Abstract
Despite substantial progress in understanding the mechanism by which expanded CTG/CAG trinucleotide repeats cause neurodegenerative diseases, little is known about the basis for repeat instability itself. By taking advantage of a novel phenomenon, we have developed a selectable assay to detect contractions of CTG/CAG triplets. When inserted into an intron in the APRT gene or the HPRT minigene, long tracts of CTG/CAG repeats (more than about 33 repeat units) are efficiently incorporated into mRNA as a new exon, thereby rendering the encoded protein nonfunctional, whereas short repeat tracts do not affect the phenotype. Therefore, contractions of long repeats can be monitored in large cell populations, by selecting for HPRT+ or APRT+ clones. Using this selectable system, we determined the frequency of spontaneous contractions and showed that treatments with DNA-damaging agents stimulate repeat contractions. The selectable system that we have developed provides a versatile tool for the analysis of CTG/CAG repeat instability in mammalian cells. We also discuss how the effect of long CTG/CAG repeat tracts on splicing may contribute to the progression of polyglutamine diseases.
A growing number of neurodegenerative diseases such as myotonic dystrophy, Huntington's disease, and spinocerebellar ataxias have been found to result from expansion of CTG/CAG trinucleotide repeats. Normal individuals generally have fewer than 30 repeats at a disease locus, whereas affected individuals may have from 30 to several thousand repeat units, depending on the disease. The mechanism by which an expanded repeat causes the characteristic features of a disease is complex and depends on the orientation of the repeat within the gene. In myotonic dystrophy, for example, the mRNA from affected individuals carries an expanded CUG repeat in its 3′ untranslated region that may contribute to disease pathogenesis by binding CUG binding proteins, which can alter the splicing of other, unrelated mRNAs (see reference 51 for a review). Additional factors such as nuclear retention of the CUG-containing transcripts (49) and hypermethylation of the adjacent chromatin (48) may also contribute to the disease phenotype. In Huntington's disease and spinocerebellar ataxias 1, 2, 3, 6, and 7, a CAG trinucleotide repeat is expanded within the coding region of the affected gene. The resulting neurodegeneration has been linked to accumulation of aberrant polyglutamine-containing proteins, which induce neuronal death (see reference 55 for a review).
The exact mechanism leading to expansion of trinucleotide repeats is unknown. It is likely to be related to the ability of repeat tracts to form unusual DNA secondary structures such as hairpins and slipped-strand DNA duplexes, which can interfere with aspects of DNA metabolism (see reference 52 for a review). Both Escherichia coli and Saccharomyces cerevisiae have been used as model systems to study the instability of CTG/CAG repeats. Virtually every process that exposes single strands of DNA destabilizes triplet repeats, including transcription (2, 45), nucleotide excision repair (35, 38), mismatch repair (21, 42, 44, 46), replication (17, 22, 30, 43), and recombination (9, 18-20, 39, 40). CTG/CAG triplet repeats also cause double-strand DNA breaks in yeast (9, 20). Similarly, studies with mammalian cells have shown that DNA replication (6, 37), mismatch repair (23, 27, 53), and proximity to CpG islands (4) contribute to destabilization of triplet repeats. Based on these studies, several models of repeat expansion have been proposed (reviewed in references 3 and 52). Small changes in repeat length may be caused by the slippage of DNA polymerases, while larger changes may result from errors of DNA repair machinery. For example, if single- or double-strand breaks are formed close to the repeat tract, flaps, hairpins, or other complex DNA structures could form at the ends, leading to errors in DNA repair. Furthermore, alternative DNA structures formed at the repeat locus might by themselves be recognized by DNA repair proteins (such as mismatch repair machinery), which could lead to aberrant processing and promote repeat expansions or contractions (reviewed in reference 47).
Understanding the mechanisms that lead to expansion of triplet repeats may allow development of approaches to prevent the lengthening of repeat tracts or even to induce contractions of the expanded repeats in order to stop the progression of neurodegenerative disorders. To search for genetic factors or therapeutic treatments that affect repeat stability, it is critical to use a sensitive assay. Assays based on inactivation of a selectable reporter gene such as URA3 or chloramphenicol acetyltransferase have been developed for yeast (34, 41) and E. coli (14). However, to achieve the ultimate goal of finding a cure for triplet repeat diseases, it is essential to find treatments that are effective in mammalian cells. Presently, trinucleotide repeat instability is assayed in mammalian cells by methods such as small-pool PCR and GeneScan, which can detect frequencies of repeat change in the range from 10−2 to 10−3, and thus lack the sensitivity of a selectable genetic assay.
In this report we describe a new genetic assay for trinucleotide repeat contractions in mammalian cells that is selective and quantitative. The assay is based on the novel finding that long CAG repeats cloned into an intron of a reporter gene disrupt correct splicing and become incorporated into mRNA, thereby inactivating the gene. Using this system, we demonstrate that aphidicolin and hydroxyurea, which affect DNA replication, and gamma irradiation, which induces DNA breaks, destabilize long repeat tracts in mammalian cells. The selectable system that we have developed provides a versatile tool for the further analysis of CTG/CAG repeat instability.
MATERIALS AND METHODS
Construction of plasmids.
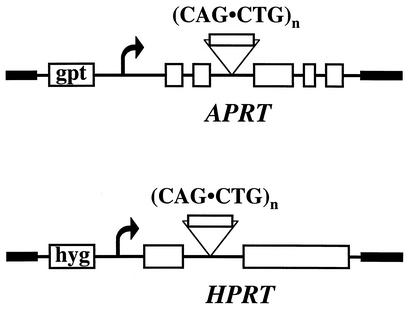
The plasmids containing CTG/CAG repeats in the APRT gene were constructed as follows: the CTG/CAG repeat tract that included 19 nucleotides of 5′ and 43 nucleotides of 3′ flanking sequences from the myotonic dystrophy locus was inserted into the polylinker in intron 2 of the APRT gene (17). CTG/CAG repeat tracts in plasmids pRW3502, pRW3504, and pRW3506 are 17, 98, and 175 repeats in length, respectively (17). The NotI sites that flank the human sequences in these plasmids were used to clone the repeats in both orientations into the NotI site in the polylinker with an adjacent FRT site in the second intron of the APRT gene in plasmid pJHW1. This series of plasmids carries an otherwise wild-type copy of the hamster APRT gene and an adjacent bacterial GPT gene (Fig. 1).
FIG. 1.
Reporter cassettes for detection of repeat instability. CTG-CAG repeat tracts of various lengths were cloned in two orientations into intron 2 of the hamster APRT gene or the single intron in the human HPRT minigene, as described in Materials and Methods.
In order to construct the plasmids containing CTG/CAG repeats of various length in the HPRT minigene, the (CTG)98 tract was subcloned into pUC19 and was propagated in E. coli to obtain length variants. The repeat tracts flanked by NotI sites were then cloned into the XbaI site within the intron of the HPRT minigene (13, 29) in pMHAd, which also contains the hygromycin resistance gene. The pMHAd plasmid consists of the KpnI-BamHI fragment from pGKhprt-mini5 (kindly provided by A. Bradley) cloned into pINd-Hygro (Invitrogen).
