Abstract
Bicoid (Bcd) is a Drosophila melanogaster morphogenetic gradient that controls embryonic patterning by activating target gene expression in a concentration-dependent manner. In this study we describe experiments to determine how different enhancers respond to Bcd distinctively, focusing on two natural Bcd-responsive enhancer elements, hunchback (hb) and knirps (kni). Our results show that, on the hb enhancer element, the amino-terminal domain of Bcd (residues 1 to 91) plays primarily an inhibitory role, whereas on the kni enhancer element this same Bcd domain plays a positive role at low protein concentrations. We further demonstrate that while the amino-terminal domain is largely dispensable for cooperative binding to the hb enhancer element, it is preferentially required for cooperative binding to the kni enhancer element. Alteration of the arrangement of Bcd binding sites in the kni enhancer element reduces the role of the amino-terminal domain in cooperative DNA binding but increases the effectiveness of the self-inhibitory function. In addition, elimination of symmetric pairs of Bcd binding sites in the kni enhancer element reduces both DNA binding and activation by Bcd. We propose that the amino-terminal domain of Bcd is an enhancer-specific switch that contributes to the protein's ability to activate different target genes in distinct manners.
Bicoid (Bcd) is a Drosophila melanogaster morphogenetic protein required for anterior patterning during early embryogenesis (8, 24). Embryos lacking Bcd fail to develop anterior structures, including the head and thorax (13). Bcd protein, which is synthesized from the maternally contributed and anteriorly localized bcd mRNA (4), is distributed in early embryos as an anterior-to-posterior gradient (11). This 489-amino-acid protein is a homeodomain-containing transcriptional activator, with its homeodomain located at residues 92 to 151 (4). An essential function of Bcd during development is to activate gap genes in a concentration-dependent manner (8). For example, while Bcd activates the head-specific gap gene orthodenticle (otd) in the most anterior part of the embryo (14, 15), it activates the expression of hunchback (hb), a gap gene required for thoracic development (30), in the anterior half of the embryo (10, 28). Another gap target gene of Bcd, knirps (kni), contains a Bcd-responsive enhancer element that can sense even lower concentrations of Bcd in the embryo (6, 26). How Bcd activates different target genes at discrete concentrations remains poorly understood, and a previously proposed simple affinity threshold model has been challenged by recent findings (see Discussion for details).
Previous studies have shown that Bcd binds DNA in a highly cooperative manner (19). Such cooperativity is likely to play an important role in facilitating the sharp on/off switches of target genes, such as hb, in response to the Bcd gradient in embryos. Cooperative DNA binding by Bcd is achieved through interactions between Bcd molecules (19), primarily relying on protein sequences outside the homeodomain (35), including the amino-terminal domain (residues 1 to 91). In particular, the homeodomain alone, which fails to self associate efficiently in vitro, has little cooperative DNA binding activity (35, 37). Experiments that used an altered-specificity mutant of another homeodomain protein, Ftz(Q50K), further suggest that protein-protein interaction mediated by Bcd sequences outside its homeodomain represents an important mechanism for selecting specific target genes for activation (36, 37).
In addition to protein-protein interaction and cooperative DNA binding, the evolutionarily conserved amino-terminal domain of Bcd also provides a self-inhibitory function (38). In particular, Bcd derivatives lacking the amino-terminal domain exhibit a greatly increased ability to activate a hb-CAT reporter gene containing the Bcd-responsive hb enhancer element in Drosophila S2 cells. A mutant protein, Bcd(A52-56), which has the critical residues 52 to 56 mutated to alanines, exhibits a dominant effect in wild-type embryos, causing a posterior shift of the fatemap and patterning defects (38). The expression of Bcd target genes and other segmentation genes is also significantly altered in embryos containing Bcd(A52-56) (38; unpublished data). These results suggest that the self-inhibitory function provided by the amino-terminal domain of Bcd is essential to embryogenesis.
In this report we demonstrate that, depending on enhancer sequences, the amino-terminal domain of Bcd is preferentially utilized for one of two distinct functions: cooperativity or self inhibition. Specifically, while cooperative DNA binding to the kni enhancer element is highly dependent on the amino-terminal domain of Bcd, this domain is largely dispensable for cooperative binding to the hb enhancer element. In contrast, while activation from the hb enhancer element is highly sensitive to the self-inhibitory function, mutations affecting this function only cause limited effects on the kni enhancer element. Our further analysis of kni enhancer derivatives suggests that enhancer structure plays a critical role in determining the characteristics in responding to Bcd. We propose that a coordinated execution of the two functions provided by the amino-terminal domain—cooperativity and self inhibition—helps define distinctive responses to the Bcd gradient.
MATERIALS AND METHODS
Plasmid construction.
Table 1 lists the plasmids used in this study. The genes encoding truncated and point mutation derivatives of Bcd were generated by a PCR-mediated mutagenesis procedure with pFY441 (38) as the template, which is based on pGEM3 and contains wild-type bcd with the hemagglutinin (HA) tag-coding sequence immediately upstream of the unique NdeI site. To construct pFY7009, the resulting PCR product was cloned into the NdeI-XbaI sites of pFY441 to replace the full-length bcd. The plasmids pFY7015 and pFY443 were described previously (38). The plasmids expressing Bcd-VP16 derivatives in yeast cells were based on AAH5 (1) with the yeast Leu2 marker. These deletion derivatives were generated by PCR. Effector plasmids expressing Bcd derivatives in S2 cells were based on pAc5.1/V5-HisC vector (Invitrogen). The kni enhancer derivatives kni(3R), 3HH, and 3TT were generated by annealing oligonucleotides and filling in by reverse transcriptase. These DNA fragments were cloned into the EcoRI-NotI sites of pBluescript KS(−) vector (Stratagene) to generate pDF501, pDF510, and pDF511. The oligonucleotides were as follows: 5′-AATTCGTACTGGCTTAGGCGATTTCGTTACGCGATTAGGGGATCAGCTTACCAGCTTAGCAGATTATCCTAGC and 5′-GGCCGCTAGGATAATCTGCTAAGCTGGTAAGCTGATCCCCTAATCGCGTAACGAAATCGCCTAAGCCAGTACG for kni(3R), 5′-CCGGAATTCGTAATCCAGGGATTACGCCATAGACAACCGGTGGACAACGTAATCCAGGGATTACGGATCCAAGTGCGC and 5′-TTTTCCTTTTGCGGCCGCGTAATCCGCGGATTACGAACGCTATGCGCACTTGGATCCGTAATCC for 3HH, and 5′-CCGGAATTCGGATTACGTTACCTAATCCCCCCATAGACAACCGGTGGACAACCGGATTACGTTATGTAATCCCGGATCCAAG and 5′-TTTTCCTTTTGCGGCCGCGGATTAGCCTACGTAATCCCGAACGCTATGCGCACTTGGATCCGGGA TTACA for 3TT. To construct pDF520, pDF521, and pDF522, the NotI (Klenow filled-in)-SalI fragments from pDF501, pDF510, and pDF511 were inserted into the XhoI (Klenow filled-in)-SalI sites of pCZ3005 (37) to replace the 250-bp natural hb enhancer.
