Abstract
The p53 and NF-κB transcription factor families are important, multifunctional regulators of the cellular response to stress. Here we have investigated the regulatory mechanisms controlling p53-dependent cell cycle arrest and cross talk with NF-κB. Upon induction of p53 in H1299 or U-2 OS cells, we observed specific repression of cyclin D1 promoter activity, correlating with a decrease in cyclin D1 protein and mRNA levels. This repression was dependent on the proximal NF-κB binding site of the cyclin D1 promoter, which has been shown to bind the p52 NF-κB subunit. p53 inhibited the expression of Bcl-3 protein, a member of the IκB family that functions as a transcriptional coactivator for p52 NF-κB and also reduced p52/Bcl-3 complex levels. Concomitant with this, p53 induced a significant increase in the association of p52 and histone deacetylase 1 (HDAC1). Importantly, p53-mediated suppression of the cyclin D1 promoter was reversed by coexpression of Bcl-3 and inhibition of p52 or deacetylase activity. p53 therefore induces a transcriptional switch in which p52/Bcl-3 activator complexes are replaced by p52/HDAC1 repressor complexes, resulting in active repression of cyclin D1 transcription. These results reveal a unique mechanism by which p53 regulates NF-κB function and cell cycle progression.
The transcriptional effects resulting from cellular stresses such as DNA damage or hypoxia are complex and involve the action of multiple transcription factors. Depending on the cell type and exact nature of the stimuli, different patterns of gene expression are established which can result in distinct cellular outcomes. For example, activation of the tumor suppressor p53 in response to DNA damage can result in either cell cycle arrest or the induction of apoptosis (25, 55). Moreover, cell cycle arrest can occur at either the G1/S or G2 phase of the cell cycle, depending on the circumstances of p53 activation (52). These apparent differences in p53 functionality can be accounted for by a variety of regulatory mechanisms. For example, posttranslational modification of p53, either by phosphorylation, acetylation, or sumoylation, can have profound effects on its behavior, influencing DNA binding, stability, and the interaction with other transcriptional regulators such as the p300 coactivator protein (25, 55).
In resting cells, p53 has a short half-life of only a few minutes as a result of the Mdm2 ubiquitin ligase, which binds directly to p53 and mediates its proteolytic degradation (25, 55). Upon stimulation, p53 rapidly becomes stabilized by different and stimulus-specific posttranslation modifications, which disrupt its interaction with Mdm2, thus extending its half-life to several hours and allowing it to become transcriptionally active (25, 55). Among the p53 downstream target genes are mdm2 itself (which results in a negative feedback loop restricting the duration of the p53 response), proapoptotic genes such as bax and PIG3, and cell cycle inhibitory proteins such as the cyclin-dependent kinase inhibitor p21 (WAF1/CIP1) (25, 55). p53 is capable both of activating and repressing transcription (62). These important antitumor effects of p53 have resulted in its being a primary target for alteration during cancer development, being mutated in approximately 50% of human tumors (22).
In addition to posttranslational modification, another potentially important determinant of p53 action is its ability to interact with or modulate the function of other heterologous transcriptional regulators in the cell, although this aspect of its regulation is not as well investigated. Recently, it has become apparent that a complex relationship exists between p53 and members of the NF-κB and IκB families of proteins and that the functions of these proteins are integrated within the cell through a variety of mechanisms. Although most typically associated with activation by inflammatory cytokines, NF-κB subunits are also activated by DNA damage and other forms of stress more commonly associated with p53 induction (41). As with p53, the activation of NF-κB can affect the cell in a variety of ways. Under some circumstances, NF-κB function can apparently antagonize p53, being proproliferative and antiapoptotic (44). Consequently, aberrantly active forms of NF-κB have been described in human tumors and transformed cell lines (6, 47). Under other circumstances, however, NF-κB activation is associated with cellular differentiation and proapoptotic functions (7, 44, 49).
Investigation of the integration of p53 and NF-κB function has largely centered on the function of the RelA(p65) subunit. Significantly, RelA was recently shown under some circumstances to be required for p53-dependent apoptosis (49). Moreover, activation of p53 was shown to result in the induction of a p50/RelA DNA-binding complex (49). In contrast, a recent report failed to note this effect but indicated that NF-κB can suppress p53 activity by inducing Mdm2 protein levels (53). These two reports also contradict each other concerning whether NF-κB is required for p53-mediated apoptosis or acts to prevent it. In addition, our laboratory, and others, had previously observed that p53 and RelA can mutually suppress, or modulate, each other's transcriptional activity in a manner consistent with competition for limiting amounts of the p300 coactivator protein (46, 57, 59). Furthermore, other mechanisms of cross talk have also been described. These include reports that the promoter of the p53 gene itself has been found to contain a functional NF-κB binding site (24, 30, 42), that p53 can inhibit NF-κB activation (29, 51), that Mdm2 expression can stimulate transcription from the relA gene promoter (19), and that p53 can bind directly to IκBα (10).
In this report, we have further investigated the integration of p53 and NF-κB function. We observed a p53-induced, NF-κB- dependent repression of the cyclin D1 promoter resulting from down regulation of Bcl-3 protein levels and increased association of p52 with histone deacetylase 1 (HDAC1). These results demonstrate that p53/NF-κB cross talk is not confined to the RelA subunit and effects on apoptosis. Instead, by influencing the activity of other family members, p53's effects on the cell cycle can in part be mediated by regulating the activity of distinct NF-κB subunits.
MATERIALS AND METHODS
Cells.
Human embryonic kidney 293 cells, human non-small-cell lung carcinoma H1299 cells, and human osteosarcoma U-2 OS cells were maintained at 5% CO2 in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin-streptomycin (Sigma), and 1% l-glutamine (Sigma). H1299 cells containing stably transfected forms of p53 were additionally grown in the presence of 400 μg of hygromycin B per ml and 400 μg of G418 per ml. Human osteosarcoma Saos-2 cells containing p53 under the control of the Tet-On-inducible system were a gift from Sonia Lain and David Lane (University of Dundee, Dundee, Scotland, United Kingdom) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% tetracycline-free fetal bovine serum (Clontech), 1% penicillin-streptomycin, 1% l-glutamine, and 1 mg of G418 per ml. H1299 cells stably expressing isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible wild-type or the serine-15-to-alanine mutant of p53 were created by using the LacSwitchTMII inducible expression system according to the manufacturer's instructions (Stratagene). Induction of p53 was achieved by treating cells with 20 to 100 μM IPTG (Melford Laboratories) for the indicated times. Where indicated, endogenous NF-κB activity was induced by tumor necrosis factor alpha (TNF-α; Sigma) at a final concentration of 10 ng/ml. Inhibition of HDAC activity by trichostatin A (TSA) (Upstate Biotechnology) was performed by incubating cells with 100 ng of TSA per ml for 16 h before harvesting.
Flow cytometric analysis of cell cycle distribution.
Adherent and detached cells were harvested, pooled, washed once in phosphate-buffered saline (PBS), and fixed in ice-cold 70% (vol/vol) ethanol in distilled water. Cells were then washed twice in PBS (plus 1% [wt/vol] bovine serum albumin) and resuspended in PBS containing 0.1% (vol/vol) Triton X-100, 50 μg of propidium iodide per ml and 50 μg of RNase A per ml. After incubation at room temperature for 20 min, cells were analyzed for cell cycle distribution with a FACS Calibur flow cytometer (fluorescence-activated cell sorter; FACS) and Cell Quest software (Becton Dickinson). Red fluorescence (585 ± 42 nm) was evaluated on a linear scale, and pulse width analysis was used to exclude cell doublets and aggregates from the analysis. Cells with a DNA content between 2N and 4N were designated as being in the G1, S, or G2/M phase of the cell cycle. The number of cells in each compartment of the cell cycle was expressed as a percentage of the total number of cells present.
Transient transfections and protein extracts.
Calcium phosphate transient transfections were performed essentially as described previously (59). All transfections contained appropriate levels of Rous sarcoma virus (RSV) control plasmid such that each dish received the same amount of DNA. Nuclear extracts were prepared as described previously (15). For whole-protein lysates, cells were washed once with PBS and resuspended in buffer A (20 mM HEPES [pH 7.6], 400 mM NaCl, 1 mM EDTA, 25% glycerol, 0.1% NP-40, 1 mM dithiothreitol). Cells were lysed by using 10 strokes of a 26-gauge syringe. Cells were incubated on ice for 30 min before centrifugation at maximum speed in an Eppendorf 5417R centrifuge for 15 min. All solutions used in protein extractions contained protease and phosphatase inhibitors, phenylmethylsulfonyl fluoride (1 mM), leupeptin (1 μg/μl), aprotinin (1 μg/μl), pepstatin A (1 μg/μl), NaF (5 mM), and Na3VO4 (500 μM).
Immunoblotting and antibodies.
Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), resolved proteins were electroblotted onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked in 10% blocking buffer (Tris-buffered saline [TBS]-0.1% Tween 20, 10% milk) (TTBS) for 30 min. The membrane was then probed with the primary antibody in 5% TTBS-milk overnight at 4°C, followed by three 10-min washes with TTBS. Incubation with the secondary antibody was performed for 1 h at room temperature followed by three 10-min washes with TTBS. Detection of proteins was achieved by using enhanced chemiluminescence (Amersham).
