Abstract
Drugs that produce covalent interstrand cross-links (ICLs) in DNA remain central to the treatment of cancer, but the cell cycle checkpoints activated by ICLs have received little attention. We have used the fission yeast, Schizosaccharomyces pombe, to elucidate the checkpoint responses to the ICL-inducing anticancer drugs nitrogen mustard and mitomycin C. First we confirmed that the repair pathways acting on ICLs in this yeast are similar to those in the main organisms studied to date (Escherichia coli, budding yeast, and mammalian cells), principally nucleotide excision repair and homologous recombination. We also identified and disrupted the S. pombe homologue of the Saccharomyces cerevisiae SNM1/PSO2 ICL repair gene and found that this activity is required for normal resistance to cross-linking agents, but not other forms of DNA damage. Survival and biochemical analysis indicated a key role for the “checkpoint Rad” family acting through the chk1-dependent DNA damage checkpoint in the ICL response. Rhp9-dependent phosphorylation of Chk1 correlates with G2 arrest following ICL induction. In cells able to bypass the G2 block, a second-cycle (S-phase) arrest was observed. Only a transient activation of the Cds1 DNA replication checkpoint factor occurs following ICL formation in wild-type cells, but this is increased and persists in G2 arrest-deficient mutants. This likely reflects the fraction of cells escaping the G2 damage checkpoint and arresting in the subsequent S phase due to ICL replication blocks. Disruption of cds1 confers increased resistance to ICLs, suggesting that this second-cycle S-phase arrest might be a lethal event.
Agents that produce covalent interstrand cross-links (ICLs) between the complementary strands of the DNA double helix remain a key component of many cancer chemotherapy regimens (36). Although drugs such as the nitrogen mustards also produce abundant DNA monoadducts, there is compelling evidence that ICLs are the critical cytotoxic lesion (44) and kill proliferating cells by disrupting essential processes such as transcription and replication.
In the best-characterized model organisms, Escherichia coli and Saccharomyces cerevisiae, the response to ICLs is known to involve both nucleotide excision repair (NER) and homologous recombination (13, 24, 35, 39, 54). In these systems experimental evidence is consistent with two main scenarios for ICL repair via a combination of these pathways. The bacterial model invokes an initial ICL “uncoupling” reaction by NER, incising the ICL on one strand only, followed by a recA-dependent recombination event into the (resected) gap created, providing a template for the subsequent excision repair of the opposing adducted strand (13, 58). For S. cerevisiae there is evidence supporting a related pathway during psoralen ICL repair but with the key difference that NER may excise the cross-linked DNA on both strands, producing a double-strand break (DSB) intermediate, which is then repaired through homologous recombination (24, 39). In addition, an accumulating body of evidence from studies of yeast and mammalian cells suggests that events during DNA replication may influence the processing of nitrogen mustard ICLs (1, 14, 35). In rapidly dividing (exponential-phase) yeast cells there is a requirement for functional homologous recombination, and DNA DSBs form at high frequency (35). Interestingly these DSBs still form in the absence of functional NER and therefore could be associated with either of the two recombination repair pathways proposed above. These DSBs most likely arise as a result of processing events at replication forks stalled by ICLs (14, 35).
To date, the cell cycle checkpoints elicited in response to interstrand cross-linking agents have received little attention (16). This is clearly of considerable importance since tumors often exhibit dysregulated checkpoint activity (18), and the response to cross-linking drugs might be influenced by this. Here we take advantage of the fission yeast, Schizosaccharomyces pombe, as a model organism to define the checkpoints involved in the ICL response. This yeast has been the subject of intense investigation, leading to a detailed understanding of its cell cycle and the checkpoints activated by DNA damage (10).
At the heart of the checkpoint response in S. pombe are the checkpoint Rad proteins (Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1) (3, 4, 19, 26, 49). Rad3 and Rad26 exist as a complex (17) which signals to downstream effectors in response to DNA damage through the lipid kinase motif (ATM-related) activity of Rad3 (33). The Rad1-Rad9-Hus1 proteins form a complex that resembles the PCNA sliding clamp that may act to generate a checkpoint signal at aberrant DNA structures (11). Rad17 belongs to a further complex which, by analogy with the S. cerevisiae and human homologues, contains the four small subunits of the replication factor C and is required for the association of Rad9-Rad1-Hus1 (9-1-1) complex proteins with DNA damage (9). The checkpoint Rad family proteins are required for both the replication checkpoint, which is activated by replication fork blocks (such as those induced by hydroxyurea [HU] treatment) preventing mitosis, and the DNA damage checkpoint, acting to arrest cells in late S or G2 following ionizing radiation treatment (10, 19, 47). In response to replication blocks, Rad3 phosphorylates and activates Cds1 (29), whereas during the damage checkpoint Rad3 phosphorylates Chk1 (46, 62); however, the biological significance of this latter event is still not understood. In addition, when Cds1 is absent, blocking DNA replication activates Chk1. This is interpreted as follows: normally, Cds1 prevents the occurrence of DNA damage in S phase when replication forks are stalled, as well as ensuring that mitosis is inhibited. In the absence of Cds1, the resulting DNA damage from stalled and/or collapsed replication activates the Chk1-dependent DNA damage checkpoint to prevent mitosis (17, 29). The kinases Cds1 and Chk1 prevent the activation of the Cdc2-Cdc13 complex through maintenance of Cdc2 Tyr15 phosphorylation (45, 48).
Given the complexities of ICL metabolism, we wished to determine if these adducts elicit replication checkpoints (since they are potent inhibitors of replication) or DNA damage checkpoints (since they are highly distorting lesions which are associated with DSBs) or whether both of these key checkpoints are utilized. We first conducted a large-scale sensitivity screen of mutants in the main DNA damage response pathways to gain an overview of the key factors involved. Functional studies confirmed and extended the findings of this screen, at both a biochemical and a physiological level. Our experiments demonstrate that the damage checkpoint in G2, mediated through Chk1, is the primary pathway activated by ICLs in fission yeast, whereas the replication checkpoint utilizing Cds1 is a less significant, secondary response, which may actually be detrimental to cell survival.