Cell lines.
For integration of CAG repeat tracts at the endogenous APRT locus in CHO cells, the XhoI site in the third exon of the APRT gene in pJHW1 plasmids carrying (CAG)98 and (CAG)175 was cleaved, filled in, and ligated. FLP recombinase-mediated site-specific integration at the FRT site in RMP34 cells, which carry a wild-type APRT gene with an FRT site in the second intron (31), was expected to generate an upstream APRT− gene that carried the XhoI site mutation and a downstream gene that was APRT− by virtue of the CAG triplet repeat. As described previously (31), this procedure generates apparent single-step replacements at a frequency of about 25%. Two such single-step replacements were isolated from transfections with (CAG)98 and (CAG)175 and were shown by sequencing to carry (CAG)95 and (CAG)61, respectively, at the expected location in intron 2 in an otherwise wild-type APRT gene. The structure of the integrated constructs was verified by Southern blotting. It is unclear at what point the changes in repeat tract length occurred, but single colonies were isolated and confirmed by DNA sequencing to contain (CAG)95 and (CAG)61.
APRT and HPRT selection assays.
Cells were plated at 5 × 105 cells per 100-mm-diameter plate in the presence of ALASA selection (50 μM azaserine, 25 μM alanosine, and 100 μM adenine) to select for APRT+ colonies or in the presence of HAT selection (0.1 mM hypoxanthine, 0.4 μM aminopterine, and 16 μM thymidine) to select for gpt+ or HPRT+ colonies. Plate contents were incubated undisturbed for 3 weeks. The colonies were then picked for analysis or stained with 1% Coomassie blue for counting.
Northern blot analysis and reverse transcriptase-PCR (RT-PCR).
Northern blot analysis was performed by using the NorthernMax-Gly kit (Ambion) according to the manufacturer's instructions. The probes for Northern analysis of APRT transcripts were two PCR products amplified from exons 2 and 3 of the APRT gene, which were subsequently labeled by random priming in the presence of [32P]dCTP.
For RT-PCR, total RNA was extracted from cells by using an RNA-Easy kit (Qiagen) and was then reverse transcribed and amplified with a Titan Single Tube RT-PCR kit (Roche). Primers that anneal within exons 2 (5′-ACCTTAAGTCCACGCATGGCGGCAAGATCG) and 3 (5′-CTTCCCTCGCTTCCGGATGAGCACACAGCC) were used for the APRT gene transcript. The following primers were used to amplify exon 2-exon 3 junctions from the human HPRT minigene: 5′-CCTTGATTTATTTTGCATACCTAATCATTATGCT and 5′-ACAATGTGATGGCCTCCCATCTCCTTCATC. These primers were designed so that they do not amplify the hamster HPRT gene from CHO cells.
PCR amplification of CTG/CAG repeat tracts and sequencing.
CTG/CAG repeat tracts were amplified by using a GC-rich PCR amplification kit (Roche). The following primer pairs that anneal within intron 2 of the APRT gene or inside exons 2 and 3 were used to amplify and characterize contraction events: primers immediately flanking the repeat tract (5′-CCTCTAGAGTCGTCCTTGTAGCCGGGAATG and 5′-GCCTGGCCGAAAGAAAGAAATGGTCTGTGATCC); primers that anneal 100 bp away from the repeat tract (5′-GAAACACCCTAGGGTCGCTGAATGTCCACC and 5′-TAGCACATGTCAGGGCTACCGAATTCGCGG); primers that anneal 200 bp away from the repeat tract (5′-TAGGAGTAGCACCTAAGATGAACTAGATGC and 5′-AGTTCAGGGTATATGTCTGGGGTCACTTCC); and primers that anneal within the exons flanking the repeat tract, i.e., exon 2 (5′-ACCTTAAGTCCACGCATGGCGGCAAGATCG) and exon 3 (5′-CTTCCCTCGCTTCCGGATGAGCACACAGCC). Following initial characterization of contracted repeats by PCR, the PCR products were cloned by using a TOPO-2 PCR cloning kit (Invitrogen) and were sequenced.
Mutagenic treatments.
Actively dividing cells were treated with 1.7 Gy of gamma irradiation or were incubated in the presence of 0.5 μg of aphidicolin/ml or 0.5 mM hydroxyurea for 12 h. The cells were then allowed to recover for 3 days. For each treatment, 2 × 107 treated cells were plated at a density of 5 × 105 cells per 100-mm-diameter dish and were incubated in the presence of APRT+ selection (ALASA) for 3 weeks and were stained with Coomassie blue. Aliquots of cells were plated on nonselective media to calculate survival and plating efficiency. The rate of formation of APRT+ clones was calculated by dividing the number of APRT+ clones by the survival rate (plated cells/surviving cells) and dividing by the number of population doublings that the cells underwent after treatment prior to applying APRT+ selection.
RESULTS
Assay system.
To examine the effect of CTG/CAG repeats on gene expression, we cloned various lengths of CTG/CAG repeats into a wild-type APRT gene as shown in Fig. 1. We chose several different lengths of CTG/CAG repeats. (CTG/CAG)17 and (CTG/CAG)25 are within the range of repeats in normal individuals and do not show instability, (CTG/CAG)28 and (CTG/CAG)32 are considered premutational because they lie on the borderline between the lengths found in normal and affected individuals, and (CTG/CAG)67 and the longer repeats are in the range found in affected individuals and are known to be unstable (52). Plasmids in which the CTG/CAG repeat was oriented so that the APRT transcript contained CUG sequences were designated CTG, and the plasmids expressing CAG sequences in the APRT transcript were designated CAG.
Effects of repeat orientation on the APRT phenotype were examined by transfection and by analysis of stable transformants. Plasmids carrying the modified APRT gene and an adjacent GPT gene (Fig. 1) were transfected into CHO RMP41 cells, which are APRT− (31), and the ratio of APRT+ to GPT+ colonies was determined (Table 1). In some cases, stable GPT+ transformants were isolated and screened by Southern blotting for single-copy integrants that carried the adjacent APRT gene, and then the ratio of APRT+ to GPT+ colonies was determined by plating the cells under appropriate selection conditions (Table 1). In both assays, all lengths of CTG repeat yielded approximately equal numbers of APRT+ and GPT+ colonies, indicating that repeats in the CTG orientation do not interfere with APRT expression, as expected from our previous results (33). By contrast, the outcome with repeats in the CAG orientation depended on the length of the tract. Plasmids with 32 or fewer repeats gave equal numbers of APRT+ and GPT+ colonies, whereas plasmids with (CAG)67, (CAG)98, or (CAG)175 gave few if any APRT+ colonies, indicating that long CAG tracts in some way kill the activity of the APRT gene.
TABLE 1.