TABLE 1.
Plasmids used in this study
| Activator or reportera | Plasmid | Note | Reference or source |
|---|---|---|---|
| Plasmids for in vitro transcription and translation | |||
| Bcd(WT) | pFY441 | SP6 promoter | 38 |
| Bed(92-489) | pFY7015 | SP6 promoter | 38 |
| Bed(1-246) | pFY443 | SP6 promoter | 38 |
| Bcd(92-246) | pFY7009 | SP6 promoter | This study |
| Plasmids for transient transfection | |||
| Bcd(WT) | pFY442 | Drosophila actin 5C promoter | 38 |
| Bcd(92-489) | pFY413 | Drosophila actin 5C promoter | 38 |
| Bcd(A52-56) | pFY436 | Drosophila actin 5C promoter | 38 |
| Bcd(A57-61) | pFY465 | Drosophila actin 5C promoter | 38 |
| Effector plasmids for yeast cells | |||
| Bcd(WT)-VP16 | pMA1226 | LEU2; yeast ADH1 promoter | 20 |
| Bcd(92-152)-VP16 | pCZ74 | LEU2; yeast ADH1 promoter | This study |
| Bcd(92-165)-VP16 | pCZ1007 | LEU2; yeast ADH1 promoter | This study |
| Bcd(92-205)-VP16 | pCZ1008 | LEU2; yeast ADH1 promoter | This study |
| Bcd(92-246)-VP16 | pCZ1010 | LEU2; yeast ADH1 promoter | This study |
| Bcd(1-246)-VP16 | pCZ71 | LEU2; yeast ADH1 promoter | This study |
| Bcd(12-246)-VP16 | pCZ1011 | LEU2; yeast ADH1 promoter | This study |
| Bcd(27-246)-VP16 | pCZ1012 | LEU2; yeast ADH1 promoter | This study |
| Bcd(42-246)-VP16 | pCZ93 | LEU2; yeast ADH1 promoter | This study |
| Bcd(1-151)-VP16 | pCZ32 | LEU2; yeast ADH1 promoter | This study |
| Bcd(1-165)-VP16 | pCZ69 | LEU2; yeast ADH1 promoter | This study |
| Bcd(1-205)-VP16 | pCZ70 | LEU2; yeast ADH1 promoter | This study |
| Reporter plasmids | |||
| hb-CAT | pCZ3005 | CAT reporter plasmid | 37 |
| kni-CAT | pCZ3006 | CAT reporter plasmid | 37 |
| kni(3R)-CAT | pDF520 | CAT reporter plasmid | This study |
| 3HH-CAT | pDF521 | CAT reporter plasmid | This study |
| 3TT-CAT | pDF522 | CAT reporter plasmid | This study |
| hb-lacZ | pMA630R | Integrating yeast reporter plasmid | 9 |
| kni-lacZ | pTA123 | Integrating yeast reporter plasmid | 37 |
WT, wild type.
Gel shift assays.
To generate the radioactively labeled probes for gel shift experiments, the DNA fragments containing the natural or artificial enhancers were isolated from the respective plasmids and were filled in with Klenow in the presence of [α-32P]dCTP. In our experiments the kni probe was isolated as an XbaI-SacI fragment from pCZ72, and the hb probe was isolated as an XhoI-XbaI fragment from pMAX1 (37). The kni enhancer derivatives kni(3R), 3HH, and 3TT were isolated as EcoRI-NotI fragments from pDF501, pDF510, and pDF511, respectively. Probes for single pairs of head-to-head (HH) and tail-to-tail (TT) sites were isolated as NotI-BamHI fragments from pDF510 and pDF511, respectively. The probes were diluted to a final concentration of 1.6 × 10−11 M. The experimental procedures and conditions for gel shift assays were described previously (37). Wild-type Bcd and its derivatives used in this assay were expressed in vitro by using TnT quick coupled transcription/translation systems (Promega). The active protein concentrations were estimated by using high concentrations (4 × 10−8 M) of a 32P-labeled consensus Bcd binding site, A1 (7). The same amounts of active Bcd proteins were used in gel shift experiments. The Molecular Dynamics PhosphorImager ImageQuant program was used for quantitative analysis.
Yeast strain and β-galactosidase liquid assays.
The yeast strain used in this study is CY26 containing the integrated hb-lacZ or kni-lacZ reporter gene (37). The Bcd-VP16 effector plasmids were introduced into yeast cells by using the lithium acetate method, and three independent transformants were assayed for β-galactosidase activities as described previously (37).
Transient transfection experiments.
Drosophila S2 cells (Invitrogen) were grown at 25°C in Schneider's Drosophila medium (GIBCO) supplemented with 10% fetal bovine serum (GIBCO). The cells were seeded in 60-mm-diameter tissue culture plates at 4 × 106/plate 24 h prior to transfection. Transfection was performed by the calcium phosphate transfection system following the protocol from GIBCO. Each plate was transfected with 1 μg of reporter plasmid, 1 μg of Copia-lacZ internal control plasmid, and the indicated amount of effector plasmids. Salmon sperm carrier DNA (GIBCO) was used to bring the total amount of DNA to 10 μg for each transfection. The cells were harvested 48 h later, and whole-cell lysates were prepared by using the lysis buffer (50 mM HEPES [pH 7.5], 300 mM NaCl, 0.5% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) following the freeze-thaw procedure (3). The transfection efficiency was determined by monitoring the β-galactosidase activity, and the amounts of lysates used in chloramphenicol acetyltransferase (CAT) and Western blotting assays were normalized accordingly. CAT assays were performed as described previously by using three independently transfected samples for each experiment (3). For Western blotting, cell lysates were separated on 10% polyacrylamide sodium dodecyl sulfate gels and were transferred to nitrocellulose membrane (Bio-Rad), which was blotted with anti-HA monoclonal antibody (HA-11; 1:500 final dilution; Babco).