MATERIALS AND METHODS
S. pombe disruption strategy for PSO2/SNM1.
The PSO2/SNM1 gene was deleted by methods described in reference 6. The KanMX6 module, conferring G418 resistance, was targeted by homologous integration in the open reading frame, 72 nucleotides upstream and 15 nucleotides downstream of the open reading frame, respectively.
Drugs.
Analytical-grade mechlorethamine (nitrogen mustard or HN2) and mitomycin C (MMC) were purchased from Sigma Chemical Co. (Poole, United Kingdom). Mononitrogen mustard (HN1 or 2-dimethylaminoethylchloride hydrochloride), 99% pure, was obtained from Aldrich (Gillingham, United Kingdom).
Sensitivity assays.
Liquid yeast extract with supplements (YES) medium was inoculated with a single colony picked from a freshly streaked (YES) stock plate and grown overnight at 28°C with vigorous shaking. Cells were counted microscopically, and only cultures with ∼2 × 107 cells/ml were used. Cells were resuspended in phosphate-buffered saline at a density of 2 × 107 cells/ml, and 2-ml aliquots were treated with the desired concentration of drug (freshly dissolved in cold sterile water) for 60 min at 28°C with vigorous shaking. Cells were harvested, washed twice with 2 ml of phosphate-buffered saline, and then diluted and plated in duplicate onto YES plates at a density giving rise to 200 colonies per plate in untreated controls. Plates were incubated for 3 days at 28°C and then scored. Any experiments giving rise to more than 250 colonies per plate in untreated controls were rejected. For the serial-dilution colony spotting assay, cells were serially diluted from 107 to 103 cells/ml and 5 μl was spotted onto plates and irradiated at the indicated doses. For cisplatin, MMC, camptothecin, and HU experiments, the plates contained the stated concentration of test agent.
Checkpoint analysis.
Synchronous cultures of G2 cells were generated on lactose gradients, as previously described (4), and divided into two samples. One was treated with drug for 20 min at 30°C, and the other was treated with water. Cells were washed with water and suspended in complete medium at 30°C. Samples were taken every 20 min and fixed in methanol. Cells were analyzed by microscopy after DAPI (4′,6′-diamidino-2-phenylindole) and Calcofluor staining as previously described (4).
Phosphorylation-dependent mobility shift of Chk1 kinase and Cds1 kinase activity.
Log-phase cultures were treated with HN2 for 20 min at 30°C, washed with water, and incubated in complete medium at 30°C. Samples were taken after 1, 2, and 3 h. To analyze Chk1 phosphorylation, protein extracts were obtained as previously described (11) and were run on 10% acrylamide gels. Triple hemagglutinin-tagged (3-HA-tagged) Chk1 protein was detected with HA monoclonal antibody (Roche). The Cds1 kinase activity experiments were performed as previously described (29).
RESULTS
S. pombe appears to repair ICLs in a manner similar to those of other eukaryotes.
Currently very little is known about the repair of ICLs in S. pombe. In order for these studies to be of relevance to other eukaryotic systems, we first screened a library of S. pombe DNA repair mutants for nitrogen mustard (HN2) sensitivity to confirm that ICLs are processed in a similar fashion in this yeast. Disruptants in all the major pathways investigated to date in S. cerevisiae and mammalian cells were available, with the exception of snm1/pso2 mutants. S. cerevisiae snm1/pso2 mutants were originally isolated on the basis of their specific sensitivity to the interstrand cross-linking agents nitrogen mustard and psoralen (23, 50, 51). Consequently we identified a putative S. pombe snm1/pso2 gene and created a disruptant, as described above (Materials and Methods). The candidate S. pombe SpSnm1/Pso2 protein has 27 and 37% identity and 42 and 55% similarity with the S. cerevisiae (ScSnm1) and human Snm1A (HsSnm1) proteins, respectively, over the entire protein sequence.
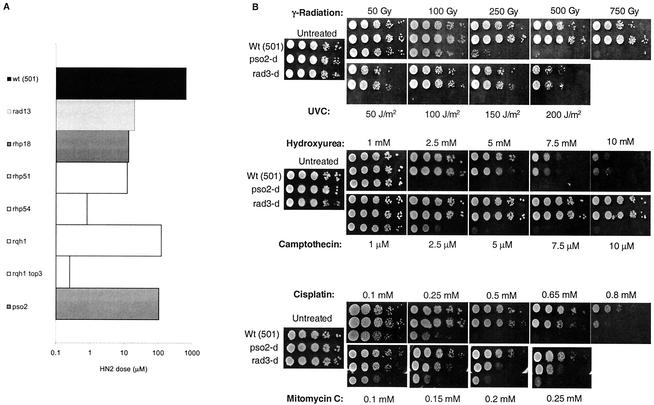
Our screen demonstrated that the repair pathways acting on ICLs in bacteria, S. cerevisiae, and mammals are largely conserved in S. pombe, and the results are summarized in Fig. 1A. Compared to the parental, repair-proficient strain (501), a rad13 mutant (S. cerevisiae Rad2 and mammalian XPG homologue [27], required for the 3′ incision during NER) was over 10-fold more sensitive. An rhp18 disruptant (S. cerevisiae and mammalian Rad18, involved in postreplication repair and damage tolerance) (60) demonstrated a similar level of sensitivity. The homologous recombination rhp51 and rhp54 mutants (S. cerevisiae and mammalian Rad51 and Rad54) (27) were also sensitive. All of these observations are consistent with those previously made for budding yeast (16, 24, 35). Combining rhp51 with a mutation in top3 (topoisomerase III) increased the HN2 sensitivity of the rhp51 mutants. Notably, top3 null mutations on their own are lethal in S. pombe and are rescued by defects in homologous recombination (43). It therefore also appears that the lethal intermediates that accumulate in rhp51-null cells during ICL processing are less efficiently resolved in the absence of Top3. The rqh1 family of RecQ helicases (related to S. cerevisiae SGS1 and the Bloom's syndrome protein) exhibit significant HN2 sensitivity. This has previously been observed for S. cerevisiae sgs1 mutants (52) and suggests that this helicase, which is important in maintaining replication fork integrity, probably by diverting unrepaired lesions into a recombinational bypass pathway (42, 55), also plays a role in the repair or tolerance of ICLs. The increased sensitivity of the rqh1 top3 mutant emphasizes the importance of top3 in the processing of ICLs and suggests that top3 may act in pathways in addition to Rqh1-dependent recombination during ICL repair. Other repair mutant strains tested included a uvde (UV damage endonuclease) disruptant and a rad2-null strain (S. cerevisiae rad27 and mammalian FEN-1 flap endonuclease) (25, 27, 64), neither of which showed any sensitivity (data not shown).
