Abstract
Smad proteins transduce transforming growth factor β (TGF-β) and bone morphogenetic protein (BMP) signals that regulate cell growth and differentiation. We have identified YY1, a transcription factor that positively or negatively regulates transcription of many genes, as a novel Smad-interacting protein. YY1 represses the induction of immediate-early genes to TGF-β and BMP, such as the plasminogen activator inhibitor 1 gene (PAI-1) and the inhibitor of differentiation/inhibitor of DNA binding 1 gene (Id-1). YY1 inhibits binding of Smads to their cognate DNA elements in vitro and blocks Smad recruitment to the Smad-binding element-rich region of the PAI-1 promoter in vivo. YY1 interacts with the conserved N-terminal Mad homology 1 domain of Smad4 and to a lesser extent with Smad1, Smad2, and Smad3. The YY1 zinc finger domain mediates the association with Smads and is necessary for the repressive effect of YY1 on Smad transcriptional activity. Moreover, downregulation of endogenous YY1 by antisense and small interfering RNA strategies results in enhanced transcriptional responses to TGF-β or BMP. Ectopic expression of YY1 inhibits, while knockdown of endogenous YY1 enhances, TGF-β- and BMP-induced cell differentiation. In contrast, overexpression or knockdown of YY1 does not affect growth inhibition induced by TGF-β or BMP. Accordingly, YY1 does not interfere with the regulation of immediate-early genes involved in the TGF-β growth-inhibitory response, the cell cycle inhibitors p15 and p21, and the proto-oncogene c-myc. In conclusion, YY1 represses Smad transcriptional activities in a gene-specific manner and thus regulates cell differentiation induced by TGF-β superfamily pathways.
The Smad proteins are major intracellular signaling effectors for transforming growth factor β (TGF-β) and bone morphogenetic protein (BMP). Receptor-regulated Smads (R-Smads) become activated via phosphorylation by the heteromeric receptor complex, oligomerize with a common effector (Co-Smad [i.e., Smad4]), and are rapidly imported to the nucleus (32, 38). In the nucleus, Smads regulate gene expression positively or negatively by binding to DNA and by interacting with DNA sequence-specific transcription factors, coactivators, and corepressors (4, 23, 32). However, little is yet known about how TGF-β-induced transcriptional control regulates cell proliferation and differentiation. Thus, it is necessary to analyze more extensively transcription factor circuits that regulate gene targets downstream of the TGF-β pathways.
This led us to identify additional transcription factors that interact with Smads. In this report we focus on the identification and functional importance of the nuclear factor Yin Yang 1 (YY1) as a Smad partner. YY1 is a nuclear factor essential for mammalian development that activates or represses gene transcription depending on the promoter context (12, 47, 52). Both the N-terminal acidic domain and the C-terminal DNA-binding zinc finger domain contribute to YY1's ability to activate and repress transcription (47, 52). In addition, YY1 cooperates and physically associates with many other transcription factors, coactivators, and corepressors, much like the Smads (52). Alternating acetylation and deacetylation mechanisms regulate the complex transcriptional properties of YY1 (58).
YY1 plays important roles in cell differentiation (47). In myoblasts, YY1 represses muscle-specific genes such as those for smooth muscle and skeletal and cardiac α-actin, whose expression is regulated by TGF-β and other extracellular stimuli (27, 31). At least in one model system, i.e., C2C12 myoblasts, differentiation to myocytes depends on the degradation of YY1, an event that relieves repression of muscle-specific genes (54). These cells present an interesting system, as their differentiation is modulated by both BMP and TGF-β. TGF-β inhibits the differentiation of mesenchymal cell types to adipocytic, myocytic, or osteocytic lineages, while BMP promotes osteoblast differentiation and inhibits myocytic differentiation (2, 18, 29, 57). Little is yet understood about the cross talk between YY1 and specific signaling pathways that modulate cell differentiation.
In this report, we analyze the physical association and functional cooperation of Smad proteins and YY1, which represses Smad transcriptional activity in a gene-specific manner. Thus, YY1 inhibits cell differentiation induced by TGF-β or BMP without affecting cell growth regulation by these factors. The ability of YY1 to regulate Smad signals provides an important example of how multifunctional nuclear integrators may define the potency and competence of TGF-β superfamily pathways that are crucial for cellular differentiation.
MATERIALS AND METHODS
Cell culture, adenoviruses, ligands, proteins, antibodies, small interfering RNAs (siRNAs), and plasmids.
Human HaCaT keratinocytes were provided by N. Fusenig, Heidelberg, Germany. Murine C2C12 myoblasts, murine mammary epithelial NMuMG cells, mink lung epithelial Mv1Lu cells, monkey kidney COS-7 cells, human embryonic kidney 293T cells, human mammary MDA-MB-468 cells, and human hepatoma HepG2 cells were obtained and cultured according to protocols from the American Type Culture Collection.
Adenoviruses expressing the control protein LacZ, constitutively active activin-like receptor kinase 5 (caALK-5), Smad4, and Smad7 were donated by K. Miyazono, Tokyo, Japan, and were amplified and titrated as previously described (41). Adenoviruses expressing green fluorescent protein (GFP) and wild-type YY1 were based on the bicistronic Adeasy viral system, which was provided by B. Vogelstein, Baltimore, Md. (20). Construction and characterization of the YY1 adenovirus will be described elsewhere (A. A. Terentiev et al., unpublished data).
Recombinant mature human TGF-β1 was provided by N. Ferrara (Genentech, Inc.), and recombinant mature BMP-6 and BMP-7 were provided by K. Sampath (Curis, Inc.). Recombinant, baculovirally expressed and purified Smad3 (phosphorylated at its C terminus by the TGF-β receptor type I) and Smad4 proteins were provided by F. M. Hoffman and A. Comer (University of Wisconsin).
Mouse monoclonal anti-YY1 (H-10) and rabbit polyclonal anti-inhibitor of differentiation/inhibitor of DNA binding 1 (anti-Id-1) (Z-8) antibodies were gifts from and rabbit anti-YY1 (C-20 and H414), rabbit anti-Smad4 (H-552), goat anti-plasminogen activator inhibitor 1 (anti-PAI-1) (C20), and mouse monoclonal anti-α-tubulin (TU-02) antibodies were purchased from Santa Cruz Biotechnology, Inc.; rabbit anti-Smad3 (LPC3) was purchased from Zymed Laboratories Inc.; mouse monoclonal anti-β-catenin (C19220) and anti-p21 (C24420) were purchased from BD Transduction Laboratories Inc.; mouse monoclonal anti-Flag antibodies (M2 and M5) were purchased from Sigma-Aldrich; and mouse monoclonal anti-Myc (9E10) and rabbit polyclonal anti-phospho-Smad1 and anti-phospho-Smad2 antibodies were produced in house.
The YY1-specific siRNA was designed based on the mouse YY1 cDNA sequence (accession number NM 009537) and corresponds to nucleotides 866 to 884 of that cDNA by numbering as nucleotide 1 the A of the ATG (start) codon of the protein-coding sequence. The siRNA design was based on the criteria of Elbashir et al. (15). The YY1 siRNA sequence is identical in the human, mouse, and Xenopus cDNAs, and BLAST searches against the human genome, the mouse genome, and the expressed sequence tag database resulted in statistically significant hits (E value of 5 × e−6) that corresponded only to YY1-related sequences. The control siRNA was a double-stranded 21-mer unrelated to the YY1 sequence. Both double-stranded RNAs were provided by Dharmacon Research, Inc.
The promoter-reporter constructs 12xCAGA-luc (9), PAI-1-Luc (p800-PAI-1-neo-luc) (1), Id1-BRE2-luc (24), c-myc-luc (pHX-luc) (7), and p15-luc (28) were gifts from J. M. Gauthier (Paris, France), D. Rifkin (New York, N.Y.), O. Korchynskyi (Amsterdam, The Netherlands), K. Miyazono (Tokyo, Japan), and X.-F. Wang (Durham, N.C.), respectively. The reporter construct p21(−143/+8)-luc has been previously described (40). The mammalian expression vectors pcDNA3 encoding constitutively active ALK-5, ALK-6, Myc6-tagged Smad3 and Smad4, and Flag-tagged Smad4 and the bacterial expression vectors pGEX-4T encoding glutathione S-transferase (GST) fusions of Smad1/2/3/4, Smad4ΔMH1, and Smad4ΔMH2 have been previously described (25, 40, 41). The mammalian expression vectors pCB6+ encoding wild-type YY1 and deletion mutants Δ2-62, Δ2-197, Δ2-273, Δ69-85, Δ92-153, Δ154-199, Δ199-273, Δ262-299, Δ296-331, Δ334-414, and Δ399-414 and the bacterial expression vector pGEX-4T encoding a GST fusion with wild-type YY1 were provided by B. Lüscher, Hannover, Germany (5). The mammalian expression vectors pcDNA3-YY1 and pcDNA3-AS YY1 encoding full-length sense and antisense YY1 mRNAs, respectively, were constructed by isolation of the YY1 cDNA from pCB6+-YY1 as an EcoRI fragment and subcloning in the correct and inverse orientations in pcDNA3 digested with EcoRI. Restriction analysis and DNA sequencing verified the orientation and integrity of the construct. The Bluescript SK-based plasmids with the rat PAI-1 cDNA, pSKPAI53 (a gift of T. D. Gelehrter, Ann Arbor, Mich.), and the human β-actin cDNA pSKhβactin (a gift of G. Mavrothalassitis, Heraklion, Greece) were used for Northern blotting probe synthesis.
Cell transfections, infections, and gene reporter assays.
Transient transfections of cells by using Lipofectamine and Lipofectamine 2000 (Invitrogen), Fugene 6 (Roche), or calcium phosphate coprecipitation were performed according to the manufacturer's protocols and as previously described (36, 40). Transient transfections of siRNA were also performed with Lipofectamine. Luciferase reporter assays were performed with the enhanced luciferase assay kit from BD PharMingen, Inc., according to the manufacturer's protocol. Adenoviral transient infections of cells, using the multiplicities of infection (MOIs) specified in the figure legends, were performed as previously described (40, 41).
In vitro and in vivo protein interaction assays.
All in vitro GST pull-down assays using endogenous cellular proteins were performed as described previously (40). For in vivo coprecipitation assays, proteins expressed in COS-7 or MDA-MB-468 cells or endogenous proteins of HaCaT cells were analyzed as described previously (40). Nuclear extracts were isolated by using the NE-PER kit (Pierce). Relative protein expression levels were quantified by using the scanning densitometric software of the Fujix BAS 2000 phosphorimager. Ratios of band intensities of the tested protein (YY1 or PAI-1) over the control protein (α-tubulin) were calculated, and the ground condition ratio was set to 1, relative to which all other conditions are expressed.
DNA affinity precipitation (DNAP) assays, EMSA, and Northern analysis.
