | YDR496C |
| 10 | Nog1 | + | + | YPL093W |
| 11, 12 | Dbp9 | + | YLR276C | |
| 12 | Spb4 | + | + | YFL002C |
| Nug1 | + | + | YER006W | |
| 13 | Arx1 | + | + | YDR101C |
| 14 | Fpr3 | + | + | YML074C |
| 15 | Ssf1 | + | YHR066W | |
| 15 | Dbp2 | + | YNL112W | |
| 15 | Nop12 | + | YOL041C | |
| 16 | Has1 | + | + | YMR290C |
| 18 | Nsa1 | + | + | YGL111W |
| 16 | Fpr4 | + | YLR449W | |
| 17 | Yp1146c | + | YPL146C | |
| 19 | Nog2 | + | + | YNR053C |
| 19, 20 | Ycr072c | + | + | YCR072C |
| 20 | Ytm1 | + | + | YOR272W |
| 20 | Nop58 | + | YOR310C | |
| 21 | Mak11 | + | YKL021C | |
| 22 | Cic1/Nsa3 | + | + | YHR052W |
| 23 | Rlp7 | + | + | YNL002C |
| 23 | Mak16 | + | YAL025C | |
| 24 | Rpf2 | + | + | YKR081C |
| 25 | Nop1 | + | + | YDL014W |
| 25 | Rrp1 | + | + | YDR087C |
| 26 | Vps75 | + | YNL246W | |
| 27 | Yil096c | + | YIL096C | |
| 27 | Rpf1 | + | YHR088W | |
| 28 | Brx1 | + | + | YOL077C |
| 29 | Nop16 | + | + | YER002W |
| 29 | Nsa2 | + | + | YER126C |
| Rlp24 | + | + | YLR009W | |
| 31 | Mrt4 | + | + | YKL009W |
| 33 | Tif6 | + | + | YPR016C |
| 30 | Nop15 | + | YNL110C | |
| 32 | Loc1 | + | YFR001W | |
| Yor252w | + | + | YOR252W |
Among the identified preribosomal proteins, Rpf1 and Ssf1 have been previously shown to associate early with pre-60S particles (10, 39) while Nog2 and Arx1 are late-associating pre-60S factors (24, 31). Since Rlp24 and Nog1 seemed to be present both in early and late pre-60S complexes, we tested the RNA composition of the isolated particles. As expected, a strong enrichment of 27SA2, 27SA3, 27SB, and 7S rRNA intermediates, but not 35S pre-rRNA, in the purified complexes was observed. We also noted an enrichment of the 5S, 5.8S, and 25S rRNA in both complexes (Fig. 2B and C). In conclusion, the protein and RNA compositions of the Nog1- and Rlp24-associated complexes revealed that these proteins are present in both early and late pre-60S particles.
Rlp24 and Nog1 are involved in the biogenesis of the 60S ribosomal subunit.
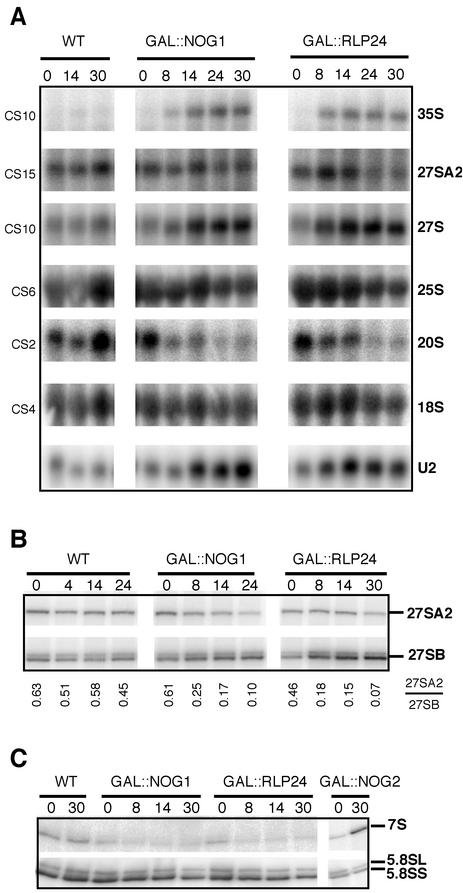
The presence of Nog1 and Rlp24 in preribosomal complexes in association with 27S and 7S rRNA intermediates did not indicate at which step of rRNA maturation these proteins are essential during the 60S ribosomal particle biogenesis. We thus investigated the effects of depleting each of these proteins on pre-rRNA steady-state levels. We used yeast strains in which the expression of NOG1 or RLP24 was under the control of the galactose-inducible GAL1 promoter. The growth rate became significantly reduced after about 8 h of culture on glucose, and this reduction was accompanied by a marked increase in the level of free 40S subunits, a concomitant decrease in the level of free 60S subunit, and the appearance of “halfmer” polysomes (data not shown) (see reference 15 for similar results on Rlp24 depletion). The 35S and 27SB intermediates in the rRNA maturation pathway were