FIG. 1.
(A) Sensitivity of S. pombe DNA repair mutants to nitrogen mustard. Results shown are the doses required to kill 10% of cells and are the means of two to three independent experiments. The repair-competent wild-type (wt) control was strain 501. (B) Serial-dilution colony spotting assay for sensitivity to a variety of DNA-damaging agents in wild-type, rad3, and pso2 strains.
The S. pombe pso2/snm1 disruptant demonstrated modest (around sixfold) sensitivity to HN2 (Fig. 1A). Since the defining phenotype of S. cerevisiae pso2/snm1 mutants is sensitivity to cross-linking agents, but not other forms of DNA damage, we tested the sensitivity of the S. pombe mutant to a range of genotoxic agents, including gamma radiation, UVC, HU, camptothecin, cisplatin, and MMC (Fig. 1B). A checkpoint-defective strain known to be highly sensitive to all of these agents (the rad3 strain) was included as a positive control in these experiments. By use of a serial-dilution colony spotting assay, modest sensitivity was observed in the pso2 strain for the cross-linking drugs (cisplatin and MMC), but not the other DNA-damaging agents, indicating that the specific ICL-processing activity of Snm1/Pso2 appears conserved between these two evolutionarily distinct yeasts.
We were concerned that HN2 sensitivity of the repair mutants in Fig. 1 might not specifically be the result of ICL-processing defects but be due to the abundant monoadducts produced by this drug (which constitute at least 95% of the total lesion load). Therefore, we tested key strains for sensitivity to a monofunctional analogue of HN2, termed HN1, capable of producing only DNA monoadducts (35). None of the strains analyzed (including rhp54, rhp51, rad13, and pso2 mutants) demonstrated any detectable sensitivity to this agent at concentrations up to 10 mM—10-fold higher than the highest dose of HN2 employed (data not shown).
Nitrogen mustard sensitivity screen of S. pombe cell cycle checkpoint mutants.
A screen of a comprehensive collection of cell cycle checkpoint mutants was undertaken (Fig. 2). The checkpoint Rad proteins all demonstrated a significant increase in sensitivity compared to the isogenic parent 501. Cells disrupted for rad3, rad26, rad1, rad9, rad17, and hus1 were all sensitive to HN2, confirming that, as for other DNA-damaging agents such as UV, ionizing radiation, and methyl methanesulfonate, this family plays a role in cell cycle integrity in response to ICLs.
FIG. 2.
Sensitivity of S. pombe mutants involved in cell cycle progression and checkpoints to nitrogen mustard. Results shown are the doses required to kill 10% of cells and are the means of two to three independent experiments. The repair-competent wild-type control (strain 501) is designated wt.
As detailed in the introduction, the downstream effectors activated by the checkpoint Rad family are defined by two main pathways, the replication response elicited through Cds1 and the damage response elicited through Chk1 (46). It is striking (Fig. 2) that cds1 mutants are resistant to HN2, compared to the isogenic parent 501 (cell killing was not observed at the highest dose used, so therefore they are at least twofold more resistant), whereas chk1-null cells are over 20-fold more sensitive. A chk1 cds1 double mutant was as sensitive as the chk1 single mutant, indicating that Cds1 does not play a role in maintaining viability in the absence of a functional DNA damage checkpoint. In addition, analysis of the rad26.T12 mutant indicated that this strain behaves indistinguishably from a cds1-null mutant. The rad26.T12 allele is specifically defective in the Cds1-dependent replication checkpoint, not the DNA damage checkpoint mediated by Chk1 (17). This result strongly supports the observation that defects in the replication checkpoint pathway confer resistance to HN2. A chk1-HA strain (where the chk gene bears a 3′ HA tag) exhibited near-wild-type sensitivity, which is significant for experimental results presented below in this report.
Other components of the checkpoint response considered included Rhp9, Mrc1, and Rad18. Mrc1 is required for the full activation of Cds1 in response to HU-induced replication stress (2, 57). However, mrc1 mutant cells were sensitive to HN2, in contrast to the resistant phenotype of the cds1-null cells, indicating that Mrc1 might play a role outside the replication checkpoint during the ICL response. The rhp9 mutant cells were also sensitive to HN2. This is consistent with the sensitivity of the chk1 mutant cells, since Rhp9 appears to be involved in the damage checkpoint, but not the replication checkpoint (63). Strikingly, a rad18-74 mutant (which is the most DNA damage sensitive of the rad18 alleles identified [59]—rad18-null mutants are inviable) was the most sensitive single mutant strain that we examined in this study. Rad18 is an SMC (structural maintenance of chromosomes) protein required to maintain cell cycle arrest in response to both UV and ionizing radiation damage (28, 59). This protein is also required for the recombinational repair of ionizing-radiation-induced DSBs (59). Hence, it is likely that the extreme sensitivity of this strain results from compound defects in both the repair of ICL-induced DSBs and the Chk1-mediated DNA damage checkpoint.
We also tested key checkpoint-defective strains for sensitivity to a monofunctional analogue of HN2 (HN1). None of the strains analyzed (including rad3, chk1, cds1, and rhp9 mutants) demonstrated any detectable sensitivity to this agent at concentrations up to 10 mM (data not shown). This confirms that the checkpoint sensitivity profiles observed specifically arise from defects in the ICL response.