Cell lysates of transiently transfected COS-7 cells were precleared with streptavidin-agarose (Sigma-Aldrich) and incubated with 30 pmol of biotinylated double-stranded DNA composed of four tandemly repeated CAGA sequence motifs in the presence of 12 μg of poly(dI-dC) for 1 h at 4°C. DNA-bound proteins were precipitated with streptavidin-agarose for 30 min at 4°C, washed, and detected by immunoblotting using enhanced chemiluminescence. The protocol is adapted from that of Nishihara et al. (39). The 4xCAGA oligonucleotide (sense strand), 5′-CAGACAGTCAGACAGTCAGACAGTCAGACAGT-3′, was biotinylated at the 5′ end, and the oligonucleotides were synthesized by Cybergene AB. The same oligonucleotide (without the biotin modification) and the adeno-associated virus P5 promoter oligonucleotide that contains the YY1 motif (48) were used in electrophoretic mobility shift assays (EMSA) with purified recombinant YY1 (produced in Escherichia coli as a GST fusion) and phosphorylated Smad3 and Smad4 proteins (produced in a baculovirus system) under conditions described previously (16, 40).
Northern blot analysis of 10 μg of total RNA isolated from infected Mv1Lu cells by using the TriZol reagent (Gibco-BRL) according to the manufacturer's protocol was performed as previously described (24). The PAI-1 cDNA probe was derived from pSKPAI53, and the β-actin probe was derived from pSKhβactin.
RT-PCR.
Total RNA was extracted from HaCaT cells with the RNeasy kit (Qiagen) and digested with DNase RQI (Promega) to remove any contaminating genomic DNA. For reverse transcription (RT), a 40-μl reaction mixture contained 1 μg of RNA, 12.5 ng of anchored oligo(dT17) primers (5′-AGCT17-3′) per μl, a 500 μM concentration of each deoxynucleoside triphosphate (dNTP), 100 ng of bovine serum albumin per μl, 10 mM dithiothreitol, 4 U of RNasin (Promega), and 200 U of SuperScript II RNase H− (Invitrogen). Reactions were carried out at 42°C for 50 min, followed by inactivation of the enzyme at 70°C for 15 min. The cDNAs were then incubated with 4 U of RNase H (Invitrogen) at 37°C for 30 min. Two-microliter aliquots of the RT reaction product were used for PCR analyses. Routinely, each PCR amplification mixture included a 50 μM concentration of each dNTP, a 0.2 μM concentration of each primer, 1.5 mM MgCl2, and 2.5 U of AmpliTaq Gold DNA polymerase. Amplification was performed in a T3 thermocycler (Biometra) with an initial denaturation step at 95°C for 7 min; 26 to 30 cycles of 30 s at 94°C, 30 s at the optimal temperature (Table 1), and 30 s at 72°C; and a final elongation step at 72°C for 5 min. Specific primers were designed according to sequences available in the data banks or published by other authors (Table 1). Primers for the glyceraldehyde 3′-phosphate dehydrogenase (GAPDH) gene were used to ascertain that an equivalent amount of cDNA was synthesized. The RT-PCR products were separated by electrophoresis on 2% agarose and stained with ethidium bromide.
TABLE 1.
Oligonucleotide primers used for RT-PCR, real-time quantitative PCR, and ChIP analyses
| Analysis | Gene | Primer sequence (strand) | Product size (bp) | Temp (°C) | PCR cycles | Reference or accession no. |
|---|---|---|---|---|---|---|
| RT-PCR | p21 | 5′-CTGCCCAAGCTCTACCTTCC-3′ (+) | 123 | 57 | 30 | 44 |
| 5′-CAGGTCCACATGGTCTTCCT-3′ (−) | ||||||
| PAI-1 | 5′-GTGGTCTGTGTCACCGTATC-3′ (+) | 440 | 59 | 30 | M16006 | |
| 5′-GTAGTTGAATCCGAGCTGCC-3′ (−) | ||||||
| c-myc | 5′-ACCCGGACGACGAGACCTTCATCA-3′ (+) | 683 | 63 | 26 | NM_002467 | |
| 5′-GGGGCTGGTGCATTTTCGGTTGTT-3′ (−) | ||||||
| GAPDH | 5′-ATCACTGCCACCCAGAAGAC-3′ (+) | 443 | 57 | 30 | 53 | |
| 5′-ATGAGGTCCACCACCCTGTT-3′ (−) | ||||||
| Real-time quantitative PCR | PAI-1 | 5′-GAGACAGGCAGCTCGGATTC-3′ (+) | 101 | 60 | 40 | M16006 |
| 5′-GGCCTCCCAAAGTGCATTAC-3′ (−) | ||||||
| GAPDH | 5′-CCCATGTTCGTCATGGGTGT-3′ (+) | 145 | 60 | 40 | 13 | |
| 5′-TGGTCATGAGTCCTTCCACGATA-3′ (−) | ||||||
| ChIP | PAI-1 | 5′-CCTCCAACCTCAGCCAGACAAG-3′ (+) | 222 | 59 | 30-31 | X13323 |
| 5′-CCCAGCCCAACAGCCACAG-3′ (−) |
Real-time quantitative RT-PCR.
DNase RQI-digested RNA from HaCaT cells was reverse transcribed as described above. PCR was performed in a total volume of 25 μl with SYBR Green PCR Master Mix (Applied Biosystems), 1 μl of cDNA, and a 300 nM concentration of each primer (Table 1). PAI-1 primers were designed with the computer program Primer Express (Applied Biosystems), using parameters recommended by the manufacturer. Reactions were carried out in an ABI-prism 7000 sequence detector (Applied Biosystems) in triplicate, using the following conditions: an initial denaturation step consisted of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Levels of PAI-1 expression in each sample were determined by using the relative standard curve method, with the GAPDH gene used as an endogenous control. After PCR, the threshold cycle (Ct value) was selected and determined for each sample. The relative quantity for each sample was calculated by using Ct values interpolated to reference curves of amplification, obtained for each set of primers by using serially diluted cDNAs. Relative amounts of DNA in each PAI-1 sample were standardized with those in GAPDH samples, and values were reported with the ground condition as a calibrator.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed with the ChIP assay kit (Upstate Inc.) according to the protocol of the manufacturer. The equivalent of ∼107 cells was used per ChIP reaction. The antibodies (5 μg) used were anti-Smad4 (H-552; Santa Cruz) and anti-Flag M5 (Sigma-Aldrich) as a negative control antibody. Genomic DNA pellets were resuspended in 12 μl of water. PCR was performed with 2 μl of immunoprecipitated material, a 0.2 μM concentration of each primer, 0.5% 2-mercaptoethanol, a 50 μM concentration of each dNTP, and 2.5 U of BioTaq DNA polymerase (Bioline) in a buffer containing 67 mM Tris-HCl (pH 8.8), 16.6 mM ammonium sulfate, 6.5 mM MgCl2, and 0.01% gelatin. Amplification was carried out as described in “RT-PCR” above, and the primer set used to amplify the distal part of the human PAI-1 promoter harboring the Smad-binding elements (SBEs) is listed in Table 1. Relative amplified DNA levels were quantified by using the densitometric software of Adobe Photoshop 6.0. Ratios of band intensities of the immunoprecipitated DNA over the input DNA were calculated, and the ground condition ratio was set to 1, relative to which all other conditions are expressed.
Thymidine incorporation assays.
Twenty-four hours after adenoviral infection or transfection with siRNAs, cells were incubated for another 24 h in 3% fetal bovine serum-containing medium prior to addition of vehicle, TGF-β1, or BMP-7. Stimulation with growth factors lasted for 18 h, during the last 3 h of which 1 μCi of [3H]thymidine (Amersham) per ml was added. Thus, thymidine incorporation was analyzed at 60 h postinfection or posttransfection and 18 h after factor addition. At this time point, cell monolayers were 70 to 80% confluent. After metabolic labeling, cells were washed twice in phosphate-buffered saline (PBS), fixed in 5% trichloroacetic acid, rinsed with water, and dehydrated in 70% ethanol, and the DNA was extracted in 0.1 M NaOH. Radioactivity was directly measured in the DNA extracts by using scintillation counting. The data are plotted as average values with standard errors of triplicate repeats for each condition per independent experiment. Each independent experiment was repeated at least twice.
Actin cytoskeleton direct fluorescence microscopy.
NMuMG cells were infected with adenoviruses, and the TGF-β1-induced epithelial-to-mesenchymal transdifferentiation (EMT) was assayed microscopically by using phase-contrast or fluorescence microscopy after staining of fixed cell preparations with tetramethylrhodamine isothiocyanate-labeled phalloidin (Sigma-Aldrich) as previously described (41). All photomicrographs were obtained in a Zeiss Axioplan microscope equipped with a Hammamatsu digital camera and Adobe Photoshop imaging software.
Preosteoblast differentiation and alkaline phosphatase (ALP) staining.
C2C12 cells were routinely incubated with 300 ng of BMP-7 per ml, and preosteoblastic differentiation was efficiently visualized at 3 to 5 days posttreatment. For cells transiently infected with the YY1 adenovirus, BMP-7 was added at 24 h postinfection and preosteoblastic differentiation was scored 3 days later. For cells transiently transfected with constitutively active ALK-6 and Smads, no BMP incubation was included and differentiation was monitored at 4 days posttransfection, reproducing results published by others (57).
To determine ALP activity histochemically, we used an adaptation of the protocol described by Yamamoto et al. (57). Cells were fixed for 10 min with 3.7% formaldehyde at room temperature. After being washed with PBS, the cells were incubated for 30 min with a mixture of 0.1 mg of naphthol AS-MX phosphate (Sigma-Aldrich) per ml, 0.5% N,N-dimethylformamide, 2 mM MgCl2, and 0.6 mg of Fast Blue BB salt (Sigma-Aldrich) per ml in 0.1 M Tris-Cl (pH 8.5) at room temperature. Stained cell preparations were washed in PBS and inspected under a Zeiss Axioplan microscope equipped with a Hammamatsu digital camera and Adobe Photoshop imaging software. For quantitative analysis of ALP activity, cells were extracted in 20 mM Tris-HCl (pH 7.4)-150 mM NaCl containing 1% Triton X-100. The enzymatic activity was determined as described by Asahina et al. (3).
RESULTS
YY1 represses both TGF-β and BMP immediate-early gene targets.
