Abstract
Insulin stimulates glucose uptake by recruiting glucose transporter 4 (GLUT4) from an intracellular pool to the cell surface through a mechanism that is dependent on phosphatidylinositol (PI) 3-kinase (PI3-K) and cortical actin remodeling. Here we test the hypothesis that insulin-dependent actin filament remodeling determines the location of insulin signaling molecules. It has been shown previously that insulin treatment of L6 myotubes leads to a rapid rearrangement of actin filaments into submembrane structures where the p85 regulatory subunit of PI3-K and organelles containing GLUT4, VAMP2, and the insulin-regulated aminopeptidase (IRAP) colocalize. We now report that insulin receptor substrate-1 and the p110α catalytic subunit of PI3-K (but not p110β) also colocalize with the actin structures. Akt-1 was also found in the remodeled actin structures, unlike another PI3-K effector, atypical protein kinase C λ. Transiently transfected green fluorescent protein (GFP)-tagged pleckstrin homology (PH) domains of general receptor for phosphoinositides-1 (GRP1) or Akt (ligands of phosphatidylinositol-3,4,5-trisphosphate [PI-3,4,5-P3]) migrated to the periphery of the live cells; in fixed cells, they were detected in the insulin-induced actin structures. These results suggest that PI-3,4,5-P3 is generated on membranes located within the actin mesh. Actin remodeling and GLUT4 externalization were blocked in cells highly expressing GFP-PH-GRP1, suggesting that PI-3,4,5-P3 is required for both phenomena. We propose that PI-3,4,5-P3 leads to actin remodeling, which in turn segregates p85α and p110α, thus localizing PI-3,4,5-P3 production on membranes trapped by the actin mesh. Insulin-stimulated actin remodeling may spatially coordinate the localized generation of PI-3,4,5-P3 and recruitment of Akt, ultimately leading to GLUT4 insertion at the plasma membrane.
Glucose uptake into skeletal muscle cells results from the redistribution of glucose transporter 4 (GLUT4) from an intracellular membrane compartment to the plasma membrane (9, 15, 32, 56). This phenomenon is abnormally diminished in type 2 diabetes, resulting in insulin resistance (46, 71). Translocation of GLUT4 to the muscle cell surface requires activation of phosphatidylinositol 3-kinase (PI3-K) (47, 56), which occurs upon binding to tyrosine-phosphorylated insulin receptor substrates (IRSs). In several cell types, such binding takes place through the Src homology 2 domains of the 85-kDa regulatory subunit (p85) of PI3-K, thereby activating the catalytic 110-kDa subunit (p110) (70). Activated p110 causes primarily the phosphorylation of phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2) to generate phosphatidylinositol-3,4,5-trisphosphate (PI-3,4,5-P3). The latter lipid recruits the serine-threonine kinase Akt/PKB via its pleckstrin homology (PH) domain for its subsequent activation via phosphorylation by phosphoinositide-dependent kinase (PDK), which also activates atypical protein kinase C ζ (aPKCζ) and -λ (11, 30, 35). PI3-K activity, membrane recruitment, and activation of Akt and aPKC have been implicated in the stimulation of GLUT4 translocation and glucose transport in skeletal muscle (3, 61) and L6 muscle cells (2, 68).
Given that PI3-K is activated not only by insulin but also by a number of extracellular factors and cues, there must be mechanisms that safeguard the specificity of each response. The possibility that distinct spatial localization of the PI3-K-dependent signal transduction pathway may be required to ensure the fidelity and specificity of insulin signaling has been previously discussed (22, 28). However, there is a controversy about the compartmentalization of the enzyme. From subcellular fractionation of control and insulin-stimulated adipocytes followed by immunodetection of p85 or in vitro PI 3-kinase activity, it was concluded that the enzyme transfers from the cytosol to microsomes of intracellular origin (6, 18, 22, 27, 37, 45, 60). However, this conclusion was challenged by the fluorescent detection at the cell periphery of insulin-stimulated adipocytes of ligands of PI-3,4,5-P3, such as green fluorescence protein (GFP)-linked PH domains of Akt or of general receptor for phosphoinositides-1 (GRP1) (16, 29, 39, 63). The discrepancy between the biochemical and immunocytological studies has not been solved but may arise from plasma membrane contamination of subcellular fractions and/or from inability of light microscopy to differentiate plasma membrane from intracellular vesicles closely apposed to the membrane. In addition, cytoskeletal elements abutting either the plasma membrane or intracellular compartments may furnish binding sites for the enzyme (6), and some of these elements, such as microfilaments, may break down upon biochemical disruption of cells. Indeed, growing evidence supports the role of the cytoskeleton in compartmentalizing signals and organelles. Moreover, insulin causes a rapid and dynamic remodeling of actin filaments into a cortical mesh, which is required for GLUT4 translocation and glucose uptake in differentiated L6 muscle cells (28, 54, 58). Within that submembrane mesh, we detected proteins characteristic of GLUT4 vesicles, including GLUT4, VAMP2, and insulin-regulated aminopeptidase (IRAP) (28, 44).
The purpose of the present study was to examine the localization of the PI3-K catalytic subunit and its major lipid product, PI-3,4,5-P3, in L6 muscle cells by applying confocal microscopy and deconvolution analysis to maximize the three-dimensional resolution. The time course of actin remodeling and the distribution of PI-3,4,5-P3 were also analyzed in living cells. We provide evidence that PI-3,4,5-P3 is formed on the plasma membrane and in structures supported by an actin scaffold. p110α (but not p110β) and Akt-1 (but not PKCλ) gathered into the actin mesh, suggesting that the actin structures spatially segregate PI3-K isoforms and signals downstream of PI-3,4,5-P3. From these results, we hypothesize that the remodeled actin structures provide a scaffold for the transmission of signals from the insulin receptor to the insulin-responsive GLUT4 compartment. Preventing the formation of the scaffold using excess ligand of PI-3,4,5-P3 (GFP-PH-GRP1) aborted GLUT4 translocation.
MATERIALS AND METHODS
Reagents.
Human insulin was purchased from Eli Lilly (Toronto, Ontario, Canada). Tetramethylrhodamine-conjugated concanavalin A; Alexa fluor 633-, rhodamine-, and Oregon Green-conjugated phalloidin; and Alexa fluor 488 (A488)-conjugated goat anti-mouse, A488-conjugated goat anti-rabbit, and A488-conjugated rabbit anti-goat antibodies were purchased from Molecular Probes (Eugene, Oreg.). Polyclonal antibody to phosphorylated Akt was purchased from Cell Signaling (Beverly, Mass.). Monoclonal antibody against myc (9E10) and polyclonal antibodies against aPKCλ (sc-1091), PKCζ (sc-7262), and Akt-1 (sc-7126); and isoform-specific antibodies against p110α (sc-1331 and sc-7174, which were raised against regions mapping the C terminus and N terminus, respectively) and p110β (sc-602 and sc-7175, which were raised against the regions mapping the C terminus and N terminus, respectively) subunits of PI3-K, as well as corresponding blocking peptides (sc-1091 P, sc-7262 P, sc-1331 P, and sc-602 P), were all obtained from Santa Cruz (Santa Cruz, Calif.). Polyclonal antibody against IRS-1 was kindly provided by Morris White (Joslin Diabetes Centre, Boston, Mass.). Plasmids containing enhanced GFP (eGFP)-tagged PH domain of GRP1 (GFP-PH-GRP1) and the eGFP-tagged PH domain of GRP1 with a single point mutation [GFP-PH(K273A)-GRP1] were kind gifts from Michael Czech (University of Massachusetts, Worcester). The plasmid encoding eGFP-tagged PH domain of Akt (GFP-PH-Akt) was a kind gift from Tobias Meyer (Stanford University, Stanford, Calif.), and the plasmid encoding eGFP was purchased from Clontech (Palo Alto, Calif.). Effectene transfection kits and plasmid DNA purification columns were purchased from Qiagen (Mississauga, Ontario, Canada).
Cell culture and transfections.
L6 muscle cells expressing c-myc epitope-tagged GLUT4 (GLUT4myc) were maintained in myoblast monolayer culture or differentiated into multinucleated myotubes as described earlier (62, 67). Myotubes were ready for experimentation 6 to 8 days postseeding. Transfection of L6 GLUT4myc myoblasts was performed in 6-well plates as described previously (44).
Fluorescence microscopy. (i) Fixed cells.
L6 GLUT4myc myotubes or myoblasts, grown on 25-mm-diameter glass coverslips, were deprived of serum for 4 h and treated with 100 nM insulin for up to 30 min at 37°C. Labeling of actin filaments with fluorophore-coupled phalloidin and immunostaining of specific antigens in fixed and permeabilized myotubes were carried out as described previously (28). It was crucial to fix the cells immediately at 4°C after insulin stimulation and to permeabilize in 0.1% (vol/vol) Triton X-100 for exactly 3 min in order to preserve actin morphology. The primary antibody dilution factors were as follows: myc, 1:150; IRS-1, 1:100; p110α, 1:250; p110β, 1:250; phospho-Akt (Ser 473), 1:250; Akt-1, 1:100; PKCζ, 1:100; and PKCλ, 1:100 in 0.1% (wt/vol) bovine serum albumin-phosphate-buffered saline. To label actin filaments simultaneously to any of the above-listed molecules, fixed and permeabilized cells were incubated for 1 h at room temperature with the fluorophore-conjugated phalloidin indicated in the figure legends (0.01 U/coverslip) during the incubation with the secondary antibody. To eliminate artifactual colocalization in these costaining experiments, two fluorophores with maximal spectral separation were chosen. As well, the staining pattern of proteins observed in the costaining experiments was compared to that following single staining using the same antibody. Additional controls for immunofluorescence experiments are described in Results.