HN2 treatment induces a G2 checkpoint.
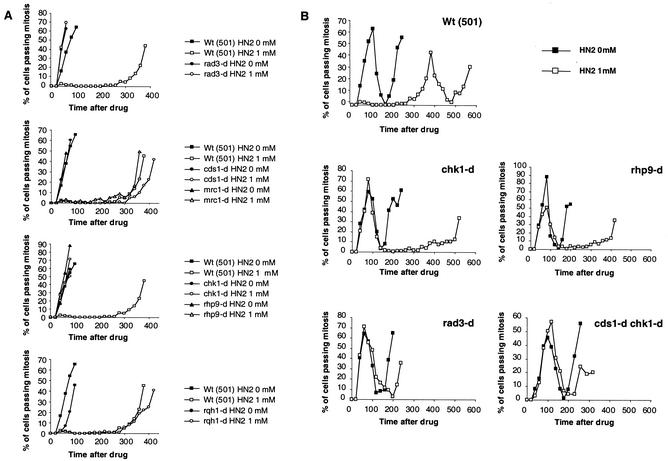
To establish if HN2 induces DNA structure-dependent checkpoint activation, as suggested by the sensitivity of checkpoint mutants, synchronous G2 cultures of cells were tested for mitotic arrest in response to treatment with HN2. In the wild-type cells (501), 1 mM HN2 treatment induced a G2 checkpoint, in a manner dependent on the rad3 checkpoint gene (Fig. 3A, top panel). The checkpoint rad pathway is separated into two subpathways: the DNA damage checkpoint, mediated by the Chk1 kinase, and the DNA replication checkpoint, mediated by the Cds1 kinase (46). The HN2-induced G2 checkpoint is independent of Cds1 and Mrc1, since cds1- and mrc1-null mutants show a normal mitotic arrest after HN2 treatment compared to the wild type (Fig. 3A, second panel from top). This observation is consistent with the absence of HN2 sensitivity in the cds1-null mutant. However, mrc1 is required for HN2 resistance. This suggests a role for Mrc1 in DNA cross-link repair, separate from its checkpoint functions, which are mediated through Cds1 (2, 57). In contrast, the HN2-induced G2 checkpoint is dependent on Chk1 and Rhp9, as mutants with null mutations in both genes fail to delay mitotic progression after HN2 treatment (Fig. 3A, third panel from top). This is in agreement with the HN2 sensitivity observed for both mutants. Finally, the G2 checkpoint induced by HN2 occurs normally in the rqh1-null mutant (Fig. 3A, bottom panel).
FIG. 3.
Checkpoint analysis in response to HN2 in different null mutants. Synchronous cultures of G2 cells were generated on lactose gradients and divided into two samples, one of which was treated with HN2 at 1 mM for 20 min. Septation index and DAPI staining were used to monitor progress through the cell cycle. (A) HN2 treatment induces mitotic arrest, and this arrest is dependent on Rad3 (top panel) and Chk1 and Rhp9 (third panel from top). In contrast the mitotic delay induced by HN2 is independent of Cds1 and Mrc1 (second panel from top) and Rqh1 (bottom panel). (B) HN2 induces a second-cell-cycle delay in the absence of a functional G2 DNA damage checkpoint. The second cell cycle is delayed in chk1- and rhp9-null mutants (middle panels) compared to the wild-type strain (top panel). The Rad3 checkpoint protein and Cds1 are required for the second-cell-cycle delay induced by HN2 (bottom panels). Wt, wild type.
We also monitored the progress of treated cells into the second cell cycle in the presence of HN2 and observed that HN2 induced a “second-cycle” delay in the absence of a functional G2 checkpoint. As shown in Fig. 3B, the second mitosis is delayed in chk1- and rhp9-null mutants compared to that in the wild-type strain (501) following HN2 treatment (Fig. 3B, top and middle panels). The second mitosis occurs at approximately 200 min in untreated cells. In wild-type cells (strain 501) treated with HN2, the first cycle is delayed until 400 min after treatment, and then the second cycle follows without a significant delay approximately 3 h later. However, in chk1- and rhp9-null mutants, where there is no first-cycle delay, the second cycle is delayed until 400 to 500 min, suggesting that cells proceed into S phase in the presence of DNA damage. This causes a block to DNA replication and subsequent activation of the DNA replication checkpoint. The replication checkpoint has previously been shown to be dependent on Cds1 and Rad3 but independent of Chk1 and Rhp9. Consistent with this, the HN2-induced second-cycle delay is dependent on Rad3 and Cds1, since it is abolished in a rad3 disruptant strain and a cds1 chk1 double disruptant (Fig. 3B, bottom panels).
We also tested the monofunctional counterpart of HN2, HN1, for checkpoint activation (Fig. 4). Consistent with the lack of sensitivity in checkpoint-deficient strains, this agent failed to elicit a G2 checkpoint or second-cycle arrest in cells treated with up to 10 mM HN1 in either a wild-type or a rad3 strain.
FIG. 4.
Checkpoint analysis in response to HN1 in different null mutants. Synchronous cultures of G2 cells were generated on lactose gradients and divided into two samples, one of which was treated with HN1 at 1 (filled circles), 2 (filled triangles), or 10 (filled squares) mM for 20 min. Septation index and DAPI staining were used to monitor progress through the cell cycle. HN1 fails to induce G2 arrest in a wild-type (Wt) strain (501). In addition, no second-cycle arrest is seen in either the wild type (501) or the rad3-d mutant.
HN2 treatment activates Chk1 and Cds1.