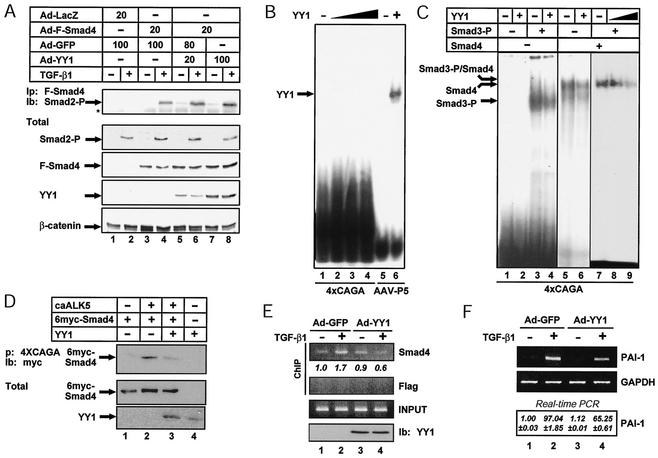
To identify novel Smad-interacting transcription factors, we performed a biochemical screen with various Smad protein domains fused to GST and a large panel of antibodies specific for known transcription factors. One of the novel interacting factors identified was YY1. Since YY1 is a transcription factor, we hypothesized that it might affect Smad-dependent regulation of gene expression. To analyze the effect of YY1 on Smad-dependent gene responses downstream of TGF-β and BMP signaling, we studied PAI-1 and Id-1, two immediate-early gene targets of these pathways (8, 9, 21, 24, 49, 50). Northern blot analysis of mink lung epithelial (Mv1Lu) cell mRNA demonstrated induction of the endogenous PAI-1 gene following treatment with TGF-β1 (Fig. 1A, lanes 1 and 2). Infection of the cells with increasing amounts of a YY1 adenovirus repressed the TGF-β1-inducible levels of PAI-1 mRNA in a dose-dependent manner (lanes 5 and 6). Control adenovirus did not interfere with induction of PAI-1 mRNA by TGF-β1 and exhibited minor effects on the basal levels of expression (Fig. 1A, lanes 7 to 10).
FIG. 1.
YY1 represses TGF-β and BMP immediate-early gene responses. (A) YY1 represses PAI-1 induction by TGF-β1. Northern analysis of PAI-1 mRNA from Mv1Lu cells mock treated (−) or treated (+) for 24 h with 10 ng of TGF-β1 per ml in the absence of YY1 (−) or in the presence of increasing doses of YY1 (Ad-YY1) (triangle; MOI of 20 or 100) or control GFP (Ad-GFP) (MOI of 20 or 100) adenovirus is shown. Sample loading and blotting efficiency was monitored with β-actin mRNA on the same blot. (B) YY1 represses Id-1 induction by TGF-β1. Immunoblot analysis of endogenous Id-1 expression in MDA-MB-468 cells infected with the indicated MOI of recombinant adenoviruses and then treated with vehicle (−) or 2.5 ng of TGF-β1 per ml (+) for 1 h is shown. Arrows mark the positions of endogenous Id-1, adenoviral YY1, adenoviral Flag-Smad4 (F-Smad4), endogenous phosphorylated Smad2 (Smad2-P), and endogenous β-catenin, which serves as a loading and blotting efficiency control. (C) YY1 represses Id-1 induction by BMP-7. Immunoblot analysis of endogenous Id-1 expression in MDA-MB-468 cells is as for panel B but in response to 300 ng of BMP-7 per ml. Infection conditions were as for panel B, and arrows mark the same proteins as in panel B except for endogenous phosphorylated Smad1 (Smad1-P) instead of Smad2. Asterisks mark nonspecific protein bands. (D) YY1 represses PAI-1 promoter induction by TGF-β1. PAI-1 (800)-luciferase assays were performed in HepG2 cells with increasing amounts of YY1 plasmid (triangle; 0.1 to 1.0 μg) in the absence (−) or presence (+) of overnight stimulation with 2.5 ng of TGF-β1 per ml. The averaged data for normalized reporter activity are shown with standard errors derived from triplicate transfections repeated at least twice independently. (E) YY1 represses the Smad-specific 12xCAGA promoter. The assays were similar to those for panel D, with the 12xCAGA-luciferase reporter in HepG2 cells after overnight stimulation with 2.5 ng of TGF-β1 per ml. Data are reported as in panel D. (F) YY1 represses the Smad-specific Id1-BRE2 promoter. The assays were similar to those for panel D, with the Id1-BRE2-luciferase reporter in C2C12 cells after overnight stimulation with 100 ng of BMP-6 per ml. Data are reported as in panel D.
In a parallel experiment, we investigated the effect of YY1 on Id-1 gene expression following induction with TGF-β1 or BMP-7. In order to simultaneously monitor Smad dependency in regulation of gene expression, we performed experiments with the Smad4-deficient human mammary epithelial cell line MDA-MB-468. Previous studies established that ectopic expression of Smad4 in these cells restores many TGF-β-dependent responses, including growth inhibition (see below) (11). While Id-1 induction by BMP (Fig. 1C) has been established (24), we also observed inducible regulation of Id-1 in response to TGF-β1 after reconstitution of Smad4 expression in these cells (Fig. 1B, lanes 3 and 4). It is worth noting that reconstitution of Smad4 expression results in significant induction of the Id-1 expression levels even in the absence of exogenously added growth factors (Fig. 1B and C, lanes 3). Ectopic expression of YY1 led to dose-dependent repression of the Smad4- and TGF-β1- or BMP-7-inducible levels of Id-1 expression (Fig. 1B and C, lanes 5 to 8). YY1 specifically targeted Id-1 gene expression, as it did not affect the TGF-β1- or BMP-7-inducible phosphorylation of endogenous Smad2 (Fig. 1B, lanes 2, 4, 6, and 8) or Smad1 (Fig. 1C, lanes 2, 4, 6, and 8), respectively, or the levels of Smad4. These findings suggest that YY1 negatively interferes with the Smad-mediated induction of PAI-1 and Id-1 gene expression.
We investigated further the role of YY1 in TGF-β and BMP signaling by transactivation assays (Fig. 1D to F). YY1 repressed the TGF-β-induced expression of the PAI-1 promoter in HepG2 cells in a dose-dependent manner (Fig. 1D), in agreement with our data on the endogenous gene in Mv1Lu cells (Fig. 1A) and in HaCaT cells (see Fig. 2F). To confirm that the YY1 effects on target promoters depended on Smads, we analyzed two synthetic promoters whose activities are dependent on Smad proteins (Fig. 1E and F). YY1 potently repressed the TGF-β-mediated induction of the 12xCAGA promoter (9), which contains 12 copies of the SBE (Fig. 1E). The repressive effect of YY1 on this promoter was detected in cell types of both epithelial (Mv1Lu and HaCaT) and mesenchymal (C2C12) origin. YY1 repressed the TGF-β-induced promoter activity, while it showed weak, if any, repression of the low, basal activity of this promoter (data not shown). Similar results were obtained with the BMP-responsive synthetic reporter Id1-BRE2, which contains a duplication of the SBE motif from the Id-1 promoter (24) (Fig. 1F). In contrast, other enhancer-promoter constructs, such as the immediate-early simian virus 40 or the cytomegalovirus promoter, which show strong constitutive activity in mammalian cells, were not repressed and occasionally were weakly induced by YY1 (data not shown). Thus, specific TGF-β and BMP immediate-early gene responses are affected dramatically by YY1, establishing YY1 as a potent repressor of Smad transcriptional activity that is mediated by strong SBE promoter elements.
FIG. 2.
YY1 inhibits the DNA-binding activity of Smads without affecting their oligomerization. (A) YY1 overexpression does not disrupt TGF-β1-induced Smad oligomers. Analysis of coimmunoprecipitation of endogenous phosphorylated Smad2 (Smad2-P) with Flag-tagged adenoviral Smad4 in lysates from infected MDA-MB-468 cells (as for Fig. 1B) after treatment with 2.5 ng of TGF-β1 per ml for 1 h is shown. Total cell extracts were immunoblotted with antibodies against endogenous phosphorylated Smad2 (Smad2-P), adenoviral Flag-tagged Smad4 (F-Smad4), YY1, and the control protein β-catenin. An asterisk marks immunoglobulin heavy-chain bands. (B) YY1 does not bind directly to the Smad-binding element. EMSA was performed with no protein (−), increasing amounts of recombinant YY1 (triangle; 125, 250, and 500 ng), or 125 ng of YY1 (+) and radiolabeled 4xCAGA and adeno-associated virus P5 promoter (AAV-P5) oligonucleotides. (C) YY1 inhibits DNA binding of Smad3 and Smad4. EMSA was performed with 250 ng (+) or 100 and 250 ng (triangle) of recombinant YY1, 100 ng of phosphorylated Smad3 (Smad3-P) (+), 300 ng of Smad4 (+), and radiolabeled 4xCAGA oligonucleotide. The exposure times in each autoradiogram were 24 h (lanes 1 to 4), 10 h (lanes 5 and 6), and 5 h (lanes 7 to 9). In panels B and C the specific band shifts are shown with arrows. (D) YY1 inhibits binding of nuclear Smad4 to SBE DNA. 4xCAGA DNA precipitation (p) assays were performed with 293T cell extracts coexpressing Myc6-Smad4 plus YY1 in the absence or presence of caALK-5. DNA precipitates were immunoblotted (Ib) with anti-Myc antibody, and immunoblots of total cell extracts are shown (Total), as marked by arrows. (E) YY1 inhibits recruitment of endogenous nuclear Smad4 to the SBE-rich area of the endogenous PAI-1 promoter. ChIP assays were performed with the indicated antibodies and PCR amplification of the distal and SBE-rich human PAI-1 promoter from HaCaT cells, infected transiently with the indicated adenoviruses (MOI of 20) and treated (+) or not (−) with 5 ng of TGF-β1 per ml for 2 h. Control ChIPs with an unrelated (Flag) antibody and 2% of the chromatin preparation, amplified without prior immunoprecipitation (INPUT), are shown. A control immunoblot (Ib) of the infected cells is also shown, demonstrating the levels of YY1 overexpression. Numbers below the top panel indicate densitometric values of the specific bands normalized over the corresponding input bands and expressed relative to the control band of lane 1, which is set to 1. (F) YY1 represses TGF-β1-induced PAI-1 mRNA levels in HaCaT cells. Semiquantitative RT-PCR assays with mRNA isolated from duplicate HaCaT cell plates for the ChIP assays demonstrate proper PAI-1 gene regulation in these cells. GAPDH serves as a control in the RT-PCRs. The bottom panel shows average expression values and standard deviations of PAI-1 mRNA levels from the same experiment and after normalization to control GAPDH mRNA levels derived from real-time quantitative PCR experiments.
YY1 does not interfere with R-Smad/Co-Smad oligomerization.
To examine the mechanism by which YY1 repressed the transcriptional activity of Smads, we tested whether YY1 could disrupt nuclear R-Smad/Co-Smad oligomers that are essential for signal transduction (Fig. 2A). To this end, we used MDA-MB-468 cells after Smad4 reconstitution and monitored endogenous Smad2 coprecipitation with the reconstituted Smad4 after stimulation with TGF-β1. Using an antibody recognizing specifically phosphorylated Smad2, we detected coprecipitation of phospho-Smad2 with Smad4 but failed to observe any effects of YY1 on the complexes between these two Smads (Fig. 2A, top panel). Adenoviral infection did not perturb in any detectable way the activation of endogenous Smad2 (detected as TGF-β-induced phosphorylation of Smad2) (Fig. 2A, second panel). YY1 did not affect the oligomeric complexes between the BMP-specific R-Smad, Smad1, and Smad4 (data not shown). We conclude that YY1 interferes with nuclear Smad activities without affecting their oligomeric status.
YY1 inhibits binding of Smads to their cognate DNA sites.
