Abstract
We previously reported that expression of the human forkhead/winged helix transcription factor, CHES1 (checkpoint suppressor 1; FOXN3), suppresses sensitivity to DNA damage and restores damage-induced G2/M arrest in checkpoint-deficient strains of Saccharomyces cerevisiae. We find that a functional glutathione S-transferase-Ches1 fusion protein binds in vivo to Sin3, a component of the S. cerevisiae Sin3/Rpd3 histone deacetylase complex. Checkpoint mutant strains with SIN3 deleted show increased resistance to UV irradiation, which is not further enhanced by CHES1 expression. Conversely, overexpression of SIN3 blocks the Ches1-mediated G2/M delay in response to DNA damage, which is consistent with Ches1 acting by inhibiting the Sin3/Rpd3 complex. Deletion of either SIN3 or RPD3 in rad9 or mec1 checkpoint mutant strains suppresses sensitivity to replication blocks and DNA damage resulting from Cdc9 ligase deficiency and UV irradiation. SIN3 or RPD3 deletions also restored G2/M arrest after DNA damage without concomitant Rad53 phosphorylation in mec1 mutant strains. This DNA damage response is absent in mad1 spindle checkpoint mutants. These data suggest that modulation of chromatin structure may regulate checkpoint responses in S. cerevisiae. Inhibition of histone deacetylation results in a DNA damage checkpoint response mediated by the spindle checkpoint pathway that compensates for loss of the primary DNA damage checkpoint pathway.
Eukaryotes contain large networks of chromatin remodeling machinery that coordinately regulate gene transcription (19). Chromatin remodeling complexes are commonly grouped into two classes based on their activity: (i) ATP-dependent chromatin remodeling complexes (45, 49) and (ii) histone acetylase and histone deacetylase (HDAC) complexes (23, 43). The second class is made up of protein complexes that modulate the status of histone acetylation, which occurs posttranslationally at lysine residues located at the amino-terminal ends of histones. Acetylation of the lysine residues neutralizes their positive charge, which interferes with the histone-DNA electrostatic interaction (15) and thereby loosens chromatin structure, allowing for a more transcriptionally competent DNA template. Conversely, removal of acetyl groups by HDAC activity restores the positive charge on the histone lysine residue and thereby reestablishes a tight interaction between the histone and DNA. Increased histone acetylation is generally associated with transcriptional activity, whereas decreased acetylation is associated with transcriptional repression.
Saccharomyces cerevisiae expresses multiple HDACs, which are encoded by SIR2, HOS1, HOS2, HOS3, HDA1, HST1 to -4, and RPD3. Rpd3 is known to associate into a large ≥2-MDa multiprotein complex that includes Sin3 (21). The Sin3/Rpd3 HDAC complex does not directly bind to DNA but interacts with other factors that target the complex to specific genes. For example, Sin3 mediates interactions with site-specific DNA-binding proteins such as Ume6 to target HDAC activity to specific promoter regions (20, 35). Microarray transcription profiles generated from rpd3Δ and sin3Δ mutants indicate that in each mutant a significant number of genes are similarly up- or down-regulated compared to wild type (3). These data, combined with the genetic evidence that Rpd3 and Sin3 regulate genes in a nonadditive manner, indicate that the proteins function together in the same regulatory pathway (41, 47).
Yeast Rpd3 most closely resembles enzymes in the human class I HDAC category, including HDAC1, HDAC2, HDAC3, and HDAC8 (5, 9, 17, 46). Human HDACs are also directed to specific promoter targets through association with an assortment of proteins, including the human homolog of the yeast Sin3 protein. These complexes in turn interact with other proteins that include several tumor suppressor genes and oncogenes involved in cell cycle progression and cancer development (9, 50). For example, HDAC1, -2, and -3 are thought to be important for regulating the G1/S transition in human cells through interaction with Rb, a tumor suppressor gene that represses transcription of E2F-regulated genes. The relationship between HDACs and Rb, along with other examples of HDAC influence on cell proliferation, clearly demonstrates the significance of histone deacetylation in the regulation of the cell cycle in human cells.
Eukaryotes also possess well-conserved pathways to monitor genomic stability and respond to DNA damage. These pathways are termed checkpoints and function to delay transitions between the different cell cycle phases in response to damaged DNA and defects in replication and chromosome segregation (14, 53). S. cerevisiae contains at least three conserved checkpoint pathways that elicit cell cycle arrest in response to genotoxic stress, and these pathways differ in both the genes required for their activity and the stimuli to which they respond. While the DNA damage checkpoint responds to insults caused by DNA-damaging agents, the replication and spindle checkpoints react to problems associated with replication and bipolar spindle-kinetochore attachment, respectively. Though these pathways respond to fundamentally different stimuli, recent studies indicate that checkpoint pathways may overlap in their response to damage (12, 26, 29). It is likely that the machinery participating in the different checkpoint responses must localize to DNA, and this localization is likely to be influenced by chromatin structure. Therefore, chromatin status may also play a key role in coordinating checkpoint responses to genotoxic stress.
We previously reported the identification of a human cDNA encoding a member of the forkhead/winged helix family of transcription factors, CHES1 (checkpoint suppressor 1; FOXN3), which can function as a high-copy suppressor of the S. cerevisiae G2/M checkpoint mutants rad9, mec1, rad24, rad53, and dun1 (33). Specifically, Ches1 confers increased survival after exposure to UV irradiation, ionizing irradiation, and methylmethane sulfonate (MMS). Furthermore, Ches1 is able to reconstitute a DNA damage-induced G2/M arrest absent in checkpoint-deficient strains, even in a mec1Δ mutant strain, which is normally nonviable. This present study was initiated to identify Ches1 in vivo binding partners. We found that a functional glutathione S-transferase (GST)-Ches1 fusion protein interacts with Sin3, a component of the Sin3/Rpd3 HDAC complex in S. cerevisiae. This led to investigation of the requirement for Sin3 in Ches1-mediated responses. Expression of CHES1 in a sin3Δ strain had no additional effect on sensitivity to DNA damage, and overexpression of SIN3 in cells with the RAD9 checkpoint gene deleted attenuated the Ches1-mediated cell cycle delay in response to DNA damage, which suggests that Ches1 functions to inhibit Sin3 activity. Deletion of either SIN3 or its associated HDAC, RPD3, in rad9 and mec1 DNA damage checkpoint mutant strains restores a DNA damage-inducible cell cycle arrest and increases survival after DNA damage. Restoration of the damage-induced arrest in the HDAC-deficient mutants requires an intact spindle checkpoint pathway, which suggests that the spindle checkpoint can act as a redundant pathway in response to DNA damage to delay cell cycle progression.