enriched in cells depleted of either Rlp24 or Nog1 (Fig. 3A and B). In these cells, we observed the accumulation of aberrant 23S intermediates that are generated by A3 cleavages directly from 35S precursors if the processing at sites A0, A1, and A2 is delayed. The 35S processing defects could explain the observed decrease in the levels of 27SA2 and 20S intermediates, which are formed downstream the processing pathway (Fig. 3A and B; Fig. 2D shows the order of processing steps). Since Rlp24 and Nog1 are not associated with 35S rRNA intermediates, the 35S accumulation is most likely indirect. This effect was previously observed in studies of a large number of mutant genes involved in 60S ribosome biogenesis (37). The 27SB rRNA species, present in the complexes associated with both Nog1 and Rlp24, accumulated compared with the preceding 27SA2 intermediate (Fig. 3B), indicating a block in 27SB processing when either Nog1 or Rlp24 was depleted. This block could explain the observed decrease of the 7S rRNA intermediate (Fig. 3C). We concluded that the absence of Rlp24 or Nog1 from the 27S-containing particles impedes 27SB processing.
FIG. 3.
Rlp24 and Nog1 are involved in 27SB pre-rRNA processing. (A) Steady-state levels of mature and precursor rRNA in strains depleted of Rlp24 or Nog1. Cells grown on galactose-containing medium were shifted to glucose for the indicated number of hours, and total RNAs were analyzed by Northern blotting after denaturing 1.2% formaldehyde agarose electrophoresis. The positions of the probes are indicated in the Fig. 2D. WT, wild type. (B) Accumulation of 27SB over 27SA2 intermediates. Following primer extension with oligonucleotide CS10 (specific for the ITS2 sequence), the changes in the ratio of 27SA2 to 27SB in cells depleted of Rlp24 and Nog1 were quantified with a PhosphorImager and the ImageQuant software (Molecular Dynamics) and are shown below the gel image. (C) Small RNAs separated on denaturing urea-5% polyacrylamide gels were analyzed by Northern blotting using a probe specific for 7S and 5.8S intermediates (CS5).
Dynamics of preribosomes in Nog1-depleted cells.
In contrast to the depletion of Nog1 or Rlp24, which leads to the accumulation of only 27SB intermediates, Nog2 depletion leads to an accumulation of 27SB and 7S intermediates (Fig. 3C shows a comparison of the effects on 7S). We have previously shown that Nog2 depletion leads to nucleoplasmic accumulation of late pre-60S particles and suggested that these late steps, by which the maturation of 7S intermediates is accomplished, could occur in the nucleoplasm (31). Any difference in the intranuclear accumulation of pre-60S particles between Rlp24- or Nog1-depleted cells and Nog2-depleted cells would support this hypothesis. Because Rlp24 depletion appeared to lead to unstable complexes (see Discussion), we chose to compare Nog1-depleted cells to Nog2-depleted cells.