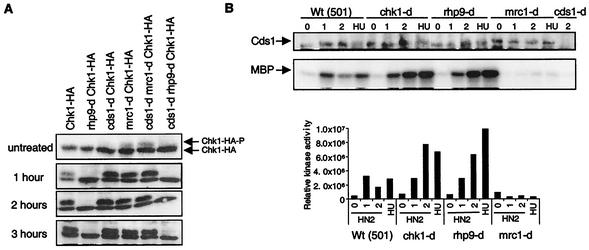
Chk1 has been shown to be phosphorylated in response to DNA damage, and this phosphorylation event generally correlates with its activation (62). To confirm that HN2 induces a Chk1-dependent G2 checkpoint, we compared Chk1 phosphorylation in asynchronous cultures, following treatment with 1 mM HN2, and in wild-type and mutant strains carrying the chk1 gene tagged with 3-HA epitopes (Fig. 5A). Chk1 is rapidly phosphorylated in response to HN2. Consistent with our analysis above and with the present literature, this Chk1 phosphorylation does not occur in the absence of Rhp9. Note, however, that the chk1 disruptants are more sensitive to HN2 than are rhp9 cells (Fig. 2), suggesting that some rhp9-independent activation of Chk1 might occur in response to ICLs. In contrast, and as expected from the literature, Cds1 and Mrc1 are not required for Chk1 phosphorylation (2, 8). This was important to determine, since sensitivities of mrc1 and cds1 mutants are not identical.
FIG. 5.
Chk1 phosphorylation and Cds1 activation in response to HN2. (A) The phosphorylation-dependent mobility shift of Chk1 kinase was assayed with different null mutants. All the strains contain a Chk1 gene tagged with 3-HA epitopes. Samples from asynchronous cultures treated with 1 mM HN2 were taken at the time indicated. Chk1 was detected with anti-HA monoclonal antibody, and arrows indicate the phosphorylation shift. HN2 induced Chk1 phosphorylation in an Rhp9-dependent but Cds1- and Mrc1-independent manner. (B) HN2 induced kinase activity in different null mutants. Samples from asynchronous cultures were taken 1 and 2 h after treatment with 1 mM HN2. Samples from asynchronous cultures treated for 1 h with 20 mM HU were used as control for the Cds1 kinase activation. The Cds1 kinase activity was detected as previously described (27). The upper panel corresponds to Cds1 protein level detection by immunoblotting, after immunoprecipitation. The lower panel corresponds to the Cds1 kinase activity assayed on the myelin basic protein substrate (MBP). The bar graph displays the Cds1 kinase activity quantified by PhosphorImager analysis, corrected for the Cds1 protein levels detected after immunoprecipitation. Wt, wild type.
The activation of the Cds1 kinase has been previously characterized, and it is known that this occurs only in S-phase cells when DNA is damaged or replication is arrested (29). We analyzed Cds1 activity following HN2 treatment to ascertain if HN2 could induce Cds1 kinase. Asynchronous cultures were treated with 1 mM HN2, and Cds1 was subsequently immunoprecipitated and assayed for kinase activity against myelin basic protein (Fig. 5B). A positive control of cells treated with 20 mM HU, a known activator of Cds1 during S phase, was also included. In the wild-type strain (501), Cds1 is transiently activated: kinase activity is increased approximately sevenfold within 1 h of HN2 exposure and subsequently declines (fourfold at 2 h following HN2 treatment). The kinase activity assayed following HN2 treatment is dependent on Cds1, as no kinase activity is detected in the absence of Cds1 (Fig. 5B). Mrc1 is essential for the activation of Cds1 in response to HU (2, 57). In agreement with this, no Cds1 induction is detected in mrc1 mutant cells (Fig. 5B). In contrast, Chk1 and Rhp9 are not required for Cds1 activation. Moreover, in chk1-d and rhp9-d mutants, 2 h following HN2 exposure, the Cds1 kinase activity remains high and does not decline at 2 h. The most probable explanation for this is the absence of HN2-induced G2 checkpoint in chk1- and rhp9-null mutants: cells progress directly into S phase with DNA damage, which leads to Cds1 and replication checkpoint activation. In the wild-type strain, the HN2-induced G2 checkpoint arrests the majority of cells in G2, preventing cells not in S phase at the time of treatment from progressing into S phase and activating Cds1. The fact that cells arrested in G2 then enter the next cycle without activating the Cds1 checkpoint clearly indicates that repair is occurring in the G2 period.
MMC induces the same checkpoint response as does HN2.
Since it is possible that the responses reported up to this point are agent specific to HN2, we repeated several key experiments with MMC as the cross-linking agent. The sensitivity profile of key repair and checkpoint mutants is the same as that observed for HN2 (Fig. 6A). The recombination rhp1 mutant, the NER mutant rad13 strain, and the RecQ helicase (rqh1) disruptant are all significantly MMC sensitive. Of the key checkpoint mutants screened, the rad3, chk1-d, and cds1-d chk1-d strains were all sensitive, whereas the cds1-d strain was somewhat more resistant than the wild type (strain 501). As for HN2, a strong G2 checkpoint was elicited in response to MMC, and this was Rad3 dependent (Fig. 6B).
FIG. 6.
(A) Serial-dilution colony spotting assay for sensitivity to MMC in representative checkpoint (upper panels) and repair (lower panels) mutants. (B) MMC treatment induces mitotic arrest, and this arrest is dependent on Rad3. Experiments were performed as described for Fig. 3. Results for two drug concentrations, 1 and 3 mM, are shown. Wt, wild type.
DISCUSSION
Before embarking upon a detailed analysis of cell cycle checkpoints involved in the S. pombe ICL response, it was important to confirm that this organism repairs ICLs by mechanisms similar to those of the eukaryotic systems studied to date (principally S. cerevisiae, and to some extent mammalian cells, in particular Chinese hamster ovary cells). The sensitivity data (Fig. 1) obtained with a panel of S. pombe DNA repair mutants indicated that this is indeed the case. Both NER (Rad13) and homologous recombination (Rhp51) are important determinants of sensitivity, as is the postreplication repair pathway governed by Rhp18. These findings are consistent with several studies of budding yeast which suggest a key role for NER in the initial incision at ICLs (24, 34) and a subsequent role for homologous recombination in providing the genetic information to complete repair through a recombination reaction, probably initiated by DSBs (16, 24, 35).