RESULTS
The amino-terminal domain of Bcd plays different roles on two enhancer elements.
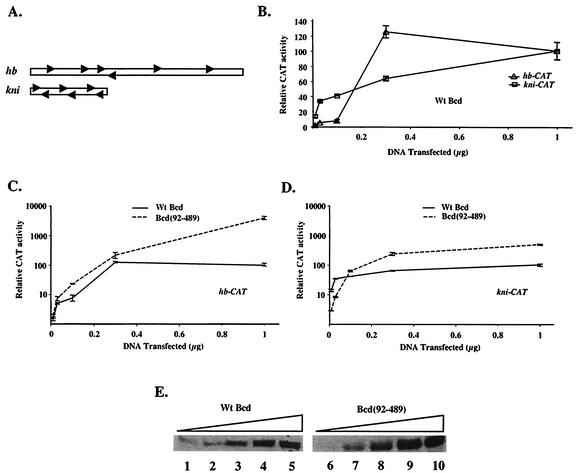
Both hb and kni are direct target genes that respond to the Bcd gradient during early embryonic development (25). The Bcd-responsive enhancer elements from these genes are sufficient to sense different Bcd concentrations in embryos (6, 10, 26, 28). Although the 250-bp hb enhancer element and the 64-bp kni enhancer element (referred to as the hb and kni enhancers hereafter) each contain six Bcd binding sites, these sites are arranged differently (Fig. 1A; also see below). We conducted a titration experiment with transiently transfected Drosophila S2 cells to determine whether such differential responses to Bcd can be recapitulated in vitro. In our experiments, the reporter genes hb-CAT and kni-CAT under the control of the respective hb and kni enhancers were cotransfected with increasing amounts of an effector plasmid expressing the wild-type Bcd protein. Our CAT assay experiments (Fig. 1B) demonstrate that the kni-CAT reporter responded to Bcd at lower concentrations than the hb-CAT reporter did, suggesting that transfection assays can reveal enhancer-specific properties of Bcd.
FIG. 1.
The amino-terminal domain of Bcd plays different roles on hb and kni enhancer elements. (A) Schematic diagrams of the 250-bp hb enhancer element and the 64-bp kni enhancer element. The Bcd binding sites are represented by arrows. (B) CAT assay results of S2 cells transfected with hb-CAT and kni-CAT reporter plasmids (1 μg) and increasing amounts of an effector plasmid expressing Bcd. Activities obtained with 1 μg of transfected effector plasmid on each reporter were arbitrarily set to 100 (fold activation was 72 and 74 for hb-CAT and kni-CAT, respectively). (C and D) CAT assay activities (in logarithmic scale) for wild-type Bcd and Bcd(92-489) on hb-CAT (C) and kni-CAT (D) reporters at different concentrations. The activities of wild-type Bcd at 1 μg of transfected effector DNA on each reporters were set to 100. (E) Representative Western blot results detecting Bcd proteins in transfected cells. For the experiments shown in this figure, the amounts of the transfected effector plasmids were 0.01, 0.03, 0.1, 0.3, and 1 μg. Wt, wild type.
To analyze the roles of the amino-terminal domain of Bcd on the hb and kni enhancers, we compared dose-response profiles for wild-type Bcd and a Bcd derivative lacking this domain [Bcd(92-489)] on both hb-CAT and kni-CAT reporter genes (Fig. 1C and D; see panel E for protein levels). Our results show that Bcd(92-489) was more active on hb-CAT than wild-type Bcd at all concentrations (Fig. 1C), confirming that the amino-terminal domain predominantly plays an inhibitory role on this reporter (38). In contrast, Bcd(92-489) had a lower activity on kni-CAT than wild-type Bcd at low concentrations (Fig. 1D), suggesting that the amino-terminal domain of Bcd plays a positive role on kni-CAT under these conditions. At high concentrations, Bcd(92-489) had a higher activity than wild-type Bcd on kni-CAT, but such an activity difference was much less significant (<4-fold) than that on hb-CAT (>40-fold). Together, these results demonstrate that the amino-terminal domain of Bcd plays distinctive roles on hb and kni enhancers.
The amino-terminal domain of Bcd is required for cooperative binding to the kni but not hb enhancer.
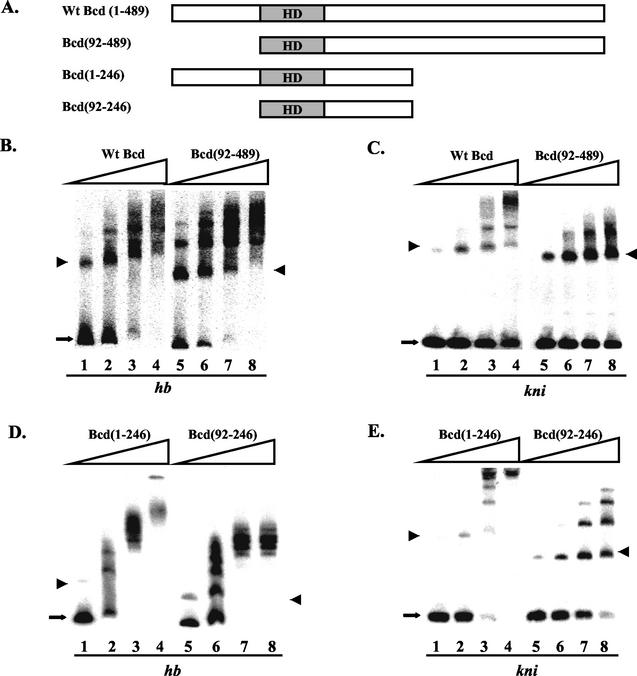
The positive effect of the amino-terminal domain of Bcd on kni-CAT expression was observed predominantly at low Bcd concentrations (Fig. 1D), suggesting that this domain may play an important role in facilitating the protein to bind the kni enhancer. To test this possibility, we conducted gel shift assays with both full-length Bcd and Bcd(92-489) on hb and kni enhancers (Fig. 2B for hb and C for kni). Our results show that wild-type Bcd can bind to both enhancer elements cooperatively (Fig. 2B and C, lanes 1 to 4). In particular, as the protein concentration increased, the monomeric complex containing a single Bcd molecule (marked with an arrowhead) was depleted to form oligomeric complexes. Interestingly, contrary to the simple affinity threshold model (see Discussion for further details), the kni enhancer did not exhibit a higher affinity for Bcd than the hb enhancer did (Fig. 2B and C, lanes 1 to 4).