Antibodies used in this report were anti-phospho-Ser-15 p53 (New England Biolabs), anti-p21 (Santa Cruz), anti-p52 monoclonal (Upstate Biotechnology), anti-p50 NF-κB (Upstate Biotechnology), anti-Bcl-3 (Santa Cruz), anti-HDAC1 (CN Biosciences), anti-cyclin A (Santa Cruz), anti-cyclin B1 (Santa Cruz), anti-cyclin E (Pharmingen), anti-β-actin (Sigma), and anti-cyclin D1 (Pharmingen). The anti-cRel, anti-RelB, anti-RelA, and anti-p100/p52 polyclonal antibodies were provided by Nancy Rice, National Cancer Institute, Frederick, Md. The anti-p53 DO1 antibody was provided by Sonia Lain and David Lane (University of Dundee).
Plasmids.
The RSV RelA (p65), p50, and c-Rel expression plasmids have been previously described (45). RSV RelB and Bcl-3 were obtained from Gary J. Nabel's laboratory (National Institutes of Health, Bethesda, Md.). The p52 expression plasmid (amino acids 1 to 405) and C-terminal p100 (amino acids 450 to 946) used in this report were generated by PCR by using the Pwo polymerase (Roche) and were subcloned into the same RSV expression plasmid backbone. Cyclin D1 and cyclin E promoter luciferase plasmids were obtained from Richard Pestell (Albert Einstein Cancer Center, Bronx, N.Y.). The 3× κB conA luciferase reporter plasmid was obtained from Ron Hay (University of St. Andrews, St. Andrews, Fife, Scotland, United Kingdom). The p21 promoter luciferase plasmid was obtained from David Lane (University of Dundee). The HDAC1 expression plasmid was obtained from Tony Kouzarides (University of Cambridge, Cambridge, United Kingdom) and has been described previously (3).
Reporter gene assays.
Cells were transfected with indicated luciferase reporter plasmids and 24 h posttransfection were treated for the indicated times with IPTG or TNF-α. Lysates were prepared by using passive lysis buffer (Promega), and luciferase assays were performed according to the manufacturer's instructions (Promega). All experiments were performed a minimum of three times before calculating means and standard deviations as shown in figures.
RNA extraction and RT-PCR.
Total RNA was extracted from H1299w/tp53 cells by using the RNeasy kit (Qiagen) following the manufacturer's instructions. Five hundred to 10 ng of RNA was used to perform reverse transcriptase PCR (RT-PCR) by using the Promega Access RT-PCR system. PCR products were resolved by agarose gel electrophoresis and visualized by staining with ethidium bromide. Primers used for Bcl-3, cyclin D1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were as follows: Bcl-3 (primer set P1), sense, 5′-TCAAGAACTGCCACAACGACAC-3′, and antisense, 5′-CTGGGGTCAGAGTCCTGGGAG-3′; Bcl-3 (primer set P2), sense, 5′-GCAGATCTTGGACTCATGAGG-3′, and antisense, same as for primer set P1; cyclin D1, sense, 5′-GACCATCCCCCTGACGGCCGAG-3′, and antisense, 5′-CCGCACGTCGGTGGGTGTGC-3′; GAPDH, sense, 5′-GGTCGTATTGGGCGCCTGGTCACC-3′, and antisense, 5′-CACACCCATGACGAACATGGGGGC-3′.
RESULTS
IPTG-dependent induction of p53 in H1299w/tp53 cells.
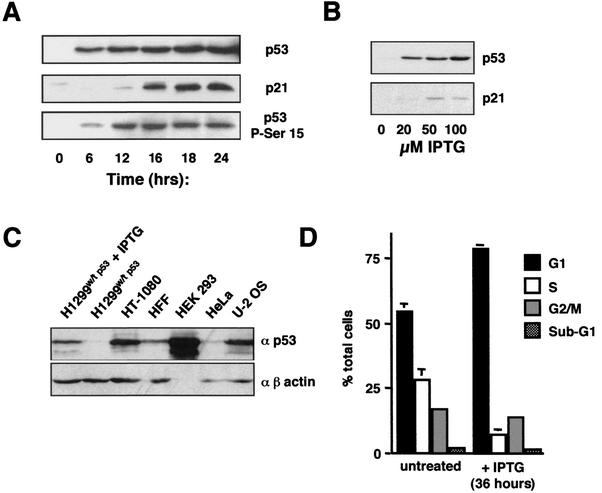
To investigate p53 function under controlled conditions, a human non-small-cell lung carcinoma cell line, H1299, expressing IPTG-inducible p53, was created by using the Stratagene LacSwitchTMII system. Wild-type H1299 cells are p53 null and in the absence of IPTG, extremely low levels of p53 expression were observed in these H1299w/tp53 cells (Fig. 1A). Upon IPTG treatment, p53 expression was observed after 6 h and increased steadily up to 24 h after treatment (Fig. 1A). Interestingly, p53 was phosphorylated at serine 15, an event that is associated with transcriptional activity (11, 16, 31). This phosphorylation lagged behind overall p53 levels, being seen at a low level at 6 h following IPTG treatment and dramatically increasing after 12 h (Fig. 1A). Consistent with IPTG-induced p53 being transcriptionally active, induction of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 was observed. Only low levels of p21 were seen at 12 h following IPTG treatment, with significant levels being seen after 16 h. Since the first transcriptional effects of p53 can be observed at 16 h, this time point was utilized for analysis in future experiments. The expression levels of p53 were also dependent on the dose of IPTG used (Fig. 1B). Comparison with cells containing endogenous wild-type p53 demonstrated that this expression was at physiological levels (Fig. 1C).
FIG. 1.
Induction of p53 expression in H1299w/tp53 cells. (A) p53 induced in H1299w/tp53 cells is phosphorylated at serine 15 and stimulates p21 expression. Western blot analysis of nuclear protein extracts showing the time course of induction of p53, p21, and phospho-serine 15-p53, following treatment with 100 μM IPTG in H1299w/tp53 cells. (B) Induction of p53 in H1299w/tp53 cells is dependent on the dose of IPTG. p53 and p21 expression was analyzed by Western blotting of nuclear protein extracts prepared from H1299w/tp53 cells following 16 h of treatment with increasing concentrations of IPTG. (C) The level of p53 induced in H1299w/tp53 is comparable to endogenous wild-type p53 levels in other cell lines. p53 expression was analyzed by Western blotting of nuclear protein extracts (15 μg) prepared from H1299w/tp53 cells following 16 h of treatment and the other indicated cell lines. (D) Induction of p53 in H1299w/tp53 cells results in a G1 cell cycle arrest. Control cells or H1299w/tp53 cells treated with 100 μM IPTG for 36 h were stained with propidium iodide and analyzed by using FACS analysis.
Induction of p53 in H1299 cells results in repression of cyclin D1.
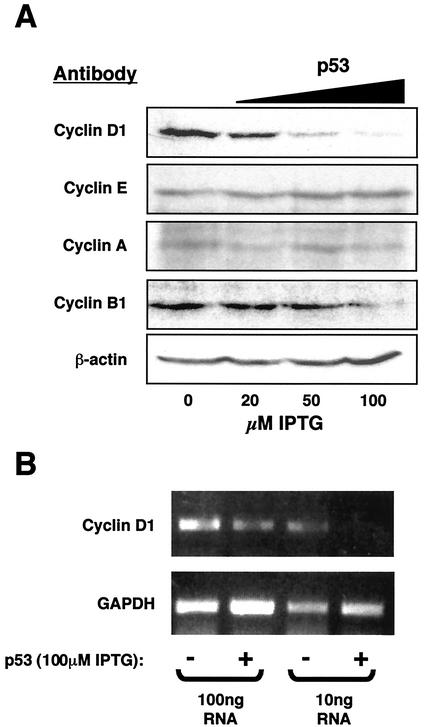
In contrast to the effects previously seen in Saos-2 cells (49), induction of p53 in the H1299 cell background, in the absence of any other stimulation, did not result in cell death. Instead, a G1 cell cycle arrest was observed after 36 h (Fig. 1D). To investigate this further, the expression levels of different cyclins were analyzed by Western blot 16 h after IPTG treatment. Significantly, of those proteins analyzed, cyclin D1 was found to be most sensitive to p53 expression levels, being repressed in a dose-dependent manner (Fig. 2A). Some repression of cyclin B1 was observed at higher levels of p53. As shown in Fig. 1, these results represent early events following p53 induction, and it would be expected that additional changes in cyclin levels would be seen at later time points. Regulation of cyclin D1 expression can occur both at the level of protein degradation and also of gene transcription (1). RT-PCR analysis demonstrated that induction of p53 also results in a significant decrease in cyclin D1 mRNA levels (Fig. 2B), suggesting that this effect, at least in part, occurred at the transcriptional level.