Effect of CAG and CTG repeats of different lengths on APRT activity
| n for APRT(CAG)n | APRT+, %a
|
n for APRT(CTG)n | APRT+, %a
|
||
|---|---|---|---|---|---|
| Transient transfection | Stable integrants | Transient transfection | Stable integrants | ||
| 17 | 91 ± 2 | 98 ± 10 | 17 | 93 ± 5 | 84 ± 10 |
| 25 | 105 ± 10 | NA | 25 | 112 ± 12 | NA |
| 32 | 91 ± 4.5 | NA | 28 | 92 ± 4.6 | NA |
| 67 | 11 ± 11 | NA | 32 | 106 ± 6.5 | NA |
| 98 | 12 ± 10 | <0.1 | 98 | 112 ± 13 | 102 ± 3.4 |
| 175 | 10 ± 11 | <0.1 | 175 | 91 ± 7.5 | 97 ± 9.6 |
Percentage of APRT+ clones from the total number of transfectants (APRT+ clones/gpt+ clones) (100). Standard errors are indicated.
To test whether the effects of long CAG repeat tracts on gene activity were specific to the APRT gene or were a general phenomenon, we cloned (CTG-CAG) repeats of various lengths into the intron in the human HPRT minigene in a plasmid that carries an adjacent hygromycin resistance gene (Fig. 1). The effect of the repeat tracts on HPRT activity was tested by transient transfection into HPRT− HT1080 human fibrosarcoma cells. As was observed with the APRT gene, CTG repeats and short CAG repeats did not interfere with HPRT gene activity, whereas longer tracts of CAG repeats strongly inhibited HPRT activity (data not shown). From these data we conclude that CAG tracts longer than about 32 repeats disrupt gene activity when inserted into an intron.
Thus, long CAG repeat tracts inserted into an intron of a reporter gene such as APRT or HPRT can provide a convenient selectable assay to monitor contractions of the repeat tracts. To verify and characterize the selectable system and to analyze treatments that affect CAG repeat stability, we inserted two different long tracts of CAG repeats—(CAG)61 and (CAG)95—into the endogenous APRT gene in CHO cells, as described in Materials and Methods. By making comparison at a single, well-characterized chromosomal locus, we sought to avoid potential difficulties in interpretation that might arise due to chromosomal context effects. As described in detail later in Results, when APRT− cells carrying (CAG)61 or (CAG)95 were subjected to selection, the APRT+ colonies that arose all contained fewer CAG triplets, validating this selective system as a tool for studying contraction of long CAG repeats. Two APRT+ colonies, which carried (CAG)24 and (CAG)31, were isolated and used along with the parental APRT− cell lines with (CAG)61 and (CAG)95 to investigate the mechanism by which long CAG repeat tracts interfere with gene expression.
Long CAG repeat tracts interfere with correct splicing of reporter genes.
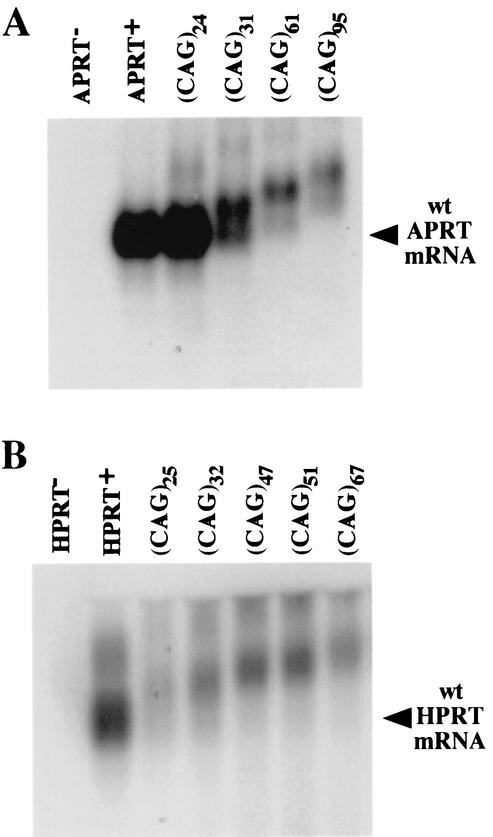
To determine the mechanism by which CAG repeat tracts interfere with gene function in our system, we analyzed mRNAs from the modified APRT and HPRT genes by Northern blotting. The APRT(CAG)24, APRT(CAG)31, APRT(CAG)61, and APRT(CAG)95 cell lines produced mRNAs that increased in size with increasing repeat length (Fig. 2A). The APRT+ cell lines, APRT(CAG)24 and APRT(CAG)31, contained an mRNA of the correct size, whereas the APRT− cell lines, APRT(CAG)61 and APRT(CAG)95, produced only an aberrant product. This corresponds well to the phenotype of the cell lines, with APRT(CAG)24 and APRT(CAG)31 being APRT+, and APRT(CAG)61 and APRT(CAG)95 being APRT−. In addition, the amount of the APRT mRNA in APRT(CAG)61 and APRT(CAG)95 cell lines was reduced approximately 10-fold from that in the wild-type APRT cell line. Transiently transfected HPRT minigene constructs carrying different lengths of CAG repeats showed the same trends (Fig. 2B).
FIG. 2.
Northern blot analysis of mRNA transcribed from APRT or HPRT genes. (A) Total RNA was extracted from an APRT− cell line carrying a deletion of the APRT gene, from wild-type (wt) APRT+ CHO cells, and from the APRT(CAG)24, APRT(CAG)31, APRT(CAG)61, and APRT(CAG)95 cell lines. (B) Total RNA was prepared from HPRT− CHO cells and from CHO cells transiently transfected with plasmids harboring the HPRT minigene alone or the HPRT minigene with insertions of CAG repeat tracts of various lengths. Equal amounts of total RNA were separated on agarose gels, transferred to a nylon membrane, and hybridized with probes to exon 2 and exon 3 of the APRT gene or to exons 1 and 2 of the HPRT minigene.
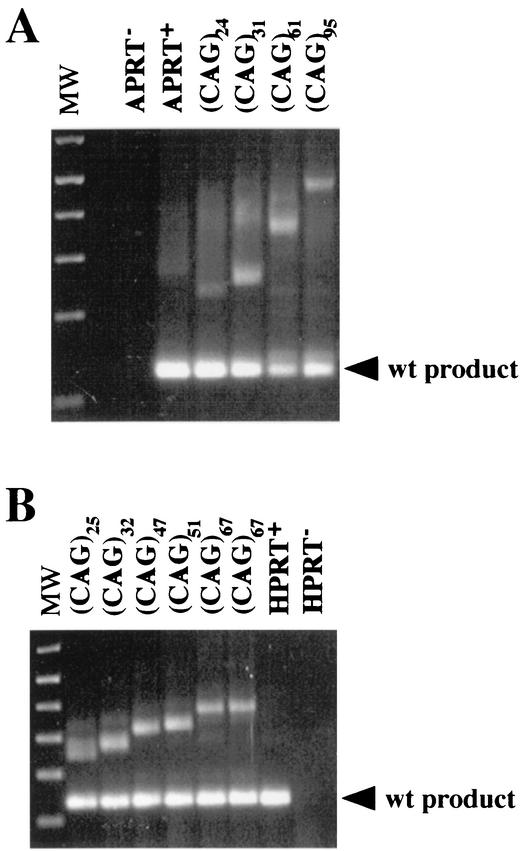
The increase in mRNA size with increasing repeat length suggested that the CAG repeats were incorporated into the mRNA instead of being spliced out. To test this possibility, we performed RT-PCR on mRNA from the APRT(CAG) cell lines (Fig. 3A), and on mRNA extracted from CHO cells transiently transfected with the HPRT(CAG) constructs (Fig. 3B). The products of RT-PCR showed increases in length similar to those seen on the Northern blots. In addition to the expanded product, all the samples yielded a second RT-PCR product corresponding in size to the correctly spliced APRT or HPRT mRNA. This normal-length mRNA was not detectable by Northern blot in cell lines with long repeat tracts (Fig. 2). We assume that efficient detection of the short RT-PCR product is caused by preferential amplification of the short fragment, because long CAG repeat tracts are difficult templates for PCR. Whatever minute amount of correctly spliced APRT mRNA is present in the cell lines with long repeats, it is not sufficient to confer an APRT+ phenotype. In contrast, in the APRT+ APRT(CAG)31 cell line, the correctly spliced product is detectable by Northern analysis (Fig. 2) and comprises approximately 30% of the APRT transcript.