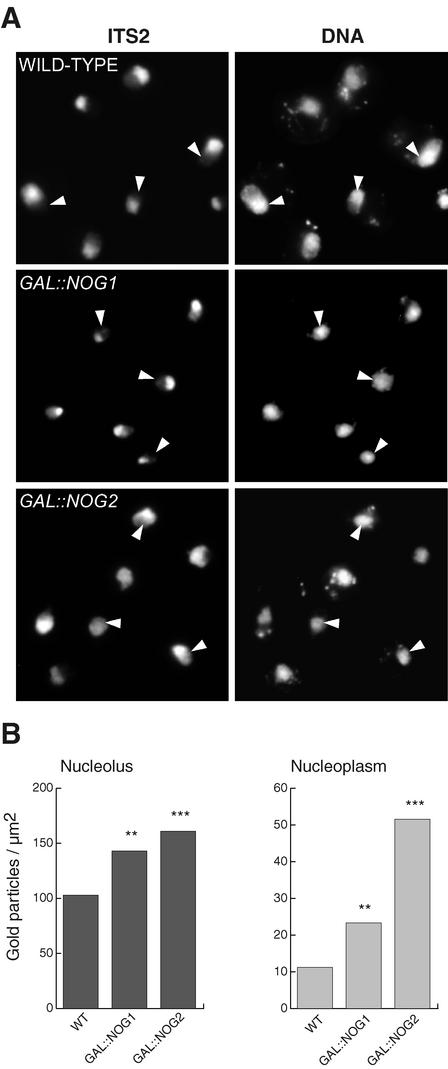
We determined the intranuclear distribution of the accumulated pre-rRNA intermediates by fluorescence in situ hybridization (FISH) and electron microscopy with probes specific to ITS2. These probes are able to detect the intermediates in the 60S processing pathway from the primary transcript to the 7S pre-rRNA. In wild-type cells, the FISH signal mostly localized to the nucleolus, with a faint labeling of the nucleoplasm (Fig. 4A). In cells depleted of Nog1 the labeling was strongly nucleolar. Consistent with what we had previously observed, the nucleoplasmic labeling was more intense in cells depleted of Nog2, in which it was sometimes impossible to distinguish the nucleolus from the rest of the nucleus. As a control, a probe complementary to the 5′ end of ETS1, which hybridizes upstream of the A0 cleavage position and which detects the 35S precursor rRNA, labeled only the nucleolus in all the strains (data not shown). Electron microscopy quantitation of pre-rRNA confirmed that, while Nog1 depletion leads to accumulation of pre-rRNA intermediates mainly in the nucleolus, Nog2 depletion leads to accumulation of these precursors in the nucleolus and nucleoplasm (Fig. 4B). Added to the observed accumulation of 7S intermediates only after Nog2 depletion, these results strongly suggest that the final steps of 7S pre-rRNA processing into 5.8S rRNA occur in the nucleoplasm. Moreover, these results also suggest that Nog1 and Rlp24 precede Nog2 function in 60S biogenesis.
FIG. 4.
In the absence of Nog1, pre-rRNAs accumulate in the nucleolus. (A) Intranuclear distribution of the preribosomal particles in Nog1- or Nog2-depleted cells was determined by FISH with a probe specific for the ITS2 sequence. DNA was stained with the DAPI (4′,6′-diamidino-2-phenylindole) fluorescent dye. Arrowheads, position of the nucleoplasm. (B) ITS2 containing pre-RNAs were detected by electron microscopy in situ hybridization in cells depleted of Nog1 or Nog2 by growth on glucose for 14 h, and the labeling densities of the nucleolus and the nucleoplasm were determined. One-to-one comparisons with the wild-type (WT) strain showed significant differences (∗∗, P < 0.01; ∗∗∗, P < 0.001). The nucleoplasmic labeling in Nog2-depleted cells is also significantly higher (P < 0.001) than that in cells lacking Nog1.
Ordered assembly of Rlp24, Nog1, and Nog2 on pre-60S ribosomal particles.
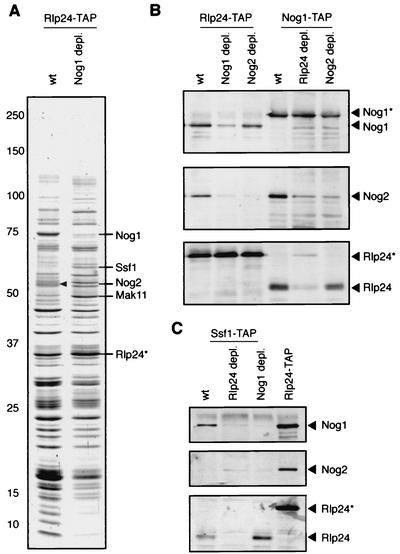
The existence of distinct, successive pre-60S complexes was suggested by the characterization of several preribosomal particles of different protein compositions (11, 24). However the factors that are essential for the coordinated association of the preribosomal proteins with nascent 60S particles are not yet known. To address this question, we took advantage of the presence of Rlp24 all along the 60S maturation pathway from the nucleolus to the cytoplasm. We used Rlp24 as a probe to measure the changes in the protein compositions of the pre-60S complexes that accumulated in cells with different mutations. We performed a series of purifications of complexes using Rlp24 or Nog1 as the bait when Rlp24, Nog1, or Nog2 was depleted. The particles purified with Rlp24 as the bait when Nog1 or Nog2 was absent showed an increase in the abundance of some preribosomal factors compared with the same complexes in wild-type cells (Fig. 5A and data not shown). In contrast, we observed a decrease in the intensities of the bands that correspond to ribosomal proteins. This decrease probably results from the depletion of the cytoplasmic fraction of the Rlp24-associated complexes (Fig. 1E).