FIG. 2.
p53 selectively down regulates cyclin D1 expression. (A) Induction of p53 in H1299w/tp53 cells represses cyclin D1 protein in a dose-dependent manner. Whole-cell lysates from H1299w/tp53 cells, treated for 16 h with increasing concentrations of IPTG, were subjected to Western blot analysis by using antibodies to cyclins A, B1, D1, E, and a β-actin control as indicated. (B) Induction of p53 in H1299w/tp53 cells represses cyclin D1 mRNA levels. RT-PCR analysis was performed by using primers specific to cyclin D1 or a GAPDH control, with total RNA prepared from H1299w/tp53 cells treated for 16 h with IPTG.
p53 represses the cyclin D1 promoter.
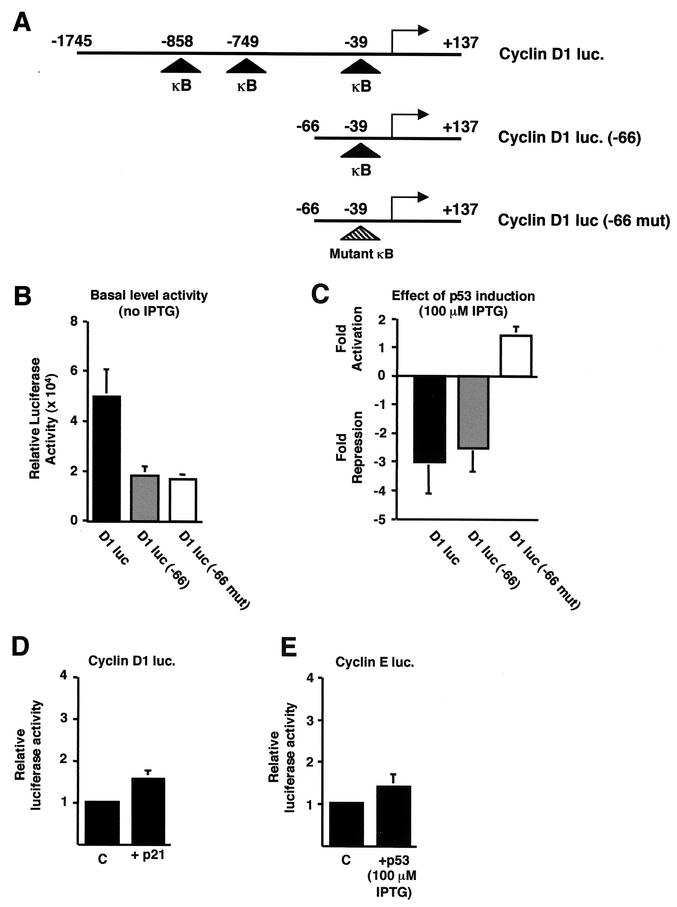
We next examined whether p53 expression regulated the activity of the cyclin D1 promoter. To perform these studies, a reporter plasmid containing the full-length cyclin D1 promoter (from −1745 to +137 relative to the start site of transcription), upstream of the luciferase gene, was transiently transfected into H1299w/tp53 cells (Fig. 3A). Cells were harvested 16 h after IPTG treatment, corresponding with the time seen for repression of endogenous cyclin D1 levels (Fig. 2). p53 expression was seen to significantly repress cyclin D1 promoter activity (Fig. 3C). To further investigate this effect, a truncation of the cyclin D1 promoter was analyzed. This reporter plasmid (containing the region from −66 to +137 relative to the start site of transcription) showed a decrease in basal activity (Fig. 3B) but was still repressed by p53 to the same extent as the full-length promoter (Fig. 3C). The cyclin D1 promoter can be regulated by many transcription factors (2, 21, 28, 33, 54, 58, 61) but does not contain any known binding sites for p53. Recently, however, it has been shown that members of the NF-κB family are also important regulators of cyclin D1 expression, with three putative κB elements being found in the promoter (21, 28, 61). Deletion to −66 removed two of these sites, but one still remained in the −66 to +137 construct (Fig. 3A). Given the previously described cross talk between p53 and NF-κB, we next investigated whether repression of cyclin D1 promoter activity might result from modulation of NF-κB. Mutation of the proximal κB element within the −66 to +137 cyclin D1 reporter plasmid abolished p53-mediated repression and actually resulted in a slight stimulation of activity (Fig. 3C). This suggested that NF-κB activity is actually required for p53-mediated repression. Since p53 induction also results in an increase in p21 protein levels (Fig. 1A), which has been shown to have numerous effects on transcription (43), we next investigated whether it might account for these effects. Cotransfection of p21 with the cyclin D1 (−1745 to +137) reporter plasmid had no significant effect, however (Fig. 3D). Furthermore, consistent with the specificity seen in Fig. 2, induction of p53 had no effect on a cyclin E promoter reporter plasmid after 16 h of IPTG treatment (Fig. 3E).
FIG.3.
p53 specifically inhibits cyclin D1 promoter activity. (A) Schematic diagram of the cyclin D1, cyclin D1 (−66), and cyclin D1 (−66 mut) luciferase reporter plasmids used. The relative positions of the previously described NF-κB binding sites are shown. luc, luciferase. (B) Relative luciferase activity of each of the cyclin D1-luciferase constructs. A quantity of 1.5 μg of each of the cyclin D1-luciferase reporter plasmids was transfected into H1299w/tp53 cells. (C) Inhibition of cyclin D1 promoter activity by p53 requires the proximal κB site. A quantity of 1.5 μg of each of the cyclin D1-luciferase reporter plasmids was transfected into H1299w/tp53 cells. p53 was induced by treatment with 100 μM IPTG, and cells were harvested 16 h after induction. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. (D) p21 does not inhibit cyclin D1 promoter activity in H1299w/tp53 cells. One and a half micrograms of cyclin D1-luciferase reporter plasmid was cotransfected with 1.5 μg of RSV-p21 or control RSV plasmid. Results are expressed as relative luciferase activity. (E) p53 does not inhibit cyclin E promoter activity in H1299w/tp53 cells. One and a half micrograms of cyclin E luciferase plasmid was transfected into H1299w/tp53 cells, and p53 expression was induced by 100 μM IPTG. Cells were harvested 16 h after IPTG treatment, and results are expressed as relative luciferase activity.
p53 repression of the cyclin D1 promoter is reversed by modulators of the NF-κB2 (p52/p100) NF-κB subunit.
Cyclin D1 promoter activity has been shown to be regulated by both p50/RelA and also p52/Bcl-3 NF-κB complexes (21, 28, 61). Bcl-3 is highly homologous to members of the IκB family, which typically bind and inhibit NF-κB subunits (44). Unlike IκB proteins, Bcl-3 is found to be constitutively present in the nucleus and is a transcriptional coactivator which can interact directly with homodimers of the p50 and p52 NF-κB subunits (8, 17, 18, 39). In H1299 cells, we did not observe any effect on cyclin D1 promoter activity, either basal or p53 repressed, upon cotransfection of a dominant-negative IκBα expression plasmid, which would be expected to more specifically target a p50/RelA complex (data not shown). Furthermore, H1299 cells lacked constitutively high levels of p50/RelA NF-κB activity, and induction of p53 did not result in any apparent significant change in NF-κB DNA binding to either the immunoglobulin/human immunodeficiency virus or H2 or proximal cyclin D1 κB elements (data not shown).
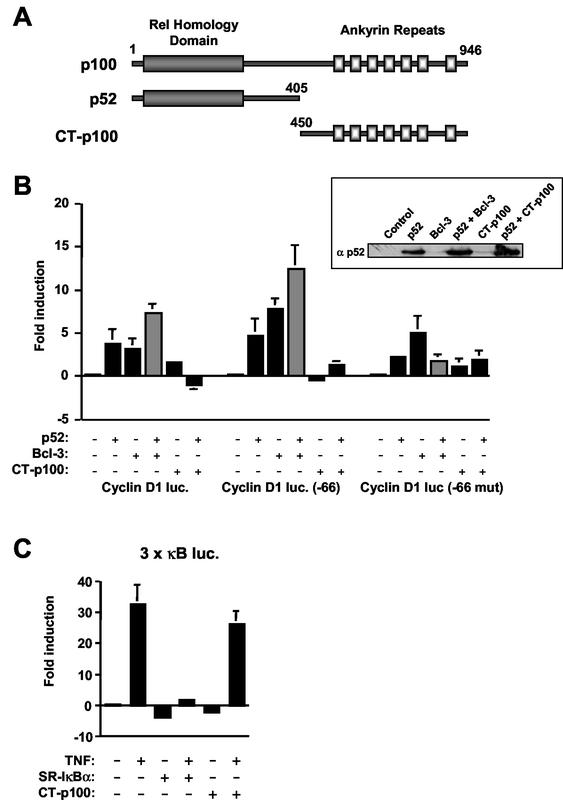
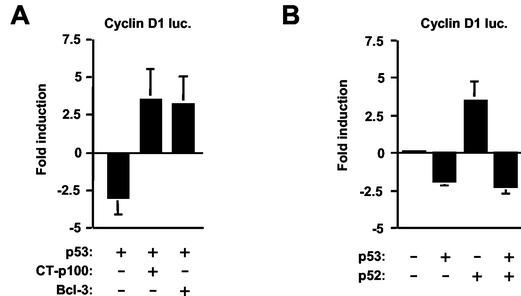
These observations suggested that other NF-κB subunits might mediate the effects of p53 through the proximal cyclin D1 κB element. We therefore investigated any possible involvement that p52 and Bcl-3 might have on p53-mediated repression. Similar to the results of Westerheide et al., coexpression of either a p52 or Bcl-3 expression plasmid, alone or in combination, resulted in stimulation of the cyclin D1 promoter (Fig. 4B) (61). Similar results were also obtained with the truncated −66 to +137 cyclin D1 promoter, and cooperative stimulation of transcription between p52 and Bcl-3 was virtually abolished by mutation of the proximal κB element (Fig. 4B). Surprisingly, significant activation by Bcl-3 alone could still be seen, suggesting that it might be able to target other factors binding this promoter region (Fig. 4B). Similarly, some residual activation by p52 alone could still be observed, although this was not affected by the CT-p100 repressor (see below), thus suggesting a nonspecific effect (Fig. 4B).