To examine whether YY1 could affect the DNA-binding properties of Smads with the SBE DNA, we performed in vitro EMSA with purified recombinant proteins and an oligonucleotide that contains concatamerized SBEs (Fig. 2B and C). First, we excluded the possibility that YY1 bound directly to the SBE (Fig. 2B). This is in agreement with the obvious sequence differences between the binding motifs of Smads (5′-CAGACAGTCTGT-3′) and YY1 (5′-[C/A]CAT-3′). In a control experiment, binding of YY1 to the adeno-associated virus P5 promoter oligonucleotide confirmed that our recombinant YY1 protein was functional and exhibited specificity in DNA-binding assays (Fig. 2B, lane 6). Furthermore, YY1 led to a reduction in the specific binding to the SBE by phosphorylated Smad3 (Fig. 2C, lanes 3 and 4), Smad4 (lanes 5 and 6), and both Smads when added together, in a dose-dependent manner (lanes 7 to 9). We then performed DNAP assays in order to confirm the in vitro EMSA results under more physiological conditions (Fig. 2D). Using a biotinylated SBE oligonucleotide, we detected efficient association of Smad4 to the DNA under conditions of stimulation with the constitutively active TGF-β type I receptor (ALK-5) (Fig. 2D, lane 2). When YY1 was coexpressed with Smad4, it inhibited Smad4 from forming nucleoprotein complexes with the SBE (lane 3).
To prove that YY1 could affect the recruitment of endogenous Smad proteins on the SBEs of a natural promoter, we performed ChIP assays for the PAI-1 promoter (Fig. 2E). As described above, this gene contains multiple SBEs in its promoter, and YY1 can repress the TGF-β-induced expression of PAI-1 mRNA and promoter activity (Fig. 1 and 2F). In HaCaT keratinocytes infected with control (GFP) or YY1 adenoviruses, we observed the detectable presence of endogenous Smad4 on the SBE-rich part of the PAI-1 promoter when chromatin was immunoprecipitated with an antibody against Smad4 (Fig. 2E, lane 1). This is agreement with the continuous shuttling of Smad4 between the cytoplasm and the nucleus and with previously published ChIP assay results for other promoters (7, 42). Stimulation of cells with TGF-β1 resulted in enhancement of Smad4 recruitment to the promoter (lane 2). Ectopic expression of YY1 inhibited the TGF-β-induced recruitment of Smad4 to the promoter (lane 4) without any major effect on the basal level of Smad4 association with the PAI-1 promoter chromatin (lane 3). The levels of ectopic YY1 expression were also determined (Fig. 2E, lanes 3 and 4, bottom panel). In a control experiment (Fig. 2F) performed with duplicate cultures of HaCaT cells infected under identical conditions as for the ChIP experiment, we confirmed that the endogenous PAI-1 gene was properly induced by TGF-β1, and this induction was reduced by ectopic YY1, reproducing the results with Mv1Lu cells (Fig. 1A). Thus, YY1 reduces the affinity of Smads for their cognate SBEs both in vitro and in vivo. This can lead to a reduction in TGF-β-induced (or BMP-induced) gene expression that is dependent on direct binding of Smads to SBEs in the regulated promoter DNA.
YY1 physically interacts with Smads in vitro and in vivo.
The negative effect of YY1 on Smad binding to DNA and recruitment to chromatin could be explained on the basis of a specific interaction between these two proteins. We examined how YY1 interacts with Smad proteins by several complementary methods. GST pull-down experiments showed that endogenous YY1 interacted strongly with the Co-Smad (Smad4), more weakly with the BMP and TGF-β R-Smads (Smad1 and Smad3, respectively), and even more weakly with Smad2, another R-Smad of the TGF-β pathway (Fig. 3A and B). Additional GST pull-down experiments demonstrated that deletion of the conserved N-terminal Mad homology 1 (MH1) domain (Smad4-ΔMH1) abolished the Smad4 interaction with YY1 to levels comparable to those for the negative GST control, whereas deletion of the conserved C-terminal MH2 domain (Smad4-ΔMH2) did not perturb the interaction in any appreciable degree (Fig. 3C). Similar results were obtained by using GST fusions of Smad1 and Smad3 deletion mutants (data not shown).
FIG. 3.
YY1 interacts physically with Smads in vitro. (A) Schematic representation of Smad proteins with their conserved MH1, the DNA-binding β-hairpin loop (black boxes), MH2, and the less conserved linker sequences. The strength of Smad protein interaction with YY1 is quantified based on the data in panels B and C. (B) Endogenous YY1 from C2C12 cells interacts with the indicated GST-Smad fusions with different relative affinities (lanes 2 to 5). A total C2C12 lysate immunoblot is shown for comparison (lane 6). Ponceau S staining of the immunoblot shows the immobilized GST-Smads and GST protein. (C) YY1 interacts with the MH1 domain of Smad4. Experiment similar to those for panel B were performed with endogenous YY1 from HaCaT cells. Immobilized GST fusion proteins are shown in a Coomassie brilliant blue (CBB) stain of the gel. In panels B and C, arrows mark the specific protein bands.
In order to address the physiological context in which such interactions occur, we performed coprecipitation assays with COS-7 cell extracts transiently expressing Smad4 in the absence or presence of the constitutively active TGF-β type I receptor ALK-5 (Fig. 4A). Smad4 was selected due to its high affinity for YY1 (Fig. 3B). Smad4 coprecipitated with endogenous YY1 both in the absence (lane 1) and in the presence of the constitutively active ALK-5 receptor (lane 2). Coprecipitation experiments performed with HaCaT cells that express Smad4 and YY1 proteins confirmed the formation of endogenous complexes between YY1 and Smad4 (Fig. 4B). The coprecipitation of Smad4 and YY1 was constitutive, and stimulation with TGF-β1 (Fig. 4B) or activated receptor (Fig. 4A) had minor effects on the interaction. To establish more firmly the constitutive nature of this interaction in vivo, we also performed coprecipitation assays using nuclear extracts from HaCaT cells (Fig. 4C) instead of total cells extracts (Fig. 4B). These experiments also confirmed the constitutive Smad4-YY1 interaction and revealed weak reduction of the complex after transient stimulation with TGF-β1. The coprecipitation was specific, since a nonspecific preimmune antiserum did not lead to detectable complexes between Smad4 and YY1 (Fig. 4C, second panel). The same endogenous protein coprecipitation was observed when the inverse immunoprecipitation with anti-YY1 antibody was followed by anti-Smad4 immunoblotting (data not shown). Under the conditions used for the endogenous coprecipitations, Smad4 exhibited significant nuclear levels in the absence and presence of TGF-β1 (Fig. 4C, third and fifth panels), which is in agreement with previous findings for the same cell line (22, 42). The TGF-β1 stimulation was robust, as it led to strong enhancement of the nuclear levels of phosphorylated Smad2 (Fig. 4C, sixth panel). These results establish the physical association of Smads and YY1 and demonstrate that formation of these protein complexes depends on Smad MH1 domain protein sequences. Furthermore, the in vivo experiments suggest that YY1 binds stably to the nuclear pool of Smad4.
FIG. 4.
YY1 and Smad4 form complexes in vivo. (A) Smad4 coimmunoprecipitates with endogenous YY1. The indicated proteins were expressed in COS-7 cells, immunoprecipitated (Ip) with anti-YY1 antibody, and immunoblotted (Ib) for Smad4 with anti-Myc antibody (top panel) or with the same anti-YY1 antibody (middle panel). The lower panel shows a total-extract immunoblot with anti-Myc for Smad4. (B) Coimmunoprecipitation of endogenous Smad4 and YY1. HaCaT total cell extracts were collected after stimulation (or not) with 10 ng of TGF-β1 per ml for 1 h, immunoprecipitated without or with specific anti-YY1 antibody, and immunoblotted with anti-Smad4 antibody (top panel). Total extracts were immunoblotted with the same anti-Smad4 antibody (middle panel) or with anti-YY1 antibody (bottom panel). (C) Coimmunoprecipitation of endogenous nuclear Smad4 and YY1. HaCaT nuclear extracts were collected after stimulation (or not) with 5 ng of TGF-β1 per ml for 2 h, immunoprecipitated (Ip) with specific anti-Smad4 antibody, and immunoblotted (Ib) with anti-YY1 antibody (top panel) or the same anti-Smad4 antibody (third panel). Immunoprecipitation with a control matched preimmune (PI) rabbit serum followed by anti-YY1 immunoblotting is shown in the second panel. Nuclear extracts were directly immunoblotted with the same anti-YY1 (fourth panel), anti-Smad4 (fifth panel), or anti-phospho-Smad2 (sixth panel) antibodies.
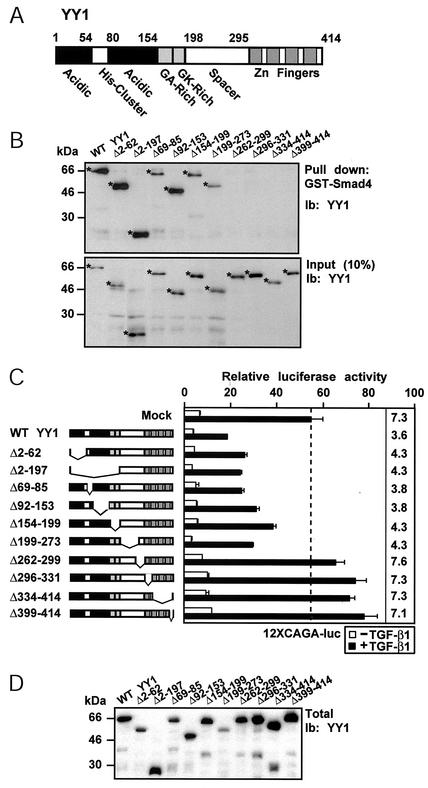
The zinc finger domain of YY1 interacts with Smad4 and is responsible for repression of Smad-dependent transcriptional activity.
YY1 consists of many distinct subdomains (Fig. 5A). The four C-terminal zinc fingers bind to DNA, whereas the N-terminal acidic domains are required for the transcriptional activity of YY1; most transcriptional partners of YY1 interact either with the zinc fingers or with the two glycine-rich subdomains (GA and GK in Fig. 5A) (52). Using a panel of internal deletion mutants of YY1 overexpressed in COS-7 cells, we performed interaction assays with Smad4 fused to GST (Fig. 5B). All deletions that span the zinc finger domain abolished the interaction of YY1 with Smad4 (Fig. 5B, Pull down), despite the proper level of expression of these mutants (Fig. 5B, Input).
FIG. 5.