MATERIALS AND METHODS
Genetic methods, strains, and plasmids.
Yeast strains used in this study are listed in Table 1. Deletion strains were prepared by standard gene replacement techniques (24). Standard procedures were used for transformations, sporulation, and tetrad dissection. All media were prepared as described elsewhere (6).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| AVL78 | MATaleu2 trp1 ura3-52 prb prc pep4-1 | V. Lundblad |
| KS197 | MATa/α cdc9-8/CDC9 rad9::HIS3/RAD9 trp1-1/TRP1 ura3-1/URA3 his3-11,3/HIS3 leu2-3,112/LEU2 ade2-1/ADE2 can1-100/CAN1 | This study |
| KS217 | Same as KS197 except for sin3::KAN/+ | This study |
| KS294 | Same as KS197 except for rpd3::KAN/+ | This study |
| KS250 | MATatrp1-1 ura3-1 his3-11,3 leu2-3,112 ade2-1 can1-100 | This study |
| KS254 | Same as KS250 except for rad9::HIS3 | This study |
| KS253 | Same as KS250 except for sin3::KAN | This study |
| KS305 | Same as KS250 except for rpd3::KAN | This study |
| KS263 | Same as KS250 except for rad9::HIS3 sin3::KAN | This study |
| KS315 | Same as KS250 except for rad9::HIS3 rpd3::KAN | This study |
| KS453 | Same as KS263 except for mad1::TRP1 | This study |
| KS485 | Same as KS315 except for mad1::TRP1 | This study |
| KS481 | Same as KS254 except mad1::TRP1 | This study |
| KS256 | Same as KS250 except for MATα cdc9-8 | This study |
| KS258 | Same as KS250 except for cdc9-8 rad9::HIS3 | This study |
| KS265 | Same as KS250 except for cdc9-8 rad9::HIS3 sin3::KAN | This study |
| KS317 | Same as KS250 except for cdc9-8 rad9::HIS3 rpd3::KAN | This study |
| KS501 | Same as KS317 except for mad1::TRP1 | This study |
| KS563 | Same as KS317 except for bub2::TRP1 | This study |
| Y669 | Same as KS250 except for mec1::HIS3 + pWJ81 (wild-type MEC1) | S. Elledge |
| KS302 | Same as Y669, except pWJ81 is replaced with pBAD070 (RNR1) | This study |
| KS300 | Same as KS302 except for sin3::KAN | This study |
| KS301 | Same as KS302 except for rpd3::KAN | This study |
| KS299 | Same as KS250 except for pBAD070 | This study |
| Y604 | Same as KS250 except for mec1-21 | S. Elledge |
| KS368 | Same as Y604 except for sin3::KAN | This study |
| KS369 | Same as Y604 except for rpd3::KAN | This study |
| KS421 | Same as Y604 except for mad1::TRP1 | This study |
| KS433 | Same as KS368 except for mad1::TRP1 | This study |
| KS427 | Same as KS368 except for bub2::TRP1 | This study |
| KS423 | Same as KS369 except for mad1::TRP1 | This study |
| KS428 | Same as KS369 except for bub2::TRP1 | This study |
The C-terminal portion of CHES1 (GenBank U68723) was obtained from the tx23 clone described by Pati et al. (33), which contains CHES1 cDNA corresponding to amino acid 292 through bp 2213 in the 3′-untranslated region fused in frame with a leader sequence derived from the ADH1 gene containing a methionine start site (ATG TCT ATC CCA GAA ACT CAA AAA GGT GTT ATC TTC TAC GAA GCT TGC). Site-directed PCR mutagenesis was used to create an XhoI site upstream of the ADH1-derived leader sequence and at bp 2214 in the CHES1 3′-untranslated region using primers 5′-AAT ATC TCG AGC TAT ACC AAG CAT AC and 5′-TCA GAA GCC TCG AGG ACT GGT GGC, respectively. This fragment was inserted at the XhoI site in the vector p424-GPD (ATCC 87357) and was named p424-GPD-CHES1. The GST-CHES1 expression construct was created by inserting the C-terminal CHES1-containing XhoI-XhoI fragment from p424-GPD-CHES1 into the XhoI site in the GST expression vector pADH-GST (provided by S. Elledge) to create pADH-GST-CHES1. The 2μm TRP-containing RNR1 expression vector pBAD 070 was a gift from the Elledge lab at Baylor College of Medicine. The CEN URA-containing SIN3 expression vector M1102 and the empty vector M1101 were provided by Z. Nawaz from Baylor College of Medicine.
GST pull-down assay.
Six 2-liter flasks containing 600 ml of selective medium were inoculated with a protease-deficient strain (AVL78) expressing either pADH-GST-CHES1 or pADH-GST empty vector (1.8-liter combined culture volume/strain) and grown to an optical density at 600 nm (OD600) of 1.0 at 30°C while shaking at 220 rpm. Cultures of each type were combined and centrifuged, and resulting pellets were washed twice with 10 ml of 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4; pH 7.3) and once with 10 ml of extraction buffer (50 mM HEPES [pH 7.3], 150 mM KOAc, 10 mM NaF, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 20 mM β-glycerol phosphate, 0.1% Triton X-100, 1× protease inhibitor cocktail [Boehringer Mannheim], and 1× phosphatase inhibitor cocktails I and II [Calbiochem]). Pellets were resuspended as a thick paste in extraction buffer and ground using a mortar and pestle under liquid nitrogen, and the resulting powder was stored at −80°C. The powder extract was thawed and centrifuged for 15 min at 5,000 × g at 4°C, and the resulting supernatant was collected and assayed for protein concentration. A 50% slurry of glutathione-Sepharose 4B (Amersham Pharmacia Biotech) was prepared according to the manufacturer's instructions and added to the crude extracts (5 ml; 20 mg/ml) at a concentration of 150 μl of slurry per 100 mg of protein and incubated for 3 h at 4°C. The Sepharose was subsequently washed with 1× PBS three times for 15 min at 4°C followed by centrifugation for 5 min at 100 × g at 4°C. The Sepharose was transferred to Micro Bio-Spin chromatography columns (Bio-Rad) and washed with 1× PBS three times by gravity flow, followed by resuspension of the beads in Western loading buffer. Samples were boiled for 10 min and centrifuged, and the supernatant was loaded onto a sodium dodecyl sulfate (SDS)-8% polyacrylamide gel for electrophoresis, followed by Coomassie staining using Bio-Safe Coomassie stain (Bio-Rad). Protein bands were sequenced by mass spectrometry at the Protein Chemistry Core Laboratory, Baylor College of Medicine.
cdc9-8 checkpoint assay.