Plasma membrane staining with tetramethylrhodamine-conjugated concanavalin A (50 μg/ml) and GLUT4myc translocation determined by cell-surface immunostaining of the myc epitope on intact myoblasts were carried out as described previously (44, 54). For conventional and confocal fluorescence microscopy, cells were examined with either an inverted Leica DM-IRE2 microscope or a Zeiss LSM 510 laser scanning confocal microscope, respectively. Acquisition parameters were adjusted to exclude saturation of the pixels. For quantification, such parameters were kept constant between the various conditions.
(iii) Live-cell studies.
For time-lapse fluorescent confocal microscopy, L6 myoblasts were seeded onto 35-mm glass bottom microwell dishes (MatTek Co., Ashland, Mass.) and transfected with GFP-PH-GRP1, GFP-PH(K273A)-GRP1, or eGFP cDNA. Sixteen hours posttransfection, the cells were serum deprived (4 h), washed and incubated with KRPH buffer (120 mM NaCl, 2.5 mM KCl, 20 mM HEPES, 1.2 mM MgSO4, 1 mM NaH2PO4, and 2 mM CaCl2) supplemented with 5 mM d-glucose, placed into a temperature-controlled chamber set at 37°C, and visualized using the Zeiss LSM 510 laser scanning confocal microscope. Images were acquired every 10 to 30 s throughout the period from 2 min prior to insulin addition to 11 min after insulin addition.
(iii) Deconvolution microscopy and image analysis.
Images obtained from several focal planes were deconvolved using the nearest-neighbor deconvolution module from Openlab software by Improvision. Volocity software by Improvision was utilized for visualization of the volume in three dimensions. To show the localization of GFP with respect to F-actin and the plasma membrane, the opacity function in Volocity was used to expose the green signal. The “sequence xyz view” function in Volocity allows us to slice the volume into three cutting planes, an xy slice, an xz slice and a yz slice. It was utilized to show the localization of GFP with respect to the plasma membrane and F-actin (see Fig. 5). To show areas of colocalization, Adobe Photoshop was used to select for yellow regions. Selected regions were copied and pasted into a new RGB channel.
FIG. 5.
PI-3,4,5-P3 is generated at the plasma membrane as well as within cellular structures formed by actin remodeling. L6 myoblasts were transiently transfected with 0.5 μg of plasmid encoding GFP-PH-GRP1 and stimulated with insulin following 4 h of serum starvation. The cells were fixed and stained for (A) F-actin using rhodamine-phalloidin or (B) the plasma membrane using tetramethylrhodamine-conjugated ConA (50 μg/ml). Optical sections of 0.2 μm from the top of the cell to the bottom were acquired and deconvolved. A cross-section of the structure was obtained (a) and analyzed for colocalization with F-actin (Ab) or the plasma membrane (Bb) by selecting for yellow regions.
(iv) Quantitation and statistical analysis.
Representative fluorescence microscopy images were quantitated with the use of NIH Image software (National Institutes of Health, Bethesda, Md.) and ImageJ for GLUT4myc translocation and GFP fluorescence, respectively. Raw data for GLUT4myc translocation were converted to reflect stimulation by insulin above basal levels relative to surface GLUT4myc in untransfected cells. Metafluor software (Universal Imaging Corp.) was used to analyze the dynamic changes in fluorescence intensity derived from the two GFP-PH-GRP1 fusion proteins in live-cell experiments (see Fig. 3B). The region of interest in the perinuclear cytosol region and the region of peripheral ruffling were determined and monitored along the entire sequence of time-lapse images. Fluorescence intensity values were then plotted over time.
FIG.3.
GFP-PH-GRP1 protein translocates from the cytosol to the periphery of the cell and concentrates in newly formed structures revealed by time-lapse live-cell microscopy. (A) L6 myoblasts were seeded on 35-mm-diameter glass bottom microwell dishes and transiently transfected with 0.5 μg of GFP-PH-GRP1 (a to c) or GFP-PH(K273A)-GRP1 (d to f), as described in Materials and Methods. The cells were then continuously visualized at 37°C by confocal fluorescent microscopy, and representative time images taken immediately prior to the addition of insulin and 2 and 10 min thereafter are presented. The complete time-lapse movie is provided at http://www.sickkids.ca/research/custom/profiles/klip.asp. Bar, 10 μm. (B) Fluorescent signal intensities in two different regions (as shown in panel A [□ and ▵, perinuclear cytosol; ○ and ⋄, peripheral structures]) within the transfected cell were obtained and quantified. To account for possible bleaching, all measurements were normalized to the signal in the nucleus within each cell. The fluorescent intensities in the perinuclear cytosol and the periphery from two representative cells expressing the wild-type and mutant PH domains were then plotted over time. The y scale represents fluorescent intensity relative to that in the nucleus. (The dotted and solid lines represent quantifications from the left and right cells in each panel, respectively.) (C) The fluorescent intensities from 10 GFP-PH-GRP1- and 19 GFP-PH(K273A)-GRP1-expressing cells at 0, 2, and 10 min following insulin stimulation were obtained. The intensities at 2 and 10 min were then expressed relative to fluorescent intensity at time zero. *, P < 0.001 compared to time zero.
All statistical analyses were carried out with the analysis of variance and post hoc analysis (Prism software).
RESULTS
Localization of p110 isoforms within the insulin-induced cortical actin mesh.
We first visualized the organization of actin labeled with fluorophore-phalloidin vis-a-vis the distribution of IRS-1 or PI3-K in L6 myotubes. In the basal state, actin formed long filamentous structures (stress fibers) aligned along the longitudinal axis of the myotubes (Fig. 1A a and b). These are best visualized in the ventral focal plane (typically 1 to 2 μm above the coverslip; Fig. 1Aa and b) but also more faintly in the dorsal focal plane (4 to 6 μm above the coverslip; Fig. 1Aa and c). In insulin-stimulated (100 nM, 10 min) myotubes, stress fibers remained visible in the ventral focal plane (Fig. 1Aa and d). However, F-actin reorganized into mesh-like structures that could be observed in the dorsal plane (Fig. 1Ae), creating dorsal extensions typically rising 2 to 3 μm above the dorsal plasma membrane of basal cells (Fig. 1Aa and f). Immunoreacted IRS-1, detected with Alexa 488-conjugated secondary antibody, appeared diffuse and punctate throughout the myoplasm in the basal state (Fig. 1Ba). Upon insulin stimulation, a significant fraction of intracellular IRS-1 was found along the newly formed dorsal actin mesh (identified by yellow regions in the dorsal extension plane in Fig. 1Bd).
FIG. 1.
IRS-1 and p110α, but not p110β, concentrate in insulin-induced actin structures. Serum-deprived (4 h) L6 myotubes were stimulated with or without 100 nM insulin for 10 min at 37°C, followed by fixation and permeabilization. (A) Images were acquired from three focal planes that are depicted in a schematic diagram (a). The ventral (V) focal plane is defined as 1 to 3 μm above the coverslip; the dorsal (D) focal plane is 4 to 6 μm above the coverslip. In response to insulin, the surface of the cell remodels into structures that protrude 2 to 3 μm above the dorsal focal plane of the cell. This is defined as the dorsal extension (DE) focal plane that is absent in basal cells. Gray scale images of the ventral surface to the dorsal surface of basal (b and c) L6 myotubes and the ventral surface to the dorsal extensions of insulin-treated (d to f) L6 myotubes stained with Oregon Green-conjugated phalloidin are shown. (B) IRS-1 (a and d) or p110 (b, c, e, and f) proteins were stained with specific polyclonal antibodies, followed by Alexa 488-conjugated secondary antibody, as described in Materials and Methods. Composite images of F-actin labeled with Alexa fluor 633-conjugated phalloidin and IRS-1 or p110 protein staining are presented (yellow indicates regions of colocalization). Immunofluorescence images for p110α (g) and p110β (h) were also acquired from cells that were stained with antibodies preincubated with a 10-fold molar excess of competitive blocking peptides. The images are representative of three experiments. Bar, 10 μm.
That insulin stimulation of L6 myotubes causes partial colocalization of the 85-kDa regulatory subunit of PI3-K with the insulin-induced actin mesh has been previously reported (28). Both isoforms of the catalytic p110 subunit (α and β) have been implicated in insulin action in various cell types (1, 26, 48). Recent studies have begun to assign specific roles to each isoform (20). To determine which p110 collects in the same region as p85α, we examined the intracellular localization of p110α and p110β in L6 myotubes using diverse isoform-specific antibodies. Under basal conditions, p110α (using an antibody directed to the C terminus) exhibited a diffuse cytosolic staining pattern, which excluded the nuclei (Fig. 1Bb). Upon stimulation with insulin, p110α markedly redistributed to regions where F-actin remodeled (Fig. 1Be). Staining with another p110α-specific antibody raised against the N terminus of p110α revealed a similar staining pattern (not shown). p110β (using an antibody directed to the C terminus) appeared diffusely distributed throughout the cytoplasm in nonstimulated cells and, in contrast to p110α, localized to the nuclei (Fig. 1Bc), consistent with recent reports (33). Further in contrast to p110α, p110β escaped relocalization into the insulin-induced actin mesh (Fig. 1Bf). A similar staining pattern was observed using another anti-p110β antibody raised against the N terminus of the protein (not shown). To further confirm the specificity of the isoform-specific antibodies, the two anti-C-terminal antibodies against each isoform were preincubated with a 10-fold molar excess of each of the two corresponding synthetic antigenic peptides. As shown in Fig. 1Bg and h, preabsorption of each antibody to its specific peptide resulted in a significant decrease in the fluorescent signal, whereas preincubation with the reciprocal peptide had no effect (not shown). Collectively, these data suggest that insulin-induced actin remodeling specifically recruited the p110α isoform but not the p110β isoform, of the catalytic subunit of PI 3-kinase.
Localization of the PI3-K effectors Akt and aPKC vis-a-vis the actin mesh.