As stated above, recombinational processing of ICLs involves the formation of DSB intermediates that are mainly repaired by Rad52-controlled events in budding yeast, with a minor role for nonhomologous end joining (NHEJ). In contrast to the repair of DSBs induced by ionizing radiation, there is also a requirement for the Pso2/Snm1 protein at a postincision (NER) stage of ICL repair (31). S. cerevisiae cells carrying a mutated pso2/snm1 allele are specifically sensitive to ICL-inducing agents but show wild-type resistance to monofunctional alkylating drugs and ionizing radiation (12, 50). In this study, we have identified the S. pombe homologue of Pso2/Snm1 and created a deletion mutant. The observation that S. pombe snm1/pso2 mutants are also modestly sensitive to a variety of cross-linking agents indicates that the function of the Snm1/Pso2 protein in DNA metabolism is at least partly conserved between these two evolutionarily distinct yeasts. This is an important observation, as mouse embryonic stem cells disrupted for the paralog SNM1 demonstrate sensitivity solely to MMC (of the cross-linking agents tested), possibly due to functional redundancy with other murine homologues (15). The Pso2/Snm1 protein is a member of the metallo-β-lactamase family of enzymes that share a hydrolytic domain similar to that of the mRNA cleavage and polyadenylation specificity factor, CPSA (5). A human gene clearly related to Pso2/Snm1, Artemis, has been identified as the mutant gene in radiation-sensitive severe combined immune deficiency (40), where the phenotype results from defects in V(D)J recombination and in the NHEJ of DSBs (30). This has been shown to arise from the failure of hairpin opening during V(D)J processing and defective processing of DNA ends during NHEJ. Artemis is a single-strand-specific 5′-to-3′ exonuclease which, when complexed with DNA-dependent protein kinase, is phosphorylated by the latter, acquiring 5′- and 3′-overhang and hairpin endonuclease activity (30). The use of model organisms such as S. cerevisiae and S. pombe, for which detailed genetic studies are possible, will be a powerful tool in further elucidating the role of the PSO2/SNM1 family in DNA repair.
Cross-links activate the DNA damage checkpoint mediated by Chk1 and Rhp9.
Cell cycle arrest is a part of the cellular response to DNA damage and is thought to ensure correct chromosome segregation in mitosis and prevent chromosome instability. Many ICL-inducing agents are used in the treatment of cancers, but little is known about the effects of ICLs on cell cycle delays. Here, we analyzed cell cycle arrest in response to ICLs. Our experiments lead us to conclude that the G2 DNA damage checkpoint mediated by Chk1 (3, 61) and Rhp9 (63) is the most biologically significant checkpoint pathway activated in response to HN2. The Cds1 (29, 41)- and Mrc1 (2, 57)-dependent DNA replication checkpoint pathway is also activated transiently, but its loss has little biological consequence in our assays. Several lines of evidence lead us to this conclusion. First, the chk1- and rhp9-null mutants are HN2 and MMC sensitive, which is not the case for the cds1-null mutant. Second, Chk1 is clearly rapidly phosphorylated in response to HN2 in a Rhp9-dependent and Cds1-Mrc1-independent manner. Third, the HN2-induced G2 checkpoint does not occur in the absence of chk1 or rhp9. Finally, Cds1 kinase activity is only transiently activated in response to HN2, suggesting that HN2 does not lead to prolonged cell accumulation within S phase. Consistent with our genetic and biochemical observations, ICLs induced by cisplatin or psoralen have been shown elsewhere to induce cell cycle arrest in S. cerevisiae G2/M but not to induce an S-phase delay (except in response to high levels of ICLs) (21, 22, 38). Together, these results suggest that replication can tolerate or bypass a certain number of ICLs.
Cells disrupted for rad3, rad26, rad1, rad9, rad17, and hus1 are all sensitive to HN2. Since we observe no cross-link-induced cell cycle arrest (HN2 or MMC) in the absence of rad3, it is likely that the arrest induced by cross-links is dependent on all checkpoint rad (10) gene products. The G2 arrest presumably represents a checkpoint that allows time for ICL repair by a combination of NER and homologous recombination. In the absence of this checkpoint, cells will initiate replication with unrepaired DNA damage, leading to DNA replication checkpoint activation mediated by Cds1 (8). This can be seen in the extended second-cycle delay observed in the absence of Chk1 and Rhp9 (Fig. 3B) and in the potentiation of the induction of Cds1 kinase activity observed in chk1- and rhp9-null mutants (Fig. 5). As the chk1 and rhp9 mutants show HN2 sensitivity compared to the wild-type and cds1-d strains, it could be speculated that ICLs are more toxic or less efficiently repaired during or after replication. In fact, as cds1-d mutants are actually more resistant to HN2 and MMC, it is possible that delaying S phase and attempting to repair ICLs in S phase are positively detrimental to cell survival.
Model for S. pombe checkpoint response to ICLs and relevance to cancer.
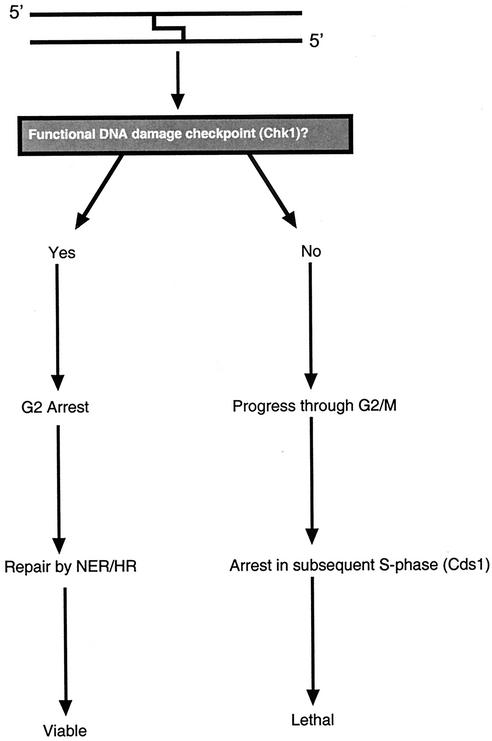
Taken together, the findings of this study allow us to suggest a basic model of the S. pombe response to ICLs (Fig. 7). In a growing population of cells, which are primarily in the G2 phase of the cell cycle, the presence of ICLs triggers the DNA damage checkpoint, requiring the full family of checkpoint Rad factors, leading to Rad3-dependent phosphorylation and activation of Chk1. The resulting arrest permits time for ICL repair through the combined action of NER and homologous recombination (24, 35). Following the completion of repair, cells reenter the normal cell cycle and pass through mitosis. In the absence of an operative damage checkpoint, cells progress through to the following S phase, where the ICLs are detected, presumably as a replication fork block. This triggers the Cds1-dependent replication checkpoint. The marked resistance of cds1-null cells to cross-linking agents suggests that this event is deleterious and might represent a terminal arrest event in at least a subset of cells.