FIG. 2.
The amino-terminal domain of Bcd is preferentially required for cooperative DNA binding to the kni enhancer element. (A) Schematic diagrams showing the Bcd proteins used in gel shift assays. HD, homeodomain. (B to E) Gel shift assay results on hb (B and D) and kni (C and E) probes with the indicated proteins. For these experiments, the DNA probe concentration was 1.6 × 10−11 M. The proteins were normalized according to their specific activity measured on a single consensus Bcd binding site. The estimated active protein concentrations were 8 × 10−11 M (lanes 1 and 5), 1.6 10−10 M (lanes 2 and 6), 3.2 × 10−10 M (lanes 3 and 7), and 6.4 × 10−10 M (lanes 4 and 8). The monomeric complexes are marked with arrowheads, and the free probes are marked with arrows. The data shown in panels B to E were used in a Scatchard analysis to estimate Hill coefficient values. The estimated values on hb and kni elements were, respectively, 1.7 and 2.4 for wild-type Bcd, 1.3 and 1.0 for Bcd(92-489), 3.1 and 3.7 for Bcd(1-246), and 2.3 and 1.8 for Bcd(92-246). Although poor resolution of protein-DNA complexes in some gels (due to protein sizes) offered only limited accuracy, these estimated Hill coefficient values show that the removal of the amino-terminal domain preferentially reduces cooperative binding to the kni enhancer element (over twofold reduction in Hill coefficient) compared to that with the hb enhancer element (less than 1.4-fold reduction). See Fig. 6C and D and the Fig. 6 legend for further details. Wt, wild type.
The N-terminally truncated derivative Bcd(92-489) exhibited contrasting behaviors on hb and kni enhancers in gel shift assays (Fig. 2B and C, lanes 5 to 8). This derivative bound cooperatively to the hb enhancer in a manner similar to that of the full-length protein (Fig. 2B, lanes 1 to 8), suggesting that the amino-terminal domain is largely dispensable for cooperative binding to the hb enhancer. However, Bcd(92-489) failed to bind to the kni enhancer in a highly cooperative manner (Fig. 2C, lanes 5 to 8). In particular, the monomeric complex (arrowhead) represented the major species of protein-DNA complexes at all protein concentrations. To further analyze the role of the amino-terminal domain of Bcd in cooperative DNA binding, we analyzed in gel shift assays two C-terminally truncated Bcd derivatives, Bcd(1-246) and Bcd(92-246), either containing or lacking the amino-terminal domain (Fig. 2A). Our gel shift results (Fig. 2D for hb, E for kni) with these proteins [lanes 1 to 4, Bcd(1-246); lanes 5 to 8, Bcd(92-246)] further confirm a preferential requirement of the amino-terminal domain for cooperative DNA binding to the kni enhancer. Hill coefficient estimates based on the data shown in Fig. 2 further suggest that removal of the amino-terminal domain of Bcd preferentially reduces cooperative binding to the kni enhancer (see the legend of Fig. 2 for details).
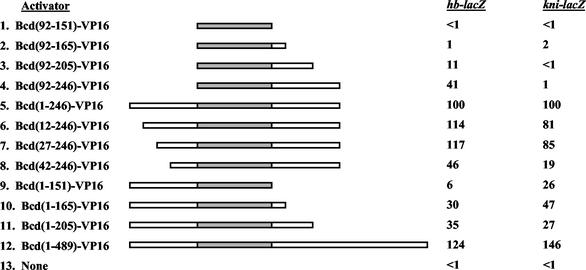
We also analyzed systematic Bcd deletion derivatives in yeast cells (Fig. 3). In this experiment, Bcd derivatives fused to the strong activation domain from VP16 (27) were compared for their abilities to activate integrated hb-lacZ and kni-lacZ reporter genes in yeast cells. Both full-length Bcd-VP16 and Bcd(1-246)-VP16 fusion proteins activated transcription from the hb-lacZ and kni-lacZ reporter genes effectively (lines 5 and 12). Therefore, all deletions were derived from the truncated protein Bcd(1-246)-VP16, with its activities on each reporter set to 100. Figure 3 shows that a derivative lacking the amino-terminal domain, Bcd(92-246)-VP16, failed to activate transcription from the kni-lacZ reporter while remaining active on hb-lacZ (line 4). In contrast, Bcd(1-151)-VP16, which lacks the Bcd sequence on the C-terminal side of its homeodomain, had a more significant reduction in activity on hb-lacZ than on kni-lacZ (line 9). The data shown in Fig. 3 further suggest that protein sequences immediately flanking the homeodomain play positive roles on kni-lacZ (compare lines 4 and 8) and hb-lacZ (compare lines 9 and 10; also compare lines 2 through 4). Interestingly, Bcd(92-151)-VP16, which contains only the homeodomain of Bcd fused to VP16, failed to efficiently activate from both reporters (line 1). Since the self-inhibitory function conferred by the amino-terminal domain of Bcd is not operative in yeast cells (38 and unpublished data) and, moreover, since all the derivatives contain the strong activation domain VP16, these results are consistent with the suggestion that different Bcd sequences outside its homeodomain play important but differential roles in cooperative binding to hb and kni enhancers.
FIG. 3.
Different domains of Bcd are required for activating transcription from hb and kni enhancers in yeast cells. Shown are relative activities of Bcd-VP16 fusion proteins on integrated hb-lacZ or kni-lacZ reporter genes in yeast cells. Only Bcd sequences are shown in schematic diagrams (not to scale), with the shaded region representing its homeodomain. The activities of Bcd(1-246)-VP16 were arbitrarily set to 100 on each reporter gene, with actual β-galactosidase activities of 576 and 91 on hb-lacZ and kni-lacZ, respectively.
The self-inhibitory function of Bcd is implemented differently on hb and kni enhancers.