The zinc finger domain of YY1 is critical for repression of Smad activity. (A) Schematic representation of the domain structure of YY1 (GA- and GK-rich, Gly/Ala- and Gly/Lys-rich domains, respectively). (B) Interaction assay of wild-type (WT) and various deletion mutants of YY1 overexpressed in COS-7 cells with semipurified GST-Smad4 fusion protein and immunoblot (Ib) detection with anti-YY1 antibody. The deletions of human YY1 amino acids are shown above each lane. The efficiency of expression and immunodetetection of YY1 proteins by immunoblotting of total lysate is shown (Input). Asterisks mark the specific YY1 protein bands. (C) Signaling assays in Mv1Lu cells with the 12xCAGA-luciferase reporter and the same panel of YY1 deletion mutants. The schematic representation of YY1 deletion mutants uses the domain conventions of panel A. Data are presented as in Fig. 1D. Basal and TGF-β-induced reporter levels are shown by open and filled bars, respectively. The dashed line indicates the TGF-β1-induced (overnight with 5 ng of TGF-β1 per ml) reporter level, and the numbers at the right indicate the respective fold induction by TGF-β1. (D) Expression levels of the YY1 deletion mutants used for panel C from duplicate transfections of COS-7 cells and immunoblot (Ib) analysis with anti-YY1 antibody.
To test the functional importance of the YY1 zinc fingers in mediating repression of Smad transcriptional activity, we examined the effect of the same YY1 deletion mutants on TGF-β-dependent induction of the 12xCAGA synthetic promoter in Mv1Lu cells (Fig. 5C). Deletion of the C-terminal zinc fingers or the juxtaposed spacer subdomain abolished repression by YY1 (mutants Δ399-414, Δ334-414, Δ296-331, and Δ262-299), which is consistent with the protein-protein interaction data of Fig. 5B. All other internal deletion mutants that retained their zinc finger domain were able to repress the induction of the 12xCAGA promoter by TGF-β1. The wild-type and truncated forms of YY1 were expressed at comparable levels (Fig. 5D). In conclusion, the combined data of interaction mapping and transcriptional repression emphasize the role of the YY1 zinc fingers as determinants of the inhibitory action of YY1 on TGF-β signaling.
YY1 knockdown renders cells more sensitive to TGF-β and BMP signaling.
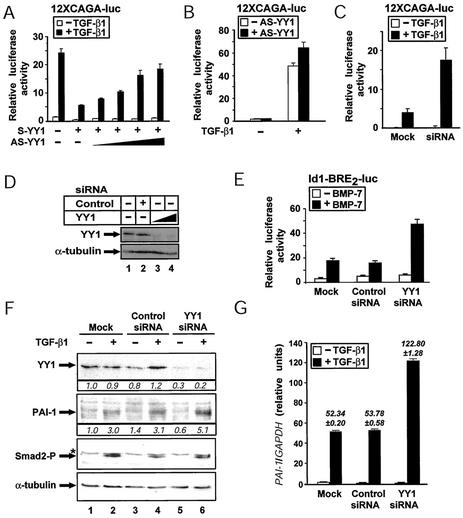
Since YY1 blocks specific TGF-β and BMP transcriptional responses when overexpressed, it was possible that endogenous YY1 could limit the cellular response to TGF-β or BMP. In order to examine this hypothesis, we constructed a full-length antisense YY1 expression vector. Such a vector was previously shown to alleviate the repressive effect of YY1 on cellular and viral gene promoters (6, 35). We tested the ability of the antisense YY1 construct to affect TGF-β-specific responses. In transient-transfection experiments with Mv1Lu cells, increasing doses of antisense YY1 could restore TGF-β-induced transcription from the 12xCAGA synthetic gene that was repressed by sense YY1 (Fig. 6A). Furthermore, in the presence of antisense YY1, the 12xCAGA reporter was more sensitive to stimulation by TGF-β1 (Fig. 6B). We conclude that the observed repression mechanism of YY1 on various TGF-β superfamily gene responses is specific to this transcription factor and that the antisense construct is able to derepress TGF-β-dependent responses.
FIG. 6.
Endogenous YY1 restricts TGF-β and BMP signaling levels. (A and B) Antisense YY1 reverses the inhibitory effect of YY1 and increases the TGF-β1 responsiveness of a Smad-dependent gene promoter in Mv1Lu cells. (A) Induction of the 12xCAGA reporter by overnight stimulation with 0.5 ng of TGF-β1 per ml in the presence of a constant amount (0.5 μg) of sense YY1 (S-YY1) and increasing amounts (0.03, 0.1, 0.3, and 1.0 μg) of antisense YY1 (AS-YY1) plasmids. (B) Increased responsiveness of the 12xCAGA reporter to TGF-β1 (0.5 ng/ml) in the absence (empty bars) or presence (full bars) of antisense YY1 (0.3 μg DNA). (C) RNAi against endogenous YY1 dramatically enhances the inducibility of the 12xCAGA-luciferase reporter by overnight stimulation with 0.5 ng of TGF-β1 per ml of HaCaT cells transfected with mock or YY1 siRNA oligonucleotides. (D) RNAi against YY1 efficiently decreases endogenous YY1 protein levels in C2C12 cells. Immunoblotting with the indicated antibodies of cell extracts from C2C12 cells transfected with no (−), an unrelated (control), or different doses of (triangle) specific YY1 siRNA oligonucleotides. (E) RNAi against YY1 dramatically enhances the inducibility of the Id1-BRE2-luciferase reporter by overnight stimulation with 300 ng of BMP-7 per ml in C2C12 cells transfected with mock, control siRNA or YY1 siRNA oligonucleotides. (F) RNAi against endogenous YY1 enhances the TGF-β1-inducible levels of endogenous PAI-1 protein. Immunoblotting of HaCaT cell extracts transfected with no (mock), control siRNA, or specific anti-YY1 siRNA oligonucleotides and treated (+) or not (−) with 2.5 ng of TGF-β1 per ml for 4 h with the indicated antibodies is shown. The asterisk marks nonspecific protein bands. The numbers below the top two panels indicate densitometric values of the specific bands normalized over the corresponding α-tubulin bands and expressed relative to the control band of lane 1, which is set to 1. (G) RNAi against endogenous YY1 enhances the inducibility of the endogenous PAI-1 gene by TGF-β1. Real-time PCR analysis of human PAI-1 mRNA from HaCaT cells transfected with no (mock), control siRNA, or specific anti-YY1 siRNA oligonucleotides and treated (+) or not (−) with 5 ng of TGF-β1 per ml for 16 h is shown. The bar graph plots average expression values and standard deviations of PAI-1 mRNA levels normalized to control GAPDH mRNA levels. The data in panels A, B, C, and E are presented as in Fig. 1D.
However, the antisense YY1 data reveal relatively weak efficiency in YY1 neutralization (Fig. 6A and B). This is in agreement with weak downregulation of endogenous YY1 protein levels (data not shown). In order to downregulate the expression of endogenous YY1 in a more efficient way, we relied on siRNA technology (14, 19). For this purpose, we designed a double-stranded RNA oligonucleotide which contains a sequence unique to the YY1 cDNA that is 100% conserved between the Xenopus, mouse, and human species. Transfection of the YY1 siRNA in C2C12 (Fig. 6D, lanes 3 and 4), HaCaT (Fig. 6F, lanes 5 and 6), and Mv1Lu (data not shown) cells, led to potent (three- to fivefold) downregulation of the endogenous YY1 protein levels. This effect was specific to the YY1 siRNA and was not observed with an independent siRNA against an unrelated gene (Fig. 6D, lane 2, and F, lanes 3 and 4). Functional assays with the TGF-β Smad-specific promoter 12xCAGA in HaCaT cells demonstrated that knockdown of the YY1 gene by siRNA resulted in a fourfold enhancement of the TGF-β1 response (Fig. 6C). A similar result was obtained when YY1 was knocked down in C2C12 cells that were stimulated with BMP-7 (Fig. 6E). The BMP Smad-specific promoter Id1-BRE2 became hypersensitive (threefold) to BMP-7 treatment under conditions of YY1 knockdown. Furthermore, when HaCaT cells transiently transfected with the YY1 siRNA were stimulated with TGF-β1, they exhibited a strong induction (fivefold) of the endogenous PAI-1 protein expression, which was significantly higher than that observed in mock-transfected cells or in cells transfected with the control siRNA (threefold) (Fig. 6F). The YY1 siRNA did not result in nonspecific downregulation of phosphorylated Smad2 levels in response to TGF-β1 or downregulation of the α-tubulin levels of the transfected cells. To verify that the observed effects on endogenous PAI-1 protein reflected regulation at the mRNA level, we repeated the siRNA transfections in HaCaT cells and monitored human PAI-1 mRNA levels by using real-time quantitative PCR assays (Fig. 6G). In this instance we also observed the same dramatic effect of YY1 knockdown on the inducibility of the PAI-1 gene by TGF-β1. The effect measured in this siRNA experiment was also YY1 specific, as mock transfection or transfection of an unrelated control siRNA did not exhibit any measurable effects on PAI-1 mRNA levels. Thus, comparison of the parallel protein (Fig. 6F) and mRNA (Fig. 6G) assays firmly establishes that endogenous YY1 knockdown potentiates the response of the PAI gene to TGF-β1 by 2.3-fold (mRNA level) and 1.6-fold (protein level). The combined data from antisense YY1 and siRNA analyses suggest that endogenous YY1 limits the capacity of various cell types to respond physiologically to TGF-β and BMP signaling pathways. Thus, YY1 may regulate threshold levels of signal transduction mediated by the Smads, presumably by associating with the endogenous nuclear pool of Smad4, and this affects specific gene expression but possibly also broader cellular responses.
YY1 blocks cellular differentiation pathways downstream of TGF-β and BMP.
We then investigated the potential role of YY1 in Smad-dependent cellular differentiation responses to TGF-β and BMP by using two well-established cell models: the EMT of mammary epithelial NMuMG cells in response to TGF-β1 (Fig. 7A) and the differentiation of myoblastic C2C12 cells to preosteoblasts in response to BMP-7 (Fig. 7B) (18, 41). NMuMG cells infected with control GFP adenovirus exhibited a robust EMT in response to TGF-β1, which is characterized by a reorganization of the actin cytoskeleton from a cortical pattern to intense stress fibers (Fig. 7A, top panels), as previously established (34, 41). Ectopic expression of YY1 resulted in reversion of the TGF-β effect, while it did not perturb the epithelial character of non-TGF-β-treated NMuMG cells. We also confirmed the inhibitory effect of YY1 against the constitutively active receptor ALK-5, which also, when provided via adenoviral transduction, leads to EMT of NMuMG cells (Fig. 7A, bottom panels). The YY1 effect thus resembles that of the inhibitory Smad7, which fully blocks the TGF-β-mediated EMT (Fig. 7A, bottom panels).
FIG. 7.
Ectopic expression of YY1 inhibits TGF-β- and BMP-induced transdifferentiation. (A) Top panels, YY1 inhibits mammary NMuMG cell EMT induced by TGF-β1. NMuMG cells were infected with Ad-GFP (MOI, 25) or Ad-YY1 (MOI, 10) adenovirus, treated for 24 h with vehicle (control) or with 2.5 ng of TGF-β1 per ml, and subjected to actin direct fluorescence microscopy. Bottom panels, YY1 inhibits NMuMG EMT induced by the TGF-β type I receptor. EMT induced by caALK-5 receptor (MOI, 60) is blocked by coinfection with the YY1 virus at an MOI of 20 (Ad-YY1) or with the Smad7 virus at an MOI of 20 (Ad-Smad7). Bar, 20 μm. (B) YY1 blocks BMP-7-induced preosteoblast differentiation of C2C12 myoblasts. In situ ALP staining of C2C12 cells treated with vehicle (control) or 300 ng of BMP-7 per ml for 3 days after infection with Ad-GFP (MOI, 20) or Ad-YY1 (MOI, 4) virus is shown. Bar, 80 μm.