FIG. 4.
Regulation of the cyclin D1 promoter by p52 and Bcl-3. (A) Schematic diagram of p100, p52, and the region of the C terminus of p100 used to construct the CT-p100 expression plasmid. (B) Effects of p52, Bcl-3, and CT-p100 expression on cyclin D1 promoter activity. One and a half micrograms of each of the indicated RSV expression plasmids was cotransfected with 1.5 μg of the indicated cyclin D1-luciferase reporter plasmids into H1299w/tp53 cells in the absence of IPTG treatment. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. Western blot analysis of p52 expression in nuclear protein extracts from comparably transfected cells is shown inset. luc, luciferase. (C) CT-p100 does not inhibit TNF-α-induced NF-κB transcriptional activity. The 3× κB-luciferase reporter plasmid (1.5 μg) was cotransfected with 1.5 μg of either the CT-p100, SR-IκBα, or control RSV expression plasmid into H1299w/tp53 cells in the absence of IPTG treatment. Luciferase activity was induced by treatment with 10 ng of TNF-α per ml, and cells were harvested 12 h later. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls.
Since endogenous p52 homodimers do not appear to be readily amenable to investigation by standard electrophoretic mobility shift assay techniques (data not shown), the role of endogenous p52 protein as a regulator of cyclin D1 promoter activity was investigated by using a plasmid expressing the C terminus of p100 (CT-p100) (Fig. 4A). Unlike other NF-κB subunits, p50 and p52 are derived by proteolytic processing of their larger precursor proteins, p105 and p100, respectively. The C-terminal ankyrin repeat domains of both p105 and p100, which are lost in p50 and p52, function as IκB-like NF-κB inhibitors (32, 50). Furthermore, expression of the C terminus of p105 has been previously shown to be able to function as a specific inhibitor of the p50 NF-κB subunit when expressed alone (32). As expected, coexpression of CT-p100 efficiently repressed p52-mediated induction of the cyclin D1 promoter (Fig. 4B) but did not affect TNF-mediated activation of an artificial NF-κB reporter containing multiple κB elements (Fig. 4C). Analysis of nuclear protein extracts from comparably transfected cells indicated that CT-p100 did not inhibit p52 expression levels (Fig. 4B). In contrast, dominant-negative IκBα was an efficient repressor of TNF-activated NF-κB, which could be expected to consist predominantly of p50/RelA complexes (Fig. 4C). These results suggested that CT-p100 is a repressor of p52 homodimers and does not inhibit the activity of p50/RelA. It cannot be ruled out that there might be effects on other members of the NF-κB family, although we find that p50 homodimers, which would be the other subunit most likely to be inhibited by CT-p100, do not regulate the cyclin D1 promoter (data not shown and reference 61).
Curiously, when expressed alone, CT-p100 did not strongly inhibit the cyclin D1 promoter (Fig. 4B). This observation is consistent with the absence of a strong effect in unstimulated H1299 cells of mutating the proximal cyclin D1 promoter κB element (Fig. 3B), suggesting either a balance between activating and inhibitory p52 complexes or that additional factors can compensate for the removal of p52. Significantly, however, expression of CT-p100 completely reversed p53-mediated repression of the cyclin D1 promoter (Fig. 5A) in a manner similar to the earlier mutation of the proximal κB element (Fig. 3C). To confirm that the effect of p53 did result from modulation of a p52/Bcl-3 complex, coexpression of Bcl-3 also resulted in complete reversal of p53-mediated repression (Fig. 5A). Interestingly, p52, despite being able to activate the cyclin D1 promoter in unstimulated cells, was unable to reverse inhibition by p53 (Fig. 5B). Taken together, these results suggested that p53-mediated repression of the cyclin D1 promoter results from its ability to modulate the activity of a p52/Bcl-3 complex.
FIG. 5.
p53-induced repression of cyclin D1 promoter activity can be prevented by Bcl-3 and the C terminus of p100. (A) Inhibition of cyclin D1 promoter activity by p53 can be reversed by expression of Bcl-3 or CT-p100. One and a half micrograms of cyclin D1-luciferase reporter plasmid was cotransfected with 1.5 μg of Bcl-3, CT-p100, or control RSV expression plasmid into H1299w/tp53 cells. p53 expression was induced by 100 μM IPTG, and cells were harvested 16 h later. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. luc, luciferase. (B) Inhibition of cyclin D1 promoter activity by p53 is not prevented by p52. One and a half micrograms of cyclin D1-luciferase reporter plasmid was cotransfected with 1.5 μg of p52 or control RSV expression plasmid into H1299w/tp53 cells. p53 expression was induced by 100 μM IPTG, and cells were harvested 16 h later. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls.
p53 expression results in down regulation of Bcl-3 protein levels in H1299w/tp53 cells.
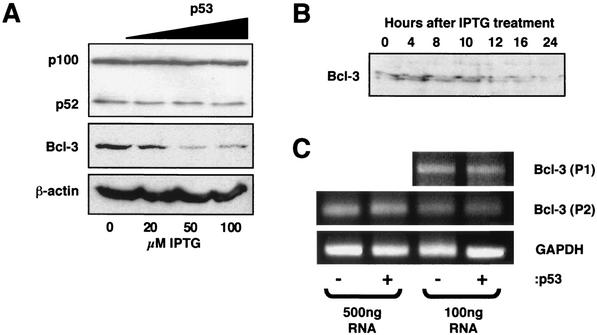
We next investigated the mechanism through which p53 is able to affect the activity of p52/Bcl-3. Western blot analysis demonstrated that IPTG induction of p53 did not alter processing of p100 to p52 (Fig. 6A). In contrast, a strong, dose-dependent down regulation of Bcl-3 protein levels was observed (Fig. 6A). A time course of p53 induction revealed that Bcl-3 repression starts at 12 h following IPTG treatment (Fig. 6B), coincident with induction of p21 protein levels (Fig. 1A), suggesting that this is an early effect of p53. Of note, as has been observed previously (9), in some extract preparations Bcl-3 runs as a doublet, probably resulting from partial dephosphorylation during extract preparation. RT-PCR analysis demonstrated that induction of p53 did not significantly affect Bcl-3 mRNA levels, however (Fig. 6C), suggesting that this effect is primarily posttranscriptional.
FIG. 6.
Induction of p53 down regulates Bcl-3 protein levels in H1299w/tp53 cells. (A) p53 represses Bcl-3 but not p100/p52 protein levels. Following 16 h of p53 induction by increasing concentrations of IPTG in H1299w/tp53 cells, whole-cell lysates were prepared and the protein levels of p100/p52, Bcl-3, and β-actin control were analyzed by Western blot. (B) Bcl-3 down regulation is evident 12 h after IPTG induction of p53. p53 was induced in H1299w/tp53 cells by 100 μM IPTG. Cells were harvested at the indicated times following IPTG treatment, and nuclear protein extracts were prepared and analyzed by Western blot for Bcl-3 levels. (C) Induction of p53 in H1299w/tp53 cells does not repress Bcl-3 mRNA levels. RT-PCR analysis was performed by using two sets of primers specific to Bcl-3 (P1 and P2) or a GAPDH control, with total RNA prepared from H1299w/tp53 cells treated for 16 h with IPTG.
p53 reduces the levels of p52/Bcl-3 complexes and induces association of p52 with HDAC1.
Consistent with the loss of Bcl-3 protein, a significant reduction in p52/Bcl-3 complexes was also observed (Fig. 7A). A number of observations suggested that p53-mediated repression of the cyclin D1 promoter did not just result from loss of Bcl-3, however. The CT-p100 construct or mutation of the proximal κB element, despite being able to reverse p53-mediated repression, had minimal effects on cyclin D1 promoter activity in unstimulated cells (Fig. 3B and 4B). These results were consistent with p52 having an active inhibitory role following p53 induction and Bcl-3 suppression. If repression of the cyclin D1 promoter and the inability of p52 to reverse this (Fig. 5B) were merely the result of removing an activator (Bcl-3), then it would not be expected that p53-mediated inhibition would be abolished by mutation of the proximal κB element or the cotransfection of CT-p100. Rather, these would mimic or enhance the p53 effect. Our results could be explained, therefore, if p52 homodimers were capable of interacting with HDACs or other transcriptional repressor complexes in H1299 cells. Interestingly, it has recently been shown that both p50 homodimers and RelA(p65) can interact with HDAC activity in unstimulated cells (4, 63). If this were also the case for p52, in the absence of p53, a balance might exist between activating p52/Bcl-3 complexes and inhibitory p52/HDAC complexes, both working through the same DNA elements. Mutation of the proximal NF-κB binding site or cotransfection of CT-p100 would have the effect of canceling out the activities of both complexes, resulting in little change. p53-mediated suppression of Bcl-3 would then tip the balance in favor of the p52/HDAC complexes, resulting in active transcriptional repression consistent with the requirement for the proximal κB element and reversal of repression by CT-p100.