FIG. 7.
Model for the ICL checkpoint response in S. pombe. HR, homologous recombination.
The role of Chk1 as the major mediator of the ICL checkpoint arrest has relevance to cancer biology, as mammalian homologues of this gene are known. Mouse embryonic stem cells disrupted for CHK1 are defective in the regulation of the G2/M DNA damage checkpoint (53, 56), and mammalian Chk1 appears to exert its major regulatory effect through the phosphorylation and inactivation of cyclin-dependent kinase following DNA damage (20, 32). Extrapolation of the results presented in our present study suggests that mammalian cells overexpressing Chk1 might be resistant to ICL-inducing agents, whereas cancers suffering dysregulation of this checkpoint might be more sensitive to these drugs. In addition, the fact that the absence of Cds1 confers resistance to HN2 in S. pombe could have some implications in chemotherapy, since some tumors harbor mutations of CHK2, the CDS1 homologue in mammalian cells. Indeed, heterozygous germ line mutations in chk2 have been identified in some patients with Li-Fraumeni syndrome, a familial cancer phenotype (7). A chk2 mutation (1100delc) leading to a truncating variant that abolishes the kinase activity results in a 2-fold increase of breast cancer risk in women and a 10-fold increase of risk in men (37). In this context, the use of ICL-inducing agents in treatment of tumors defective for CHK2 may have a reduced therapeutic index. While the yeast checkpoints represent a simple view of events in higher organisms, our study points to a potential pivotal role for Chk1 and Cds1-Chk2 in the response to cross-linking anticancer agents.
Acknowledgments
We thank J. Murray for kindly sharing unpublished strains.
This work was supported, in part, by EC grant FIGH-CT-1999-0010 (to A.M.C.) and was also partly supported by Cancer Research UK Programme grant SP2000/0402 (to J.A.H.). L.J.B. was supported by a Cancer Research UK studentship, and P.J.M. was supported by a Royal Society University Research fellowship.
REFERENCES
- 1.Akkari, Y. M., R. L. Bateman, C. A. Reifsteck, S. B. Olson, and M. Grompe. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 20:8283-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 3.al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.al-Khodairy, F., E. Fotou, K. S. Sheldrick, D. J. Griffiths, A. R. Lehmann, and A. M. Carr. 1994. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell 5:147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind, L., D. R. Walker, and E. V. Koonin. 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27:1223-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 7.Bell, D. W., J. M. Varley, T. E. Szydlo, D. H. Kang, D. C. Wahrer, K. E. Shannon, M. Lubratovich, S. J. Verselis, K. J. Isselbacher, J. F. Fraumeni, J. M. Birch, F. P. Li, J. E. Garber, and D. A. Haber. 1999. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528-2531. [DOI] [PubMed] [Google Scholar]
- 8.Brondello, J. M., M. N. Boddy, B. Furnari, and P. Russell. 1999. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell. Biol. 19:4262-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caspari, T., and A. M. Carr. 2002. Checkpoints: how to flag up double-strand breaks. Curr. Biol. 12:R105-R107. [DOI] [PubMed] [Google Scholar]
- 10.Caspari, T., and A. M. Carr. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81:173-181. [DOI] [PubMed] [Google Scholar]
- 11.Caspari, T., M. Dahlen, G. Kanter-Smoler, H. D. Lindsay, K. Hofmann, K. Papadimitriou, P. Sunnerhagen, and A. M. Carr. 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20:1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassier, C., and E. Moustacchi. 1981. Mutagenesis induced by mono- and bi-functional alkylating agents in yeast mutants sensitive to photo-addition of furocoumarins (pso). Mutat. Res. 84:37-47. [DOI] [PubMed] [Google Scholar]
- 13.Cole, R. S., and R. R. Sinden. 1975. Repair of cross-linked DNA in Escherichia coli. Basic Life Sci. 5B:487-495. [DOI] [PubMed] [Google Scholar]
- 14.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dronkert, M. L., J. de Wit, M. Boeve, M. L. Vasconcelos, H. van Steeg, T. L. Tan, J. H. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 20:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dronkert, M. L., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486:217-247. [DOI] [PubMed] [Google Scholar]
- 17.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 18.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 19.Enoch, T., A. M. Carr, and P. Nurse. 1992. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6:2035-2046. [DOI] [PubMed] [Google Scholar]
- 20.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O'Connor, and H. Piwnica-Worms. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275:5600-5605. [DOI] [PubMed] [Google Scholar]
- 21.Grossmann, K. F., J. C. Brown, and R. E. Moses. 1999. Cisplatin DNA cross-links do not inhibit S-phase and cause only a G2/M arrest in Saccharomyces cerevisiae. Mutat. Res. 434:29-39. [DOI] [PubMed] [Google Scholar]
- 22.Grossmann, K. F., A. M. Ward, and R. E. Moses. 2000. Saccharomyces cerevisiae lacking Snm1, Rev3 or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment. Mutat. Res. 461:1-13. [DOI] [PubMed] [Google Scholar]
- 23.Henriques, J. A., and E. Moustacchi. 1980. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 95:273-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jachymczyk, W. J., R. C. von Borstel, M. R. Mowat, and P. J. Hastings. 1981. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol. Gen. Genet. 182:196-205. [DOI] [PubMed] [Google Scholar]
- 25.Kaur, B., A. M. Avery, and P. W. Doetsch. 1998. Expression, purification, and characterization of ultraviolet DNA endonuclease from Schizosaccharomyces pombe. Biochemistry 37:11599-11604. [DOI] [PubMed] [Google Scholar]