In the C2C12 cell model, BMP-7 treatment of cells infected with the GFP adenovirus resulted in differentiation to preosteoblasts with intense ALP (an early differentiation marker) staining (Fig. 7B), much like in control uninfected cell cultures (data not shown). Infection of C2C12 cells with the YY1 adenovirus fully inhibited the differentiation to ALP-positive cells. We therefore conclude that YY1 efficiently blocks two separate differentiation responses elicited by TGF-β and BMP, i.e., the EMT and preosteoblastic differentiation, respectively.
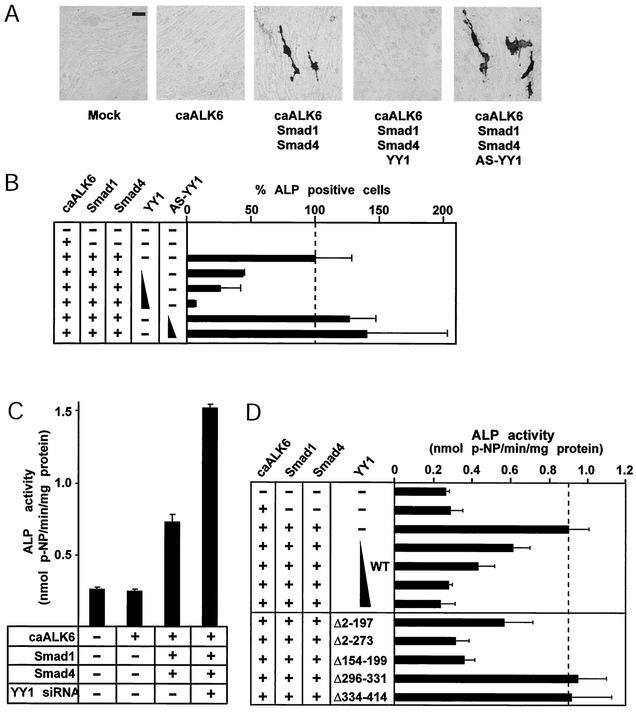
Smad-induced osteogenic differentiation is repressed by ectopic YY1 and enhanced by knockdown of YY1.
In order to confirm that the YY1-mediated block of preosteoblastic differentiation of C2C12 cells involves the Smad pathway, we set up conditions of differentiation that depend exclusively on the transient expression of exogenous Smads (Fig. 8). Constitutively active BMP type IB receptor (ALK-6) failed to induce preosteoblastic differentiation due to the low signal achieved by transient expression of this receptor. However, when the receptor was combined with Smad1 and Smad4, significant numbers of ALP-positive cells were observed (Fig. 8A). Cotransfection of YY1 with the Smads and the activated receptor resulted in a dose-dependent decrease of ALP-positive cells (Fig. 8A and B) and ALP enzymatic activity (Fig. 8D). Thus, YY1 overexpression inhibits the Smad-mediated induction of preosteoblastic differentiation.
FIG. 8.
Smad-dependent myoblast-to-preosteoblast differentiation is regulated by YY1. (A) ALP staining of C2C12 cells transiently transfected with the indicated plasmids for caALK-6, Smad1, Smad4, and sense and antisense (AS) YY1. Representative photomicrographs are shown. Bar, 80 μm. (B) Bar graph of the C2C12 differentiation data from panel A, expressing the relative number of ALP-positive cells compared to the total number of cells counted in the whole transfected culture. Values are averages from duplicate cell cultures with standard errors. The percentage of ALP-positive cells detected after cotransfection with caALK-6, Smad1, and Smad4 was set at 100% (dashed line), relative to which all other conditions are plotted. Triangles represent 0.01, 0.1, and 0.3 μg of transfected YY1 and 0.1 and 0.3 μg of AS-YY1 plasmid DNA. (C) RNAi against endogenous YY1 enhances the preosteoblastic differentiation of C2C12 cells in response to BMP Smads. ALP enzymatic activity measured in cell extracts of transfected C2C12 cells with the indicated expression vectors in the absence (−) or presence (+) of YY1 siRNA oligonucleotides is shown. Average values of triplicate transfected-extract measurements with standard errors are expressed as nanomoles of released p-nitrophenol (p-NP) per minute of reaction per milligram of total protein in the extract. (D) C2C12 preosteoblastic differentiation is not altered by YY1 mutants that fail to interact with Smads. ALP enzymatic activity measured in transfected C2C12 cells with the indicated expression vectors is shown. The data are expressed as in panel C. The triangle represents 0.03, 0.1, 0.3, and 0.5 μg of transfected YY1 plasmid DNA, and the YY1 deletion mutants (0.1 μg each) are listed as in Fig. 4. The dashed line corresponds to the specific activity of the receptor-plus-Smad condition.
In order to examine whether endogenous YY1 could limit the differentiation response to BMP-7, we used the antisense YY1 expression vector and siRNA against YY1 (Fig. 8B and C). When antisense YY1 was expressed in C2C12 cells, the efficiency of ALP-positive C2C12 cell differentiation was enhanced (Fig. 8A and B). The effect of endogenous YY1 knockdown on C2C12 osteoblastic differentiation induced by BMP-7 was even more dramatic (Fig. 8C), as the siRNA approach was more efficient in suppressing YY1 expression in this cell line (Fig. 6D). Thus, C2C12 cells became hypersensitive to BMP-7 treatment or BMP receptor and Smad expression under conditions of endogenous YY1 knockdown. We conclude that YY1 plays a role in regulating the threshold levels of BMP signaling that lead to preosteoblastic differentiation.
Finally, in order to examine whether the ability of YY1 to inhibit BMP-induced C2C12 differentiation depended on the specificity of interaction with the Smads, we repeated the C2C12 differentiation assays by using some of the YY1 deletion mutants shown in Fig. 5 (Fig. 8D). Preosteoblastic differentiation of C2C12 cells was repressed by all mutants that interacted with Smad4 (Δ2-197 and Δ154-199) but remained unaltered by those mutants that failed to interact with Smad4 (Δ296-331 and Δ334-414). In addition, mutant Δ2-273, which retains only the zinc finger domain of YY1, also exhibited similar repression against C2C12 differentiation. Thus, for YY1 to block BMP Smad-mediated osteoblastic differentiation of C2C12 cells, the zinc finger domain of YY1 is necessary and sufficient.
Ectopic expression of YY1 fails to interfere with growth inhibition induced by TGF-β or BMP.
Since differentiation responses to TGF-β or BMP are sensitive to the repressor levels of YY1, we wanted to test whether this was the case for the growth response of recipient cells. In most cell types, TGF-β exhibits an antiproliferative response, which is most characteristic in epithelial cells as their cell cycle is arrested in the early G1 phase (33, 51). To examine the effect of YY1 on the growth-inhibitory pathway downstream of TGF-β, we analyzed three independent epithelial cell lines that are potently growth inhibited by TGF-β, i.e., Mv1Lu, HaCaT, and NMuMG cells (Fig. 9A). Under conditions of 70 to 95% adenoviral infection efficiency and high levels of ectopic expression of YY1 (Fig. 9C), YY1 did not significantly affect the growth-inhibitory response to TGF-β1 as determined by incorporation of radiolabeled thymidine. In control experiments, an adenovirus expressing the inhibitory Smad7, which shuts down the TGF-β pathway at the level of receptors and Smads (23, 32, 38), efficiently blocked this response in all cell lines tested (Fig. 9A). It is worth noting that in NMuMG cells, the same dose of the YY1 adenovirus failed to interfere with the growth-inhibitory response to TGF-β1 (Fig. 9A), yet it was competent to modulate transdifferentiation of these cells (Fig. 7A). Furthermore, YY1 did not interfere with the growth response of MDA-MB-468 cells to either TGF-β1 or BMP-7, in the absence or presence of Smad4 (Fig. 9B), indicating that YY1 failed to antagonize the Smad-dependent reduction in DNA replication induced by both TGF-β1 and BMP-7. Finally, the growth-inhibitory response of HaCaT cells to TGF-β1 was not affected by the YY1 siRNA (Fig. 9D), while the same degree of endogenous YY1 knockdown in HaCaT cells effectively enhanced SBE-mediated transcriptional responses (Fig. 6C, F, and G). We conclude that while YY1 can repress the Smad-mediated induction of certain genes by TGF-β and BMP, it fails to interfere with Smad-dependent TGF-β- and BMP-induced inhibition of epithelial cell proliferation.
FIG. 9.
YY1 does not block the growth-inhibitory pathway downstream of TGF-β or BMP. (A) Dose-response growth inhibition assays with TGF-β-sensitive Mv1Lu, NMuMG, and HaCaT cells either not infected (CTL) or infected with Ad-GFP (MOI of 25), Ad-YY1 (MOI of 20), or Ad-Smad7 (MOI of 20) (S7) virus. (B) Dose-response growth inhibition assays with nonresponsive MDA-MB-468 cells infected as for panel A, except that Ad-YY1 was at an MOI of 5 and sensitivity to TGF-β1 and BMP-7 was restored by infection with Ad-Smad4 (MOI of 40) (S4). In panels A and B, the percentage of radiolabeled thymidine incorporated after TGF-β1 or BMP-7 treatment is graphed in a semilogarithmic plot relative to the level of vehicle-treated cells (set to 100%). (C) Representative immunoblot of endogenous and adenovirally expressed YY1 levels in infected NMuMG cells. An arrow marks the YY1 band, and input viral MOIs are shown above each lane. (D) Endogenous YY1 knockdown by siRNA does not affect growth inhibition by TGF-β1. Thymidine incorporation assays with HaCaT cells transiently transfected with mock or YY1 siRNA and stimulated with TGF-β1 are presented as described above.
YY1 fails to repress the TGF-β-mediated regulation of the p15, p21, and c-myc genes.