Strains containing the indicated plasmids were grown to log phase in selective medium at 25°C and then diluted with rich medium (5-ml final volume) to a final OD600 of 0.25 and further grown at 25°C in log phase (OD600, ∼0.7), followed by a shift to 37°C for 4 h. Samples (0.5 ml) were briefly sonicated, fixed with 70% ethanol for 1 h at 4°C, and resuspended in TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0). Samples were counted by inspection for the fraction of large-budded cells as previously described (38).
Survival assays. (i) cdc9-8 temperature sensitivity assays
Logarithmically growing cells were fivefold diluted and spotted onto rich or selective medium, followed by incubation at the indicated temperatures.
(ii) UV irradiation.
Logarithmically growing cells were diluted, briefly sonicated, and plated on rich medium. Plates were irradiated at the indicated doses using a Spectronics UV cross-linker, followed by incubation at 30°C for 48 to 72 h. Colonies were counted, and percent survival compared to that in the unirradiated control was determined.
(iii) HU.
Logarithmically growing cells were fivefold diluted and spotted onto rich medium supplemented with either 10 mM or 50 mM hydroxyurea (HU; Sigma) and incubated at 30°C. Colony formation was assessed at 48 to 72 h.
Effect of DNA damage on the release of G2/M-arrested cells.
The effect of UV irradiation on release from G2/M arrest was examined using a modified protocol of Pati et al. (33). Briefly, logarithmically growing cultures were transferred to rich medium and incubated at 30°C while shaking at 220 rpm. Fifteen milliliters of each culture (OD600 ≈ 0.4) was pelleted and resuspended in 5 ml of rich medium containing nocodazole (10 mg/ml in dimethyl sulfoxide; Sigma) to a final concentration of 10 μg/ml. After 2.5 to 3.0 h of incubation with rotation at 30°C, greater than 90% of cells were arrested with large-budded morphology. The cells were pelleted and plated in duplicate, one of which was UV irradiated at a dose of 10 J/m2 and another which served as a mock-irradiated control. The cells were immediately harvested from the plates, washed twice with rich medium, and recovered in 5 ml of rich medium. These cultures were incubated with rotation at 30°C, and 1-ml samples were collected at 20-min intervals. The samples were briefly sonicated, fixed with 70% ethanol for 1 h at 4°C, and resuspended in TE buffer. Samples were counted for the fraction of large-budded cells as previously described (38) and for nuclear morphology by staining with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) as described elsewhere (1).
Rad53 phosphorylation assay.
Logarithmically growing cultures were arrested with nocodazole, and 4 × 108 to 6 × 108 cells were split and plated in duplicate as above, one of which was UV irradiated at a dose of 15 J/m2 and another that served as a mock-irradiated control. The cells were immediately harvested from the plates and recovered in 10 ml of rich medium, followed by incubation at 30°C while shaking at 220 rpm. Ten-milliliter samples were collected at 10 min, pelleted, and immediately frozen under liquid nitrogen and stored at −80°C. Pellets were thawed, and protein extracts for Western analysis were prepared using the trichloroacetic acid method (32). Protein extracts were resolved by electrophoresis on an SDS-8% polyacrylamide gel and transferred to polyvinylidene difluoride blotting membrane (Millipore). Rad53 was detected using an anti-Rad53 polyclonal antibody (provided by S. Elledge; 1:300 dilution in Tris-buffered saline with 0.1% Tween 20 and 10 mg of bovine serum albumin/ml), followed by a peroxidase-labeled donkey anti-rabbit antibody (Amersham Pharmacia Biotech) with visualization by using the ECL chemiluminescence reagent (Amersham Pharmacia Biotech).
RESULTS
Sin3 interacts with a GST-Ches1 fusion protein in vivo.
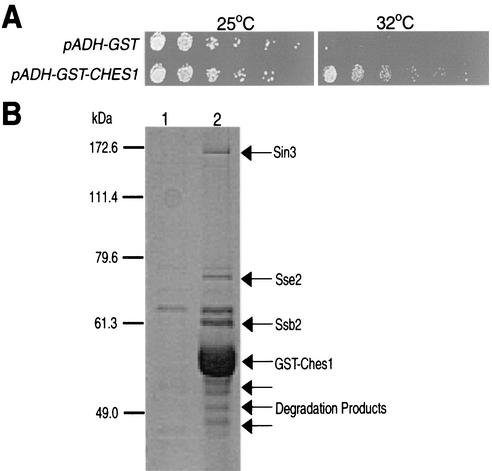
CHES1 was originally identified in a genetic screen for human cDNAs capable of restoring viability at the nonpermissive temperature (32°C) in cells containing the temperature-sensitive allele of DNA ligase (cdc9-8) and a second mutation in the checkpoint genes MEC1 or RAD9. The C-terminal portion of Ches1, not including the forkhead/winged-helix DNA-binding domain, is sufficient for suppression of all checkpoint phenotypes assayed (33). Therefore, we created a construct that directed expression in yeast of C-terminal Ches1 fused to GST in order to identify S. cerevisiae proteins that interact with the Ches1 protein. We tested the functionality of this construct by testing for growth at 32°C of cdc9-8 rad9Δ strains containing either the pADH-GST empty vector or the pADH-GST-CHES1 construct (Fig. 1A). Strains containing the GST vector did not grow at the nonpermissive temperature; however, strains containing the GST-CHES1 construct did grow at 32°C, similar to those containing the original CHES1 construct, demonstrating that the CHES1 fusion protein folds and functions appropriately.
FIG. 1.
Sin3 interacts in vivo with a GST-Ches1 fusion protein. (A) Fivefold serial dilutions of the mutant strain cdc9-8 rad9Δ carrying multicopy plasmids expressing either GST (pADH-GST) or the GST-Ches1 fusion protein (pADH-GST-CHES1) were incubated at either 25 or 32°C. (B) Protein lysates prepared from cells (AVL78) expressing either GST alone (lane 1) or GST-Ches1 (lane 2) were incubated with glutathione-bound Sepharose, and the resulting purified material was separated on an SDS-8% polyacrylamide gel electrophoresis gel and Coomassie stained.
Yeast protein extracts were prepared from cultures of a protease-deficient strain (AVL78) expressing either GST-Ches1 or GST alone, followed by incubation with glutathione-bound Sepharose beads (Fig. 1B). Sepharose-bound proteins were visualized by SDS-polyacrylamide gel electrophoresis and Coomassie staining, and potential Ches1-interacting proteins were isolated and sequenced by mass spectrometry. GST-Ches1 specifically pulled down two members of the Hsp70 chaperonin family, Sse2 and Ssb2 (8), and Sin3, which is a 174.8-kDa protein that is part of a large multiprotein HDAC complex containing Rpd3 (21).