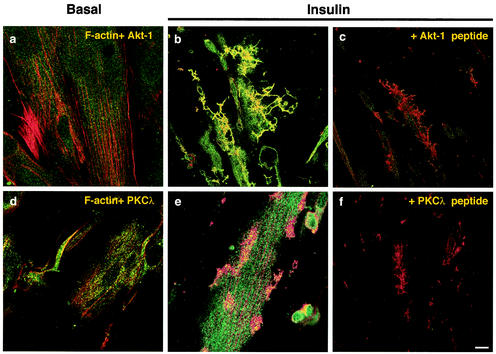
PI3-K activation leads to the stimulation of the serine/threonine kinases Akt and aPKCζ/λ. Both enzymes have been implicated in insulin-stimulated GLUT4 recruitment to the cell surface of L6 muscle cells (2, 68). Thus, it was important to assess the cellular localization of these targets of PI3-K vis-a-vis the actin mesh that had segregated distinct PI3-K isoforms. L6 myotubes were left untreated or stimulated with insulin, and subsequently, F-actin and the serine/threonine kinases were labeled with rhodamine-phalloidin and isoform-specific antibody coupled to Alexa 488-conjugated secondary antibody, respectively. Under unstimulated conditions, the Akt-1 signal was largely cytosolic and diffuse (Fig. 2a). Insulin stimulation caused a clear enhancement in the Akt-1 signal colocalizing with insulin-induced actin structures seen at the dorsal extension plane (yellow regions in Fig. 2b). Using an antibody against phospho-Akt produced a similar result (not shown). The antibody against PKCλ behaved in marked contrast to anti-Akt-1. In the basal state, PKCλ was diffusely distributed throughout the myoplasm and did not colocalize with actin (Fig. 2d). Upon insulin stimulation, the fluorescent signal of PKCλ remained cytosolic and did not concentrate significantly with the newly formed actin structures (Fig. 2e). Preincubation of the antibodies with a 10-fold molar excess of corresponding blocking peptides resulted in a complete reduction in the fluorescent signal of Akt-1 (Fig. 2c) and PKCλ (Fig. 2f). Like PKCλ, immunolocalized PKCζ also escaped colocalization with the remodeled actin structures following insulin stimulation (not shown). Therefore, while a portion of one PI3-K effector, Akt, redistributed to the actin structures, the other effector, aPKC, was largely excluded from this localization.
FIG. 2.
The PI3-K effector, Akt-1, but not aPKC, colocalizes with insulin-induced actin structures. Serum-deprived (4 h) L6 myotubes were untreated (a and d) or stimulated with 100 nM insulin for 10 min at 37°C (b, c, e, and f), followed by fixation and permeabilization. F-actin was labeled with rhodamine-conjugated phalloidin. Akt-1 (a to c) or PKCλ (d to f) proteins were stained with specific polyclonal antibodies, followed by Alexa 488-conjugated secondary antibodies, as described in Materials and Methods. Competitive inhibition of Akt-1 (c) and PKCλ (f) using a 10-fold molar excess of blocking peptides is also shown. Yellow regions indicate regions of remodeled actin filaments with colocalized Akt-1 or PKCλ protein. Shown are images of the dorsal plane of basal cells and dorsal extension planes of insulin-stimulated cells, as defined in the legend for panel a of Fig. 1A. The images are representative of four experiments. Bar, 10 μm.
PI-3,4,5-P3 production within the actin mesh.
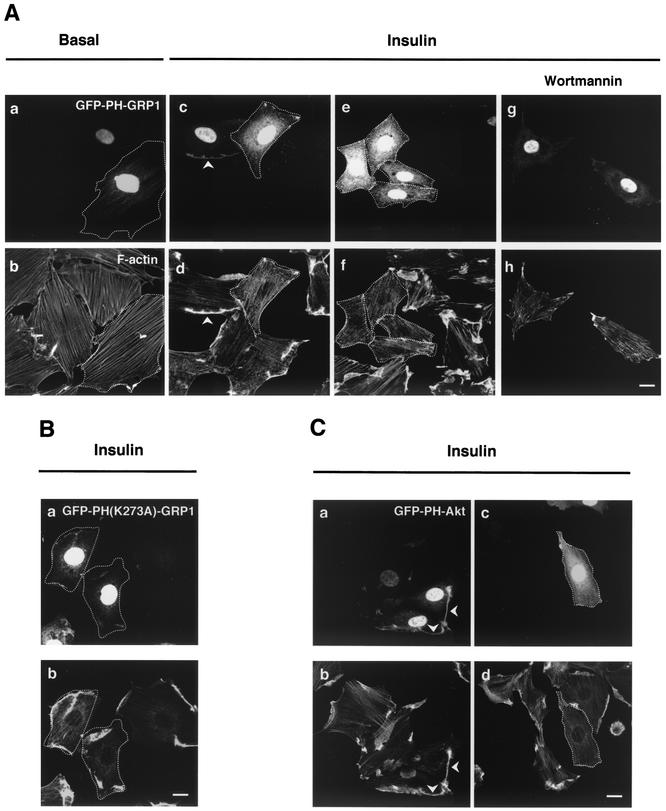
The colocalization of IRS-1, p85α and p110α with the insulin-induced actin mesh suggested the possibility that PI3-K was active in this region. Substrates for such an activity could conceivably be provided by the plasma membrane above the actin structures and/or membrane vesicles entrapped by the actin mesh, which contain GLUT4, IRAP, and VAMP-2 (28, 44). To visualize whether PI-3,4,5-P3 is generated in these regions, we used a fluorescent indicator consisting of eGFP fused to the PH domain of the ADP-ribosylating factor exchange factor GRP1. PH-GRP1 has a higher affinity (Kd, ∼25 nM) for PI-3,4,5-P3 than for other phosphoinositides in vitro (29), making it an ideal marker to detect production of PI-3,4,5-P3 (43). A similar construct harboring a single point mutation (K273A) in the PH domain of GRP1 that results in loss of PI-3,4,5-P3 binding capacity (29) was used for control experiments. GFP-PH-GRP1 cDNA was transiently transfected into L6 myoblasts. Myoblasts are far more amenable to transfection than myotubes, and transcript expression can be detected within 16 h. Actin filaments also reorganize in response to insulin in myoblasts (44), albeit the structures are not as prominent as in the much larger myotubes. To gain information on the dynamic localization of PI-3,4,5-P3, we analyzed the distribution of GFP-PH-GRP1 in live cells by time-lapse confocal microscopy. GFP-PH-GRP1 was concentrated in the nucleus, and a faint signal spread throughout the cytosol (Fig. 3Aa). Stimulation with insulin caused a rapid (∼2 min) translocation of GFP-PH-GRP1 from the perinuclear cytosol to peripheral thread-like structures (Fig. 3Ab). Ten minutes after insulin stimulation, the GFP-PH-GRP1 signal appeared to reorganize and preferentially localize in distinct peripheral structures (Fig. 3Ac). To verify that the cellular dynamics of the GFP-PH-GRP1 fusion protein in response to insulin depended upon its binding to PI-3,4,5-P3, a similar analysis was performed using the mutant GFP-PH(K273A)-GRP1 (Fig. 3Ad to f). As previously described for GFP-PH-GRP1 (39), this mutant fusion protein also displayed nuclear localization, suggesting that this feature is independent of PI-3,4,5-P3 binding. Yet, the changes observed both in the perinuclear cytosol and in the cell periphery were minor compared to those observed with the wild-type PH domain. The insulin-stimulated cellular relocalization of the GFP-PH-GRP1 in comparison to that of the mutated protein can be more readily appreciated in the time-lapse movies (http://www.sickkids.ca/research/custom/profiles/klip.asp). The dynamics of the fluorescence intensity over time of GFP-PH-GRP1 or GFP-PH(K273A)-GRP1 in the perinuclear cytosol and in the peripheral region of two representative cells each is shown in Fig. 3B. The fluorescence intensities at the perinuclear cytosol and the peripheral regions at 2 and 10 min following insulin stimulation were determined from 10 and 19 cells expressing GFP-PH-GRP1 and GFP-PH(K273A)-GRP1, respectively. In cells expressing GFP-PH-GRP1, the fluorescence intensity at the perinuclear cytosol was reduced by 70% within 2 min of insulin stimulation. Such a reduction was absent in cells expressing the mutant construct (Fig. 3B and C). This reduction in the perinuclear cytosolic region at 2 min was accompanied by a 2.8-fold increase in fluorescent intensity in the peripheral region of the GFP-PH-GRP1-expressing cells compared to only a 1.25-fold increase in GFP-PH(K273)A-GRP1-expressing cells. Ten minutes after insulin stimulation, the fluorescent signal in the perinuclear region of GFP-PH-GRP1-expressing cells seemed to regain some of its intensity. In contrast, a progressive increase in GFP-PH-GRP1 fluorescent signal was observed at the cell periphery, reaching an eightfold intensity compared to that at time zero. A much smaller effect (1.6-fold) was observed in cells expressing the mutant construct. Using this tool, it appears that insulin causes PI-3,4,5-P3 production at the cell periphery and induces unique localized sites of further PI-3,4,5-P3 accumulation.
In order to assess the possibility that these localized sites of PI-3,4,5-P3 accumulation are related to the insulin-induced actin structures described in Fig. 1, the above experiments with live cells were complemented by analysis of the distribution of GFP-PH-GFP1 in fixed cells vis-a-vis the localization of actin filaments. Consistent with the above results and other studies using fixed cells (39), GFP-PH-GRP1 was largely located in the nucleus (Fig. 4 a, c, and e) and faintly throughout the cytosol (Fig. 4a). Upon insulin stimulation, actin remodeled into structures at the dorsal surface of the myoblasts (Fig. 4f). Compared to myotubes, these structures created smaller dorsal extensions, making the difference between the dorsal and dorsal extension planes defined in Fig. 1A less pronounced. Such structures were absent from either the dorsal or ventral surfaces of unstimulated cells (Fig. 4b and d). Interestingly, insulin stimulation led to the preferential appearance of GFP-PH-GRP1 in regions of the myoblasts where actin remodeling occurred (Fig. 4e to f). This finding suggested the possibility that PI-3,4,5-P3 was generated within the actin structures, consistent with the insulin-stimulated localization of p110α within the actin mesh (Fig. 1Be).