In order to explain why YY1 failed to interfere with growth-inhibitory responses, we hypothesized that YY1 may be acting as a repressor of Smad-dependent transcriptional activities in a gene-specific manner. For this reason, we examined the effect of YY1 overexpression on three critical gene targets of TGF-β that mediate cell cycle arrest, i.e., the cell cycle inhibitors p15 and p21 and the proto-oncogene c-myc (51) (Fig. 10). The p15 and p21 genes are induced, whereas c-myc is repressed, by TGF-β1 in a Smad-dependent manner (7, 17, 40). Infection of HaCaT cells with increasing doses of the YY1 adenovirus and RT-PCR analysis of endogenous p21 mRNA levels showed the same degree of p21 inducibility by TGF-β1 as in cells infected with the GFP adenovirus (Fig. 10A). The same result was obtained when endogenous p21 protein levels in response to TGF-β1 were measured in HaCaT cells (Fig. 10B). The inability of YY1 to repress expression of the p15 gene in HaCaT cells was also confirmed (data not shown). Furthermore, analysis of p21 and p15 promoter activities in response to TGF-β1 indicated a two- to threefold induction, respectively (Fig. 10C and D). YY1 coexpression increased the basal activities of both promoters, and the responsiveness to TGF-β1 remained intact (p21) or was weakly reduced (p15). When endogenous c-myc mRNA was analyzed in the same experiments, we observed downregulation of c-myc expression induced by TGF-β1, which was even more enhanced in the presence of ectopic YY1 (Fig. 10E). Analysis of the c-myc promoter activity in response to TGF-β1 indicated the expected repression, and YY1 overexpression did not alter the response to TGF-β1 in any measurable manner while significantly increasing the basal promoter levels (Fig. 10F). The combined data for p15, p21, and c-myc expression and promoter analysis indicate that the responsiveness of these three genes to TGF-β cannot be neutralized by the repressor YY1, unlike the case for the PAI-1 and Id-1 genes. Based on the critical roles that c-Myc, p15, and p21 play in eliciting cell cycle arrest of epithelial cells in response to TGF-β, we suggest that these genes may explain why YY1 fails to interfere with the growth-inhibitory response to TGF-β.
FIG. 10.
YY1 does not block the regulation of p15, p21, and c-myc gene expression by TGF-β. (A) TGF-β1 induces p21 mRNA accumulation in the presence of high exogenous YY1 levels. Semiquantitative RT-PCR assays were performed with mRNA isolated from HaCaT cells infected with adenoviruses expressing GFP (Ad-GFP) (MOI of 200) or YY1 (Ad-YY1) (MOIs of 20 and 100) and treated overnight with vehicle (−) or 2.5 ng of TGF-β1 per ml (+). p21-specific PCR is shown along with GAPDH-specific PCR, which serves as a control. Reactions performed in the absence of reverse transcriptase (−RT) or in the presence of water (H2O) instead of cDNA are included. An immunoblot (Ib) of the infected cells is also shown, demonstrating the dose-dependent levels of YY1 overexpression. (B) p21 protein induction by TGF-β1 is not altered by YY1 overexpression. Immunoblot analysis of HaCaT cells infected with the indicated MOI of adenoviruses, treated overnight with vehicle (−) or with 5 ng of TGF-β1 per ml and analyzed with the indicated antibodies is shown. (C) YY1 does not repress the p21 promoter. p21(−143/+8)-luciferase assays were performed in HaCaT cells transiently transfected with pcDNA3 (−) or pcDNA3-YY1 (triangle; 250, 500, and 750 ng) and treated with vehicle (−) or 2.5 ng of TGF-β1 per ml (+) for 20 h. Data are represented as in Fig. 1D, and the fold induction by TGF-β1 is indicated above the brackets. (D) YY1 does not repress the p15 promoter. p15-luciferase assays were performed in HaCaT cells transiently transfected with pcDNA3 (−) or pcDNA3-YY1 (triangle; 250, 500, and 750 ng) and treated with vehicle (−) or 5 ng of TGF-β1 per ml (+) for 20 h. Data are represented as in Fig. 1D, and the fold induction by TGF-β1 is indicated above the brackets. (E) YY1 does not interfere with c-myc repression by TGF-β1. Semiquantitative RT-PCR assays were performed with mRNA isolated from HaCaT cells infected with adenoviruses expressing GFP (MOI of 200) or YY1 (MOI of 100) and treated overnight with vehicle (−) or 5 ng of TGF-β1 per ml. c-myc- and control GAPDH-specific PCRs are shown. A YY1 immunoblot in HaCaT transfectants duplicate to those used for the RT-PCR assays is also shown. (F) YY1 does not repress the c-myc promoter. c-Myc (pHX)-luciferase assays were performed in HaCaT cells transiently transfected with pcDNA3 (−) or pcDNA3-YY1 (+) and treated with vehicle (−) or 5 ng of TGF-β1 per ml (+) for 20 h. Data are represented as in Fig. 1D, and the fold repression by TGF-β1 is indicated above the brackets. A YY1 immunoblot in HaCaT transfectants duplicate to those used for the luciferase assays is also shown.
DISCUSSION
Among the various transcription factors that associate with Smads, a small group, the proto-oncogenes c-ski and sno-N, exhibit broad negative regulation of most TGF-β superfamily responses (30). Another group of Smad-interacting corepressors, TGIF and Tob, selectively inactivate the TGF-β and BMP pathways, respectively (55, 59). Here we report the first repressor of TGF-β- and BMP-specific Smad pathways that exhibits functional and target gene selectivity.
YY1 can repress specific gene targets of both TGF-β and BMP pathways that depend on Smad binding to SBE-like DNA, such as PAI-1 and Id-1 (Fig. 1). Both of these genes contain multiple SBEs scattered in their promoter regions, many of which contribute to the maximal induction of the genes by the TGF-β/BMP/Smad signal (9, 24). Thus, multiple Smad complexes with the promoter DNA may be critical for proper gene induction, and YY1 appears to be able to interfere with this Smad-dependent transcriptional mechanism. On the other hand, YY1 does not affect the responsiveness of genes such as p15, p21, and c-myc to TGF-β (Fig. 10). For the Smad-mediated regulation of these three promoters, Smads need to form complexes with other transcription factors, such as Sp1, Miz-1, or p107/E2F4/5, that associate with the responsive sites on the promoter (7, 17, 40, 45). Neither the p15, the p21, nor the c-myc gene contains multiple SBEs scattered in the responsive areas of its promoter. Furthermore, the p21 promoter can be induced by Smad3 and Smad4 mutants that are defective in SBE binding (40). Consequently, YY1 cannot effectively block the Smad-dependent regulation of these genes.
Alternatively, the repressive effect of YY1 may depend on its ability to be recruited to the target promoter independent of Smad proteins. Among the promoters we have analyzed, only c-myc has been previously reported to be regulated by YY1 (43), and a number of observations favor a model of YY1 interference with the Smad pathway via a protein-protein-dependent mechanism. In vitro EMSA with recombinant Smad and YY1 proteins showed an inhibitory activity for YY1 against Smad-SBE binding (Fig. 2C). This occurred in the absence of YY1 binding to the same DNA sequences (Fig. 2B). Furthermore, this effect mimicked rather well the in vivo inhibition of Smad-SBE binding (Fig. 2D) or Smad-PAI-1 chromatin recruitment (Fig. 2E), suggesting that nuclear complexes between Smads and YY1 may be sufficient for repression of Smad-dependent and SBE-containing gene promoters. Finally, YY1 and Smads interact via their respective DNA-binding domains (Fig. 3 and 5), and thus it is possible that the YY1-Smad complexes are distinct from YY1-DNA complexes at other genomic sites. Since in this study, only a limited set of responsive genes and promoters was tested, a large-scale analysis of gene expression downstream of TGF-β and BMP signaling in the presence of YY1 is an important step to undertake in the future. This may reveal additional and more complex mechanisms of cross talk between these two protein factors.
YY1 interacts with the conserved N-terminal MH1 domain of Smad4, more weakly with the MH1 domains of Smad1 and Smad3, and even more weakly with Smad2 (Fig. 3). This selective interaction with Smad4 distinguishes YY1 from all other known repressors of Smads, which preferably associate with the C-terminal MH2 domain or the MH1 domain of an R-Smad (4). The MH1 domain provides specificity for DNA binding and nuclear localization to the Smads (25, 26, 46). In agreement with these conclusions, Smad2, whose MH1 domain cannot associate with DNA or with importins (25, 56), interacts with YY1 very weakly (Fig. 3). The selectivity for the MH1 domain is of functional importance, as all presently known Smad-interacting transcription factors that bind to the MH1 domain are DNA-binding factors (23). YY1 is also a DNA-binding protein (47), yet it can repress Smad transcriptional activity without binding to DNA itself (Fig. 2B and C). Rather, it appears that the DNA-binding domain of YY1, which is also known to be important for its transcriptional repressor activity, is responsible for the negative regulation of the Smad pathway through conformational changes that interfere with Smad binding to DNA (Fig. 2). Since this domain of YY1 is multifunctional and is also regulated by alternating acetylation-deacetylation (58), it is possible that more complex mechanisms may govern the repressive effect of YY1 on the Smad pathway. Efforts to correlate the repression mediated by YY1 on Smads via inhibition of coactivators of the CBP family or via recruitment of histone deacetylases were not successful (data not shown).
The apparent specificity of YY1 in antagonizing cellular differentiation (Fig. 7 and 8) but not proliferative responses (Fig. 9) to TGF-β family ligands correlates with the selectivity that YY1 exhibits by repressing only specific gene targets of the Smad pathway (Fig. 1, 2, and 10). This hypothesis is consistent with the complex transcriptional functions of YY1 that depend on the promoter and cellular context. The distinct antagonism that YY1 exhibits towards cell differentiation regulated by TGF-β or BMP is consistent with the previously described involvement of YY1 in cellular differentiation (47). A novel action of YY1 in EMT and preosteoblastic differentiation is established here, in addition to the role of YY1 as a negative regulator of myocyte differentiation (52). The negative effect of YY1 against TGF-β-induced EMT (Fig. 7A) and the neutral effect of YY1 on cell growth (Fig. 9) suggest that YY1 may act as an antitumor agent with respect to the action of TGF-β in cancer progression. TGF-β is known to act as a tumor suppressor due to its growth-inhibitory effects on many cell types but also as an enhancer of tumor progression based on its ability to induce EMT, tumor cell invasiveness, immune cell suppression, and angiogenesis (10, 37). YY1 interferes with one of the protumorigenic actions of TGF-β, EMT, yet it leaves a tumor-suppressing action (growth inhibition) intact, which would make it a suitable therapeutic target in cancer cases with a strong contribution of the TGF-β pathway.
It is attractive to suggest that during TGF-β or BMP signaling, the activity of YY1 is regulated negatively to allow efficient activation of gene targets that lead to cell differentiation. However, we have so far been unable to obtain convincing evidence that TGF-β superfamily signaling leads to regulation of YY1 protein levels (data not shown). The antisense and siRNA experiments provide evidence that YY1 may define threshold levels of TGF-β or BMP signaling (Fig. 6 and 8). The association of YY1 with specific nuclear Smad complexes, and in particular the nuclear Smad4 pool, may offer the cell a mechanism to titrate TGF-β or BMP signaling towards specific gene targets. According to this model, Smad4 plays a crucial role in defining the strength and/or length of the nuclear signal induced by TGF-β family members.
In summary, the identification of YY1 as a Smad-interacting protein that negatively regulates TGF-β superfamily signal transduction leading to cellular differentiation opens novel ground for future analysis of the complex mechanisms that integrate diverse extracellular signals to choices of cellular fate.