FIG. 5.
Nog1 association with pre-60S complexes depends on Rlp24 and is a prerequisite for Nog2 assembly. (A) Early pre-60S complexes accumulate in the absence of Nog1. Rlp24-associated complexes were purified from wild-type (wt) or Nog1-depleted cells (GAL1::NOG1 cells shifted to glucose for 14 h), and the proteins were separated on a 5 to 20% polyacrylamide gel and stained with colloidal Coomassie blue. Asterisks (A to C), positions of the tagged proteins used as baits. The proteins are the same as those in Fig. 2A for comparison. Arrowhead, the Nog2 band identified by mass spectrometry. (B) The role of Nog1 and Rlp24 in Nog2 assembly on pre-60S particles was confirmed by immunoblots with tandem affinity-purified complexes and specific antibodies. Cells were shifted to glucose for 14 h to deplete Nog1 or Rlp24 and for 18 h to deplete Nog2. (C) The requirement of Nog1 for Rlp24 assembly on pre-60S particles was tested by using Ssf1-TAP purified complexes and immunoblots in wild-type cells or after Nog1 or Rlp24 depletion for 14 h.
Particularly interesting was the strong increase in the intensities of the bands containing Ssf1 and Mak11 when Nog1 was depleted (Fig. 5A). Since the absence of Nog1 blocks 27SB processing and since Ssf1 leaves the pre-60S particles before the generation of the 7S pre-rRNA (10), we concluded that in these mutant cells the Rlp24-associated complex mirrors the composition of early 27S-containing particles. Moreover, Nog2 is absent from these particles (Fig. 5A and B), suggesting that Nog1 function is required for Nog2 assembly and that Ssf1 leaves the pre-60S particles before Nog2 association (see below). We tested the presence of Nog2 and Rlp24 in Ssf1-associated complexes with specific antibodies (Fig. 5C). Nog2 is absent from these complexes, in contrast to Rlp24, which was enriched when Nog1 was depleted.
Altogether these results allowed us to conclude that (i) Ssf1 leaves pre-60S particles before or concomitantly with Nog2 association, (ii) Nog2 is not required for stable association of Nog1 or Rlp24 with pre-60S particles, and (iii) the presence of Nog1 is an absolute requirement for the later addition of Nog2 to 27SB-containing pre-60S particles but Nog1 is not required for the addition of many other preribosomal proteins such as Ssf1, Mak11, and Rlp24.
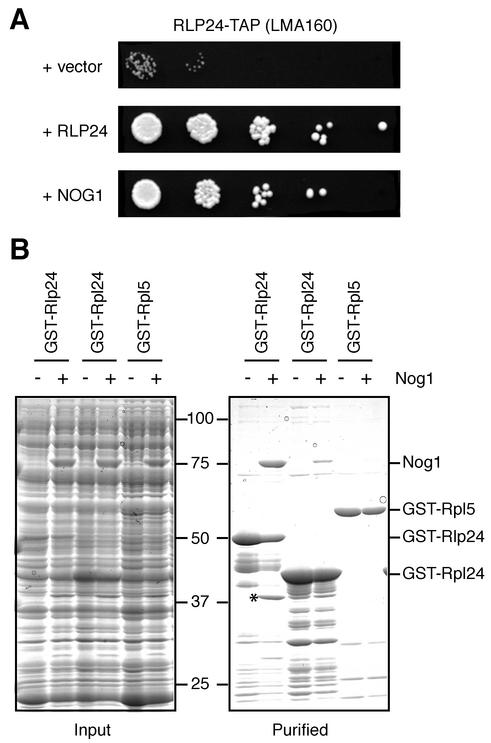
The study of the composition of the pre-60S complexes in different mutant cells could provide information only on global physical associations of proteins inside the preribosomal particles. Since we were interested in understanding the mechanisms responsible for the ordered assembly of proteins on preribosomes, we searched for functional partners of Rlp24. Taking advantage of the slow-growth phenotype of the strain expressing C-terminally TAP-tagged Rlp24, we performed a high-copy-number suppressor screen which identified genes that rescued the slow-growth phenotype of this strain when overexpressed. In addition to clones expressing Rlp24, two independent plasmids carrying the sequence corresponding to the complete NOG1 gene were selected (the smallest insert encompassed the region from 212 bp upstream of the initiation codon to 250 bp downstream of the stop codon) (Fig. 6A). This interaction between the NOG1 and RLP24 genes suggested that the two proteins might directly interact. Strong indications for a direct physical interaction also came from a two-hybrid genomic screen using Nog1 as bait that specifically selected Rlp24 as a prey (M. Fromont-Racine, J. C. Rain, and P. Legrain, unpublished data). To confirm these results in vitro, we tested whether recombinant Nog1 and Rlp24 produced in E. coli interact. In pull-down experiments, Nog1 copurified with a GST-Rlp24 fusion when expressed in E. coli. A smaller amount of Nog1 was purified when the related GST-Rpl24 protein was used, but no Nog1 could be detected in similar experiments when GST-Rpl5 (Fig. 6B) or GST alone (not shown) was used. In addition, the Rlp24-Nog1 interaction was resistant to an RNase A treatment (not shown). We conclude that Nog1 directly and specifically interacts with Rlp24.