FIG. 4.
Preferential generation of PI-3,4,5-P3 in association with actin structures detected by low-level expression of GFP-PH-GRP. L6 myoblasts were transiently transfected with 0.5 μg of GFP-PH-GRP1 cDNA for 16 h. Cells were serum deprived (4 h) and then stimulated with 100 nM insulin for 10 min at 37°C. After this period, the cells were fixed, permeabilized, and stained for F-actin (rhodamine-phalloidin). For the basal condition, images from both the ventral focal plane (near the bottom of the cell) (a and b) and the dorsal plane (near the top of the cell) (c and d) were obtained using a confocal microscope. For insulin-stimulated cells, images from the dorsal focal planes were obtained (e and f). Closed arrowheads indicate regions of remodeled actin filaments and localization of GFP-PH-GRP1 protein in insulin-stimulated cells. The images are representative of at least four experiments. Bar, 10 μm.
Additional spatial analysis of PI-3,4,5-P3 at the plasma membrane.
Some of the confocal fluorescent signal of GFP-PH-GRP1 colocalizing with the actin structure could potentially arise from optical bleeding of an intense signal on the cytosolic leaflet of the plasma membrane. To better ascertain the localization of PI-3,4,5-P3 production, we applied deconvolution analysis to the confocal fluorescent images obtained from several focal planes. Optical sections of 0.2 μm were taken from the bottom to the top of a typical myoblast expressing GFP-PH-GRP1 and analyzed with Openlab deconvolution software. A cross-sectional view of the deconvolved insulin-induced actin structure (Fig. 5Aa) reveals a significant colocalization of the GFP-PH-GRP1 signal with F-actin (Fig. 5Ab). The above result illustrates that PI-3,4,5-P3 produced in response to insulin is found in intracellular regions colocalizing with the remodeled cortical actin mesh. These results do not rule out, however, the possibility that PI-3,4,5-P3 could also be generated on the cell membrane. To examine this possibility, insulin-stimulated myoblasts expressing GFP-PH-GRP1 were reacted with tetramethylrhodamine-conjugated concanavalin A (Fig. 5B). Concanavalin A specifically binds to carbohydrate moieties and has been widely used to label the plasma membrane of intact cells. A cross-section revealed tetramethylrhodamine-concanavalin A signal (red) in the periphery of the structure, which was absent from the cytosol (Fig. 5Ba). Conversely, the GFP-PH-GRP1 signal (green) was largely intracellular, although a discernible signal colocalized with the plasma membrane (yellow signal and Fig. 5Bb). Together, these results suggest that in addition to PI-3,4,5-P3 produced in endomembranes within the actin structure (Fig. 5), PI-3,4,5-P3 is generated along the plasma membrane in response to insulin.
To obtain spatial detail on the localization of GFP-PH-GRP1 vis-a-vis the insulin-induced actin structures, the deconvolved images of this construct and rhodamine-conjugated phalloidin were volume rendered to obtain a three-dimensional reconstruction of the cell. The F-actin signal (red) was more prominent towards the top of the cell compared to the GFP-PH-GRP1 signal (green), which was more intense at the middle and base of the remodeled actin structure (Fig. 6a). Electronic reduction of the red signal revealed a green signal spanning the entire structure that was more intense near the middle and below the peak of the actin (red signal) structure (Fig. 6b). To assess the possible contribution of a cytosolic signal in the region, we analyzed optical sections of 0.2 μm from cells with comparable expression of GFP-PH(K273A)-GRP1. As before, a more intense F-actin signal was observed towards the top of the cell (Fig. 6c). Electronic removal of this red signal revealed only a low-reaching GFP-PH(K273A)-GRP1 (green) signal near the bottom of the actin structure (Fig. 6d). This relatively small abundance of GFP-PH(K273A)-GRP1 within the actin structure contrasts with GFP-PH-GRP1, which spanned throughout the actin structure. A similar result was obtained from cells expressing eGFP only (not shown). This result suggests that the contribution of cytosolic GFP-PH-GRP1 to the signal within the actin structures is minor. Therefore, most of the GFP-PH-GRP1 signal within the actin structure appears to originate from specifically concentrated PI-3,4,5-P3, potentially attached to vesicles gathered by the structures.
FIG. 6.
GFP-PH-GRP1 protein localizes within and below the peak of the actin structure assessed by deconvolution analysis. L6 myoblasts were transiently transfected with 0.5 μg of plasmid encoding GFP-PH-GRP1 (a and b) or GFP-PH(K273A)-GRP1 (c and d), stimulated with insulin, and stained for F-actin as described in the legend for Fig. 4. Images from several focal planes (optical sections of 0.2 μm) were obtained, deconvolved, and volume rendered to obtain a three-dimensional reconstruction of the cell. The red signal (F-actin) is reduced to reveal the green signal (GFP) below (b and d). Bar, 10 μm.
PI-3,4,5-P3 is required for insulin-dependent formation of the cortical actin mesh.
The localization of PI3-K and its lipid product near or within the actin mesh raises the question of whether PI3-K is required for the formation of such a mesh. Hence, the susceptibility of the actin structure to wortmannin was first assessed. Pretreatment with wortmannin (100 nM, 20 min) followed by insulin stimulation prevented actin remodeling and, as expected, also precluded generation of PI-3,4,5-P3 (Fig. 7Ag, and h versus c and d). This result suggests that PI3-K activity is required for actin mesh formation but does not identify whether PI-3,4,5-P3 is the required product. An approach to answer this question was afforded by cells with high levels of expression of GFP-PH-GRP1. Low-level-expressing cells were defined as those with a relatively low nuclear fluorescent signal, which allowed clear discernment of dense structures within the nucleus. High-level-expressing cells were defined as those in which the nuclear signal of GFP-PH-GRP1 was saturated under the same acquisition settings (see the cells outlined in Fig. 7Aa, c, and e). In highly expressing cells, insulin-stimulated phosphorylation of extracellular signal-regulated kinase (ERK) was maintained (not shown), suggesting that PI-3,4,5-P3-independent pathways were not affected. Strikingly, in cells expressing high levels of GFP-PH-GRP1, stimulation with insulin did not lead to any actin remodeling (Fig. 7Ab, d, and f). In contrast, cells expressing high levels of the mutant GFP-PH(K273A)-GRP1 (see the outlined cell in Fig. 7Ba) responded to the hormone by mounting a subcortical actin mesh akin to that in untransfected cells (Fig. 7Bb). In a similar set of studies, we used the PH domain of Akt fused to eGFP (GFP-PH-Akt) as an alternative method to detect PI-3,4,5-P3 within the cells. While GFP-PH-Akt can detect PI-3,4,5-P3 levels, it has also been shown to bind to PI-3,4-P2 (13, 14). Nonetheless, as was the case of GFP-PH-GRP1, the GFP-PH-Akt protein colocalized with remodeled actin structures following insulin stimulation in L6 cells expressing low levels of the construct (Fig. 7Ca and b). In cells highly expressing GFP-PH-Akt (see the outlined cell in Fig. 7C c), the insulin-induced actin remodeling was abolished (Fig. 7C d). A likely interpretation of these results is that highly expressed GFP-PH-GRP1 or GFP-PH-Akt sequesters PI-3,4,5-P3, thereby blocking signals leading to actin remodeling. The high expression of GFP-PH-GRP1 also abolished the localized foci of phosphorylated endogenous Akt, although it did not visibly reduce the overall cytosolic signal of phosphorylated Akt (not shown). This result confirms the significance of localized changes in PI-3,4,5-P3 availability for downstream signaling to GLUT4 (see below).
FIG. 7.
High-level expression of GFP-PH-GRP1 or wortmannin pretreatment inhibits insulin-stimulated actin remodeling. L6 myoblasts transiently transfected with 0.5 μg of GFP-PH-GRP1 (A) or GFP-PH(K273A)-GRP1 (B) or GFP-PH-Akt (C) cDNA were stimulated with (Ac to h; Ba and b; and Ca to d) or without (Aa and b) 100 nM insulin for 10 min at 37°C. L6 myoblasts transfected with GFP-PH-GRP1 were also exposed to 100 nM wortmannin for 20 min prior to and during insulin stimulation (A, g and h). After this period, the cells were fixed, permeabilized, and stained for F-actin (rhodamine-phalloidin). Arrowheads indicate regions of remodeled actin filaments and localization of GFP-tagged protein in insulin-stimulated cells. Cells highly expressing the constructs are outlined. The images are representative of three experiments. Bar, 10 μm.
PI-3,4,5-P3 production is required for GLUT4myc translocation to the cell surface.
It is well established that PI3-K activity is essential for the insulin-dependent externalization of GLUT4 in muscle and adipose cells (4, 8, 56). However, the role of different PI3-K lipid products in this process is unclear. High-level expression of GFP-PH-GRP1 provided a strategy to approach this question. The L6 myoblasts used in this study express high levels of myc-tagged GLUT4 (21, 67). The presence of the myc epitope in the first exofacial loop of GLUT4 allows for the direct detection of GLUT4 insertion in the plasma membrane in unpermeabilized myoblasts by cell surface immunostaining. The GLUT4myc staining in unstimulated cells was unaffected by the different levels of GFP-PH-GRP1 expression (Fig. 8a to d). Strikingly, the insulin-dependent appearance of GLUT4myc staining on the cell surface was reduced in cells transfected with high levels of GFP-PH-GRP1 (Fig. 8g and h) while the density of GLUT4myc staining in lower-expressing cells did not appear to be reduced (Fig. 8e and f). The surface density of GLUT4myc staining was quantified in order to compare the insulin-dependent GLUT4myc translocation in untransfected cells and adjacent cells transfected with GFP-PH-GRP1 in the same optical field. In cells expressing high levels of GFP-PH-GRP1, cell surface GLUT4myc staining in response to insulin was reduced by 80% ± 4% compared to only a 26% ± 4% reduction in lower-expressing cells. The decrease in insulin-stimulated GLUT4myc translocation in high-GFP-PH-GRP1-expressing cells was not associated with a decrease in total GLUT4myc expression (1.07 ± 0.06 times that of untransfected cells). High-level expression of the GFP-PH(K273A)-GRP1 had no effect on insulin-stimulated cell surface GLUT4myc (results not shown). These results suggest that PI-3,4,5-P3 is a key contributor to GLUT4 translocation.