FIG. 7.
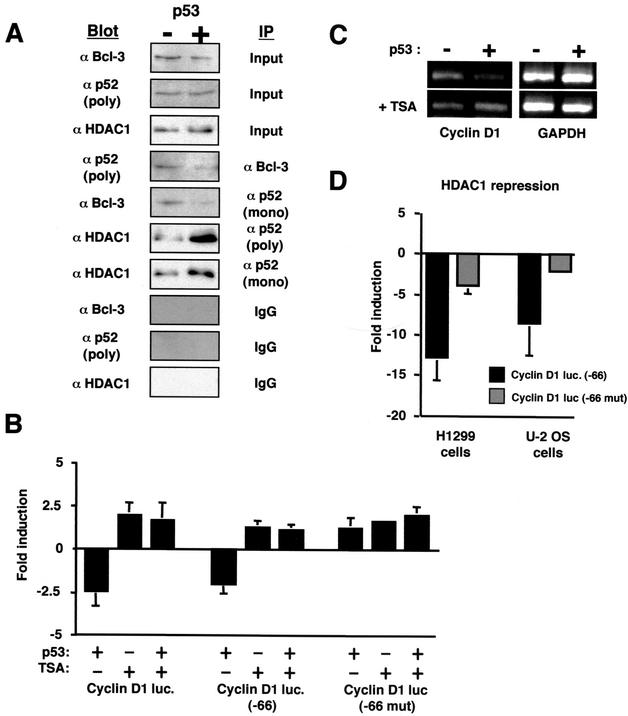
Induction of p53 reduces p52/Bcl-3 complexes and increases p52 association with HDAC1. (A) Modulation of p52 complex formation by p53. Following 16 h of p53 induction by 100 μM IPTG in H1299w/tp53 cells, nuclear protein extracts were prepared. Immunoprecipitation experiments were then performed with either a monoclonal or polyclonal antibody to p52, Bcl-3 antibody, or an immunoglobulin G control, from 100 μg of protein extract. The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Samples of input material are shown (10 μg). (B) Inhibition of HDAC activity abolishes p53-mediated repression of the cyclin D1 promoter. One and a half micrograms of each of the cyclin D1, cyclin D1 (−66), and cyclin D1 (−66 mut) luciferase reporter plasmids was transfected into H1299w/tp53 cells. p53 was induced by treatment with 100 μM IPTG, and cells were harvested 16 h after induction. Cells were treated with 100 ng of TSA per ml for 16 h before harvesting as indicated. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. luc, luciferase. (C) Inhibition of HDAC activity abolishes p53-mediated repression of endogenous cyclin D1. RT-PCR analysis was performed by using primers specific to cyclin D1 or a GAPDH control, with total RNA (100 ng) prepared from H1299w/tp53 cells treated for 16 h with IPTG. Cells were treated with 100 ng of TSA per ml for 16 h before harvesting as indicated. (D) HDAC1 represses the cyclin D1 promoter in a κB element-dependent manner. The cyclin D1 (−66) or cyclin D1 (−66 mut) luciferase reporter plasmid (1.5 μg of each) was transfected into H1299w/tp53 or U-2 OS cells together with 1 μg of HDAC1 expression plasmid. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls.
We therefore investigated whether p52 was associated with HDAC1. Immunoprecipitation of p52 with two distinct antibodies revealed that not only did endogenous p52 associate with HDAC1 in untreated cells but also that this interaction increased significantly upon p53 induction (Fig. 7A). This change did not reflect an alteration in intrinsic HDAC1 levels, which were unaffected upon induction of p53 (Fig. 7A). We next investigated whether HDAC activity was required for p53-mediated inhibition of the cyclin D1 promoter. Treatment with TSA, an inhibitor of deacetylase activity, abolished p53-mediated inhibition of both the full-length and truncated versions of the cyclin D1 promoter (Fig. 7B). Moreover, TSA treatment also abolished p53 inhibition of endogenous cyclin D1 mRNA levels (Fig. 7C). Consistent with these effects, transfection of an HDAC1 expression plasmid strongly inhibited the full-length and truncated cyclin D1 promoters in both H1299 and U-2 OS cells (Fig. 7D and data not shown). Mutation of the proximal κB element again strongly reduced this repression, consistent with effects mediated through p52 binding to this site (Fig. 7D). These results indicated therefore that induction of p53 results in loss of activating p52/Bcl-3 complexes (through inhibition of Bcl-3 levels) and an increase in inhibitory p52/HDAC1 complexes. This observation explains that requirement of the proximal κB element for p53-mediated repression and the reversal of this repression by the CT-p100 inhibitor.
Repression of the cyclin D1 promoter and Bcl-3 protein requires p53 phosphorylation on serine 15.
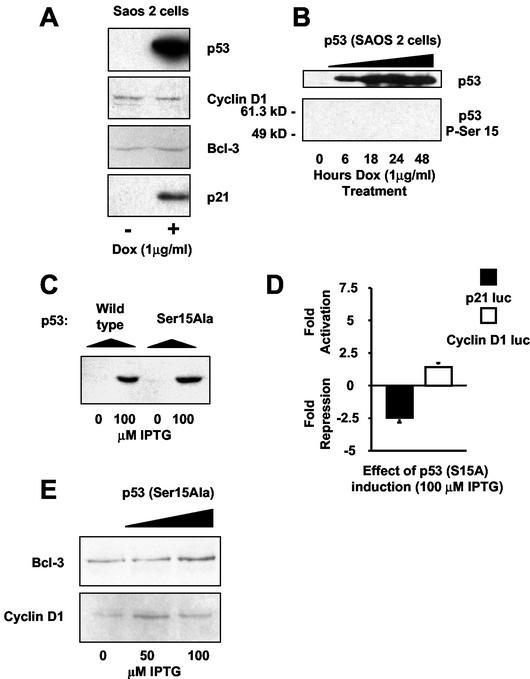
We next investigated whether Bcl-3 repression was seen in other cell lines expressing p53. In contrast to the results seen in H1299w/tp53 cells, tetracycline-regulated p53 in a Saos-2 stable cell line did not result in down regulation of cyclin D1 or Bcl-3 protein levels (Fig. 8A). We had earlier observed that H1299-cell p53 was phosphorylated at serine 15 (Fig. 1A). In contrast, Saos-2-cell p53 was not detectably phosphorylated at this residue (Fig. 8B). While there are many other possible mechanisms that could account for the cell type specificity seen for Bcl-3 and cyclin D1 repression, this suggested that serine 15 phosphorylation might account, at least in part, for some of these differences. This result also suggests that other mechanisms can compensate for lack of serine 15 phosphorylation since p53 still induces p21 and apoptosis in Saos-2 cells.
FIG. 8.
Down regulation of Bcl-3 and cyclin D1 by p53 in H1299 cells is dependent on phosphorylation of p53 at serine 15. (A) p53 expression in Saos-2 cells does not result in either the down regulation of Bcl-3 or cyclin D1. Saos-2 cells containing p53 under the control of the Tet-On-inducible system were harvested after 24 h of doxycycline treatment, and nuclear protein extracts were prepared. Western blot analysis of p53, p21, cyclin D1, and Bcl-3 was performed on p53-induced and control-treated samples. (B) p53 expressed in Saos-2 cells, in the absence of additional stimulation, is not phosphorylated at serine 15. Saos-2 cells containing p53 under the control of the Tet-On-inducible system were harvested at the indicated times following doxycycline treatment, and nuclear protein extracts were prepared. p53 and phospho-serine 15 p53 levels were then determined by Western blot analysis. (C) p53 is expressed to similar levels in both H1299w/tp53 and H1299S15A cells. Equivalent levels of protein from nuclear protein extracts prepared from H1299w/tp53 and H1299S15A cells treated with IPTG for 16 h were analyzed for p53 levels by Western blot. (D) Ser15Ala mutant p53 is unable to induce p21-luciferase activity or inhibit cyclin D1 promoter activity. One and a half micrograms of either p21-luciferase or cyclin D1-luciferase reporter plasmid was transfected into H1299/mutp53 cells. Cells were harvested following 16 h of IPTG treatment. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. luc, luciferase. (E) Ser15Ala mutant p53 fails to down regulate Bcl-3 or cyclin D1 protein levels. Following 16 h of p53 induction by increasing concentrations of IPTG in H1299S15A cells, whole-cell lysates were prepared, and the protein levels of Bcl-3 and cyclin D1 were analyzed by Western blot.