Acknowledgments
We thank O. Korchynskyi for stimulating discussions and for performing the experiments for Fig. 1F and T. Imamura for advice on the DNAP assays. We thank Santa Cruz Biotechnology, K. Sampath, N. Ferrara, F. M. Hoffman, A. Comer, S. Itoh, B. Lüscher, M. Austen, T. D. Gelehrter, X.-F. Wang, G. Mavrothalassitis, J.-M. Gauthier, D. Rifkin, K. Miyazono, B. Vogelstein, and N. Fusenig for valuable reagents.
This research was funded in part by grants from the Human Frontier Science Program (to A.M.) and the Dutch Cancer Society (to P.T.D.) (NKI 2000-2217) and by a STINT grant from the Swedish Foundation for International Cooperation in Research and Higher Education (to C.-H.H.). U.V. was supported by a postdoctoral fellowship from the French Association pour la Recherche sur le Cancer.
K. Kurisaki, A. Kurisaki, and U. Valcourt contributed equally to this work.
REFERENCES
- 1.Abe, M., J. G. Harpel, C. N. Metz, I. Nunes, D. J. Loskutoff, and D. B. Rifkin. 1994. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276-284. [DOI] [PubMed] [Google Scholar]
- 2.Alliston, T., L. Choy, P. Ducy, G. Karsenty, and R. Derynck. 2001. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 20:2254-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahina, I., T. K. Sampath, I. Nishimura, and P. V. Hauschka. 1993. Human osteogenic protein-1 induces both chondroblastic and osteoblastic differentiation of osteoprogenitor cells derived from newborn rat calvaria. J. Cell Biol. 123:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attisano, L., and J. L. Wrana. 2000. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Austen, M., B. Lüscher, and J. M. Lüscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 6.Breslin, M. B., and W. V. Vedeckis. 1998. The human glucocorticoid receptor promoter upstream sequences contain binding sites for the ubiquitous transcription factor, Yin Yang 1. J. Steroid Biochem. Mol. Biol. 67:369-381. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massagué. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 8.Datta, P. K., M. C. Blake, and H. L. Moses. 2000. Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-β-induced physical and functional interactions between smads and Sp1. J. Biol. Chem. 275:40014-40019. [DOI] [PubMed] [Google Scholar]
- 9.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J.-M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 11.de Winter, J. P., B. A. Roelen, P. ten Dijke, B. van der Burg, and A. J. van den Eijnden-van Raaij. 1997. DPC4 (SMAD4) mediates transforming growth factor-β1 (TGF-β1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene 14:1891-1899. [DOI] [PubMed] [Google Scholar]
- 12.Donohoe, M. E., X. Zhang, L. McGinnis, J. Biggers, E. Li, and Y. Shi. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dotsch, J., A. Harmjanz, H. Christiansen, J. Hanze, F. Lampert, and W. Rascher. 2000. Gene expression of neuronal nitric oxide synthase and adrenomedullin in human neuroblastoma using real-time PCR. Int. J. Cancer 88:172-175. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 16.Ericsson, J., A. Usheva, and P. A. Edwards. 1999. YY1 is a negative regulator of transcription of three sterol regulatory element-binding protein-responsive genes. J. Biol. Chem. 274:14508-14513. [DOI] [PubMed] [Google Scholar]
- 17.Feng, X.-H., X. Lin, and R. Derynck. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-β. EMBO J. 19:5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii, M., K. Takeda, T. Imamura, H. Aoki, T. K. Sampath, S. Enomoto, M. Kawabata, M. Kato, H. Ichijo, and K. Miyazono. 1999. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell 10:3801-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harborth, J., S. M. Elbashir, K. Bechert, T. Tuschl, and K. Weber. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114:4557-4565. [DOI] [PubMed] [Google Scholar]
- 20.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua, X., X. Liu, D. O. Ansari, and H. F. Lodish. 1998. Synergistic cooperation of TFE3 and smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 12:3084-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inman, G. J., F. J. Nicolas, and C. S. Hill. 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell 10:283-294. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, S., F. Itoh, M. J. Goumans, and P. ten Dijke. 2000. Signaling of transforming growth factor-β family members through Smad proteins. Eur. J. Biochem. 267:6954-6967. [DOI] [PubMed] [Google Scholar]
- 24.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277:4883-4891. [DOI] [PubMed] [Google Scholar]
- 25.Kurisaki, A., S. Kose, Y. Yoneda, C.-H. Heldin, and A. Moustakas. 2001. Transforming growth factor-β induces nuclear import of Smad3 in an importin-β1 and Ran-dependent manner. Mol. Biol. Cell 12:1079-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusanagi, K., M. Kawabata, H. K. Mishima, and K. Miyazono. 2001. α-Helix 2 in the amino-terminal mad homology 1 domain is responsible for specific DNA binding of Smad3. J. Biol. Chem. 276:28155-28163. [DOI] [PubMed] [Google Scholar]
- 27.Lee, T. C., Y. Zhang, and R. J. Schwartz. 1994. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9:1047-1052. [PubMed] [Google Scholar]
- 28.Li, J. M., M. A. Nichols, S. Chandrasekharan, Y. Xiong, and X.-F. Wang. 1995. Transforming growth factor β activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J. Biol. Chem. 270:26750-26753. [DOI] [PubMed] [Google Scholar]
- 29.Liu, D., B. L. Black, and R. Derynck. 2001. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 15:2950-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, X., Y. Sun, R. A. Weinberg, and H. F. Lodish. 2001. Ski/Sno and TGF-β signaling. Cytokine Growth Factor Rev. 12:1-8. [DOI] [PubMed] [Google Scholar]
- 31.MacLellan, W. R., T. C. Lee, R. J. Schwartz, and M. D. Schneider. 1994. Transforming growth factor-β response elements of the skeletal α-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J. Biol. Chem. 269:16754-16760. [PubMed] [Google Scholar]
- 32.Massagué, J. 2000. How cells read TGF-β signals. Nat. Rev. Mol. Cell. Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 33.Massagué, J., S. W. Blain, and R. S. Lo. 2000. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 34.Miettinen, P. J., R. Ebner, A. R. Lopez, and R. Derynck. 1994. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127:2021-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montalvo, E. A., M. Cottam, S. Hill, and Y. J. Wang. 1995. YY1 binds to and regulates cis-acting negative elements in the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:4158-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morén, A., S. Itoh, A. Moustakas, P. ten Dijke, and C.-H. Heldin. 2000. Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene 19:4396-4404. [DOI] [PubMed] [Google Scholar]
- 37.Moustakas, A., K. Pardali, A. Gaal, and C.-H. Heldin. 2002. Mechanisms of TGF-β signaling in regulation of cell growth and differentiation. Immunol. Lett. 82:85-91. [DOI] [PubMed] [Google Scholar]
- 38.Moustakas, A., S. Souchelnytskyi, and C.-H. Heldin. 2001. Smad regulation in TGF-β signal transduction. J. Cell Sci. 114:4359-4369. [DOI] [PubMed] [Google Scholar]
- 39.Nishihara, A., J. Hanai, T. Imamura, K. Miyazono, and M. Kawabata. 1999. E1A inhibits transforming growth factor-β signaling through binding to Smad proteins. J. Biol. Chem. 274:28716-28723. [DOI] [PubMed] [Google Scholar]
- 40.Pardali, K., A. Kurisaki, A. Morén, P. ten Dijke, D. Kardassis, and A. Moustakas. 2000. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-β. J. Biol. Chem. 275:29244-29256. [DOI] [PubMed] [Google Scholar]
- 41.Piek, E., A. Moustakas, A. Kurisaki, C.-H. Heldin, and P. ten Dijke. 1999. TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 112:4557-4568. [DOI] [PubMed] [Google Scholar]
- 42.Pierreux, C. E., F. J. Nicolas, and C. S. Hill. 2000. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20:9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riggs, K. J., S. Saleque, K. K. Wong, K. T. Merrell, J. S. Lee, Y. Shi, and K. Calame. 1993. Yin-yang 1 activates the c-myc promoter. Mol. Cell Biol. 13:7487-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schonherr, E., B. Levkau, L. Schaefer, H. Kresse, and K. Walsh. 2001. Decorin-mediated signal transduction in endothelial cells. Involvement of Akt/protein kinase B in up-regulation of p21(WAF1/CIP1) but not p27(KIP1). J. Biol. Chem. 276:40687-40692. [DOI] [PubMed] [Google Scholar]
- 45.Seoane, J., C. Pouponnot, P. Staller, M. Schader, M. Eilers, and J. Massagué. 2001. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400-408. [DOI] [PubMed] [Google Scholar]
- 46.Shi, Y. 2001. Structural insights on Smad function in TGFβ signaling. Bioessays 23:223-232. [DOI] [PubMed] [Google Scholar]
- 47.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed]
- 48.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 49.Song, C. Z., T. E. Siok, and T. D. Gelehrter. 1998. Smad4/DPC4 and Smad3 mediate transforming growth factor-β (TGF-β) signaling through direct binding to a novel TGF-β-responsive element in the human plasminogen activator inhibitor-1 promoter. J. Biol. Chem. 273:29287-29290. [DOI] [PubMed] [Google Scholar]
- 50.Stroschein, S. L., W. Wang, and K. Luo. 1999. Cooperative binding of Smad proteins to two adjacent DNA elements in the plasminogen activator inhibitor-1 promoter mediates transforming growth factor β-induced smad-dependent transcriptional activation. J. Biol. Chem. 274:9431-9441. [DOI] [PubMed] [Google Scholar]
- 51.ten Dijke, P., M. J. Goumans, F. Itoh, and S. Itoh. 2002. Regulation of cell proliferation by Smad proteins. J. Cell Physiol. 191:1-16. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 53.Valcourt, U., J. Gouttenoire, A. Moustakas, D. Herbage, and F. Mallein-Gerin. 2002. Functions of transforming growth factor-β family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J. Biol. Chem. 277:33545-33558. [DOI] [PubMed] [Google Scholar]
- 54.Walowitz, J. L., M. E. Bradley, S. Chen, and T. Lee. 1998. Proteolytic regulation of the zinc finger transcription factor YY1, a repressor of muscle-restricted gene expression. J. Biol. Chem. 273:6656-6661. [DOI] [PubMed] [Google Scholar]
- 55.Wotton, D., R. S. Lo, S. Lee, and J. Massagué. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 56.Yagi, K., D. Goto, T. Hamamoto, S. Takenoshita, M. Kato, and K. Miyazono. 1999. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 274:703-709. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto, N., S. Akiyama, T. Katagiri, M. Namiki, T. Kurokawa, and T. Suda. 1997. Smad1 and Smad5 act downstream of intracellular signalings of BMP-2 that inhibits myogenic differentiation and induces osteoblast differentiation in C2C12 myoblasts. Biochem. Biophys. Res. Commun. 238:574-580. [DOI] [PubMed] [Google Scholar]
- 58.Yao, Y. L., W. M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida, Y., S. Tanaka, H. Umemori, O. Minowa, M. Usui, N. Ikematsu, E. Hosoda, T. Imamura, J. Kuno, T. Yamashita, K. Miyazono, M. Noda, T. Noda, and T. Yamamoto. 2000. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103:1085-1097. [DOI] [PubMed] [Google Scholar]