FIG. 8.
High levels of GFP-PH-GRP1 inhibit insulin-stimulated GLUT4 translocation. L6 myoblasts transiently transfected with 0.5 μg of GFP-PH-GRP1 cDNA were serum deprived (4 h) and then stimulated with (e to h) or without (a to d) 100 nM insulin for 30 min at 37°C. After this period, the cells were left intact and processed for detection of GLUT4myc at the cell surface with anti-myc antibody followed by Cy3-conjugated goat anti-mouse antibody. For each condition, top and bottom panels show the same optical field of cells. Closed arrowheads indicate the positions of cells expressing low levels of GFP-PH-GRP1, and the open arrowheads indicate the position of a cell highly expressing the protein. The images are representative of three experiments. Bar, 10 μm.
DISCUSSION
Insulin initiates a signaling cascade that leads to various metabolic and mitogenic effects in specific target tissues. Actin filaments are required to preserve the integrity of the mitogenic signals, specifically the SOS-Grb2 interaction (55). Most, if not all, of the metabolic actions of insulin require PI3-K activation. This profuse array of responses suggests the need to segregate PI3-K spatially and temporally in order to guide individual responses. Recent studies analyzing the subcellular localization of PI3-K have relied largely on subcellular fractionation to separate the cytosol from the plasma membrane and light microsomes (6, 22). Those studies consistently found the p85 subunit or IRS-dependent PI3-K activity in the cytosol of resting cells and in the cytosol and low-density microsomes of insulin-stimulated cells. In contrast, after platelet-derived growth factor stimulation, a portion of the p85 purified with the plasma membrane (37, 45). These studies naturally concluded that insulin selectively causes migration of PI3-K from within the cytosol to the low-density microsome (LDM). LDM is a heterogeneous fraction containing not only membranes but also diverse filaments (6, 66); other membrane fractions can also bring down attached and copurifying filaments. Hence, subcellular fractionation has limitations in resolving the nature of the association of IRS-1/p85 with the isolated membranes. Immunofluorescence studies would add resolution to this question, but detection of PI3-K subunits by this means has so far been lacking. Instead, previous studies have analyzed the localization of GFP-PH domains with affinity for PI3Ps (39, 42, 63) and found that GFP-PH-Akt localized in the cell perimeter upon insulin stimulation (16). Acknowledging that the GFP-PH-Akt detects PI3-K products, it was tempting to relate the localization of the products to that of the enzyme. Those studies prompted suggestions that the isolated LDM might have been contaminated with the plasma membrane and/or with vesicles lying closely apposed to the cell surface. Thus, the question of the subcellular localization of PI3-K requires further examination.
Insulin-induced F-actin remodeling segregates PI 3-kinase isoforms.
In the present study, we used confocal fluorescence microscopy to locate two isoforms of the catalytic subunits of class I PI3-K and to examine the effect of insulin on such distribution. Specific antibodies with selectivity towards IRS-1, p110α, or p110β were found to be reliable for immunofluorescence detection and were thus used as probes for their corresponding enzymes. The localization of these proteins was determined in GLUT4myc myotubes arising from multiple fusion events among myoblasts. The myotubes, measuring several hundred micrometers in length and about 10 μm in height, present a large cytoplasmic domain devoid of sarcomeric cross-striations. Instead, filaments of actin span the length of the myotubes. We have previously reported that insulin stimulation leads to the reorganization of cortical actin filaments into a mesh running from the cell surface (which consequently develops ruffles) through about half of the interior of the cell (54).
Strikingly, our results revealed that like IRS-1, the catalytic p110α subunits of PI3-K, but not p110β, colocalized with the actin mesh formed in response to insulin (Fig. 1). We have previously observed that p85α is distributed as punctae in the cytosol and redistributes into the actin structures following insulin stimulation (28). Therefore, it is conceivable that the binding partner of p110α in this locale is p85α. The differential behaviors of the p110 isoforms underscores that the association with the actin cytoskeleton does not arise from optical artifacts potentially associated with actin remodeling, although direct interaction of p110α and p85α with F-actin has not been demonstrated. Regardless of the mechanism for isoform-specific localization, F-actin-dependent differential localization of PI3-K isoforms may have implications for downstream insulin signal transduction, as Akt, but not aPKCs, also colocalized with insulin-induced actin structures. In conclusion, IRS-1 and isoforms of the regulatory and catalytic subunits of PI3-K relocate to regions on or within the remodeled actin mesh in response to insulin.
Cellular loci of PI-3,4,5-P3 production defined by actin remodeling.
The above results prompt the question of whether the p110 subunit isoforms of PI3-K gathered into the insulin-induced actin mesh are catalytically active and where the products are located. Previous studies had used the PH domain of GRP1 fused to eGFP to track the generation and localization of PI-3,4,5-P3 in response to insulin stimulation (16, 39), but they lacked sufficient resolution to separate signals emanating from the membrane from those potentially within the adjacent cytosol. Laser confocal microscopy was not always employed. Here we analyzed the localization of the PI-3,4,5-P3 ligand GFP-PH-GRP1 by two approaches: confocal fluorescence imaging of live cells in real time (Fig. 3) and confocal immunofluorescence coupled to deconvolution analysis of fixed cells in which actin filaments were concomitantly labeled with rhodamine-phalloidin (Fig. 5). Deconvolution analysis was used to further reduce the out-of-focus fluorescence obtained by confocal microscopy, thus enhancing the Z resolution. Both approaches revealed the segregation of GFP-PH-GRP1 signals along thread-like elements highly reminiscent of actin structures. The results further show that GFP-PH-GRP1 can be detected along the plasma membrane and in regions several micrometers deep contained within the actin mesh (Fig. 5 and 6). To further strengthen the spatial localization of PI-3,4,5-P3 vis-a-vis the actin cytoskeleton, we used the PH domain of Akt fused to eGFP in addition to GFP-PH-GRP1. Our results show that GFP-PH-Akt also colocalizes with insulin-stimulated actin structures (Fig. 7C). During the process of revision of this paper, Tengholm and Meyer (51) reported the need for a threshold of 3′-phosphoinositides (detected via CFP-PH-Akt1) at the plasma membrane of 3T3-L1 adipocytes for GLUT4 translocation. The use of evanescent wave microscopy and the reliance on the PH domain of Akt precluded complete spatial analysis of PI-3,4,5-P3 production. Our results complement the conclusions of that study by offering whole-cell spatial detail and assign a specific role to the PI-3,4,5-P3 lipid product in actin remodeling and GLUT4 translocation (see below).
The GFP-PH-GRP1 signal within the actin mesh is likely to emanate from PI-3,4,5-P3 present in membranes located in this region. Indeed, we have previously detected three membrane-bound proteins colocalizing with the actin mesh (VAMP2, GLUT4, and IRAP), while other membrane proteins were excluded (insulin receptor and VAMP3/cellubrevin) (44, 54), suggesting that only a subset of intracellular membranes gather in the insulin-induced actin structures. We hypothesize that the actin mesh formed in response to insulin constitutes a pull mechanism that segregates a subpopulation of vesicles containing GLUT4 and VAMP2. PI3-K would then produce local foci of PI-3,4,5-P3. In addition, the enzyme also generates PI-3,4,5-P3 on the cytosolic leaflet of the plasma membrane (Fig. 5B). To our knowledge, this is the first visualization of such dual localization of PI-3,4,5-P3.
Actin mesh formation requires PI3-K input.
The above results suggest that insulin causes actin remodeling leading to segregation of PI3-K isoforms and generation of PI3-K products within the actin mesh. Interestingly, formation of the actin mesh was itself regulated by PI3-K. This is borne out from the complete inhibition of actin remodeling in the presence of wortmannin (57, 59) and the abolition of this phenomenon in myoblasts expressing either the dominant-negative mutant Δp85 (28, 57, 66) or, as shown here, high levels of GFP-PH-GRP1 or GFP-PH-Akt (Fig. 7). One potential interpretation of our data is that a positive-feedback loop operates whereby PI3-K is required for actin remodeling, which in turn gathers the activated enzyme towards membranes, where production of PI-3,4,5-P3 is essentially localized. A feed-forward loop involving PI3-K was recently found to operate in neutrophils, where PI-3,4,5-P3 at the leading edge stimulated its own additional accumulation (65, 69). Further experimentation is required to prove the presence of a similar mechanism in muscle cells.
The PI3-K dependence of insulin-induced actin remodeling in muscle cells is in stark contrast to the recently reported increase in actin dynamics in 3T3-L1 adipocytes (24, 25). In those cells, wortmannin failed to prevent the increase in a cortical actin band which appears to be controlled by N-Wasp via the small G protein TC10 (5, 23, 24). Conversely, actin mesh formation in muscle cells is independent of TC10 (L. JeBailey and A. Klip, unpublished observations) and instead requires the small G protein Rac (28). These results underscore the cell type-specific differences in both the architecture and dynamics of actin and the insulin-dependent signals governing it.
Functional consequences of segregating PI-3,4,5-P3 within the actin mesh on downstream signals.