Deletion of SIN3 or RPD3 suppresses rad9Δ DNA damage sensitivity.
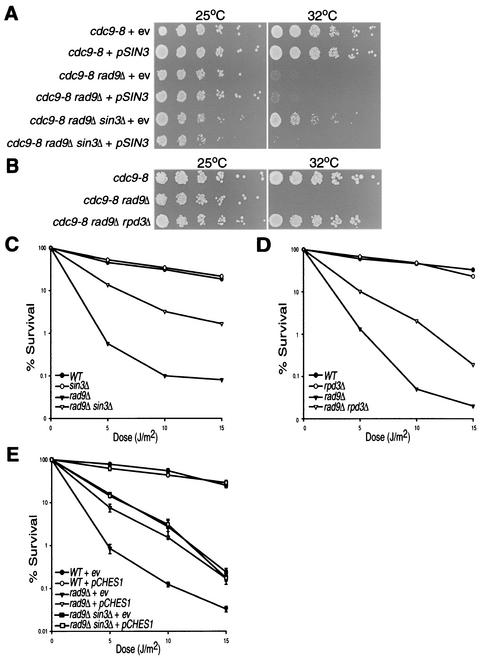
The results of the GST pull-down assays suggested that Sin3 and its related HDAC, Rpd3, may play a role in the response to DNA damage. We created cdc9-8 rad9Δ sin3Δ triple mutant strains transformed with either a low-copy-number SIN3 expression plasmid or empty vector and assayed for growth at 32°C (Fig. 2A). As expected, cdc9-8 rad9Δ cells did not grow at 32°C, compared to cdc9-8 cells with an intact damage checkpoint pathway. Interestingly, deletion of SIN3 allowed growth at 32°C, while the reintroduction of SIN3 on a plasmid complemented the phenotype, thus preventing growth at 32°C. Since Sin3 and Rpd3 are both required for Rpd3-mediated HDAC activity, we also tested the ability of a cdc9-8 rad9Δ rpd3Δ triple mutant strain to grow at the nonpermissive temperature. Similar to the SIN3 deletion, disruption of RPD3 in the cdc9-8 rad9Δ background restored growth at 32°C, compared to cells containing a wild-type RPD3 allele (Fig. 2B).
FIG. 2.
Deletion of SIN3 or RPD3 suppresses cdc9-8 rad9Δ temperature sensitivity and rad9Δ UV sensitivity. (A) Fivefold serial dilutions of cdc9-8 mutant strains containing wild-type or mutant alleles of RAD9 and SIN3 incubated at either 25 or 32°C. Each strain carries low-copy-number plasmids expressing either empty vector (ev) or wild-type Sin3 (pSIN3). (B) Fivefold serial dilutions of cdc9-8 mutant strains containing wild-type or mutant alleles of RAD9 and RPD3 were incubated at either 25 or 32°C. (C to E) Colony survival assays following UV irradiation of yeast containing wild-type or mutant alleles of RAD9 and SIN3 (C), wild-type or mutant alleles of RAD9 and RPD3 (D), or wild-type or mutant alleles of RAD9 and SIN3 (E) transformed with either a high-copy-number CHES1 expression vector, p424-GPD-CHES1 (pCHES1), or the corresponding empty vector, p424-GPD (ev).
To determine whether deletion of SIN3 or RPD3 could compensate for the loss of RAD9 function in response to other forms of DNA damage, we performed colony survival assays to assess whether deletion of either SIN3 or RPD3 could suppress the UV sensitivity of a rad9Δ mutant (Fig. 2C and D). Deletion of SIN3 or RPD3 had no significant effect on UV sensitivity in cells with wild-type RAD9. However, deletion of SIN3 or RPD3 in the rad9Δ background reproducibly suppressed the UV sensitivity phenotype compared to rad9Δ mutants containing wild-type alleles of SIN3 and RPD3.
Ches1 acts through inhibition of the Sin3/Rpd3 complex.
Based on the results of the GST pull-down assay and the HDAC mutants' phenotypes, we hypothesized that Ches1 acted by inhibition of Sin3/Rpd3 activity. To further test this hypothesis we assayed the ability of Ches1 to suppress the DNA damage response in a strain with SIN3 deleted and, conversely, whether overexpression of SIN3 would inhibit Ches1 function.
We performed UV colony survival assays using rad9Δ and rad9Δ sin3Δ mutant strains containing either a CHES1 expression construct or the comparable empty vector (Fig. 2E). As previously reported by Pati et al. (33), expression of CHES1 in rad9Δ cells suppressed their sensitivity to UV-induced damage compared to rad9Δ cells carrying the empty vector. As shown above, deleting SIN3 in the rad9Δ background suppressed UV sensitivity to a similar degree, and expression of CHES1 in a rad9Δ sin3Δ strain did not further enhance survival compared to the rad9Δ sin3Δ strain carrying the empty vector.
We previously reported that expression of CHES1 in checkpoint-deficient yeast strains restored a G2/M delay in response to DNA damage (33). We determined whether overexpression of SIN3 would inhibit this G2/M delay by monitoring the ability of cdc9-8 rad9Δ mutant strains to arrest at G2/M following a 4-h incubation at 37°C when transformed with vectors expressing CHES1 and/or SIN3 (Table 2). A cdc9-8 strain wild type for RAD9 arrested at 83.4%, while only 44.2% of cdc9-8 rad9Δ double mutant cells maintained the large-budded morphology. As expected, expression of CHES1 partially restored G2/M arrest in cdc9-8 rad9Δ cells to 62.0%. Cotransformation of a low-copy expression vector containing SIN3 with the CHES1 expression vector resulted in a decrease in the percentage of large-budded cells to 47.7%, which indicates that increased expression of SIN3 attenuates Ches1's activity. These combined data further support the conclusion that Ches1 acts through inhibition of Sin3/Rpd3 HDAC activity, resulting in increased survival from DNA damage and G2/M arrest in Rad9-deficient strains.
TABLE 2.
Cell cycle arrest of strains containing a temperature-sensitive DNA ligase mutation, cdc9-8, after 4 h at 37°C
| Genotype | Plasmidsa | % Large-budded cellsb |
|---|---|---|
| cdc9-8 RAD9 | p424-GPD (ev), pM1101 (ev) | 83.4 ± 0.6 |
| cdc9-8 rad9Δ | p424-GPD (ev), pM1101 (ev) | 44.2 ± 0.4 |
| cdc9-8 rad9Δ | p424-GPD-CHES1, pM1101 (ev) | 62.0 ± 0.6 |
| cdc9-8 rad9Δ | p424-GPD-CHES1, pM1102 (SIN3) | 47.7 ± 1.7 |
ev, empty vector.
Mean ± standard error of the mean.