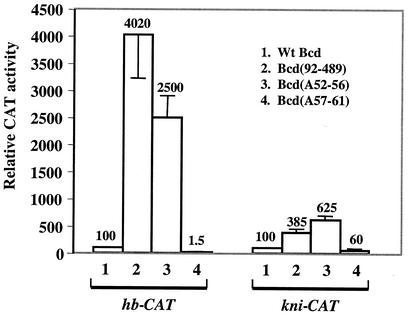
The experiments described thus far relied on either full-length or truncated Bcd derivatives. Our recent systematic analysis of the self-inhibitory function has generated point mutations within the amino-terminal domain that either disrupt or strengthen the self-inhibitory function (38 and unpublished data). In particular, Bcd(A52-56), which has residues 52 to 56 of Bcd changed to alanines, disrupts the self-inhibitory function, and results in an increased ability to activate the hb-CAT reporter (38). In contrast, Bcd(A57-61), which contains the neighboring five residues converted to alanines, further strengthens the self-inhibitory function, resulting in a virtually inactive protein on the hb-CAT reporter.
To further determine whether the self-inhibitory function provided by the amino-terminal domain of Bcd is differentially implemented on different enhancers, we took advantage of these point mutation derivatives. We performed transient transfection assays by using high protein concentrations to specifically compare the effectiveness of the self-inhibitory function on the hb-CAT and kni-CAT reporters. Figure 4 shows that, while Bcd(A52-56) was 25 times more active than wild-type Bcd on hb-CAT, the difference was only 6.25-fold on kni-CAT (lane 3). In addition, while Bcd(A57-61) was virtually inactive on hb-CAT, its activity on kni-CAT was reduced only by 40% (lane 4). As shown in Fig. 1C and D, at high protein concentrations the activity difference between full-length Bcd and the N-terminally truncated derivative Bcd(92-489) was also much greater on hb-CAT (>40-fold) than on kni-CAT (<4-fold) (Fig. 4, lane 2). Together these results demonstrate that the self-inhibitory function is more robust on hb-CAT than on kni-CAT. Interestingly, Bcd(92-489) was more active on hb-CAT than Bcd(A52-56), but the opposite was true on kni-CAT; this difference is consistent with the finding that the amino-terminal domain is preferentially required for cooperative binding to the kni enhancer (Fig. 2).
FIG. 4.
The self-inhibitory function of Bcd is differentially implemented on hb and kni enhancers. (A) CAT assay results from S2 cells transfected with 1 μg of hb-CAT or kni-CAT reporter plasmid and 1 μg of effector plasmid expressing wild-type Bcd (lane 1), Bcd(92-489) (lane 2), Bcd(A52-56) (lane 3), or Bcd(A57-61) (lane 4). The activities of wild-type Bcd on each reporter were set to 100. The data for the hb-CAT reporter, except for that of Bcd(A57-61), are from Zhao et al. (38). Wt, wild type.
Bcd binding site arrangements influence their dependence on the amino-terminal domain for cooperative DNA binding.
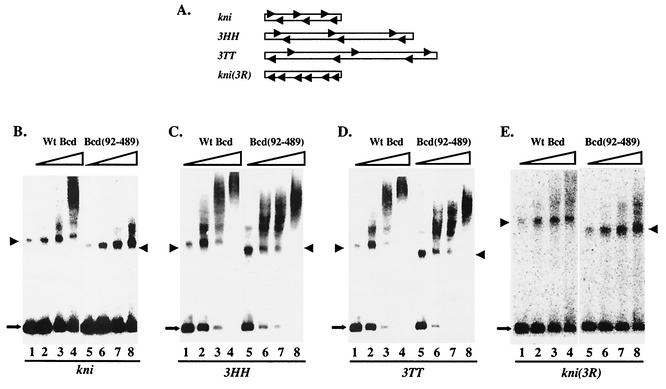
Previous binding site selection experiments have isolated from random DNA sequences symmetric head-to-head (HH) and tail-to-tail (TT) sites, suggesting that Bcd can preferentially recognize symmetric pairs of DNA sites that are separated by defined, short spacing (34). An examination of the hb and kni enhancers (Fig. 1A) reveals that, while all the Bcd binding sites in the 64-bp kni enhancer are arranged symmetrically similar to the selected patterns, those in the 250-bp hb enhancer are dispersed with only one centrally located head-to-head pair. Both enhancers have six Bcd binding sites. To determine whether the arrangements of Bcd binding sites can influence the dependence on the amino-terminal domain of Bcd for cooperative DNA binding, we generated three enhancer derivatives (Fig. 5A). In kni(3R), the orientations of three of the Bcd binding sites were reversed, eliminating all symmetric pairs of sites while maintaining the same number and spacing of the sites. Two other enhancers, 3HH and 3TT, contain three closely spaced head-to-head and tail-to-tail pairs, respectively, that are separated by long spacing (25 bp).
FIG. 5.
Both spacing and orientation of Bcd binding sites affect the role of the amino-terminal domain in cooperative DNA binding. (A) Schematic diagrams of the enhancer derivatives. (B to E) Gel shift assay results on kni (B), 3HH (C), 3TT (D), and kni(3R) (E) with either wild-type Bcd or Bcd(92-489). The estimated active protein concentrations were 8 × 10−11 M (lanes 1 and 5), 1.6 × 10−10 M (lanes 2 and 6), 3.2 × 10−10 M (lanes 3 and 7), and 6.4 × 10−10 M (lanes 4 and 8). The DNA probe concentration was 1.5 × 10−11 M. The monomeric complexes are marked with arrowheads. Estimated Hill coefficient values on 3HH and 3TT were, respectively, 3.4 and 3.2 for wild-type Bcd and 2.7 and 1.7 for Bcd(92-489). See the legend to Fig. 2 for further details. Wt, wild type.
Figure 5 shows our gel shift results on these three enhancer derivatives (and wild-type kni enhancer as a control), with full-length Bcd and Bcd(92-489). These results reveal the following findings. First, both full-length Bcd (lanes 1 to 4) and Bcd(92-489) (lanes 5 to 8) bound cooperatively to enhancer derivatives 3HH (Fig. 5C) and 3TT (Fig. 5D), suggesting that increased spacing between the symmetric pairs of Bcd binding sites reduces the dependence on the amino-terminal domain of Bcd for cooperative DNA binding (also see the legend to Fig. 5). Second, wild-type Bcd could no longer bind cooperatively to kni(3R) (Fig. 5E, lanes 1 to 4). Removing the amino-terminal domain did not further reduce DNA binding, suggesting that the amino-terminal domain plays little role in binding to kni(3R) (Fig. 5E, lanes 5 to 8). Together these results show that the role of the amino-terminal domain in cooperative binding to the kni enhancer is determined by the special arrangements of the Bcd binding sites in this enhancer. They also suggest that, when the Bcd binding sites are tightly packed, such as in the kni enhancer, cooperative DNA binding by wild-type Bcd requires relatively rigid arrangements of these sites. This is in contrast to the Bcd binding sites that are separated by long spacing, where there is no such rigid requirement of orientation or spacing (34).