FIG. 6.
Rlp24 and Nog1 genetically and physically interact. (A) The Rlp24-TAP strain was transformed with a high-copy-number LEU2 vector (pFL46S) carrying no insert or inserts corresponding to Nog1 or Rlp24. The rescue of the slow-growth phenotype was tested by spotting transformants on minimal-medium plates in 10−1-dilution steps. The plates were incubated at 25°C for 4 days. (B) Total protein extracts from bacteria expressing GST-Rlp24, GST-Rpl24B, and GST-Rpl5 were incubated with extracts containing Nog1 and then with glutathione-Sepharose beads (Pharmacia). After extensive washing, the bound proteins were eluted under denaturing conditions and separated by electrophoresis. The Coomassie blue-stained gels representing the input mixture of bacterial protein extracts (left) and the purified proteins (right) are shown side by side for comparison. A band of about 38 kDa that copurifies with GST-Rlp24 only in the presence of Nog1 (∗) is a C-terminal Nog1 fragment (matching peptides between amino acids 361 and 599 as determined by MALDI-TOF mass spectrometry and assignment of peptide masses to the Nog1 sequence).
Altogether these results show that Rlp24 plays an early role in pre-60S particle assembly and guides the association of Nog1 with these complexes and that later assembly of Nog2 is dependent on the preassembly of both Nog1 and Rlp24.
DISCUSSION
Ordered assembly of proteins during 60S ribosomal subunit biogenesis.
Ribosome biogenesis in eukaryotes involves coordinated assembly of a large number of protein factors on, and their dissociation from, dynamic ribonucleoprotein complexes that contain precursors of the rRNAs. More than 120 preribosomal factors, many of them of previously unknown function, were identified by their presence in intermediate complexes purified by affinity methods (3, 8, 10, 13-15, 17, 24, 31). At least 60 proteins are physically associated with pre-60S complexes in their maturation from the nucleolus to the cytoplasm. Multiple changes in the protein compositions of these complexes are likely to occur in correlation with the rRNA processing steps. However, knowledge of the global protein composition of the preribosomal complexes is not sufficient to define either the order of assembly and dissociation of these factors or their roles in the progression from one preribosomal particle to another. Indirect information about these assembly events comes from localization studies and the analysis of pre-rRNA associated with different preribosomal factors. For example, late nucleoplasmic pre-60S complexes associated with Sda1 consist of apparently mature rRNA, ribosomal proteins, and a subset of preribosomal factors (24). In contrast, early nucleolar pre-60S particles with which most of the known preribosomal factors associate cannot be investigated on the basis of protein localization.
A valuable tool for the study of the early maturation events is the identification of rRNA precursors associated with different pre-60S proteins (3, 10, 15, 24, 31, 39). For example, Ssf1 is a protein involved in early steps of pre-rRNA processing since it is present in 27SA2- and 27SB-containing particles (10, 39). In contrast, other proteins, such as Nog2, associate with 27SB and 7S rRNA intermediates (31). Preribosomal factors are thus commonly associated with multiple preribosomal complexes that contain successive rRNA intermediates.