To investigate whether serine 15 phosphorylation of p53 might be required for repression of Bcl-3 and cyclin D1, we utilized another stable cell line, H1299S15A, created at the same time as the H1299w/tp53 cells, which contained an IPTG-inducible p53 with a serine-15-to-alanine mutation (p53S15A). p53S15A protein levels were induced to the same extent as in wild-type p53 (Fig. 8C). p53S15A also repressed the activity of a p21 promoter containing reporter plasmid (Fig. 8D), failed to induce endogenous p21 protein, and did not undergo a G1 cell cycle arrest (data not shown). The former observation might indicate that while the S15A mutation inhibits the formation of p53/coactivator complexes, p53/corepressor complexes remain intact. This possibility was not pursued any further, however. In contrast to the H1299w/tp53 cells, IPTG treatment of H1299S15A cells failed to repress the cyclin D1 promoter or cyclin D1 and Bcl-3 protein (Fig. 8D and E). In fact, a slight activation of both the cyclin D1 promoter and Bcl-3 protein levels was observed, although this was not reflected by a change in cyclin D1 protein. Inhibition of Bcl-3 protein levels and consequent repression of cyclin D1 promoter activity and protein therefore require specific posttranslational modification of p53 at serine 15.
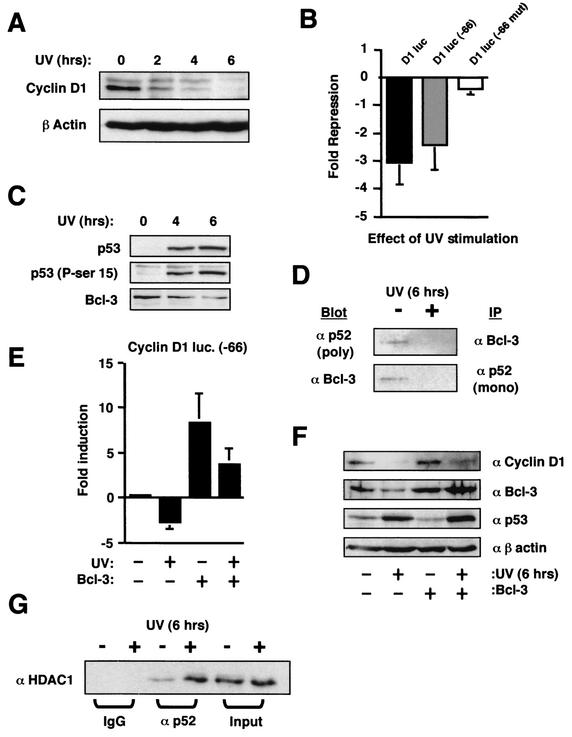
UV light stimulation of U-2 OS cells also results in repression of cyclin D1, loss of Bcl-3, and an increase in p52/HDAC1 complexes.
These results had all been obtained in p53-null cell lines into which p53 had been reintroduced under the control of an inducible promoter. While this allowed us to assess the direct contribution of p53 to these effects, it was important to demonstrate that similar regulation of p52 and the cyclin D1 promoter occurred in a cell line in which endogenous p53 was induced by an activating stimulus. We therefore investigated the effect of stimulating the human osteosarcoma U-2 OS cell line, which contains wild-type p53, with UV light. It was not possible to demonstrate a cell cycle effect of UV stimulation in these cells, since they underwent apoptosis before any significant accumulation in the G1 phase could be observed (data not shown). p53's effects on the cell cycle occur concomitantly with effects of apoptosis, however (discussed in reference 56), and these cells still provided a suitable model system to investigate the effects of endogenous p53 activation on p52 and cyclin D1 promoter activity. UV stimulation resulted in strong repression of cyclin D1 protein levels (Fig. 9A). Furthermore, UV also mimicked p53 induction in H1299w/tp53 cells by inhibiting the cyclin D1 promoter in a manner dependent upon the proximal κB element (Fig. 9B). As expected, UV stimulation induced both p53 protein and its phosphorylation on serine 15 (Fig. 9C). Significantly, as in the H1299w/tp53 cells, Bcl-3 protein levels were also suppressed (Fig. 9C), resulting in a significant decrease in p52/Bcl-3 complexes (Fig. 9D). As also seen previously in the H1299w/tp53 cells, Bcl-3 expression has a strong stimulatory effect on the cyclin D1 promoter and reversed UV-induced repression (Fig. 9E). A similar effect was also seen on endogenous cyclin D1 protein levels, where Bcl-3 expression partially recovered the loss of protein seen upon UV treatment (Fig. 9F). Since the transfection efficiency of these cells is typically less than 50% (data not shown), a more significant recovery would be unlikely to be observed by using these techniques. Finally, and also consistent with the effects seen in H1299w/tp53 cells, a significant increase in the association of p52 with HDAC1 followed UV treatment (Fig. 9G). Therefore, p52 undergoes the same transcriptional switch from activator to repressor upon induction of endogenous p53 in response to UV light.
FIG.9.
Effects of UV stimulation on cyclin D1 and p52. (A) UV treatment down regulates cyclin D1 protein levels. U-2 OS cells were treated with 40-J/m2 UV radiation for the indicated times. Whole-cell lysates were prepared and immunoblotted for cyclin D1 and a β-actin control as indicated. (B) UV treatment inhibits the cyclin D1 promoter in a manner dependent upon the proximal κB element. One and a half micrograms of each of the cyclin D1 (−66) and cyclin D1 (−66 mut) luciferase reporter plasmids were transfected into U-2 OS cells. Cells were treated with 40-J/m2 UV radiation for 6 h as indicated. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. luc, luciferase. (C) UV treatment induces Ser-15 phosphorylated endogenous p53 and down regulates Bcl-3 levels. U-2 OS cells were treated with 40-J/m2 UV radiation for the indicated times. Nuclear protein extracts were prepared and immunoblotted for p53, phospho-Ser-15-modified p53, and Bcl-3 as indicated. (D) UV treatment results in loss of p52/Bcl-3 complexes. U-2 OS cells were either left untreated or stimulated with 40-J/m2 UV radiation for 6 h, and nuclear protein extracts were prepared. p52 or Bcl-3 was then immunoprecipitated from 100 μg of protein extract. The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted for for the reciprocal protein, as indicated. A sample of input material is shown (10 μg). (E) Inhibition of cyclin D1 promoter activity following UV treatment can bereversed by expression of Bcl-3. One and a half micrograms of cyclin D1-luciferase reporter plasmid was cotransfected with 1.5 μg of Bcl-3 or control RSV expression plasmid into U-2 OS cells. Cells were treated with 40-J/m2 UV radiation for 6 h as indicated. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. (F) Down regulation of cyclin D1 protein levels following UV treatment can be reversed by expression of Bcl-3. U-2 OS cells were transfected with 1.5 μg of Bcl-3 or control RSV expression plasmid. After 36 h, cells were treated with 40-J/m2 UV radiation for 6 h as indicated. Whole-cell lysates were then prepared and analyzed by immunoblotting with the indicated antibodies. (G) UV treatment increases the association of p52 with HDAC1. U-2 OS cells were either left untreated or stimulated with 40-J/m2 UV radiation for 6 h, and nuclear protein extracts were prepared. p52 was then immunoprecipitated with the polyclonal antibody, together with an immunoglobulin G control, from 100 μg of protein extract. The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted for HDAC1. A sample of input material is shown (10 μg).
DISCUSSION
In this report we demonstrate that induction of p53 can result in inhibition of the cyclin D1 promoter through down regulation of Bcl-3 protein levels, p52/Bcl-3 complexes, and an increase in p52 association with HDAC1. Cyclin D1 has previously been found to be an indirect target of p53. For example, the p53-inducible protein PC3 was shown to repress cyclin D1 transcription in NIH 3T3 cells (20). Whether PC3 can regulate Bcl-3 is unknown. In contrast, other reports have indicated that p53 can actually induce cyclin D1 expression, possibly as a consequence of p21 expression (12). Our results are consistent with this apparent cell type specificity, since we show that down regulation of Bcl-3 and subsequent repression of cyclin D1 levels require phosphorylation of p53 at serine 15. Serine 15 phosphorylation has been shown to promote the interaction of p53 with the p300 coactivator protein and to be required for its transcriptional activity (11, 16, 31). These observations imply that down regulation of Bcl-3 by p53 requires the action of a directly inducible p53 target gene. An obvious candidate would be Mdm2, which is known to function as an E3 ubiquitin ligase (55). We have been unable to detect an interaction between Mdm2 and Bcl-3, however (data not shown). Another candidate is a protein termed BRCA1-associated RING domain (BARD1). BARD1 interacts with the breast cancer susceptibility protein BRCA1 (26) and the BRCA1/BARD1 complex functions as an E3 ubiquitin ligase (5). BARD1 has also been shown to associate both with p53 and with a Bcl-3 complex (14, 27). Furthermore, BARD1 is induced by genotoxic stimuli that also induce p53 (27). It is possible that p53 might facilitate BARD1-dependent proteolytic degradation of Bcl-3. p53 induction can also activate signaling pathways (49). It is possible, therefore, that p53 induces Bcl-3 phosphorylation, targeting it for degradation in a manner analogous to other members of the IκB family. The precise mechanism controlling down regulation of Bcl-3 will require further investigation, however.