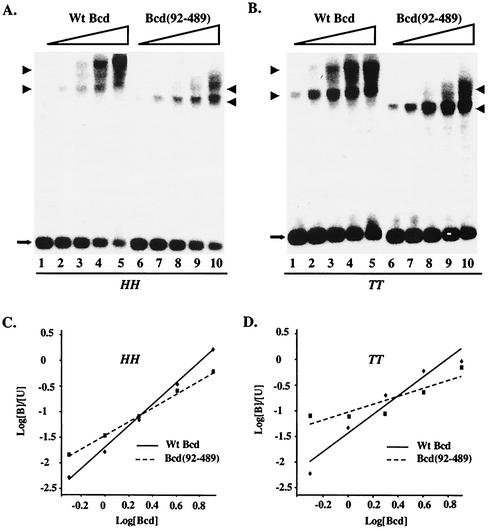
On enhancers that contain multiple Bcd binding sites, the Bcd-DNA complexes were poorly resolved in gel shift assays (Fig. 2 and 5). In contrast, the monomeric and dimeric Bcd complexes could be readily resolved on DNA probes containing only two Bcd binding sites (Fig. 6). This permitted a more accurate and quantitative analysis of the role of the amino-terminal domain in cooperative DNA binding. Specifically, we determined Hill coefficient values for full-length Bcd and Bcd(92-489) on single pairs of closely spaced HH (Fig. 6A and C) and TT (Fig. 6B and D) binding sites. Wild-type Bcd bound to both HH and TT pairs cooperatively, with an estimated Hill coefficient of 1.88 ± 0.28 and 1.48 ± 0.25, respectively. In contrast, Bcd(92-489) had a significantly reduced but measurable cooperativity on the HH pair, with an estimated Hill coefficient of 1.40 ± 0.30. This same Bcd derivative became completely noncooperative on the TT pair, with an estimated Hill coefficient of 0.66 ± 0.10, which is similar to the value (0.8) on a single Bcd binding site. These results further demonstrate that the amino-terminal domain plays an important role on closely spaced symmetric pairs of Bcd binding sites, particularly the TT pair.
FIG. 6.
The amino-terminal domain of Bcd plays an important role in cooperative binding to symmetric pairs of Bcd binding sites. (A and B) Gel shift experiments on single pairs of closely spaced HH (A) and TT (B) sites with wild-type Bcd (lanes 1 to 5) and Bcd(92-489) (lanes 6 to 10). The monomeric and dimeric complexes are marked with arrowheads, and the free probes are marked with arrows. (C and D) Hill coefficient measurements. The Scatchard plots shown here were based on the data shown in panels A and B. Hill coefficient values given in the text were averages of three independent gel shift assays. The estimated active proteins were 0.3 × 10−10, 0.6 × 10−10, 1.2 × 10−10, 2.4 × 10−10, and 4.8 × 10−10 M, expressed in arbitrary units of 0.5, 1, 2, 4, and 8, respectively. [B] and [U] represent bound and unbound probe concentrations as described previously (34). Wt, wild type.
Bcd binding site arrangements influence the effectiveness of the self-inhibitory function of Bcd.
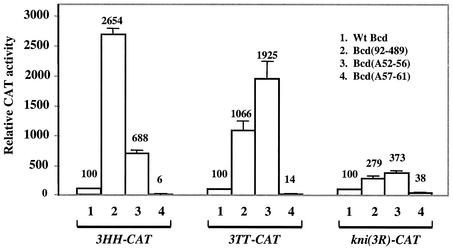
Our analysis of Bcd activities on hb and kni enhancers suggests that the amino-terminal domain is preferentially used for self inhibition and cooperativity, respectively. This further suggests that cooperative DNA binding mediated by the amino-terminal domain may preclude (or hinder) this domain from being utilized effectively for self inhibition. To further test this possibility, we compared the effectiveness of the self-inhibitory function on the artificial enhancers described above. Since both 3TT and 3HH had a reduced dependence on the amino-terminal domain for cooperative DNA binding, a prediction of the preclusion model is that the self-inhibitory function would become more robust on these enhancers than on the kni enhancer. For these experiments, we analyzed the activities of wild-type Bcd, Bcd(92-489), Bcd(A52-56), and Bcd(A57-61) on these enhancers at high protein concentrations to specifically compare the effectiveness of the self-inhibitory function.
The results shown in Fig. 7 demonstrate that both 3HH-CAT and 3TT-CAT reporters became more sensitive to the self-inhibitory function of Bcd. In particular, Bcd(A57-61) had much lower activities on both 3TT-CAT and 3HH-CAT than on kni-CAT (compare lane 4 of Fig. 7 with Fig. 4). In addition, both Bcd(A52-56) and Bcd(92-489) exhibited higher activities on 3TT-CAT and 3HH-CAT than on the kni-CAT reporter (Fig. 7, lanes 2 and 3). Our results also reveal a difference between 3HH-CAT and 3TT-CAT in their responses to the A52-56 mutation and removal of the entire amino-terminal domain (Fig. 7, lanes 2 and 3). This difference is consistent with the finding that cooperative binding to the TT pair is more dependent on the amino-terminal domain than to the HH pair (Fig. 6). Finally, although the responses to the mutations affecting the self-inhibitory function on the kni(3R)-CAT reporter were not dramatically altered compared to those of kni-CAT, all activities were significantly reduced on knic(3R)-CAT (27%) (see also reference 6). These results are consistent with the finding that Bcd fails to bind cooperatively to kni(3R) (Fig. 5), demonstrating that reversion of the orientations of Bcd binding sites in the kni enhancer alters its characteristics in responding to Bcd.
FIG. 7.
Bcd binding site arrangements influence the effectiveness of the self-inhibitory function. Shown are CAT assay results on 3TT-CAT, 3HH-CAT, and kni(3R)-CAT by using Bcd derivatives at high concentrations (1 μg of transfected effector plasmid). The Bcd proteins used in these experiments were wild-type Bcd (lane 1), Bcd(92-489) (lane 2), Bcd(A52-56) (lane 3), and Bcd(A57-61) (lane 4).