Deletion of SIN3 or RPD3 suppresses mec1 mutant strain phenotypes.
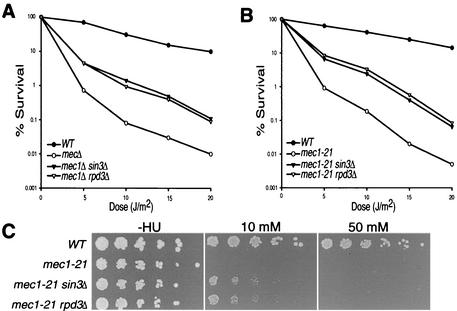
Rad9 mediates the DNA damage response through the Mec1 kinase-dependent checkpoint pathway. Therefore, we tested the effect of deleting SIN3 or RPD3 in two mec1 mutant strain sets: (i) a mec1Δ strain set transformed with a high-copy RNR1 expression vector, which suppresses mec1Δ lethality with no apparent effect on the DNA damage checkpoint response (10, 52); and (ii) a strain set containing the viable, checkpoint-deficient allele mec1-21. As expected, mec1Δ and mec1-21 cells displayed greatly reduced viability after exposure to UV irradiation compared to wild type (Fig. 3A and B). Deletion of SIN3 or RPD3 in both mec1 backgrounds reproducibly suppressed UV sensitivity to a similar degree as that seen in the rad9Δ colony survival experiments.
FIG. 3.
Deletion of SIN3 or RPD3 suppresses mec1 UV and HU sensitivity. (A and B) Colony survival assays of yeast containing wild-type or mutant alleles of SIN3 or RPD3 in combination with MEC1 or mec1Δ (A) and MEC1 or mec1-21 (B) were UV irradiated and colony survival was measured. (C) Fivefold serial dilutions of cells containing wild-type or mutant alleles of MEC1, SIN3, or RPD3 plated on medium supplemented with 0, 10, or 50 mM HU and incubated at 30°C.
In contrast to rad9Δ strains, mec1 mutant strains are also defective in the regulation of the checkpoint response to replication stress (51), which can be elicited through inhibiting ribonucleotide reductase by exposure to HU. We sought to determine whether deletion of SIN3 or RPD3 had any effect on HU sensitivity in a Mec1-deficient mutant background (Fig. 3C). As expected, mec1-21 cells did not grow when plated onto HU-containing medium. However, mec1-21 sin3Δ and mec1-21 rpd3Δ cells displayed growth on medium containing low HU concentrations (10 mM) but not on medium with higher HU concentrations (50 mM). We also observed the same response in cells harboring the mec1Δ allele (data not shown). Thus, deletion of SIN3 or RPD3 moderately suppresses the HU sensitivity exhibited by mec1 mutants.
Deletion of SIN3 or RPD3 restores the DNA damage-induced G2/M delay in checkpoint-deficient strains.
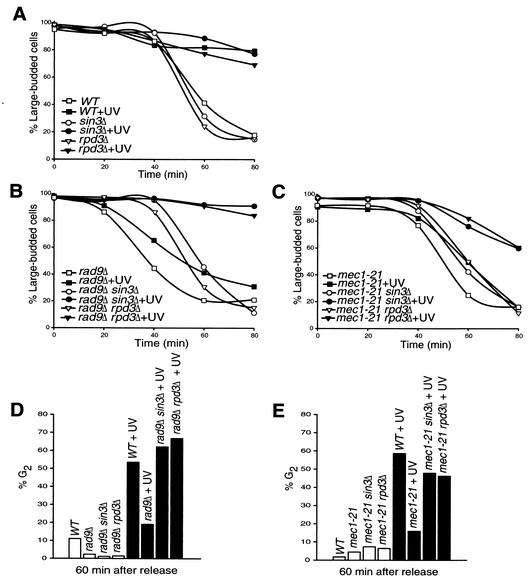
Deletion of SIN3 or RPD3 could suppress sensitivity of checkpoint mutants by making them less prone to DNA damage, by increasing the efficiency of repair, and/or by restoring checkpoint arrest. Therefore, we specifically asked whether deletion of SIN3 or RPD3 restores the G2/M delay of rad9Δ and mec1-21 mutants after damage. Cells containing mutations in either RAD9 or MEC1 combined with wild-type or mutant alleles of SIN3 and RPD3 were synchronized in G2 using nocodazole, UV irradiated (10 J/m2), and then released into nocodazole-free medium to monitor cell cycle progression by morphology and DAPI staining (Fig. 4). In unirradiated cells, deletion of RPD3 or SIN3 had no effect on the kinetics of G2/M release compared to that in wild-type cells. Irradiated wild-type, sin3Δ, and rpd3Δ cells remained arrested in G2/M, as evidenced by retention of large-budded morphology and concentration of DNA at the bud-neck region. As expected, rad9Δ and mec1-21 mutants lost the DNA damage checkpoint response and had similar kinetics of exit from G2/M with or without irradiation; however, rad9Δ mutant cells exited G2/M earlier than all other strains. Interestingly, deletion of either SIN3 or RPD3 in both the rad9Δ and mec1-21 backgrounds reconstituted the UV irradiation-induced G2/M delay to wild-type levels. These data indicate that deletion of SIN3 or RPD3 restores the G2/M delay induced by DNA damage in rad9 and mec1 checkpoint mutants.
FIG. 4.
Deletion of SIN3 or RPD3 restores the DNA damage-induced G2/M delay. Cultures of the indicated strains were nocodazole arrested, mock (open symbols) or UV (filled symbols) irradiated (10 J/m2), and released into rich medium for sample collection at the indicated time points. (A) Assessment of strains wild type or mutant for SIN3 or RPD3 for large-budded morphology. (B and C) Assessment of strains wild type or mutant for SIN3 or RPD3 containing rad9Δ (B) or mec1-21 (C) for large-budded morphology. (D and E) Strains wild type or mutant for SIN3 or RPD3 containing rad9Δ (D) or mec1-21 (E) assessed for DAPI staining 60 min after release from nocodazole arrest.
Loss of SIN3 or RPD3 does not restore checkpoint signaling through Rad53.