PI-3,4,5-P3 binds to the PH domains of Akt and of its upstream activator, PDK. Therefore, it was conceivable that membranes containing PI-3,4,5-P3 and lodged within the actin mesh might attract Akt. Indeed, upon insulin stimulation, Akt was found along with the newly formed actin mesh (Fig. 2). This Akt was phosphorylated (results not shown), suggesting the possibility that PI-3,4,5-P3 trapped in the mesh binds the enzyme, which in turn becomes phosphorylated by PDK potentially bound to the lipid vesicles. Alternatively, Akt may become phosphorylated in other loci, including the plasma membrane, and subsequently migrate to the lipid vesicles trapped in the actin mesh, potentially to phosphorylate target proteins. The present study does not differentiate between these possibilities but suggests that Akt does not bind directly to actin, insofar as Akt did not colocalize with actin stress fibers in the basal state. While colocalizing with the actin mesh in the insulin-stimulated state, it is likely that Akt binds to lipid vesicles within this structure rather than to actin itself. This is because the enzyme was soluble in ice-cold Triton X-100, whereas cytoskeletal elements sediment under these conditions (results not shown). The hypothesis that remodeled actin couples PI3-K to Akt is also borne out from studies using cytochalasin D, which inhibits actin dynamics. This drug reduced the response of Akt to insulin (42;K. Yaworsky and A. Klip, unpublished observations) in the face of normal activation of PI3-K (42, 58). These results suggest that actin dynamics is instrumental in the coupling of activated PI3-K to Akt in actin-generated loci. The converse scenario, that Akt lies upstream of actin reorganization, is deemed unlikely because transfection of a dominant-negative mutant of Akt into L6 myoblasts did not affect insulin-stimulated actin remodeling (68).
Collectively, the results presented buttress the concept that the insulin-induced actin mesh serves as a scaffold that segregates foci of selective isoforms of PI3-K, membrane vesicles containing selective membrane proteins, PI-3,4,5-P3, and specific downstream signaling molecules such as activated Akt. The importance of localized Akt phosphorylation is further suggested by the observation that high expression of the GFP-PH-GRP1 (but not the mutant) prevented actin remodeling (Fig. 7) as well as localized Akt phosphorylation (not shown) and GLUT4 translocation (Fig. 8).
Consistent with the scenario that localized activation of PI3-K within the actin mesh is critical for downstream signaling to GLUT4, several in vivo models of insulin resistance show reduced Akt activity and diminished GLUT4 translocation in the absence of alterations in overall insulin-stimulated PI3-K activity. Where examined, PI3-K was found to be mistargeted, presumably precluding the activation of Akt at the proper location (17, 38, 52). Moreover, in at least two cellular models of insulin resistance, insulin-dependent actin remodeling is altered (53, 54). Alterations in the cytoskeleton have also been proposed to occur in insulin-resistant 3T3-L1 adipocytes, leading to IRS-1 release from cytoskeletal complexes associated with membranes (7). Hence, actin remodeling may play a central role controlling the fidelity of insulin signaling, which may go awry in insulin resistance.
Our work also illustrates a selective topological segregation of Akt vis-a-vis another downstream effector of PI3-K, the aPKC. The latter enzyme did not localize to the actin mesh (Fig. 2), although it is activated by insulin in a wortmannin-sensitive manner in these cells (49). One possibility to explain the differential behavior of Akt and aPKC is that only the former binds to PI-3,4,5-P3. Indeed, aPKC lacks a bona fide PH domain recognizing this lipid, although it has the potential to bind phosphoinositides (50). It is conceivable, then, that an actin-independent PI3-K signaling complex (perhaps involving p110β) may trigger the activation of aPKC. Further resolution by electron microscopy is required to establish more precisely the spatial correspondence of the putative actin scaffold to insulin-signaling molecules and membrane vesicles.
PI-3,4,5-P3 is required for GLUT4 translocation.
The above results suggest a model whereby PI-3,4,5-P3 is generated on vesicles contained in the actin mesh that also harbor GLUT4 and VAMP2, attracting and activating Akt and promoting GLUT4 translocation. Several observations support this model. First, high-level expression of GFP-PH-GRP1 abolishes both actin remodeling and GLUT4 translocation (Fig. 8), suggesting a requirement of PI-3,4,5-P3 in GLUT4 translocation. Second, depolymerization of F-actin with cytochalasin D precludes the subsequent copurification of PI3-K with GLUT4-containing membranes (66). Third, dominant-negative Akt and Akt-inhibiting peptides prevent GLUT4 translocation in myoblasts and 3T3-L1 adipocytes (19, 68). Fourth, Akt promotes the interendosomal transit of GLUT4 within myoblasts, conceivably leading to the sorting of specialized GLUT4 vesicles (12). Intracellular GLUT4 compartments have been identified as a target for Akt in rat adipocytes (10, 31). Whether the actin mesh-segregated pools of PI-3,4,5-P3 and Akt are those required for GLUT4 translocation remains to be determined.
The above results were not geared to test whether there is a cause-effect relationship between actin remodeling and GLUT4 translocation. Those two events have instead been linked in several studies using a diverse array of strategies interfering with actin remodeling (23, 24, 28, 40, 41, 54, 58, 66-68). However, these strategies interfere with all types of actin dynamics, falling short from linking a specific structure such as the mesh observed in myoblasts and myotubes or the cortical band observed in 3T3-L1 adipocytes. In any case, our study suggests the involvement of PI-3,4,5-P3 in both actin remodeling and GLUT4 translocation, possibly with preference over PI-3,4-P2, which binds GFP-PH-GRP1 only weakly (29). These results are in agreement with the observed reduction in GLUT4 translocation in cells expressing PTEN (phosphatase and tensin homolog) (34, 36) or the Src homology 2-containing inositol 5′-phosphatase 2 (64). Our results were also not geared to test the participation of aPKC in either actin remodeling or GLUT4 translocation. It is conceivable that this enzyme participates in other selective steps in GLUT4 translocation, such as insertion into the plasma membrane. Indeed, a recent study suggests that aPKC can phosphorylate cellular VAMP2, a protein important for GLUT4 vesicle incorporation into the plasma membrane (2a).
In conclusion, our findings suggest the following role for PI3-K and its lipid product in insulin-dependent GLUT4 translocation in L6 muscle cells. Generation of PI-3,4,5-P3 is required to trigger cortical actin remodeling, which may then concentrate the p85α/p110α PI3-K isoform to increase the local content of PI-3,4,5-P3. This in turn creates localized sites for attachment of Akt in endomembranes gathered in the remodeled actin mesh. In this manner, PI-3,4,5-P3 could potentially be involved in a positive-feedback loop that further increases localized production of PI-3,4,5-P3 and Akt activation.
Acknowledgments
This study was supported by a grant from the Canadian Institutes of Health Research (MT7307) to A. Klip. N. Patel was supported by a Banting & Best Diabetes Centre-Novo Nordisk Studentship Award. A. Rudich is the recipient of the Albert Renold Career Development Award from the European Foundation for the Study of Diabetes. Z. Khayat was supported by a Canadian Institutes of Health Research Doctoral Student Award.
We are grateful to Morris White, Michael Czech, and Tobias Meyer for generously supplying IRS-1 antibody, GFP-PH-GRP1, GFP-PH-(K273A)-GRP1, and GFP-PH-Akt cDNAs used in this study.