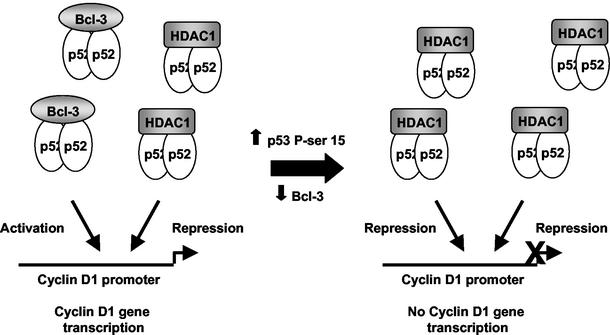
Inhibition of the cyclin D1 promoter by p53 through the proximal κB element does not merely result from the removal of an activator protein (Bcl-3). Repression by p53 required the proximal κB element (Fig. 3C) and was inhibited by the C terminus of p100 (CT-p100) (Fig. 5A). If p53 inhibited cyclin D1 through a passive mechanism, that is, by just removing an activator complex, then these experiments would have been expected to mimic the effect of p53 induction. Instead, these observations all indicated that p53 induced active repression of the cyclin D1 promoter. Consistent with this, we found that p52 can associate with the transcriptional repressor HDAC1 and that this interaction increases significantly upon p53 induction (Fig. 7A). This effect is also observed in U-2 OS cells upon UV stimulation (Fig. 9G), demonstrating that this is not an effect limited to H1299 cells and artificial induction of p53 protein. An active role for this p52/HDAC1 complex is indicated by the reversal of p53-mediated inhibition of the cyclin D1 promoter by the deacetylase inhibitor TSA (Fig. 7B). The p52/HDAC interaction can be observed in untreated cells (Fig. 7A and 9G), and mutation of the proximal cyclin D1 κB element or coexpression of CT-p100 has a minimal effect on promoter activity in the absence of p53 (Fig. 3B and Fig. 4B). We propose, therefore, that in untreated H1299 cells, there is a balance between activator p52/Bcl-3 complexes and repressor p52/HDAC complexes. p53 induction results in a transcriptional switch, whereby loss of Bcl-3 and an increase in p52/HDAC1 complexes results in repression of the cyclin D1 promoter (Fig. 10). We consider it likely, however, that in different cell types, the balance between repressor and activator p52 complexes will vary, with p52/Bcl-3 complexes dominating in many instances, resulting in positive regulation of cyclin D1 regulation.
FIG. 10.
Schematic diagram illustrating the effects of p53 induction on p52 function and cyclin D1 promoter activity. In unstimulated cells, p52 can associate with both Bcl-3 and HDAC1. Upon p53 induction, Bcl-3 is down regulated and p52 associates predominantly with HDAC1, resulting in the repression of the cyclin D1 promoter. It is likely that, dependent upon the cell type, the ratio of p52/Bcl-3 to p52/HDAC1 complexes will vary, resulting in differing levels of NF-κB-dependent transcription.
Inhibition of cyclin D1 and Bcl-3 levels is consistent with the tumor suppressor activity of p53 since both are proto-oncogenes. Cyclin D1 functions in the mid-G1 phase of the cell cycle where, in conjunction with its cyclin-dependent kinase partners CDK4 and CDK6, it acts to inhibit the retinoblastoma tumor suppressor (23). Consistent with this proliferative function, cyclin D1 is found to be overexpressed in most human breast cancers (60). In addition, cyclin D1 can have other CDK-independent functions such as binding to and stimulating the activity of the estrogen receptor (38, 64). Similarly, Bcl-3 has also been shown to have proproliferative functions and can, under some circumstances, inhibit apoptosis (37, 40, 48). Bcl-3 is up regulated in some cases of human B-cell chronic lymphocytic leukemia as a result of its proximity to the breakpoint junction of the t (14, 19) translocation (34, 35). Furthermore, and consistent with its role as a physiologically significant regulator of cyclin D1, it has also been reported to have elevated levels in human breast cancers (13). Although the physiological target genes for p50 and p52 NF-κB homodimers are less well known than those for the classical p50/RelA NF-κB heterodimer, it is likely that they regulate many other promoters. In addition, Bcl-3 has been found to function as a coactivator for the AP-1 and RXR transcription factors (36, 37). By repressing Bcl-3 levels and increasing the relative levels of p52/HDAC1 complexes, p53 is likely to affect the expression of a number of other genes in addition to cyclin D1, and this mechanism could potentially therefore represent an important component of the p53 tumor suppressor pathway.
Acknowledgments
We thank Sonia Lain and David Lane for providing many reagents and for their help and assistance. We are also grateful to Rosie Clarke for help with FACS analysis; Richard Pestell for providing the cyclin D1 reporter plasmids; Julian Blow, Michelle Garrett, and Stefan Roberts for their critical reading of the manuscript; Shreeram Sathya; Nicola Wiechens; and all the members of the N.D.P. laboratory for their help and assistance.
N.D.P. is funded by a Royal Society University Fellowship, and S.R. is funded by a grant from Cancer Research UK.
REFERENCES
- 1.Agami, R., and R. Bernards. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55-66. [DOI] [PubMed] [Google Scholar]
- 2.Albanese, C., M. D'Amico, A. T. Reutens, M. F. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundaram, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclin D1 gene by the EPA-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, L. A., and N. D. Perkins. 2002. The large subunit of replication factor C interacts with the histone deacetylase, HDAC1. J. Biol. Chem. 277:29550-29554. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner, B. P., S. D. Westerheide, and A. S. Baldwin. 2001. The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer, R., and T. Ludwig. 2002. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12:86-91. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin, A. S. 2001. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J. Clin. Investig. 107:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkett, M., and T. D. Gilmore. 1999. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18:6910-6924. [DOI] [PubMed] [Google Scholar]
- 8.Bours, V., G. Franzoso, V. Azarenko, S. Park, T. Kanno, K. Brown, and U. Siebenlist. 1993. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell 72:729-739. [DOI] [PubMed] [Google Scholar]
- 9.Bundy, D. L., and T. W. McKeithan. 1997. Diverse effects of BCL3 phosphorylation on its modulation of NF-κB p52 homodimer binding to DNA. J. Biol. Chem. 272:33132-33139. [DOI] [PubMed] [Google Scholar]
- 10.Chang, N. S. 2002. The non-ankyrin C terminus of IκB α physically interacts with p53 in vivo and dissociates in response to apoptotic stress, hypoxia, DNA damage, and transforming growth factor-β 1-mediated growth suppression. J. Biol. Chem. 277:10323-10331. [DOI] [PubMed] [Google Scholar]
- 11.Chao, C., S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2000. Phosphorylation of murine p53 at Ser-18 regulates the p53 responses to DNA damage. Proc. Natl. Acad. Sci. USA 97:11936-11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X. B., J. Bargonetti, and C. Prives. 1995. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 55:4257-4263. [PubMed] [Google Scholar]
- 13.Cogswell, P. C., D. C. Guttridge, W. K. Funkhouser, and A. S. Baldwin. 2000. Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 19:1123-1131. [DOI] [PubMed] [Google Scholar]
- 14.Dechend, R., F. Hirano, K. Lehmann, V. Heissmeyer, S. Ansieau, F. G. Wulczyn, C. Scheidereit, and A. Leutz. 1999. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene 18:3316-3323. [DOI] [PubMed] [Google Scholar]
- 15.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumaz, N., and D. W. Meek. 1999. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18:7002-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franzoso, G., V. Bours, S. Park, M. Tomita-Yamaguchi, K. Kelly, and U. Siebenlist. 1992. The candidate oncoprotein Bcl-3 is an antagonist of P50/NF-κB-mediated inhibition. Nature 359:339-342. [DOI] [PubMed] [Google Scholar]
- 18.Fujita, T., G. P. Nolan, H. C. Liou, M. L. Scott, and D. Baltimore. 1993. The candidate protooncogene Bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev. 7:1354-1363. [DOI] [PubMed] [Google Scholar]
- 19.Gu, L. B., H. W. Findley, and M. X. Zhou. 2002. MDM2 induces NF-κB/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood 99:3367-3375. [DOI] [PubMed] [Google Scholar]
- 20.Guardavaccaro, D., G. Corrente, F. Covone, L. Micheli, I. D'Agnano, G. Starace, M. Caruso, and F. Tirone. 2000. Arrest of G1-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol. Cell. Biol. 20:1797-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttridge, D. C., C. Albanese, J. Y. Reuther, R. G. Pestell, and A. S. Baldwin. 1999. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 23.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 24.Hellin, A. C., P. Calmant, J. Gielen, V. Bours, and M. P. Merville. 1998. Nuclear factor-κB-dependent regulation of p53 gene expression induced by daunomycin genotoxic drug. Oncogene 16:1187-1195. [DOI] [PubMed] [Google Scholar]