The approaches described above cannot distinguish different preribosomal complexes that are found in the same cellular compartment and that contain similar rRNA precursors. We investigated the changes in protein composition that define novel preribosomal intermediates by the isolation and characterization of complexes blocked at a particular maturation stage. This approach, combined with phenotypic analysis of novel preribosomal factors Rlp24 and Nog1, allowed us to define a model for pre-60S assembly in the nucleolus. In this model, presented in Fig. 7, an order of association and dissociation for Ssf1, Mak11, Rlp24, Nog1, and Nog2 on preribosomal particles is presented. The results that support the key elements of this model are summarized below.
FIG. 7.
A model for sequential protein assembly and intracellular trafficking of pre-60S particles. Early pre-60S particles formed in the nucleolus contain Mak11, Ssf1, and Rlp24. The arrival of Nog1 is probably concomitant with Mak11 dissociation. After 27SA2-to-27SB processing, Nog2 associates with pre-60S concomitantly or after Ssf1 has left the particles. Late complexes containing Nog1, Nog2, and Rlp24 are transported to the nucleoplasm, where Nog1 and Nog2 dissociate, apparently before export to the cytoplasm. Finally, Rlp24 dissociates from cytoplasmic pre-60S particles and is likely to be exchanged for its ribosomal homologue, Rpl24.
Rlp24 ensures the docking of Nog1 to pre-60S particles.
We show here that Rlp24, a protein similar to the ribosomal protein L24e, is associated with pre-60S complexes all along the 60S biogenesis pathway from its assembly in the nucleolus to late cytoplasmic maturation steps. Rlp24 is located not only in the nucleus, like the majority of the known preribosomal proteins, but also in the cytoplasm (Fig. 1B, bottom). Rlp24 is neither free nor present in the polysomal fractions but is exclusively associated with large ribonucleoprotein complexes (Fig. 1C and data not shown). The cytoplasmic fraction of Rlp24 was lost when ribosome maturation and export were impaired (Fig. 1D to F), suggesting that Rlp24 is transported from the nucleus to the cytoplasm in association with the nascent ribosomes and that the cytoplasmic fraction of Rlp24 is associated with pre-60S particles. In Rlp24 complexes isolated by to tandem affinity purification we found both early preribosomal factors such as Ssf1 (10) and late preribosomal factors such as Arx1 (24) (Fig. 2A and Table 1). Moreover, the rRNA precursors 27SA2, 27SA3, 27SB, and 7S, as well as the mature rRNAs of the 60S ribosomal subunits, are enriched in complexes purified in association with Rlp24 (Fig. 2B to D). In conclusion, Rlp24 is a “core” preribosomal protein associated with most, if not all, of the intermediates that are generated during 60S ribosomal subunit biogenesis and follows the pre-60S precursors from their formation in the nucleolus until after export to the cytoplasm.
Several lines of evidence point to essential roles for Rlp24 in pre-60S stability and assembly. Rlp24 may be required for early pre-60S assembly because we observed a reduction in the abundance of complexes associated with Ssf1 or Nog1 when Rlp24 was depleted (not shown). Rlp24, already present in early, 27SA2-containing complexes, may ensure the specific docking of Nog1 to these pre-60S particles. In support to this view, we demonstrate here that Nog1 and Rlp24 not only interact directly (Fig. 6B) but also interact genetically (Fig. 6A). This interaction may explain the similar effects of Rlp24 and Nog1 depletion in blocking pre-rRNA processing at the level of the 27SB intermediate (Fig. 3). Direct protein-protein interactions in ribosome biogenesis by the isolation of multiprotein subcomplexes in wild-type cells (14) or after destabilization of pre-rRNAs (15) have been recently reported although the role of these interactions remains poorly understood.
Nog1 is essential for Nog2 association with the preribosomes.
Our data on the Nog1 depletion phenotype and the analysis of Nog1 localization and the associated pre-60S complexes showed that, while being very similar to Rlp24, Nog1 follows the pre-60S particle maturation on a shorter time frame. It dissociates from pre-60S particles in the nucleus or shortly after export to the cytoplasm, as demonstrated by its apparently exclusive nuclear localization (27, 30) (data not shown). Nog1 depletion does not affect the assembly and the stability of early pre-60S complexes, as demonstrated by the purification of Rlp24- and Ssf1-associated pre-60S particles from Nog1-depleted cells (Fig. 5A and C and data not shown). However in the absence of Nog1, later assembly of Nog2 is impaired (Fig. 5B), providing the first direct evidence that the association of late preribosomal factors is dependent on the presence of earlier factors on the particles (Fig. 7).