FIG. 3.
RT-PCR analysis of APRT and HPRT mRNA. (A) Total RNA was extracted from CHO cell lines carrying a deletion of the APRT gene, the wild-type (wt) APRT, and APRT genes with insertions of CAG repeats. (B) Total RNA was extracted from CHO cells transiently transfected with the control plasmid, a plasmid containing the intact human HPRT minigene, and a set of plasmids containing CAG repeat tracts of various lengths cloned into the intron of the HPRT minigene. Junctions between exons 2 and 3 of the hamster APRT gene and junctions between exons 2 and 3 of the human HPRT minigene were amplified by RT-PCR and were separated on agarose gels. The lower band in each lane corresponds to the product expected for correct splicing of exons 2 and 3. Upper bands correspond to transcripts containing the CAG exon incorporated between exons 2 and 3. MW, molecular weight.
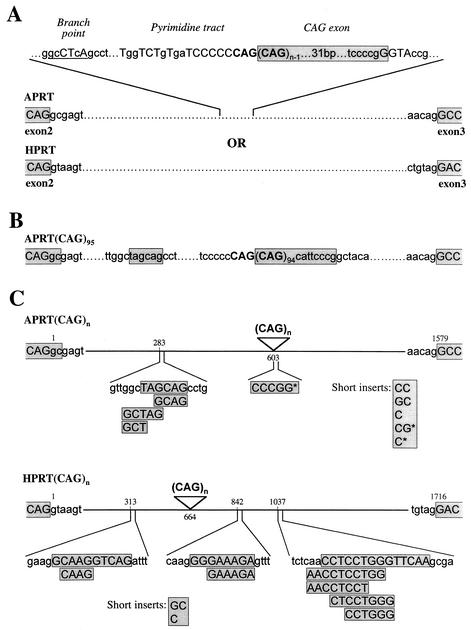
To determine the exact structure of the aberrant transcripts, we cloned and sequenced RT-PCR products from the APRT and HPRT mRNAs (Fig. 4). The majority of the products from the lower bands contained a normal junction between the adjacent exons, as expected (Table 2). Analysis of the products from the upper, expanded RT-PCR band confirmed that the CAG repeat was incorporated into the mRNA (Table 2). In the majority of cases the “CAG exon” began with a CAG triplet, extended through the repeat tract, and included 38 bases of 3′ flanking sequences (Fig. 4A). In one case the CAG exon had an alternative 3′ end, which included eight bases of 3′ flanking sequences, and it was flanked by a short stretch of nucleotides from the adjacent intron (Fig. 4B). Reasonable matches to the splicing consensus sequences surround the CAG exon (Fig. 4A). Because a small degree of variability is introduced into the repeat length by propagation of the cell lines and by PCR amplification (33), it is difficult to know the exact 5′ boundary of the CAG exon. We assume that one CAG repeat provided the consensus AG that defines the 3′ end of the adjacent intron.
FIG. 4.
Sequence analysis of APRT(CAG)n and HPRT(CAG)n mRNAs. RT-PCR products from the APRT(CAG)n and HPRT(CAG)n genes shown in Fig. 2 were cloned and sequenced. For each RT-PCR, the lower and upper bands were excised from a gel and were cloned individually. Shaded boxes indicate sequences that are included in the mRNA. (A) A typical splicing product containing the CAG repeat tract incorporated into APRT or HPRT mRNA is shown. Putative splicing signals for the CAG exon are indicated; capital letters indicate nucleotides that match the consensus for the branch point, the pyrimidine tract, and the splice sites. (B) Splicing product containing a CAG exon with an atypical 3′ splice site and a short insertion upstream of the CAG exon. (C) A summary of various insertions incorporated between the exon borders in the atypical splicing products. Each splicing product contained only one such insertion. For all the insertions, homology can be found within the intronic sequences and the position of the homology is indicated. Insertions that are too short for homology to be uniquely defined are designated short inserts and are shown in a separate box. In the APRT gene, most of the insertions begin with a GC dinucleotide, incorporation of which may have been caused by a shift in the exon boundary. Insertions in the APRT mRNA that do not begin with GC are marked with an asterisk. The CCCGG insert in APRT mRNA matches two sequences in the 3′ flanking region of the CAG repeat tract (see panels A and B).
TABLE 2.
Analysis of RT-PCR products from APRT and HPRT mRNA
| Colony | Lower band results
|
Upper band results
|
||||
|---|---|---|---|---|---|---|
| No. analyzed | No. typical | No. atypical | No. analyzed | No. with CAG exon | No. atypical | |
| APRT | 100 | 100 | 0 | |||
| APRT(CAG)24 | 3 | 3 | 0 | 3 | 3 | 0 |
| APRT(CAG)31 | 3 | 3 | 0 | 3 | 3 | 0 |
| APRT(CAG)61 | 100 | 96 | 4 | 10 | 10 | 0 |
| APRT(CAG)95 | 100 | 94 | 6 | 30 | 29 | 1 |
| HPRT | 100 | 100 | 0 | |||
| HPRT(CAG)25 | 3 | 3 | 0 | 3 | 3 | 0 |
| HPRT(CAG)32 | 3 | 3 | 0 | 3 | 3 | 0 |
| HPRT(CAG)47 | 30 | 29 | 1 | 10 | 10 | 0 |
| HPRT(CAG)51 | 30 | 29 | 1 | 10 | 10 | 0 |
| HPRT(CAG)67 | 130 | 121 | 9 | 40 | 40 | 0 |
Among the RT-PCR products derived from cell lines with long CAG repeats, we observed a small fraction of atypical products (Table 2). These mRNAs carried small insertions between the adjacent exons. In some cases an insertion was accompanied by incorporation of the CAG tract, whereas the other atypical products were obtained from the lower RT-PCR band (Fig. 3) and did not contain the CAG tract. The sequences corresponding to the insertions can be found within the intron either upstream or downstream of the CAG tract and appear to be clustered (Fig. 4C). In the case of the APRT gene, 8 out of 11 inserts began with GC, which is the dinucleotide immediately flanking exon 2. In these cases it may be that the exon boundary has been shifted. Formation of such aberrant splicing products suggests that the presence of long CAG repeat tracts in the gene may interfere with correct splicing, even where the repeat tract itself is not included in the final product.