REFERENCES
- 1.Asano, T., A. Kanda, H. Katagiri, M. Nawano, T. Ogihara, K. Inukai, M. Anai, Y. Fukushima, Y. Yazaki, M. Kikuchi, R. Hooshmand-Rad, C. H. Heldin, Y. Oka, and M. Funaki. 2000. p110beta is up-regulated during differentiation of 3T3-L1 cells and contributes to the highly insulin-responsive glucose transport activity. J. Biol. Chem. 275:17671-17676. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, G., M. L. Standaert, L. Galloway, J. Moscat, and R. V. Farese. 1997. Evidence for involvement of protein kinase C (PKC)-zeta and noninvolvement of diacylglycerol-sensitive PKCs in insulin-stimulated glucose transport in L6 myotubes. Endocrinology 138:4721-4731. [DOI] [PubMed] [Google Scholar]
- 2a.Braiman, L., A. Alt, T. Kuroki, M. Ohba, A. Bak, T. Tennenbaum, and S. R. Sampson.2001. Activation of protein kinase Cζ induces serine phosphorylation of VAMP2 in the GLUT4 compartment and increases glucose transport in skeletal muscle. Mol. Cell. Biol. 21:7852-7861. [DOI] [PMC free article] [PubMed]
- 3.Brozinick, J. T., Jr., and M. J. Birnbaum. 1998. Insulin, but not contraction, activates Akt/PKB in isolated rat skeletal muscle. J. Biol. Chem. 273:14679-14682. [DOI] [PubMed] [Google Scholar]
- 4.Cheatham, B., C. J. Vlahos, L. Cheatham, L. Wang, J. Blenis, and C. R. Kahn. 1994. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol. Cell. Biol. 14:4902-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang, S. H., C. A. Baumann, M. Kanzaki, D. C. Thurmond, R. T. Watson, C. L. Neudauer, I. G. Macara, J. E. Pessin, and A. R. Saltiel. 2001. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature 410:944-948. [DOI] [PubMed] [Google Scholar]
- 6.Clark, S. F., S. Martin, A. J. Carozzi, M. M. Hill, and D. E. James. 1998. Intracellular localization of phosphatidylinositide 3-kinase and insulin receptor substrate-1 in adipocytes: potential involvement of a membrane skeleton. J. Cell Biol. 140:1211-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, S. F., J. C. Molero, and D. E. James. 2000. Release of insulin receptor substrate proteins from an intracellular complex coincides with the development of insulin resistance. J. Biol. Chem. 275:3819-3826. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, J. F., P. W. Young, K. Yonezawa, M. Kasuga, and G. D. Holman. 1994. Inhibition of the translocation of GLUT1 and GLUT4 in 3T3-L1 cells by the phosphatidylinositol 3-kinase inhibitor, wortmannin. Biochem. J. 300:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douen, A. G., T. Ramlal, S. Rastogi, P. J. Bilan, G. D. Cartee, M. Vranic, J. O. Holloszy, and A. Klip. 1990. Exercise induces recruitment of the “insulin-responsive glucose transporter.” Evidence for distinct intracellular insulin- and exercise-recruitable transporter pools in skeletal muscle. J. Biol. Chem. 265:13427-13430. [PubMed] [Google Scholar]
- 10.Ducluzeau, P. H., L. M. Fletcher, G. I. Welsh, and J. M. Tavare. 2002. Functional consequence of targeting protein kinase B/Akt to GLUT4 vesicles. J. Cell Sci. 115:2857-2866. [DOI] [PubMed] [Google Scholar]
- 11.Egawa, K., H. Maegawa, K. Shi, T. Nakamura, T. Obata, T. Yoshizaki, K. Morino, S. Shimizu, Y. Nishio, E. Suzuki, and A. Kashiwagi. 2002. Membrane localization of 3-phosphoinositide-dependent protein kinase-1 stimulates activities of Akt and atypical PKC, but does not stimulate glucose transport and glycogen synthesis in 3T3-L1 adipocytes. J. Biol. Chem. 277:38863-38869. [DOI] [PubMed] [Google Scholar]
- 12.Foster, L. J., D. Li, V. K. Randhawa, and A. Klip. 2001. Insulin accelerates inter-endosomal GLUT4 traffic via phosphatidylinositol 3-kinase and protein kinase B. J. Biol. Chem. 276:44212-44221. [DOI] [PubMed] [Google Scholar]
- 13.Franke, T. F., D. R. Kaplan, L. C. Cantley, and A. Toker. 1997. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275:665-668. [DOI] [PubMed] [Google Scholar]
- 14.Frech, M., M. Andjelkovic, E. Ingley, K. K. Reddy, J. R. Falck, and B. A. Hemmings. 1997. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J. Biol. Chem. 272:8474-8481. [DOI] [PubMed] [Google Scholar]
- 15.Goodyear, L. J., M. F. Hirshman, R. Napoli, J. Calles, J. F. Markuns, O. Ljungqvist, and E. S. Horton. 1996. Glucose ingestion causes GLUT4 translocation in human skeletal muscle. Diabetes 45:1051-1056. [DOI] [PubMed] [Google Scholar]
- 16.Gray, A., J. Van Der Kaay, and C. P. Downes. 1999. The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem. J. 344(Pt 3):929-936. [PMC free article] [PubMed] [Google Scholar]
- 17.Heart, E., W. S. Choi, and C. K. Sung. 2000. Glucosamine-induced insulin resistance in 3T3-L1 adipocytes. Am. J. Physiol. 278:E103-E112. [DOI] [PubMed] [Google Scholar]
- 18.Heller-Harrison, R. A., M. Morin, A. Guilherme, and M. P. Czech. 1996. Insulin-mediated targeting of phosphatidylinositol 3-kinase to GLUT4-containing vesicles. J. Biol. Chem. 271:10200-10204. [DOI] [PubMed] [Google Scholar]
- 19.Hill, M. M., S. F. Clark, D. F. Tucker, M. J. Birnbaum, D. E. James, and S. L. Macaulay. 1999. A role for protein kinase Bbeta/Akt2 in insulin-stimulated GLUT4 translocation in adipocytes. Mol. Cell. Biol. 19:7771-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooshmand-Rad, R., L. Hajkova, P. Klint, R. Karlsson, B. Vanhaesebroeck, L. Claesson-Welsh, and C. H. Heldin. 2000. The PI 3-kinase isoforms p110(alpha) and p110(beta) have differential roles in PDGF- and insulin-mediated signaling. J. Cell Sci. 113(Pt. 2):207-214. [DOI] [PubMed] [Google Scholar]
- 21.Huang, C., R. Somwar, N. Patel, W. Niu, D. Torok, and A. Klip. 2002. Sustained exposure of L6 myotubes to high glucose and insulin decreases insulin-stimulated GLUT4 translocation but upregulates GLUT4 activity. Diabetes 51:2090-2098. [DOI] [PubMed] [Google Scholar]
- 22.Inoue, G., B. Cheatham, R. Emkey, and C. R. Kahn. 1998. Dynamics of insulin signaling in 3T3-L1 adipocytes. Differential compartmentalization and trafficking of insulin receptor substrate (IRS)-1 and IRS-2. J. Biol. Chem. 273:11548-11555. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, Z. Y., A. Chawla, A. Bose, M. Way, and M. P. Czech. 2002. A phosphatidylinositol 3-kinase-independent insulin signaling pathway to N-WASP/Arp2/3/F-actin required for GLUT4 glucose transporter recycling. J. Biol. Chem. 277:509-515. [DOI] [PubMed] [Google Scholar]
- 24.Kanzaki, M., and J. E. Pessin. 2001. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J. Biol. Chem. 276:42436-42444. [DOI] [PubMed] [Google Scholar]
- 25.Kanzaki, M., R. T. Watson, A. H. Khan, and J. E. Pessin. 2001. Insulin stimulates actin comet tails on intracellular GLUT4-containing compartments in differentiated 3T3L1 adipocytes. J. Biol. Chem. 276:49331-49336. [DOI] [PubMed] [Google Scholar]
- 26.Katagiri, H., T. Asano, H. Ishihara, K. Inukai, Y. Shibasaki, M. Kikuchi, Y. Yazaki, and Y. Oka. 1996. Overexpression of catalytic subunit p110alpha of phosphatidylinositol 3- kinase increases glucose transport activity with translocation of glucose transporters in 3T3-L1 adipocytes. J. Biol. Chem. 271:16987-16990. [DOI] [PubMed] [Google Scholar]
- 27.Kelly, K. L., and N. B. Ruderman. 1993. Insulin-stimulated phosphatidylinositol 3-kinase. Association with a 185-kDa tyrosine-phosphorylated protein (IRS-1) and localization in a low density membrane vesicle. J. Biol. Chem. 268:4391-4398. [PubMed] [Google Scholar]
- 28.Khayat, Z. A., P. Tong, K. Yaworsky, R. J. Bloch, and A. Klip. 2000. Insulin-induced actin filament remodeling colocalizes actin with phosphatidylinositol 3-kinase and GLUT4 in L6 myotubes. J. Cell Sci. 113(Pt. 2):279-290. [DOI] [PubMed] [Google Scholar]
- 29.Klarlund, J. K., W. Tsiaras, J. J. Holik, A. Chawla, and M. P. Czech. 2000. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J. Biol. Chem. 275:32816-32821. [DOI] [PubMed] [Google Scholar]
- 30.Klippel, A., W. M. Kavanaugh, D. Pot, and L. T. Williams. 1997. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol. Cell. Biol. 17:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kupriyanova, T. A., and K. V. Kandror. 1999. Akt-2 binds to Glut4-containing vesicles and phosphorylates their component proteins in response to insulin. J. Biol. Chem. 274:1458-1464. [DOI] [PubMed] [Google Scholar]
- 32.Marette, A., E. Burdett, A. Douen, M. Vranic, and A. Klip. 1992. Insulin induces the translocation of GLUT4 from a unique intracellular organelle to transverse tubules in rat skeletal muscle. Diabetes 41:1562-1569. [DOI] [PubMed] [Google Scholar]
- 33.Martelli, A. M., P. Borgatti, R. Bortul, M. Manfredini, L. Massari, S. Capitani, and L. M. Neri. 2000. Phosphatidylinositol 3-kinase translocates to the nucleus of osteoblast-like MC3T3-E1 cells in response to insulin-like growth factor I and platelet-derived growth factor but not to the proapoptotic cytokine tumor necrosis factor alpha. J. Bone Miner. Res. 15:1716-1730. [DOI] [PubMed] [Google Scholar]
- 34.Mosser, V. A., Y. Li, and M. J. Quon. 2001. PTEN does not modulate GLUT4 translocation in rat adipose cells under physiological conditions. Biochem. Biophys. Res. Commun. 288:1011-1017. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi, H., K. A. Brewer, and J. H. Exton. 1993. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 268:13-16. [PubMed] [Google Scholar]
- 36.Nakashima, N., P. M. Sharma, T. Imamura, R. Bookstein, and J. M. Olefsky. 2000. The tumor suppressor PTEN negatively regulates insulin signaling in 3T3-L1 adipocytes. J. Biol. Chem. 275:12889-12895. [DOI] [PubMed] [Google Scholar]