The same pre-rRNA-containing complexes differ according to their protein compositions.
Until now, the existence of several pre-60S complexes was predicted on the basis of the knowledge of the well-ordered steps of rRNA maturation. Careful examination of the compositions of the complexes isolated in association with Ssf1, Rlp24, and Nog1 in wild-type versus mutant cells revealed that the presence of a few preribosomal proteins in these particles is mutually exclusive. Mak11, a protein that is essential for 60S ribosomal subunit formation (25), is present in both Rlp24 (Fig. 2A) and Ssf1 (not shown) complexes. Moreover, this protein is clearly enriched in both Rlp24 and Ssf1 complexes after Nog1 depletion (Fig. 5A and data not shown). We conclude that Mak11 is transiently present on early pre-60S particles and leaves these complexes before, or shortly after, Nog1 association (Fig. 7). In contrast, Ssf1 was found on intermediate particles together with Nog1 in complexes containing 27SA2 and 27SB precursors (10) (Fig. 5C).
Ssf1 has been described as an early-acting pre-60S factor on the basis of its nucleolar localization, the pre-rRNA composition of its associated complex, and its depletion phenotype (10). We show here that the presence of Nog2 and Ssf1 in pre-60S complexes seems to be mutually exclusive (Fig. 7) even if both these proteins are associated with 27SB pre-rRNAs (10, 31; C. Saveanu and M. Fromont-Racine, unpublished results). Nog2 was not identified as a component of Ssf1 complexes (10, 13), and we could not detect Nog2 in Ssf1 complexes when using specific antibodies (Fig. 5C). Ssf1 is an early-acting pre-60S factor (10) that is clearly enriched in the Rlp24-associated complexes blocked at the level of 27SB processing (Fig. 5A and 3, which shows the effects of Nog1 depletion on the steady-state levels of rRNA intermediates). Nog2, which we previously reported as a late-acting preribosomal protein (31), was absent from the blocked complexes when Nog1 was depleted (Fig. 5B). We conclude that Ssf1 participates in the assembly of 27SA2 particles and dissociates from 27SB particles independently of Nog1. It is only after Ssf1 dissociation and in the presence of Nog1 that Nog2 becomes associated with 27SB particles. This is the first direct evidence for the existence of distinct pre-60S complexes which differ only by their protein compositions and not by the presence of different pre-rRNAs. At least two 27SB-containing pre-60S particles exist, an early one containing Ssf1 and a later one containing Nog2.
A function for Rlp24 in the cytoplasm?
Rlp24 has a sequence very similar to that of the cytoplasmic Rpl24 ribosomal protein, and Rlp24-like sequences are present in all eukaryotes. In Archaea, there is only one Rpl24e protein that is more similar to the preribosomal Rlp24 than to the ribosomal protein Rpl24 (Fig. 1). The N-terminal region of Rlp24 is highly conserved and contains a putative zinc finger motif of the form C-X2-C-X21-C-X2-C. The four cysteines of this motif are conserved in all Rlp24-like sequences, including those of Archaea, whereas the second and the third cysteines are not conserved in Rpl24-like sequences. The C-terminal part of Rlp24 is more divergent and is not present in the Archaea sequences. These observations suggest that a gene for an ancient Rpl24/Rlp24 ribosomal protein duplicated early during eukaryotic evolution.
The structure of the large 50S subunit from the halophilic archaean Haloarcula marismortui reveals that Rpl24e is located at the interface with the small ribosomal subunit (2). Since only one Rpl24e is present per ribosome, we speculate that, in eukaryotes, Rlp24 and Rpl24 successively occupy the same position on the ribosome and interact directly with the rRNA. In support to this model we show here that, when ribosome assembly and export are impaired, Rpl24 remains cytoplasmic, in contrast to ribosomal proteins such as Rpl25, which accumulates in the nucleus under the same conditions (Fig. 1D and F). Moreover, Rpl24 was shown to assemble late in the cytoplasm with the nascent ribosomes (20). Since Rlp24 dissociates from pre-60S particles in the cytoplasm (Fig. 1B), the exchange with Rpl24 might take place in late cytoplasmic pre-60S particles. The factor that triggers Rlp24 dissociation from pre-60S particles does not seem to be the cytoplasmic Rpl24 itself since Rlp24 localization and its sedimentation profile on sucrose gradients are not affected by the double deletion of RPL24A and RPL24B (Saveanu and Fromont-Racine, unpublished results). Candidates for Rlp24 recycling factors are cytoplasmic pre-60S-associated factors such as Lsg1 (24) and cytoplasmic GTPases such as Efl1, a protein that was shown to be involved in Tif6 recycling (32).