In summary, Northern and RT-PCR analyses have demonstrated that CAG repeat tracts are recognized as exons and are incorporated into mRNA. The CAG exon shifts the reading frame, preventing synthesis of functional APRT or HPRT enzymes. Stop codons in the new reading frame presumably trigger nonsense-mediated decay, which may account for the lower-than-normal levels of mRNAs carrying the CAG exon (Fig. 2). The efficiency of splicing apparently depends on the length of the CAG repeat tract, with short tracts of repeats generating sufficient normal mRNA to confer an APRT+ or HPRT+ phenotype on cells.
Analysis of spontaneous contraction events recovered by using the APRT+ selection assay.
To determine the frequency of spontaneous contractions, we used the APRT(CAG)61 and APRT(CAG)95 cell lines, which are APRT−. Cells were propagated under APRT− selection conditions, which kill APRT+ cells, in order to eliminate preexisting contraction events from the cell population. Then the cells were grown for 3 days (approximately 2.5 population doublings) without selection, plated under APRT+ selection, and kept in the presence of APRT+ selection until visible APRT+ colonies had formed. The colonies were counted to determine the rate of formation of APRT+ cells. For the APRT(CAG)61 cell line, the rate of reversion to APRT+ was 3.1 × 10−6 ± 0.9 × 10−6 per cell division, and for the APRT(CAG)95 cell line the rate was 1.4 × 10−6 ± 0.4 × 10−6 per cell division. To examine the nature of the events that lead to the APRT+ phenotype, we isolated individual APRT+ colonies, amplified the CAG region by PCR, and sequenced across the repeat tract. The majority of the colonies [18 of 18 from APRT(CAG)61 and 9 of 16 from APRT(CAG)95] contained a contracted CAG repeat tract, with no changes to the flanking sequences (Table 3). The CAG repeat tracts in these colonies ranged from 4 to 33 repeats, indicating that the maximum number of CAG repeats compatible with APRT activity is 33.
TABLE 3.
Analysis of sequences around CAG repeats in APRT+ clones derived from APRT(CAG)61 and APRT(CAG)95 cell lines
| APRT(CAG)61 values | APRT(CAG)95 values
|
||||||
|---|---|---|---|---|---|---|---|
| Clone no. | 5′ flanking region | CAG no. | 3′ flanking region | Clone no. | 5′ flanking region | CAG no. | 3′ flanking region |
| 1 | Intact | 10 | Intact | 1 | Intact | 5 | Intact |
| 2 | Intact | 15 | Intact | 2 | Intact | 12 | Intact |
| 3 | Intact | 17 | Intact | 3 | Intact | 15 | Intact |
| 4 | Intact | 19 | Intact | 4 | Intact | 16 | Intact |
| 5 | Intact | 21 | Intact | 5 | Intact | 17 | Intact |
| 6 | Intact | 21 | Intact | 6 | Intact | 21 | Intact |
| 7 | Intact | 22 | Intact | 7 | Intact | 24 | Intact |
| 8 | Intact | 25 | Intact | 8 | Intact | 25 | Intact |
| 9 | Intact | 27 | Intact | 9 | Intact | 31 | Intact |
| 10 | Intact | 28 | Intact | 10 | Δ106 | 0 | Δ63 |
| 11 | Intact | 29 | Intact | 11 | Δ9 | 4 | Intact |
| 12 | Intact | 30 | Intact | 12 | Δ143 | 6 | Intact |
| 13 | Intact | 30 | Intact | 13 | Intact | 31 | Δ133 |
| 14 | Intact | 30 | Intact | 14 | Intact | 44 | Δ70 |
| 15 | Intact | 31 | Intact | 15 | Intact | 53 | Δ62 |
| 16 | Intact | 32 | Intact | 16 | Intact | 64 | Δ324 |
| 17 | Intact | 33 | Intact | ||||
| 18 | Intact | 33 | Intact | ||||
Several colonies (7 of 16) derived from the APRT(CAG)95 cell line contained deletions that extended from the CAG repeats into one or both flanking regions (Table 3). Three of the deletions retained CAG repeat tracts longer than 33 repeats; they are presumably APRT+ because they have deleted sequences critical for splicing. Interestingly, no deletions were recovered from the APRT(CAG)61 cell line, suggesting that longer repeat tracts may have a higher tendency to induce rearrangements of the adjacent DNA. These results demonstrate that the selectable assay system that we have developed can be effectively used to quantify repeat instability in mammalian cells.
Effect of gamma irradiation and DNA replication inhibitors on CTG/CAG repeats.
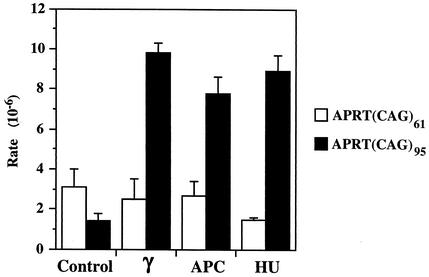
Studies with E. coli and yeast have suggested that double-strand breaks and stalled replication forks contribute to the instability of triplet repeats (9, 17, 20, 22, 30, 43). We have tested the effect of these factors on triplet repeat stability in mammalian cells by using the APRT(CAG)61 and APRT(CAG)95 cell lines. Cells were treated with 1.7 Gy of gamma irradiation to induce double-strand breaks or were incubated in the presence of 0.5 μg of aphidicolin/ml or 0.5 mM hydroxyurea for 12 h to disrupt replication forks. Following these treatments, cells were allowed to recover for 3 days and were plated under APRT+ selection, and the rates of APRT+ colony formation were determined (Fig. 5). Gamma irradiation, aphidicolin, and hydroxyurea increased the rate of APRT+ colony formation by the APRT(CAG)95 cell line by five- to sevenfold. In contrast, no stimulation of the rate was observed with the APRT(CAG)61 cell line, suggesting that the (CAG)61 repeat tract is not long enough to be destabilized effectively by these treatments. Stimulation of repeat contractions in the APRT(CAG)95 cell line by gamma irradiation and DNA replication inhibitors supports current models, which assert that DNA breaks and stalled replication forks contribute to repeat instability, and also shows that this cell line can be used as a sensitive tool to search for potential therapeutic agents for triplet repeat diseases in mammalian cells.
FIG. 5.
Effect of hydroxyurea, aphidicolin, and gamma irradiation on the rate of CAG repeat contractions in APRT(CAG)61 and APRT(CAG)95 cell lines. Cells were treated with hydroxyurea (HU), aphidicolin (APC), and gamma irradiation (γ) as described in Materials and Methods and were incubated in the presence of APRT+ selection, and the number of APRT+ clones was scored. Approximately 107 cells were used for each treatment. All the treatments were repeated at least three times, and error bars represent the standard deviation.
DISCUSSION
Novel assay for repeat instability in mammalian cells.