- 37.Nave, B. T., R. J. Haigh, A. C. Hayward, K. Siddle, and P. R. Shepherd. 1996. Compartment-specific regulation of phosphoinositide 3-kinase by platelet-derived growth factor and insulin in 3T3-L1 adipocytes. Biochem. J. 318:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawano, M., K. Ueta, A. Oku, K. Arakawa, A. Saito, M. Funaki, M. Anai, M. Kikuchi, Y. Oka, and T. Asano. 1999. Hyperglycemia impairs the insulin signaling step between PI 3-kinase and Akt/PKB activations in ZDF rat liver. Biochem. Biophys. Res. Commun. 266:252-256. [DOI] [PubMed] [Google Scholar]
- 39.Oatey, P. B., K. Venkateswarlu, A. G. Williams, L. M. Fletcher, E. J. Foulstone, P. J. Cullen, and J. M. Tavare. 1999. Confocal imaging of the subcellular distribution of phosphatidylinositol 3,4,5-trisphosphate in insulin- and PDGF-stimulated 3T3-L1 adipocytes. Biochem. J. 344(Pt. 2):511-518. [PMC free article] [PubMed] [Google Scholar]
- 40.Omata, W., H. Shibata, L. Li, K. Takata, and I. Kojima. 2000. Actin filaments play a critical role in insulin-induced exocytotic recruitment but not in endocytosis of GLUT4 in isolated rat adipocytes. Biochem. J. 346(Pt. 2):321-328. [PMC free article] [PubMed] [Google Scholar]
- 41.Patki, V., J. Buxton, A. Chawla, L. Lifshitz, K. Fogarty, W. Carrington, R. Tuft, and S. Corvera. 2001. Insulin action on GLUT4 traffic visualized in single 3T3-l1 adipocytes by using ultra-fast microscopy. Mol. Biol. Cell 12:129-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peyrollier, K., E. Hajduch, A. Gray, G. J. Litherland, A. R. Prescott, N. R. Leslie, and H. S. Hundal. 2000. A role for the actin cytoskeleton in the hormonal and growth-factor-mediated activation of protein kinase B. Biochem. J. 352(Pt. 3):617-622. [PMC free article] [PubMed] [Google Scholar]
- 43.Rameh, L. E., A. Arvidsson, K. L. Carraway III, A. D. Couvillon, G. Rathbun, A. Crompton, B. VanRenterghem, M. P. Czech, K. S. Ravichandran, S. J. Burakoff, D. S. Wang, C. S. Chen, and L. C. Cantley. 1997. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem. 272:22059-22066. [DOI] [PubMed] [Google Scholar]
- 44.Randhawa, V. K., P. J. Bilan, Z. A. Khayat, N. Daneman, Z. Liu, T. Ramlal, A. Volchuk, X. R. Peng, T. Coppola, R. Regazzi, W. S. Trimble, and A. Klip. 2000. VAMP2, but not VAMP3/cellubrevin, mediates insulin-dependent incorporation of GLUT4 into the plasma membrane of L6 myoblasts. Mol. Biol. Cell 11:2403-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricort, J. M., J. F. Tanti, E. Van Obberghen, and Y. Le Marchand-Brustel. 1996. Different effects of insulin and platelet-derived growth factor on phosphatidylinositol 3-kinase at the subcellular level in 3T3-L1 adipocytes. A possible explanation for their specific effects on glucose transport. Eur. J. Biochem. 239:17-22. [DOI] [PubMed] [Google Scholar]
- 46.Ryder, J. W., J. Yang, D. Galuska, J. Rincon, M. Bjornholm, A. Krook, S. Lund, O. Pedersen, H. Wallberg-Henriksson, J. R. Zierath, and G. D. Holman. 2000. Use of a novel impermeable biotinylated photolabeling reagent to assess insulin- and hypoxia-stimulated cell surface GLUT4 content in skeletal muscle from type 2 diabetic patients. Diabetes 49:647-654. [DOI] [PubMed] [Google Scholar]
- 47.Shepherd, P. R., D. J. Withers, and K. Siddle. 1998. Phosphoinositide 3-kinase: the key switch mechanism in insulin signaling. Biochem. J. 333:471-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddhanta, U., J. McIlroy, A. Shah, Y. Zhang, and J. M. Backer. 1998. Distinct roles for the p110alpha and hVPS34 phosphatidylinositol 3′-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J. Cell Biol. 143:1647-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somwar, R., W. Niu, D. Y. Kim, G. Sweeney, V. K. Randhawa, C. Huang, T. Ramlal, and A. Klip. 2001. Differential effects of phosphatidylinositol 3-kinase inhibition on intracellular signals regulating GLUT4 translocation and glucose transport. J. Biol. Chem. 276:46079-46087. [DOI] [PubMed] [Google Scholar]
- 50.Standaert, M. L., L. Galloway, P. Karnam, G. Bandyopadhyay, J. Moscat, and R. V. Farese. 1997. Protein kinase C-zeta as a downstream effector of phosphatidylinositol 3-kinase during insulin stimulation in rat adipocytes. Potential role in glucose transport. J. Biol. Chem. 272:30075-30082. [DOI] [PubMed] [Google Scholar]
- 51.Tengholm, A., and T. Meyer. 2002. A PI3-kinase signaling code for insulin-triggered insertion of glucose transporters into the plasma membrane. Curr. Biol. 12:1871-1876. [DOI] [PubMed] [Google Scholar]
- 52.Tirosh, A., R. Potashnik, N. Bashan, and A. Rudich. 1999. Oxidative stress disrupts insulin-induced cellular redistribution of insulin receptor substrate-1 and phosphatidylinositol 3-kinase in 3T3-L1 adipocytes. A putative cellular mechanism for impaired protein kinase B activation and GLUT4 translocation. J. Biol. Chem. 274:10595-10602. [DOI] [PubMed] [Google Scholar]
- 53.Tong, P., Z. A. Khayat, C. S. Chan, and A. Klip. 2001. A mechanism for the impairment of insulin action by high levels of insulin and glucose in L6 rat skeletal muscle cells. Diabetes 50(Suppl. 2):A507. [Google Scholar]
- 54.Tong, P., Z. A. Khayat, C. Huang, N. Patel, A. Ueyama, and A. Klip. 2001. Insulin-induced cortical actin remodeling promotes GLUT4 insertion at muscle cell membrane ruffles. J. Clin. Investig. 108:371-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsakiridis, T., A. Bergman, R. Somwar, C. Taha, K. Aktories, T. F. Cruz, A. Klip, and G. P. Downey. 1998. Actin filaments facilitate insulin activation of the src and collagen homologous/mitogen-activated protein kinase pathway leading to DNA synthesis and c-fos expression. J. Biol. Chem. 273:28322-28331. [DOI] [PubMed] [Google Scholar]
- 56.Tsakiridis, T., H. E. McDowell, T. Walker, C. P. Downes, H. S. Hundal, M. Vranic, and A. Klip. 1995. Multiple roles of phosphatidylinositol 3-kinase in regulation of glucose transport, amino acid transport, and glucose transporters in L6 skeletal muscle cells. Endocrinology 136:4315-4322. [DOI] [PubMed] [Google Scholar]
- 57.Tsakiridis, T., P. Tong, B. Matthews, E. Tsiani, P. J. Bilan, A. Klip, and G. P. Downey. 1999. Role of the actin cytoskeleton in insulin action. Microsc. Res. Tech. 47:79-92. [DOI] [PubMed] [Google Scholar]
- 58.Tsakiridis, T., M. Vranic, and A. Klip. 1994. Disassembly of the actin network inhibits insulin-dependent stimulation of glucose transport and prevents recruitment of glucose transporters to the plasma membrane. J. Biol. Chem. 269:29934-29942. [PubMed] [Google Scholar]
- 59.Tsakiridis, T., M. Vranic, and A. Klip. 1995. Phosphatidylinositol 3-kinase and the actin network are not required for the stimulation of glucose transport caused by mitochondrial uncoupling: comparison with insulin action. Biochem. J. 309:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsuji, Y., Y. Kaburagi, Y. Terauchi, S. Satoh, N. Kubota, H. Tamemoto, F. B. Kraemer, H. Sekihara, S. Aizawa, Y. Akanuma, K. Tobe, S. Kimura, and T. Kadowaki. 2001. Subcellular localization of insulin receptor substrate family proteins associated with phosphatidylinositol 3-kinase activity and alterations in lipolysis in primary mouse adipocytes from IRS-1 null mice. Diabetes 50:1455-1463. [DOI] [PubMed] [Google Scholar]
- 61.Turinsky, J., and A. Damrau-Abney. 1999. Akt kinases and 2-deoxyglucose uptake in rat skeletal muscles in vivo: study with insulin and exercise. Am. J. Physiol. 276:R277-R282. [DOI] [PubMed] [Google Scholar]
- 62.Ueyama, A., K. L. Yaworsky, Q. Wang, Y. Ebina, and A. Klip. 1999. GLUT-4myc ectopic expression in L6 myoblasts generates a GLUT-4-specific pool conferring insulin sensitivity. Am. J. Physiol. 277:E572-E578. [DOI] [PubMed] [Google Scholar]
- 63.Venkateswarlu, K., P. B. Oatey, J. M. Tavare, and P. J. Cullen. 1998. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr. Biol. 8:463-466. [DOI] [PubMed] [Google Scholar]
- 64.Wada, T., T. Sasaoka, M. Funaki, H. Hori, S. Murakami, M. Ishiki, T. Haruta, T. Asano, W. Ogawa, H. Ishihara, and M. Kobayashi. 2001. Overexpression of SH2-containing inositol phosphatase 2 results in negative regulation of insulin-induced metabolic actions in 3T3-L1 adipocytes via its 5′-phosphatase catalytic activity. Mol. Cell. Biol. 21:1633-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, F., P. Herzmark, O. D. Weiner, S. Srinivasan, G. Servant, and H. R. Bourne. 2002. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 4:513-518. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Q., P. J. Bilan, T. Tsakiridis, A. Hinek, and A. Klip. 1998. Actin filaments participate in the relocalization of phosphatidylinositol 3-kinase to glucose transporter-containing compartments and in the stimulation of glucose uptake in 3T3-L1 adipocytes. Biochem. J. 331:917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Q., Z. Khayat, K. Kishi, Y. Ebina, and A. Klip. 1998. GLUT4 translocation by insulin in intact muscle cells: detection by a fast and quantitative assay. FEBS Lett. 427:193-197. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Q., R. Somwar, P. J. Bilan, Z. Liu, J. Jin, J. R. Woodgett, and A. Klip. 1999. Protein kinase B/Akt participates in GLUT4 translocation by insulin in L6 myoblasts. Mol. Cell. Biol. 19:4008-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiner, O. D., P. O. Neilsen, G. D. Prestwich, M. W. Kirschner, L. C. Cantley, and H. R. Bourne. 2002. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 4:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White, M. F., and C. R. Kahn. 1994. The insulin signaling system. J. Biol. Chem. 269:1-4. [PubMed] [Google Scholar]
- 71.Zierath, J. R., L. He, A. Guma, E. Odegoard Wahlstrom, A. Klip, and H. Wallberg-Henriksson. 1996. Insulin action on glucose transport and plasma membrane GLUT4 content in skeletal muscle from patients with NIDDM. Diabetologia 39:1180-1189. [DOI] [PubMed] [Google Scholar]