We show here that, unlike Rpl24, Rlp24 is involved in a direct, specific interaction with Nog1 (Fig. 6). This interaction is likely to be essential for the assembly of functional pre-60S particles. Strikingly, the equivalent of ribosomal protein L24 from plants specifically interacts with TAV, a viral translational regulator (26). It is thus likely that Rpl24e proteins developed specific protein interactions with factors that are specialized in ribosome biogenesis or translation. While the role of Rlp24 in the maturation of the ribosomes is essential (Fig. 3), Rpl24 is not essential in yeast and has a role in translation (9).
In conclusion, this study reveals the existence of several preribosomal complexes of different protein compositions associated with identical pre-rRNA intermediates. Our analysis shed light on the assembly and dissociation of some pre-60S factors and allowed us to show that specific protein-protein interactions could mediate the assembly events. Applied systematically, this approach should allow the complete characterization of distinct successive complexes that define the ribosome biogenesis pathway in eukaryotes, as well as the roles of preribosomal proteins in guiding the assembly and dissociation of other factors.
Acknowledgments
We thank L. Decourty for performing the Rlp24 high-copy-number suppressor screen and J.-C. Rain (Hybrigenics, Paris, France) for providing the results of a Nog1 two-hybrid screen. We thank P. Lenormand for his assistance in mass spectrometry experiments and D. Montean for the generation of polyclonal antibodies. We are grateful to P. Legrain (Hybrigenics) for critical reading of the manuscript. We thank B. Seraphin (CGM-CNRS, Gif-sur-Yvette, France) for providing the plasmid pBS1479, A. Johnson for sharing data prior to publication and for providing the Nmd3 plasmids, and E. Hurt (Biochemie-Zentrum, Heidelberg, Germany) for the plasmid expressing the Rpl25-eGFP reporter gene. The SC1101 strain was obtained from EUROSCARF and has been constructed by CellZome AG (Heidelberg, Germany.
This work was supported by a grant of the European Union, the RNOMICS network (QLG2-CT-2001-01554). C.S. received financial support from Groupe d'Intérêt Publique-Aventis.
REFERENCES
- 1.Bachellerie, J. P., B. Michot, M. Nicoloso, A. Balakin, J. Ni, and M. J. Fournier. 1995. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci. 20:261-264. [DOI] [PubMed] [Google Scholar]
- 2.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 4.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand, E., F. Houser-Scott, A. Kendall, R. H. Singer, and D. R. Engelke. 1998. Nucleolar localization of early tRNA processing. Genes Dev. 12:2463-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonangelino, C. J., E. M. Chavez, and J. S. Bonifacino. 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2486-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugeron, M. C., and P. Linder. 1998. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4:566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dresios, J., I. L. Derkatch, S. W. Liebman, and D. Synetos. 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39:7236-7244. [DOI] [PubMed] [Google Scholar]
- 10.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 11.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 12.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 14.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 15.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 16.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, M. Tyers, J. S. Andersen, C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, and A. I. Lamond. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 18.Hurt, E., S. Hannus, B. Schmelzl, D. Lau, D. Tollervey, and G. Simos. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144:389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine, D. L., and D. Tollervey. 1998. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochemical Sci. 23:383-388. [DOI] [PubMed] [Google Scholar]
- 23.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 24.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtake, Y., and R. B. Wickner. 1995. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol. Cell. Biol. 15:2772-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, H. S., A. Himmelbach, K. S. Browning, T. Hohn, and L. A. Ryabova. 2001. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106:723-733. [DOI] [PubMed] [Google Scholar]
- 27.Park, J. H., B. C. Jensen, C. T. Kifer, and M. Parsons. 2001. A novel nucleolar G-protein conserved in eukaryotes. J. Cell Sci. 114:173-185. [DOI] [PubMed] [Google Scholar]
- 28.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 29.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 30.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapman, J., J. Retel, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 36.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412-1416. [PubMed] [Google Scholar]
- 37.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 38.Warner, J. R. 1974. The assembly of ribosomes in eukaryotes, p. 461-488. In M. Nomura, A. Tissieres, and P. Lengyel (ed.), Ribosomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Wehner, K. A., and S. J. Baserga. 2002. The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]