In this paper, we describe the first selectable assay for monitoring triplet repeat instability in mammalian cells. The reporter cassette consists of a selectable gene, either APRT or HPRT containing an insertion of CTG/CAG repeats in an intron. Because long CAG tracts interfere with normal splicing, triplet repeat contraction events can be monitored by the appearance of APRT+ or HPRT+ colonies. The main advantage of this selectable assay over the PCR-based assays such as small-pool PCR and GeneScan is its high sensitivity. Using this assay, we have detected contraction events that occurred at rates down to 10−6, which is the rate of spontaneous contractions in our cell lines, rather than the sensitivity limit of the assay. In contrast, PCR-based assays provide reproducible results only at frequencies down to 10−3, because rare events cannot be reliably detected against the background of unchanged molecules. The sensitivity of the selectable assay allows measurement of small differences in repeat instability that may result from the normal process of DNA replication, DNA repair, and recombination or from genetic and environmental perturbations.
We have demonstrated that our assay can be used with two different selectable genes, APRT and HPRT, in two different cell types: hamster CHO and HT1080 human fibrosarcoma cells. This selection system offers the possibility of analyzing triplet repeat stability in various cell types by using a common assay in order to study and compare cell lines with specific mutations or cell lines isolated from patients. The only requirement for a cell line is that it be either APRT− or HPRT−. Because the HPRT gene is located on the X chromosome, HPRT− variants can be readily selected from cell lines that carry a single functional copy of the X chromosome.
We have used this assay at the APRT locus in CHO cells to monitor contractions of repeat tracts from 95 or 61 repeats to 33 or fewer; that is, contractions of a minimum of 62 or 28 repeats, respectively. Since there seems to be a fairly sharp cutoff at 33 repeats for the APRT+ phenotype, it should be possible to construct parental cell lines with shorter repeat tracts in order to detect small contractions or with longer repeat tracts to assay larger ones. Contractions of at least 30 or more repeats, as detected by our present assay system, are perhaps the most relevant for studies designed to search for possible therapeutic agents. Only the treatments that induce substantial repeat contractions offer the possibility of a cure for patients whose genomes contain pathological expansions of triplet repeats.
The present selection assay is designed to detect contractions of CAG repeats, which represent an important avenue of research into treatment of triplet repeat diseases. Of equal importance, however, are the processes that lead to expansion of repeat tracts in the first place. Because APRT and HPRT genes can be selected in either direction, it should be possible, in principle, to design selection assays for expansion. For example, it may be feasible to use the APRT(CAG)33 cell line, which is APRT+, to select for expansions to longer repeat tracts, which are APRT−. Fortuitously, the crossover point in this assay—33 repeats—is at the upper range of normal in the progression of the human disease and is perhaps ideally suited for studying the initial instability that leads to the disease state.
Effect of CAG repeat tracts on splicing.
We describe here a novel property of CTG/CAG repeat tracts from the myotonic dystrophy locus, which serves as the basis for our selectable assay. The CTG/CAG repeat tracts in the CAG orientation behave like exons, forcing themselves into the mRNA and interfering with correct splicing. Many exons are known to contain auxiliary splicing elements called exonic splicing enhancers (ESEs) (reviewed in reference 1). ESEs usually associate with introns containing weak flanking splice sites, and function to promote utilization of adjacent splice sites (reviewed in references 10 and 26). The majority of ESEs that have been identified are purine-rich repeats. Remarkably, a CA-rich motif has also been found to function as an ESE and to promote splicing in vivo and in vitro (7, 11, 25, 50, 54). We propose, therefore, that CAG repeat tracts can function as ESEs and that the longer the repeat tract is, the stronger is the ESE signal that it provides. The possibility that CAG repeat tracts act as ESEs is supported by examination of the putative splicing signals flanking the CAG exon. The putative 3′ and 5′ splicing signals (Fig. 4A) are derived from the natural sequences at the myotonic dystrophy locus, although they are oriented opposite to the direction of transcription. The 3′ splice site signal UGGUCUGUGAUCCCCCCAG↓C resembles the consensus (Y)∼15NYAG↓G, as does the 5′ splice site signal (GG↓GUACCG versus the consensus AG↓GURAGU), but neither is a close match, and thus, they are likely to be weak. Indeed an alternative, presumably weaker 5′ splice site, CG↓GCTACA, was found among the RT-PCR products. It is unclear what sequence is used for the branch point; however, the sequence CTCAGC, which resembles the branch point consensus CTRAYY, would position the critical A residue 40 nucleotides upstream of the 3′ splice site, well within the usual range for such signals (36). These observations are consistent with the idea that tracts of CAG repeats can enhance splicing in the absence of canonical splice sites and promote utilization of cryptic splice sites in the adjacent sequences. We do not know how the presence of long CAG tracts leads to the appearance of spliced products with shifted exon boundaries or extra nucleotides in addition to or in place of the CAG tract itself (Fig. 4C). We can speculate that long CAG repeat tracts disrupt the normal course of splicing, resulting in various aberrant products.
The effect of CAG repeat tracts on splicing may contribute to the etiology of trinucleotide repeat disorders. In the case of the myotonic dystrophy gene, the repeat tract is in the CTG orientation (52), which does not interfere with splicing. It is possible, however, that the presence of the repeat may affect the splicing of adjacent genes that are transcribed in the opposite direction. In the polyglutamine disorders, such as Huntington's disease and spinocerebellar ataxias 1, 2, 3, 6, and 7, the CAG repeat is expanded within an exon (reviewed in reference 55). Based on our results that a CAG tract can interfere with normal splicing and recruit various cryptic sequences as splice sites, we speculate that an expanded CAG tract within a functional exon may also promote utilization of aberrant splice sites. Alterations in the splicing of the adjacent downstream intron could lead to frameshifts and formation of truncated proteins containing polyglutamine tracts. Indeed, for huntingtin, atrophin-1, and the androgen receptor, the neurotoxicity of the expanded CAG repeat has been attributed to the formation of truncated polyglutamine-containing proteins (5, 15, 16, 24, 28, 32). The truncated versions of huntingtin and the androgen receptor have been detected in cells containing expanded CAG tracts but not in the normal cells (8, 12, 24, 32). Furthermore, overexpression of full-length huntingtin, atrophin-1, and the androgen receptor in tissue culture cells revealed that these proteins have predominantly cytoplasmic and/or perinuclear localization with some propensity to aggregate. In contrast, nuclear localization and aggregation, which are the hallmarks of the toxicity of polyglutamine proteins, have been observed only when truncated peptides harboring the expanded polyglutamine tracts were overexpressed (5, 15, 16, 24, 28, 32). It has been suggested that some processing of the full-length polyglutamine protein takes place in the cells, which liberates the toxic peptide (55). Based on the results presented here, we propose an alternative explanation: that the toxic peptides are produced by aberrant splicing of CAG-containing exons, which creates premature termination codons.
Mechanisms of repeat contraction.
The rates of spontaneous repeat contractions leading to the APRT+ phenotype were 3.1 × 10−6 ± 0.9 × 10−6 and 1.4 × 10−6 ± 0.4 × 10−6 per cell generation for the (CAG)61 and (CAG)95 cell lines, respectively. It is somewhat surprising that the longer repeat tract appears to be more stable. In our system, however, in order to convert to the APRT+ phenotype, (CAG)61 has to contract by 28 repeats, while (CAG)95 has to contract by at least 62 repeats. Therefore, the lower rates of APRT+ colony formation in the (CAG)95 cell line may be due to a lower rate of large contractions.