DISCUSSION
The experiments described in this report reveal that the amino-terminal domain of Bcd executes its cooperative DNA binding and self-inhibitory functions in a coordinated manner. On the hb enhancer this domain plays little role in cooperative DNA binding (Fig. 2B and D) but exhibits a potent self-inhibitory function (Fig. 4). In contrast, this domain is preferentially required for cooperative binding to the kni enhancer (Fig. 2C and E) but has a muted self-inhibitory function (Fig. 4). Our experiments further suggest that differential utilization of these two functions is reflective of the structural differences between the hb and kni enhancers. Although both enhancers have a comparable number of Bcd binding sites, they are arranged differently (Fig. 1A). The Bcd binding sites in the kni enhancer form tightly spaced symmetric pairs, whereas those in the hb enhancer are dispersed and predominantly arranged in tandem with only one closely spaced symmetric pair. Alternations of the arrangement of the Bcd binding sites in the kni enhancer dramatically changed the role of the amino-terminal domain in transcription control (Fig. 5C and D and 7). In addition, elimination of symmetric arrangements of Bcd sites significantly reduced both DNA binding (Fig. 5E) and activation by Bcd. These results demonstrate that enhancers are not merely docking sites for transcription factors; rather, their structure can dramatically influence the behaviors of activators.
Both cooperative DNA binding and self inhibition involve protein-protein interactions (19, 35, 38), suggesting that the amino-terminal domain of Bcd is engaged in distinct protein-protein interaction events on different enhancers. Our results suggest that the engagement of the amino-terminal domain in cooperative DNA binding on the kni enhancer likely hinders its availability for self inhibition. Tight spacing between the Bcd binding sites may also reduce the effectiveness of the self-inhibitory function. We presently do not know precisely how the amino-terminal domain of Bcd facilitates cooperative DNA binding. While this domain is strictly required for a single pair of closely spaced TT sites, its role on a single pair of closely spaced HH sites, though important, is somewhat less critical (Fig. 6). It is possible that the amino-terminal domains of two Bcd molecules are engaged in a direct homodimeric interaction. It is also possible that this domain of one Bcd molecule interacts with a different domain of another Bcd molecule on DNA. Our previous protein-protein interaction experiments suggest that the amino-terminal domain alone does not self associate efficiently without the homeodomain (35). In this context it is relevant that cooperative DNA binding can be achieved on the hb enhancer even without the amino-terminal domain. Bcd contains multiple self-association domains (35) which play differential roles on distinct enhancers (Fig. 3). Regardless, our findings suggest that Bcd can accommodate and coordinate multiple interaction events that involve different parts of the protein, particularly its amino-terminal domain. Interestingly, the amino-terminal domain of Bcd also contains a motif that can interact with the translation initiation factor eIF4E and participates in translation repression of caudal (cad) mRNA (23), further underscoring the importance of this evolutionarily conserved domain.
One of the fundamental biological questions concerning the action of Bcd is how different target genes respond to distinct thresholds of the Bcd gradient in embryos. It was previously proposed that the affinity of Bcd binding sites within an enhancer dictates the concentration of Bcd required for activation in embryos (12, 14). However, this simple affinity threshold model has been challenged recently (36). In particular, the assumption of this model—the consensus A-type sites have higher Bcd affinity than the nonconsensus X-type sites—has been proven incorrect. Biochemical studies have shown that Bcd binds to a consensus A1 site and a nonconsensus X1 site with comparable affinities (7, 19). In addition, the kni enhancer, despite its ability to respond to lower Bcd concentrations, does not exhibit any higher Bcd affinity than the hb enhancer (Fig. 2). A more recent study also suggests that the concentration of Bcd in embryos, though important, is unlikely to be the only determinant for defining the target gene responses to the Bcd gradient (17). Our characterizations of the self-inhibitory function in embryos suggest that the activation potential of Bcd may play an important role in controlling target gene expression. In particular, Bcd(A52-56), which has a stronger activation potential, can exert a dominant effect in wild-type embryos, causing a posterior shift of the target genes (38). These results suggest that a stronger Bcd can activate transcription at a lower concentration (or with fewer DNA-bound molecules) in embryos, a concept that has been proposed previously for various activators (2, 5, 18, 20, 29). We propose that the muted self-inhibitory function of Bcd on the kni enhancer contributes to the protein's ability to activate transcription at low concentrations.
Many homeodomain proteins have been shown to cooperate specifically with their DNA binding partners (16, 21, 22, 31-33). Although no such partner proteins have been identified to specifically help Bcd to select its target genes, this possibility cannot be formally ruled out at this time. If such proteins do exist, they could potentially play important roles in facilitating Bcd to act distinctively on different enhancers. In this context, it is interesting that the Bcd-responsive kni enhancer element used in this study contains binding sites for the Caudal (Cad) protein, although shorter kni enhancer fragments without these Cad binding sites can similarly respond to low concentrations of Bcd in embryos (26). The fact that the experiments described in this report use only Bcd-responsive enhancer elements in Drosophila cells indicates that Bcd may have an intrinsic ability to activate different target genes distinctively. These studies demonstrate that even relatively simple enhancers can have profound effects in influencing activator behaviors. Understanding precisely how complex enhancers orchestrate in vivo the individual actions of different transcription factors represents a rewarding challenge for molecular biologists.
Acknowledgments
We thank members of our laboratory for discussions and technical assistance. We are particularly grateful to Cindy Bachurski and Dihua Huang for transient transfection assays and Vrushank Dave for gel shift assays.
This work was supported in part by NIH and AHA grants (to J.M.).