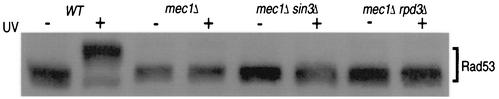
Our results indicate that loss of Sin3 or Rpd3 activity restores the cell cycle delay in response to DNA damage independently of RAD9 and MEC1. This may be due to activation of an alternative checkpoint pathway or to activation of proteins downstream of RAD9 and MEC1. Activation of the Rad9/Mec1 DNA damage checkpoint leads to Rad53 phosphorylation (37); therefore, we assayed whether DNA damage-induced Rad53 phosphorylation is restored in checkpoint-deficient sin3Δ or rpd3Δ mutant strains. We treated cultures as described above by arresting cells in nocodazole, followed by exposure to UV irradiation. Cells were subsequently released into nocodazole-free medium, and samples were collected at 0 (mock-irradiated group) and 10 min. We tested this response in strain backgrounds containing the rad9Δ, mec1-21, or mec1Δ allele. As previously published, rad9Δ and mec1-21 mutants retained some damage-induced Rad53 phosphorylation (reference 37 and data not shown), but this phosphorylation is absent in mec1Δ strains. As expected, Rad53 phosphorylation was visible in wild-type protein extracts at 10 min following release compared to the mock-irradiated sample, and the Rad53 mobility shift was not detected in mec1Δ protein extracts (Fig. 5). Under conditions in which deleting SIN3 or RPD3 restores G2/M delay, there was no evidence for Rad53 phosphorylation in the mec1Δ mutant background. This indicates that the G2/M delay seen with loss of Sin3/Rpd3 HDAC activity does not require activation of Rad53 by phosphorylation.
FIG. 5.
sin3Δ- or rpd3Δ-dependent restoration of the damage-induced G2/M delay does not require Rad53 phosphorylation. Cultures of the indicated strains were nocodazole arrested, mock (−) or UV (+) irradiated, and released into rich medium for sample collection at 10 min. Lysates were analyzed by Western blotting using an anti-Rad53 polyclonal antibody.
sin3Δ- or rpd3Δ-mediated activity is dependent on the spindle checkpoint.
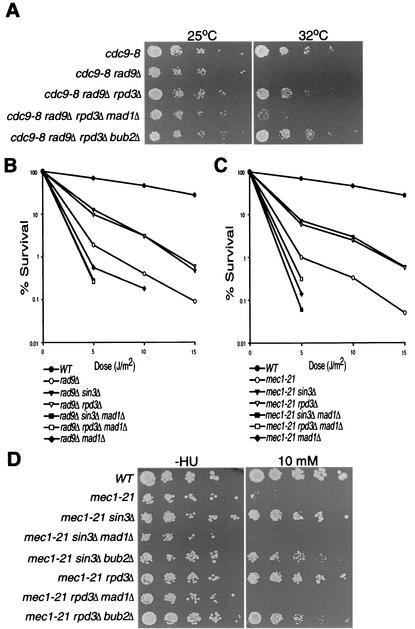
The spindle checkpoint monitors bipolar attachment of the mitotic spindle to kinetochores in order to promote faithful chromosomal segregation during mitosis (2). Defects in spindle-kinetochore attachment activate the checkpoint, which ultimately inhibits the anaphase-promoting complex, leading to cell cycle arrest at the metaphase-to-anaphase transition. In S. cerevisiae, cells arrested via the spindle checkpoint appear as large-budded cells, with DNA concentrated at the bud-neck region, similar to cells arrested by the DNA damage checkpoint. Therefore, we sought to determine whether the observed sin3Δ- and rpd3Δ-dependent DNA damage responses require an intact spindle checkpoint by deleting MAD1, a required component of the spindle checkpoint. Strains with spindle checkpoint genes deleted do not arrest in nocodazole; therefore, we determined the impact of the mad1Δ allele on survival to DNA damage in strains with or without SIN3 and RPD3. As described above, deleting RPD3 in a cdc9-8 rad9Δ background restores growth at the nonpermissive temperature of 32°C. Deleting MAD1 in both the cdc9-8 rad9Δ sin3Δ and cdc9-8 rad9Δ rpd3Δ backgrounds lowered the maximum permissive temperature back to 25°C, similar to the cdc9-8 rad9Δ double mutant (Fig. 6A and data not shown). We also examined the effect of deleting BUB2, which acts on a separate branch of the spindle checkpoint that regulates exit from mitosis (13). Unlike deletion of MAD1, deletion of BUB2 did not alter the growth at 32°C of a cdc9-8 rad9Δ mutant strain.
FIG. 6.
The spindle checkpoint pathway is required for sin3Δ- and rpd3Δ-dependent activity. (A) Fivefold serial dilutions of cdc9-8 mutant strains containing wild-type or mutant alleles of RAD9, RPD3, MAD1, and BUB2 were incubated at either 25 or 32°C. (B and C) Colony survival assays of yeast containing wild-type or mutant alleles of RAD9, SIN3, RPD3, and MAD1 (B) or MEC1, SIN3, RPD3, and MAD1 (C) were UV irradiated, and colony survival was measured. (D) Fivefold serial dilutions of cells containing wild-type or mutant alleles of MEC1, SIN3, RPD3, MAD1, and BUB2 plated on medium supplemented with 0 or 10 mM HU and incubated at 30°C.
Our data indicate that the spindle checkpoint is required for the sin3Δ- and rpd3Δ-dependent growth advantage in a cdc9-8 rad9Δ background; therefore, we also tested whether deletion of MAD1 would reverse the increase in survival to UV exhibited by Sin3- or Rpd3-deficient checkpoint mutant strains. We performed colony survival assays in rad9Δ and mec1-21 checkpoint-deficient strains containing either wild-type or mutant alleles of SIN3, RPD3, and MAD1 (Fig. 6B and C). While deleting SIN3 or RPD3 suppressed UV sensitivity in both the rad9Δ and mec1-21 mutant backgrounds (as described above), disruption of MAD1 consistently inhibited this response by decreasing the percentage of surviving cells to levels below those exhibited by the rad9Δ and mec1-21 mutants. Furthermore, the percent survival exhibited by the rad9Δ sin3Δ (or rpd3Δ) mad1Δ and mec1-21 sin3Δ (or rpd3Δ) mad1Δ triple mutants was similar to that in the rad9Δ mad1Δ and mec1-21 mad1Δ mutant strains.
We also tested whether the spindle checkpoint is necessary for the sin3Δ- and rpd3Δ-dependent HU sensitivity suppression in a mec1-21 strain background. While mec1-21 sin3Δ and mec1-21 rpd3Δ double mutants display an increase in survival to low HU concentrations (as described above), mec1-21 sin3Δ mad1Δ and mec1-21 rpd3Δ mad1Δ triple mutants fail to grow on medium containing 10 mM HU (Fig. 6D). Similar to the cdc9-8 temperature sensitivity assay, deleting BUB2 in the mec1-21 sin3Δ and mec1-21 rpd3Δ mutant backgrounds had little or no effect on the sin3Δ- and rpd3Δ-dependent growth advantage on HU. These combined data indicate that the MAD1-dependent spindle checkpoint pathway is required for the sin3Δ- and rpd3Δ-dependent suppression of sensitivity to DNA damage and replication stress in checkpoint-deficient strains.