The difference in rates is paralleled by a difference in the types of contraction events observed in the (CAG)61 and (CAG)95 cell lines. While all the colonies obtained from the (CAG)61 cell line were “clean” contractions involving only the CAG repeats, about 44% of the events from the (CAG)95 cell line carried deletions that extended into the DNA flanking the repeat tract. This observation suggests that the longer contractions in the (CAG)95 cell line may be generated in part by a mechanism different from the one that gives rise to the shorter contractions in the (CAG)61 cell line. For example, it could be that contraction of (CAG)61 repeats occurs mainly by formation of a hairpin followed by replication slippage, which leaves the flanking regions intact. In addition to this mechanism, it may be that larger hairpins formed by (CAG)95 repeats are recognized by DNA repair enzymes and are resolved via some kind of nonhomologous recombination event that involves DNA breakage and promotes deletions in the flanking sequences. Whatever the mechanism, it is important to note that both types of events—pure contractions and contractions with flanking deletions—have been observed in human patients suffering from triplet repeat diseases (see reference 33 for discussion).
Chemotherapeutic approaches to the treatment of triplet repeat diseases.
Treatments with aphidicolin, hydroxyurea, and gamma irradiation induced contractions in (CAG)95 but not in (CAG)61. As discussed above, repeat contractions in (CAG)61 and (CAG)95 cell lines may occur, in part, by different mechanisms, with longer contractions being dependent on nonhomologous recombination events. It would be consistent, therefore, that gamma irradiation-induced DNA breaks, which are repaired via a recombination pathway, have stronger effects on large contractions in (CAG)95. Similarly, stalled replication forks induced by aphidicolin and hydroxyurea may be likelier to resolve by a recombination event if they involve a long hairpin in (CAG)95. In terms of chemotherapy, the observation that longer repeats show a greater increase in the rate of contractions following mutagenic treatment is very promising. The very long repeat tracts found in myotonic dystrophy, fragile X syndrome, and Friedreich ataxia may be especially sensitive to this kind of therapy.
The results presented in this report show that chemical treatments can induce contractions of trinucleotide repeat tracts, suggesting that chemotherapeutic approaches may be applicable to trinucleotide repeat diseases. The five- to sevenfold stimulation of repeat contractions that we observed with aphidicolin, hydroxyurea, and gamma irradiation is still too low to have any therapeutic significance. In addition, nonspecific mutagens like the reagents used here are likely to induce mutations in many other genomic sites. However, since trinucleotide repeat tracts form unusual DNA structures, it seems feasible that drugs can be found that specifically interact with long repeat tracts and promote high rates of contraction events. We believe that the selectable assay for triplet repeat contractions, reported here, will greatly facilitate the search for such drugs.
Acknowledgments
We thank Fung Chan for excellent technical help and Robert Wells and members of his laboratory for help and advice.
V.G. and A.S. contributed equally to this research.
This work was supported by a Human Frontier of Science postdoctoral fellowship to V.G. and an NIH grant (GM38219) and a Muscular Dystrophy Association grant to J.H.W.
REFERENCES
- 1.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 2.Bowater, R. P., A. Jaworski, J. E. Larson, P. Parniewski, and R. D. Wells. 1997. Transcription increases the deletion frequency of long CTG·CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 25:2861-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowater, R. P., and R. D. Wells. 2001. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 66:159-202. [DOI] [PubMed] [Google Scholar]
- 4.Brock, G. J. R., N. H. Anderson, and D. G. Monckton. 1999. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 8:1061-1067. [DOI] [PubMed] [Google Scholar]
- 5.Butler, R., P. N. Leigh, M. J. McPhaul, and J. M. Gallo. 1998. Truncated forms of the androgen receptor are associated with polyglutamine expansion in X-linked spinal and bulbar muscular atrophy. Hum. Mol. Genet. 7:121-127. [DOI] [PubMed] [Google Scholar]
- 6.Cleary, J. D., K. Nichol, Y. H. Wang, and C. E. Pearson. 2002. Evidence of cis-acting factors in replication-mediated repeat instability in primate cells. Nat. Genet. 31:37-46. [DOI] [PubMed] [Google Scholar]
- 7.Coulter, L. R., M. A. Landree, and T. A. Cooper. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiFiglia, M., E. Sapp, K. O. Chase, S. W. Davies, G. P. Bates, J. P. Vonsattel, and N. Aronin. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990-1993. [DOI] [PubMed] [Google Scholar]
- 9.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279:853-856. [DOI] [PubMed] [Google Scholar]
- 10.Fu, X.-D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 11.Gersappe, A., and D. Pintel. 1999. CA- and purine-rich elements form a novel bipartite exon enhancer which governs inclusion of the minute virus of mice NS2-specific exon in both singly and doubly spliced mRNAs. Mol. Cell. Biol. 19:364-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg, Y. P., D. W. Nicholson, D. M. Rasper, M. A. Kalchman, H. B. Koide, R. K. Graham, M. Bromm, P. Kazemi-Esfarjani, N. A. Thornberry, J. P. Vaillancourt, and M. R. Hayden. 1996. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat. Genet. 13:442-449. [DOI] [PubMed] [Google Scholar]
- 13.Harriman, G. R., A. Bradley, S. Das, P. Rogers-Fani, and A. C. Davis. 1996. IgA class switch in I alpha exon-deficient mice. Role of germline transcription in class switch recombination. J. Clin. Investig. 97:477-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashem, V. I., W. A. Rosche, and R. R. Sinden. 2002. Genetic assays for measuring rates of (CAG)·(CTG) repeat instability in Escherichia coli. Mutat. Res. 2002:25-37. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi, S., R. Koide, T. Shimohata, M. Yamada, Y. Hayashi, H. Takano, H. Date, M. Oyake, T. Sato, A. Sato, S. Egawa, T. Ikeuchi, H. Tanaka, R. Nakano, T. K., I. Hozumi, T. Inuzuka, H. Takahashi, and S. Tsuji. 1998. Suppression of aggregate formation and apoptosis by transglutaminase inhibitors in cells expressing truncated DRPLA protein with an expanded polyglutamine stretch. Nat. Genet. 18:111-117. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, H., M. Yamaguchi, S. Sugai, Y. Aze, S. Narumiya, and A. Kakizuka. 1996. Expanded polyglutamine in the Machado-Joseph disease protein induces cell death in vitro and in vivo. Nat. Genet. 13:196-202. [DOI] [PubMed] [Google Scholar]
- 17.Iyer, R. R., A. Pluciennik, W. A. Rosche, R. R. Sinden, and R. D. Wells. 2000. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in E. coli. J. Biol. Chem. 275:2174-2181. [DOI] [PubMed] [Google Scholar]