- 25.Hupp, T. R., D. P. Lane, and K. L. Ball. 2000. Strategies for manipulating the p53 pathway in the treatment of human cancer. Biochem. J. 352:1-17. [PMC free article] [PubMed] [Google Scholar]
- 26.Irminger-Finger, I., and W. C. Leung. 2002. BRCA1-dependent and independent functions of BARD1. Int. J. Biochem. Cell Biol. 34:582-587. [DOI] [PubMed] [Google Scholar]
- 27.Irminger-Finger, I., W. C. Leung, J. Li, M. Dubois-Dauphin, J. Harb, A. Feki, C. E. Jefford, J. V. Soriano, M. Jaconi, R. Montesano, and K. H. Krause. 2001. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 8:1255-1266. [DOI] [PubMed] [Google Scholar]
- 28.Joyce, D., B. Bouzahzah, M. F. Fu, C. Albanese, M. D'Amico, J. Steer, J. U. Klein, R. J. Lee, J. E. Segall, J. K. Westwick, C. J. Der, and R. G. Pestell. 1999. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-κB-dependent pathway. J. Biol. Chem. 274:25245-25249. [DOI] [PubMed] [Google Scholar]
- 29.Kawai, H., Y. Yamada, M. Tatsuka, O. Niwa, K. Yamamoto, and F. Suzuki. 1999. Down-regulation of nuclear factor κB is required for p53-dependent apoptosis in X-ray-irradiated mouse lymphoma cells and thymocytes. Cancer Res. 59:6038-6041. [PubMed] [Google Scholar]
- 30.Kirch, H. C., S. Flaswinkel, H. Rumpf, D. Brockmann, and H. Esche. 1999. Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-κB and Myc/Max. Oncogene 18:2728-2738. [DOI] [PubMed] [Google Scholar]
- 31.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 32.Liou, H. C., G. P. Nolan, S. Ghosh, T. Fujita, and D. Baltimore. 1992. The NF-κB p50 precursor, p105, contains an internal I κB-like inhibitor that preferentially inhibits p50. EMBO J. 11:3003-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumura, I., T. Kitamura, H. Wakao, H. Tanaka, K. Hashimoto, C. Albanese, J. Downward, R. G. Pestell, and Y. Kanakura. 1999. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18:1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeithan, T. W., G. S. Takimoto, H. Ohno, V. S. Bjorling, R. Morgan, B. K. Hecht, I. Dube, A. A. Sandberg, and J. D. Rowley. 1997. BCL3 rearrangements and t(14;19) in chronic lymphocytic leukemia and other B-cell malignancies: a molecular and cytogenetic study. Genes Chromosomes Cancer 20:64-72. [PubMed] [Google Scholar]
- 35.Michaux, L., C. Mecucci, M. Stul, I. Wlodarska, J. M. Hernandez, P. Meeus, J. L. Michaux, J. M. Scheiff, H. Noel, A. Louwagie, A. Criel, M. Boogaerts, A. Van Orshoven, J. J. Cassiman, and H. Van Den Berghe. 1996. BCL3 rearrangement and t(14;19)(q32;q13) in lymphoproliferative disorders. Genes Chromosomes Cancer 15:38-47. [DOI] [PubMed] [Google Scholar]
- 36.Na, S. Y., H. S. Choi, J. W. Kim, D. S. Na, and J. W. Lee. 1998. Bcl3, an IκB protein, as a novel transcription coactivator of the retinoid X receptor. J. Biol. Chem. 273:30933-30938. [DOI] [PubMed] [Google Scholar]
- 37.Na, S. Y., J. E. Choi, H. J. Kim, B. H. Jhun, Y. C. Lee, and J. W. Lee. 1999. Bcl3, an IκB protein, stimulates activating protein-1 transactivation and cellular proliferation. J. Biol. Chem. 274:28491-28496. [DOI] [PubMed] [Google Scholar]
- 38.Neuman, E., M. H. Ladha, N. Lin, T. M. Upton, S. J. Miller, J. DiRenzo, R. G. Pestell, P. W. Hinds, S. F. Dowdy, M. Brown, and M. E. Ewen. 1997. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol. Cell. Biol. 17:5338-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolan, G. P., T. Fujita, K. Bhatia, C. Huppi, H. C. Liou, M. L. Scott, and D. Baltimore. 1993. The Bcl-3 protooncogene encodes a nuclear IκB-like molecule that preferentially interacts with NF-κB p50 and p52 in a phosphorylation-dependent manner. Mol. Cell. Biol. 13:3557-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong, S. T., M. L. Hackbarth, L. C. Degenstein, D. A. Baunoch, J. Anastasi, and T. W. McKeithan. 1998. Lymphadenopathy, splenomegaly, and altered immunoglobulin production in BCL3 transgenic mice. Oncogene 16:2333-2343. [DOI] [PubMed] [Google Scholar]
- 41.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 42.Pei, X. H., Y. Nakanishi, K. Takayama, F. Bai, and N. Hara. 1999. Benzo[a]pyrene activates the human p53 gene through induction of nuclear factor κB activity. J. Biol. Chem. 274:35240-35246. [DOI] [PubMed] [Google Scholar]
- 43.Perkins, N. D. 2002. Not just a CDK inhibitor: regulation of transcription by p21WAF1/CIP1/SDI1. Cell Cycle 1:39-41. [PubMed] [Google Scholar]
- 44.Perkins, N. D. 2000. The Rel/NF-κB family: friend and foe. Trends Biochem. Sci. 25:434-440. [DOI] [PubMed] [Google Scholar]
- 45.Perkins, N. D., R. M. Schmid, C. S. Duckett, K. Leung, N. R. Rice, and G. J. Nabel. 1992. Distinct combinations of NF-κB subunits determine the specificity of transcriptional activation. Proc. Natl. Acad. Sci. USA 89:1529-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravi, R., B. Mookerjee, Y. van Hensbergen, G. C. Bedi, A. Giordano, W. S. El-Deiry, E. J. Fuchs, and A. Bedi. 1998. p53-mediated repression of nuclear factor-κB RelA via the transcriptional integrator p300. Cancer Res. 58:4531-4536. [PubMed] [Google Scholar]
- 47.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18:6938-6947. [DOI] [PubMed] [Google Scholar]
- 48.Rebollo, A., L. Dumoutier, J.-C. Renauld, A. Zaballos, V. Ayllón, and C. Martínez-A. 2000. Bcl-3 expression promotes cell survival following interleukin-4 deprivation and is controlled by AP1 and AP1-like transcription factors. Mol. Cell. Biol. 20:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-κB in p53-mediated programmed cell death. Nature 404:892-897. [DOI] [PubMed] [Google Scholar]
- 50.Scheinman, R. I., A. A. Beg, and A. S. Baldwin, Jr. 1993. NF-κB p100 (Lyt-10) is a component of H2TF1 and can function as an IκB-like molecule. Mol. Cell. Biol. 13:6089-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao, J., T. Fujiwara, Y. Kadowaki, T. Fukazawa, T. Waku, T. Itoshima, T. Yamatsuji, M. Nishizaki, J. A. Roth, and N. Tanaka. 2000. Overexpression of the wild-type p53 gene inhibits NF-κB activity and synergizes with aspirin to induce apoptosis in human colon cancer cells. Oncogene 19:726-736. [DOI] [PubMed] [Google Scholar]
- 52.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20:1803-1815. [DOI] [PubMed] [Google Scholar]
- 53.Tergaonkar, V., M. Pando, O. Vafa, G. Wahl, and I. Verma. 2002. p53 stabilization is decreased upon NFκB activation: a role for NFκB in acquisition of resistance to chemotherapy. Cancer Cell 1:493-503. [DOI] [PubMed] [Google Scholar]
- 54.Tetsu, O., and F. McCormick. 1999. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 55.Vousden, K. H. 2002. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta Rev. Cancer 1602:47-59. [DOI] [PubMed] [Google Scholar]
- 56.Vousden, K. H. 2002. Switching from life to death: the Miz-ing link between Myc and p53. Cancer Cell 2:351-352. [DOI] [PubMed] [Google Scholar]
- 57.Wadgaonkar, R., K. M. Phelps, Z. Haque, A. J. Williams, E. S. Silverman, and T. Collins. 1999. CREB-binding protein is a nuclear integrator of nuclear factor-κB and p53 signaling. J. Biol. Chem. 274:1879-1882. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe, G., C. Albanese, R. J. Lee, A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webster, G. A., and N. D. Perkins. 1999. Transcriptional cross talk between NF-κB and p53. Mol. Cell. Biol. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinstat-Saslow, D., M. J. Merino, R. E. Manrow, J. A. Lawrence, R. F. Bluth, K. D. Wittenbel, J. F. Simpson, D. L. Page, and P. S. Steeg. 1995. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat. Med. 1:1257-1260. [DOI] [PubMed] [Google Scholar]
- 61.Westerheide, S. D., M. W. Mayo, V. Anest, J. L. Hanson, and A. S. Baldwin, Jr. 2001. The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G1 transition. Mol. Cell. Biol. 21:8428-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao, R. B., K. Gish, M. Murphy, Y. X. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong, H. H., M. J. May, E. Jimi, and S. Ghosh. 2002. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9:625-636. [DOI] [PubMed] [Google Scholar]
- 64.Zwijsen, R. M. L., E. Wientjens, R. Klompmaker, J. van der Sman, R. Bernards, and R. J. Michalides. 1997. CDK-independent activation of estrogen receptor by cyclin D1. Cell 88:405-415. [DOI] [PubMed] [Google Scholar]