DISCUSSION
We initiated this study to better understand the mechanism by which Ches1 suppresses checkpoint phenotypes in S. cerevisiae checkpoint mutants. We found that the Ches1 protein interacts with Sin3, a member of the S. cerevisiae Sin3/Rpd3 HDAC complex. We further demonstrated that Ches1 does not suppress the DNA damage response in sin3 mutant strains, and overexpression of SIN3 blocks Ches1-mediated G2/M arrest after DNA damage. Thus, Ches1 appears to function by inhibition of Sin3 function. This inhibition may be the result of competition for binding between the high-level expression of the heterologous human Ches1 protein and endogenous yeast proteins.
These findings also led us to determine the role of Sin3 and its associated HDAC, Rpd3, in the response to DNA damage. Loss of Sin3/Rpd3 activity by deletion of either the SIN3 or RPD3 gene in checkpoint mutant strains increased survival in response to DNA damage and replication blocks and restored an appropriate cell cycle delay.
Prior work has demonstrated that chromatin status influences the S. cerevisiae cell cycle. In particular, acetylation of H3 and H4 histone tails is necessary for transit through the G2/M cell cycle phase in the absence of damage (16, 25, 27, 30). Alteration of centromeric structure by mutation of NPS1, a component of the RSC chromatin-remodeling complex, also results in a permanent G2/M arrest that is dependent on the spindle checkpoint (44). In contrast, we observed no significant G2/M delay in Sin3/Rpd3-deficient strains in the absence of damage. Our data indicate that disrupting the Sin3/Rpd3 deacetylase restores a G2/M cell cycle arrest, but this response occurs only after the cells are exposed to DNA damage.
There are several potential models to explain how the sin3Δ- or rpd3Δ-dependent G2/M delay is activated in response to damage. It is likely that chromatin must be remodeled in response to DNA damage in order for the damage signal to be initiated or for the damage to be processed. Much evidence suggests that checkpoint proteins must load onto damaged DNA to initiate and carry out checkpoint activities. A recent study (54) reported the association of the human replication factor C-related Rad17 protein with the PCNA-related Rad1-Rad9-Hus1 protein complex at sites of DNA damage in vivo. Additionally, two reports (22, 28) demonstrated that components of the S. cerevisiae DNA damage checkpoint complex, including Mec1, Ddc1, and Ddc2, localize to a region proximal to a single double-stranded break. An interaction between the S. cerevisiae chromatin assembly factor, Asf1, and Rad53 has also recently been described (18). Rad53 localizes to chromatin in a manner dependent on the DNA helicase Sgs1, which functions in the S-phase checkpoint pathway (11). In human cells, HDAC1 interacts with the checkpoint proteins hRad9 and hHus1, possibly to regulate the G2/M checkpoint (7). Thus, depletion of Sin3/Rpd3 activity may directly promote a more accessible chromatin state and may partially obviate the need for Mec1 and Rad9 in response to DNA damage.
An alternative but not exclusive explanation for these data is that depleting HDAC activity could result in the specific misregulation of genes affecting G2/M progression and DNA damage checkpoint activity. Microarray analysis of strains with SIN3 or RPD3 deleted showed a greater-than-twofold up- and down-regulation of approximately 170 and 265 transcripts, respectively, many of which function during meiosis (3). Inhibition of the HDAC complex permits the expression of many meiotic genes in mitotic cells (4, 42, 47, 48). However, analysis of the sin3 and rpd3 mutant microarray data sets revealed no significant change in expression levels of those genes playing a primary role in the spindle checkpoint, including MAD1-3, BUB1, BUB3, and MPS1 (3).
The DNA damage-induced arrest exhibited in the SIN3 or RPD3 mutant strains is dependent on the Mad1-containing branch of the spindle checkpoint pathway. The spindle checkpoint elicits preanaphase arrest through inhibition of the anaphase-promoting complex in cells whose replicated chromosomes have failed to form proper attachments to the mitotic spindle (2). Three recent studies have reported potential functions of the spindle checkpoint pathways in response to DNA damage or replication blocks. Maringele and Lydall (26) demonstrated that the cell cycle arrest exhibited by yku70Δ mutants (which accumulate telomeric single-stranded DNA) is dependent on the DNA damage checkpoint genes CHK1, MEC1, and RAD9, as well as the spindle checkpoint gene MAD2. A second study also found a contribution of the spindle checkpoint to cell cycle arrest in response to MMS and HU when assayed in strains deficient for the DNA damage and replication checkpoints (12). In this latter study, spindle checkpoint mutants containing an intact DNA damage checkpoint exhibited a wild-type response to HU and MMS; therefore, the spindle pathway may not play a major role in the response to DNA damage when the MEC1-dependent pathway is active.
Most recently, a role for the spindle checkpoint in responding to DNA damage in some human cells has been reported (29). Extensive chromosomal damage revealed a mitotic arrest independent of p53, and this delay was present when cells were treated with inhibitors of the ATM kinase. Immunofluorescence microscopy indicated the presence of Mad2 at kinetochores, suggesting activation of the spindle checkpoint, and inhibition of Mad2 by microinjection of a dominant-negative form of Mad2 cells alleviated the damage-induced arrest, thus allowing cells to enter anaphase. Those authors propose that the arrest exhibited by these cells is likely due to the spindle checkpoint's response to damaged kinetochores, which when disrupted can no longer attach to the mitotic spindle.
In the present work we demonstrate a similar response of S. cerevisiae to DNA damage. Using specific mutants of the ATM-related Mec1 kinase, we demonstrate that the delay is independent of this pathway. Furthermore, the deletion of SIN3 or RPD3 potentiates the DNA damage response, resulting in substantial resistance to DNA damage and a wild-type cell cycle arrest even at relatively low doses of UV irradiation. Involvement of the spindle checkpoint suggests that the damage response may be acting through damaged kinetochores or, alternatively, there may be decreased tension elicited by damaged chromosomes with altered chromatin structure. Silverstein et al. (40) recently uncovered a direct role for the Schizosaccharomyces pombe Sin3 homolog, PstI, in regulating centromere acetylation levels through association with the Clr6 HDAC. Mutants lacking PstI exhibit reduced levels of gene silencing near the centromere and display a variety of phenotypes consistent with altered centromere and kinetochore structure, including increased sensitivity to the microtubule-destabilizing agent thiabendazole and defective centromeric sister chromatid cohesion.