- 18.Jakupciak, J. P., and R. D. Wells. 2000. Gene conversion (recombination) mediates expansions of CTG/CAG repeats. J. Biol. Chem. 275:40003-40013. [DOI] [PubMed] [Google Scholar]
- 19.Jakupciak, J. P., and R. D. Wells. 1999. Genetic instabilities in CTG/CAG repeats occur by recombination. J. Biol. Chem. 274:23468-23479. [DOI] [PubMed] [Google Scholar]
- 20.Jankowski, C., F. Nasar, and D. K. Nag. 2000. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl. Acad. Sci. USA 97:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaworski, A., W. A. Rosche, R. Gellibolian, S. Kang, M. Shimizu, R. P. Bowater, R. R. Sinden, and R. D. Wells. 1995. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl. Acad. Sci. USA 92:11019-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, S., A. Jaworski, K. Ohshima, and R. D. Wells. 1995. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10:213-218. [DOI] [PubMed] [Google Scholar]
- 23.Kovtun, I. V., and C. T. McMurray. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27:407-411. [DOI] [PubMed] [Google Scholar]
- 24.Lunkes, A., and J. L. Mandel. 1998. A cellular model that recapitulates major pathogenic steps of Huntington's disease. Hum. Mol. Genet. 7:1355-1361. [DOI] [PubMed] [Google Scholar]
- 25.Lynch, K. W., and T. Maniatis. 1995. Synergistic interaction between two distinct elements of a regulated splicing enhancer. Genes Dev. 9:284-293. [DOI] [PubMed] [Google Scholar]
- 26.Manley, J., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 27.Manley, K., T. L. Shirley, L. Flaherty, and A. Messer. 1999. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 23:471-473. [DOI] [PubMed] [Google Scholar]
- 28.Martindale, D., A. Hackam, A. Wieczorek, L. Ellerby, C. Wellington, K. McCutcheon, R. Singaraja, P. Kazemi-Esfarjani, R. Devon, S. U. Kim, D. E. Bredesen, F. Tufaro, and M. R. Hayden. 1998. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat. Genet. 18:150-154. [DOI] [PubMed] [Google Scholar]
- 29.Matzuk, M. M., M. J. Finegold, J. G. Su, A. J. Hsueh, and A. Bradley. 1992. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 30.Maurer, D. J., B. L. O'Callaghan, and D. M. Livingston. 1996. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:6617-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrihew, R. V., R. G. Sargent, and J. H. Wilson. 1995. Efficient modification of the APRT gene by FLP/FRT site-specific targeting. Somat. Cell Mol. Genet. 21:299-307. [DOI] [PubMed] [Google Scholar]
- 32.Merry, D. E., Y. Kobayashi, C. K. Bailey, A. A. Taye, and K. H. Fischbeck. 1998. Cleavage, aggregation and toxicity of the expanded androgen receptor in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 7:693-701. [DOI] [PubMed] [Google Scholar]
- 33.Meservy, J. M., R. G. Sargent, R. R. Iyer, F. Chan, G. J. McKenzie, R. D. Wells, and J. H. Wilson. 2003. Long CTG tracks from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol. Cell. Biol. 23:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miret, J. J., L. Pessoa-Brandao, and R. S. Lahue. 1998. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:12438-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oussatcheva, E. A., V. I. Hashem, Y. Zou, R. R. Sinden, and V. N. Potaman. 2001. Involvement of the nucleotide excision repair protein UvrA in instability of CAG·CTG repeat sequences in Escherichia coli. J. Biol. Chem. 276:30878-30884. [DOI] [PubMed] [Google Scholar]
- 36.Padgett, R. A., P. J. Grabowski, M. M. Konarska, S. Seiler, and P. A. Sharp. 1986. Splicing of messenger RNA precursors. Annu. Rev. Biochem. 55:1119-1150. [DOI] [PubMed] [Google Scholar]
- 37.Panigrahi, G. B., J. D. Cleary, and C. E. Pearson. 2002. In vitro (CTG)·(CAG) expansions and deletions by human cell extracts. J. Biol. Chem. 277:13926-13934. [DOI] [PubMed] [Google Scholar]
- 38.Parniewski, P., A. Bacolla, A. Jaworski, and R. D. Wells. 1999. Nucleotide excision repair affects the stability of long transcribed (CTG·CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 27:616-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluciennik, A., R. R. Iyer, M. Napierala, J. E. Larson, M. Filutowicz, and R. D. Wells. 2002. Long CTG·CAG repeats from myotonic dystrophy are preferred sites for intermolecular recombination. J. Biol. Chem. 277:34074-34086. [DOI] [PubMed] [Google Scholar]
- 40.Richard, G.-F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richard, G. F., B. Dujon, and J. E. Haber. 1999. Double-strand break repair can lead to high frequencies of deletions within short CAG/CTG trinucleotide repeats. Mol. Gen. Genet. 261:871-882. [DOI] [PubMed] [Google Scholar]
- 42.Rolfsmeier, M. L., M. J. Dixon, and R. S. Lahue. 2000. Mismatch repair blocks expansions of interrupted trinucleotide repeats in yeast. Mol. Cell 6:1501-1507. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar, P. S., H. C. Chang, F. B. Boudi, and S. Reddy. 1998. CTG repeats show bimodal amplification in E. coli. Cell 95:531-540. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, K. H., C. M. Abbot, and D. R. Leach. 2000. Two opposing effects of mismatch repair on CTG repeat instability in Escherichia coli. Mol. Microbiol. 35:463-471. [DOI] [PubMed] [Google Scholar]
- 45.Schumacher, S., I. Pinet, and M. Bichara. 2001. Modulation of transcription reveals a new mechanism of triplet repeat instability in Escherichia coli. J. Mol. Biol. 307:39-49. [DOI] [PubMed] [Google Scholar]
- 46.Schweitzer, J. K., and D. M. Livingston. 1997. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum. Mol. Genet. 6:349-355. [DOI] [PubMed] [Google Scholar]
- 47.Sinden, R. R. 2001. Neurodegenerative diseases. Origins of instability. Nature 411:757-758. [DOI] [PubMed] [Google Scholar]
- 48.Steinbach, P., D. Glaser, W. Vogel, M. Wolf, and S. Schwemmle. 1998. The DMPK gene of severely affected myotonic dystrophy patients is hypermethylated proximal to the largely expanded CTG repeat. Am. J. Hum. Genet. 62:278-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taneja, K. L., M. McCurrach, M. Schalling, D. Housman, and R. H. Singer. 1995. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128:995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian, M., and T. Maniatis. 1992. Positive control of pre-mRNA splicing in vitro. Science 256:237-240. [DOI] [PubMed] [Google Scholar]
- 51.Timchenko, L. T. 1999. Myotonic dystrophy: the role of RNA CUG triplet repeats. Am. J. Hum. Genet. 64:360-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usdin, K., and E. Grabczyk. 2000. DNA repeat expansions and human disease. Cell. Mol. Life Sci. 57:914-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van den Broek, W. J., M. R. Nelen, D. G. Wansink, M. M. Coerwinkel, H. te Riele, P. Groenen, and B. Wieringa. 2002. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11:191-198. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Z., H. M. Hoffman, and P. J. Grabowski. 1995. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA 1:21-35. [PMC free article] [PubMed] [Google Scholar]
- 55.Zoghbi, H. Y., and H. T. Orr. 2000. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23:217-247. [DOI] [PubMed] [Google Scholar]