REFERENCES
- 1.Ammerer, G. 1983. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 101:192-201. [DOI] [PubMed] [Google Scholar]
- 2.Arnosti, D. N., S. Barolo, M. Levine, and S. Small. 1996. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122:205-214. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Berleth, T., M. Burri, G. Thoma, D. Bopp, S. Richstein, G. Frigerio, M. Noll, and C. Nüsslein-Volhard. 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7:1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunker, C. A., and R. E. Kingston. 1996. Activation domain-mediated enhancement of activator binding to chromatin in mammalian cells. Proc. Natl. Acad. Sci. USA 93:10820-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burz, D. S., R. Pivera-Pomar, H. Jackle, and S. D. Hanes. 1998. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 17:5998-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dave, V., C. Zhao, F. Yang, C. S. Tung, and J. Ma. 2000. Reprogrammable recognition codes in Bicoid homeodomain-DNA interaction. Mol. Cell. Biol. 20:7673-7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driever, W. 1992. The Bicoid morphogen: concentration dependent transcriptional activation of zygotic target genes during early Drosophila development, p. 1221-1250. In S. L. McKnight and K. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Driever, W., J. Ma, C. Nusslein-Volhard, and M. Ptashne. 1989. Rescue of bicoid mutant Drosophila embryos by Bicoid fusion proteins containing heterologous activating sequences. Nature 342:149-154. [DOI] [PubMed] [Google Scholar]
- 10.Driever, W., and C. Nüsslein-Volhard. 1989. Bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337:138-143. [DOI] [PubMed] [Google Scholar]
- 11.Driever, W., and C. Nüsslein-Volhard. 1988. A gradient of bicoid protein in Drosophila embryos. Cell 54:83-93. [DOI] [PubMed] [Google Scholar]
- 12.Driever, W., G. Thoma, and C. Nüsslein-Volhard. 1989. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding site for the bicoid morphogen. Nature 340:363-367. [DOI] [PubMed] [Google Scholar]
- 13.Frohnhöfer, H. G., and C. Nüsslein-Volhard. 1986. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature 324:120-125. [Google Scholar]
- 14.Gao, Q., and R. Finkelstein. 1998. Targeting gene expression to the head: the Drosophila orthodenticle gene is a direct target of the Bicoid morphogen. Development 125:4185-4193. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Q., Y. Wang, and R. Finkelstein. 1996. Orthodenticle regulation during embryonic head development in Drosophila. Mech. Dev. 56:3-15. [DOI] [PubMed] [Google Scholar]
- 16.Guichet, A., J. W. Copeland, M. Erdelyi, D. Hlousek, P. Zavorszky, J. Ho, S. Brown, A. Percival-Smith, H. M. Krause, and A. Ephrussi. 1997. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385:548-552. [DOI] [PubMed] [Google Scholar]
- 17.Houchmandzadeh, B., E. Wieschaus, and S. Leibler. 2002. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature 415:798-802. [DOI] [PubMed] [Google Scholar]
- 18.Lehman, A. M., K. B. Ellwood, B. E. Middleton, and M. Carey. 1998. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J. Biol. Chem. 273:932-939. [DOI] [PubMed] [Google Scholar]
- 19.Ma, X., D. Yuan, K. Diepold, T. Scarborough, and J. Ma. 1996. The Drosophila morphogenetic protein Bicoid binds DNA cooperatively. Development 122:1195-1206. [DOI] [PubMed] [Google Scholar]
- 20.Ma, X., D. Yuan, T. Scarborough, and J. Ma. 1999. Contributions to gene activation by multiple functions of Bicoid. Biochem. J. 338:447-455. [PMC free article] [PubMed] [Google Scholar]
- 21.Mak, A., and A. D. Johnson. 1993. The carboxy-terminal tail of the homeodomain protein α2 is required for function with a second homeodomain protein. Genes Dev. 7:1862-1870. [DOI] [PubMed] [Google Scholar]
- 22.Mann, R. S., and S. K. Chan. 1996. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 12:258-262. [DOI] [PubMed] [Google Scholar]
- 23.Niessing, D., S. Blanke, and H. Jackle. 2002. Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes Dev. 16:2576-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nusslein-Volhard, C. 1991. Determination of the embryonic axis of Drosophila. Development 1(Suppl.):1-10. [PubMed] [Google Scholar]
- 25.Rivera-Pomar, R., and H. Jackle. 1996. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 12:478-483. [DOI] [PubMed] [Google Scholar]
- 26.Rivera-Pomar, R., X. Lu, H. Taubert, N. Perrimon, and H. Jackle. 1995. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature 376:253-256. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335:563-564. [DOI] [PubMed] [Google Scholar]
- 28.Struhl, G., K. Struhl, and P. Macdonald. 1989. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57:1259-1273. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, M. 1996. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc. Natl. Acad. Sci. USA 93:4311-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tautz, D., R. Lehmann, H. Schnürch, R. Schuh, E. Seifert, A. Kienlin, K. Jones, and H. Jäckle. 1987. Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature 327:383-389. [Google Scholar]
- 31.Vershon, A. K., and A. D. Johnson. 1993. A short, disordered protein region mediates interactions between the homeodomain of the yeast α2 protein and the MCM1 protein. Cell 72:105-112. [DOI] [PubMed] [Google Scholar]
- 32.Xue, D., Y. Tu, and M. Chalfie. 1993. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science 261:1324-1328. [DOI] [PubMed] [Google Scholar]
- 33.Yu, Y., W. Li, K. Su, M. Yussa, W. Han, N. Perrimon, and L. Pick. 1997. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature 385:552-555. [DOI] [PubMed] [Google Scholar]
- 34.Yuan, D., X. Ma, and J. Ma. 1999. Recognition of multiple patterns of DNA sites by Drosophila homeodomain protein Bicoid. J. Biochem. 125:809-817. [DOI] [PubMed] [Google Scholar]
- 35.Yuan, D., X. Ma, and J. Ma. 1996. Sequences outside the homeodomain of Bicoid are required for protein-protein interaction. J. Biol. Chem. 271:21660-21665. [PubMed] [Google Scholar]
- 36.Zhao, C., V. Dave, D. Fu, A. York, and J. Ma. Insights into the molecular functions of the Drosophila morphogenetic protein Bicoid. In A. Gayathri (ed.), Recent developments in molecular and cellular biology, vol. 3, in press. Research Signpost, Kerala, India.
- 37.Zhao, C., V. Dave, F. Yang, T. Scarborough, and J. Ma. 2000. Target selectivity of Bicoid is dependent on non-consensus site recognition and protein-protein interaction. Mol. Cell. Biol. 20:8112-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, C., A. York, F. Yang, D. J. Forsthoefel, V. Dave, D. Fu, D. Zhang, M. S. Corado, S. Small, M. A. Seeger, and J. Ma. 2002. The activity of the Drosophila morphogenetic protein Bicoid is inhibited by a domain located outside its homeodomain. Development 129:1669-1680. [DOI] [PubMed] [Google Scholar]