- 26.Kostrub, C. F., K. Knudsen, S. Subramani, and T. Enoch. 1998. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 17:2055-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann, A. R. 1996. Molecular biology of DNA repair in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 363:147-161. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann, A. R., M. Walicka, D. J. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15:7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray, F. Osman, N. Walworth, and A. M. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12:382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 31.Magana-Schwencke, N., J. A. Henriques, R. Chanet, and E. Moustacchi. 1982. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA 79:1722-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 33.Martinho, R. G., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra, A. M. Carr, and N. J. Bentley. 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17:7239-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHugh, P. J., R. D. Gill, R. Waters, and J. A. Hartley. 1999. Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res. 27:3259-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McHugh, P. J., W. R. Sones, and J. A. Hartley. 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHugh, P. J., V. J. Spanswick, and J. A. Hartley. 2001. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2:483-490. [DOI] [PubMed] [Google Scholar]
- 37.Meijers-Heijboer, H., A. van den Ouweland, J. Klijn, M. Wasielewski, A. de Snoo, R. Oldenburg, A. Hollestelle, M. Houben, E. Crepin, M. van Veghel-Plandsoen, F. Elstrodt, C. van Duijn, C. Bartels, C. Meijers, M. Schutte, L. McGuffog, D. Thompson, D. Easton, N. Sodha, S. Seal, R. Barfoot, J. Mangion, J. Chang-Claude, D. Eccles, R. Eeles, D. G. Evans, R. Houlston, V. Murday, S. Narod, T. Peretz, J. Peto, C. Phelan, H. X. Zhang, C. Szabo, P. Devilee, D. Goldgar, P. A. Futreal, K. L. Nathanson, B. Weber, N. Rahman, and M. R. Stratton. 2002. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet. 31:55-59. [DOI] [PubMed] [Google Scholar]
- 38.Meniel, V., N. Magana-Schwencke, D. Averbeck, and R. Waters. 1997. Preferential incision of interstrand crosslinks induced by 8-methoxypsoralen plus UVA in yeast during the cell cycle. Mutat. Res. 384:23-32. [DOI] [PubMed] [Google Scholar]
- 39.Miller, R. D., L. Prakash, and S. Prakash. 1982. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol. Cell. Biol. 2:939-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 41.Murakami, H., and H. Okayama. 1995. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature 374:817-819. [DOI] [PubMed] [Google Scholar]
- 42.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oakley, T. J., A. Goodwin, R. K. Chakraverty, and I. D. Hickson. 2002. Inactivation of homologous recombination suppresses defects in topoisomerase III-deficient mutants. DNA Repair (Amsterdam) 1:463-482. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor, P. M., and K. W. Kohn. 1990. Comparative pharmacokinetics of DNA lesion formation and removal following treatment of L1210 cells with nitrogen mustards. Cancer Commun. 2:387-394. [DOI] [PubMed] [Google Scholar]
- 45.Rhind, N., B. Furnari, and P. Russell. 1997. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 11:504-511. [DOI] [PubMed] [Google Scholar]
- 46.Rhind, N., and P. Russell. 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113:3889-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhind, N., and P. Russell. 1998. Mitotic DNA damage and replication checkpoints in yeast. Curr. Opin. Cell Biol. 10:749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhind, N., and P. Russell. 1998. Tyrosine phosphorylation of Cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol. 18:3782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowley, R., S. Subramani, and P. G. Young. 1992. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 11:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruhland, A., E. Haase, W. Siede, and M. Brendel. 1981. Isolation of yeast mutants sensitive to the bifunctional alkylating agent nitrogen mustard. Mol. Gen. Genet. 181:346-351. [DOI] [PubMed] [Google Scholar]
- 51.Ruhland, A., M. Kircher, F. Wilborn, and M. Brendel. 1981. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat. Res. 91:457-462. [DOI] [PubMed] [Google Scholar]
- 52.Saffi, J., V. R. Pereira, and J. A. Henriques. 2000. Importance of the Sgs1 helicase activity in DNA repair of Saccharomyces cerevisiae. Curr. Genet. 37:75-78. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez, Y., C. Wong, R. S. Thoma, R. Richman, Z. Wu, H. Piwnica-Worms, and S. J. Elledge. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497-1501. [DOI] [PubMed] [Google Scholar]
- 54.Sladek, F. M., M. M. Munn, W. D. Rupp, and P. Howard-Flanders. 1989. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 264:6755-6765. [PubMed] [Google Scholar]
- 55.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takai, H., K. Tominaga, N. Motoyama, Y. A. Minamishima, H. Nagahama, T. Tsukiyama, K. Ikeda, K. Nakayama, and M. Nakanishi. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 14:1439-1447. [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3:966-972. [DOI] [PubMed] [Google Scholar]
- 58.Van Houten, B., H. Gamper, S. R. Holbrook, J. E. Hearst, and A. Sancar. 1986. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. USA 83:8077-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10:2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verkade, H. M., T. Teli, L. V. Laursen, J. M. Murray, and M. J. O'Connell. 2001. A homologue of the Rad18 postreplication repair gene is required for DNA damage responses throughout the fission yeast cell cycle. Mol. Genet. Genomics 265:993-1003. [DOI] [PubMed] [Google Scholar]
- 61.Walworth, N., S. Davey, and D. Beach. 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363:368-371. [DOI] [PubMed] [Google Scholar]
- 62.Walworth, N. C., and R. Bernards. 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271:353-356. [DOI] [PubMed] [Google Scholar]
- 63.Willson, J., S. Wilson, N. Warr, and F. Z. Watts. 1997. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 25:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yonemasu, R., S. J. McCready, J. M. Murray, F. Osman, M. Takao, K. Yamamoto, A. R. Lehmann, and A. Yasui. 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]