Many reports implicate HDAC activity as being involved in cell cycle progression and cancer development (9). The complicated nature of the Rb/E2F pathway provides many ways in which genetic defects can impede Rb-dependent repression, and this pathway is targeted in many tumor types (39). Not all the transformed mammalian cell lines tested by Mikhailov exhibited a DNA damage-dependent anaphase arrest, and these differences may reflect whether they contain dysregulation of histone deacetylation. Studies using HDAC inhibitors further support the possibility that checkpoint pathways monitor acetylation status in human cells. For example, certain HDAC inhibitors cause cell cycle arrest in some tumor cell lines (31, 36). One study (34) reported that the HDAC inhibitor azelaic bishydroxamic acid (ABHA) activates a novel G2 cell cycle arrest in normal human cells, and this checkpoint is absent in some ABHA-sensitive tumor cell lines.
HDAC inhibitors are being assessed for anticancer activity in clinical trials. One confounding factor in studies using pharmacologic means to inhibit HDACs is that the resulting phenotype may be secondary to other cellular targets of these agents. Our genetic studies in yeast directly demonstrate that deletion of an HDAC complex restores a DNA damage-induced G2/M arrest in checkpoint-deficient strains that is dependent on an intact spindle checkpoint. These data suggest that HDAC inhibitors may cause a paradoxical increase in survival to genotoxic agents when used in cancer cells containing mutations in genes performing functions analogous to RAD9 and MEC1, including BRCA1 and ATM, respectively.
Acknowledgments
We extend thanks to Debananda Pati, Jim Huang, and Lisa Wang for technical support and helpful discussions. We also thank Zafar Nawaz, Steve Elledge, and Vicki Lundblad for advice, yeast strains, and other reagents.
This work was supported by grant DAMD17-98-1-8023 to K.L.S. from the U.S. Army and grant RO1-GM57426 to S.E.P. from the National Institutes of Health.
REFERENCES
- 1.Adams, A. E., and J. R. Pringle. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowdish, K. S., and A. P. Mitchell. 1993. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2172-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buggy, J. J., M. L. Sideris, P. Mak, D. D. Lorimer, B. McIntosh, and J. M. Clark. 2000. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J. 350:199-205. [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Cai, R. L., Y. Yan-Neale, M. A. Cueto, H. Xu, and D. Cohen. 2000. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275:27909-27916. [DOI] [PubMed] [Google Scholar]
- 8.Craig, E. A., B. D. Gambill, and R. J. Nelson. 1993. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol. Rev. 57:402-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 10.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 12.Garber, P. M., and J. Rine. 2002. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161:521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, R. D., and D. J. Burke. 2000. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 10:154-158. [DOI] [PubMed] [Google Scholar]
- 14.Hartwell, L. H., and T. A. Weinert. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629-634. [DOI] [PubMed] [Google Scholar]
- 15.Hong, L., G. P. Schroth, H. R. Matthews, P. Yau, and E. M. Bradbury. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268:305-314. [PubMed] [Google Scholar]
- 16.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, E., Z. Chen, T. Fredrickson, Y. Zhu, R. Kirkpatrick, G. F. Zhang, K. Johanson, C. M. Sung, R. Liu, and J. Winkler. 2000. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem. 275:15254-15264. [DOI] [PubMed] [Google Scholar]
- 18.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadonaga, J. T. 1998. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92:307-313. [DOI] [PubMed] [Google Scholar]
- 20.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 21.Kasten, M. M., S. Dorland, and D. J. Stillman. 1997. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol. Cell. Biol. 17:4852-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 24.Longtine, M. S., A. McKenzie I I I, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 25.Magnaghi-Jaulin, L., S. Ait-Si-Ali, and A. Harel-Bellan. 2000. Histone acetylation and the control of the cell cycle. Prog. Cell Cycle Res. 4:41-47. [DOI] [PubMed] [Google Scholar]
- 26.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Megee, P. C., B. A. Morgan, and M. M. Smith. 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9:1716-1727. [DOI] [PubMed] [Google Scholar]
- 28.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikhailov, A., R. W. Cole, and C. L. Rieder. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797-1806. [DOI] [PubMed] [Google Scholar]
- 30.Morgan, B. A., B. A. Mittman, and M. M. Smith. 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 11:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima, H., Y. B. Kim, H. Terano, M. Yoshida, and S. Horinouchi. 1998. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp. Cell. Res. 241:126-133. [DOI] [PubMed] [Google Scholar]
- 32.Paciotti, V., G. Lucchini, P. Plevani, and M. P. Longhese. 1998. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 17:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pati, D., C. Keller, M. Groudine, and S. E. Plon. 1997. Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol. Cell. Biol. 17:3037-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu, L., A. Burgess, D. P. Fairlie, H. Leonard, P. G. Parsons, and B. G. Gabrielli. 2000. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol. Biol. Cell 11:2069-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 36.Sambucetti, L. C., D. D. Fischer, S. Zabludoff, P. O. Kwon, H. Chamberlin, N. Trogani, H. Xu, and D. Cohen. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J. Biol. Chem. 274:34940-34947. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 38.Schiestl, R. H., P. Reynolds, S. Prakash, and L. Prakash. 1989. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol. Cell. Biol. 9:1882-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sellers, W. R., and W. G. Kaelin, Jr. 1997. Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 15:3301-3312. [DOI] [PubMed] [Google Scholar]
- 40.Silverstein, R. A., W. Richardson, H. Levin, R. Allshire, and K. Ekwall. 2003. A new role for the transcriptional corepressor SIN3; regulation of centromeres. Curr. Biol. 13:68-72. [DOI] [PubMed] [Google Scholar]
- 41.Stillman, D. J., S. Dorland, and Y. Yu. 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics 136:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strich, R., M. R. Slater, and R. E. Esposito. 1989. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc. Natl. Acad. Sci. USA 86:10018-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 44.Tsuchiya, E., T. Hosotani, and T. Miyakawa. 1998. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 26:3286-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukiyama, T., and C. Wu. 1997. Chromatin remodeling and transcription. Curr. Opin. Genet. Dev. 7:182-191. [DOI] [PubMed] [Google Scholar]
- 46.Van den Wyngaert, I., W. de Vries, A. Kremer, J. Neefs, P. Verhasselt, W. H. Luyten, and S. U. Kass. 2000. Cloning and characterization of human histone deacetylase 8. FEBS Lett. 478:77-83. [DOI] [PubMed] [Google Scholar]
- 47.Vidal, M., and R. F. Gaber. 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:6317-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidal, M., R. Strich, R. E. Esposito, and R. F. Gaber. 1991. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol. Cell. Biol. 11:6306-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wade, P. A. 2001. Transcriptional control at regulatory checkpoints by histone deacetylases: molecular connections between cancer and chromatin. Hum. Mol. Genet. 10:693-698. [DOI] [PubMed] [Google Scholar]
- 51.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 54.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]