Abstract
Most eukaryotic cells contain nearly equimolar amounts of nucleosomes and H1 linker histones. Despite their abundance and the potential functional specialization of H1 subtypes in multicellular organisms, gene inactivation studies have failed to reveal essential functions for linker histones in vivo. Moreover, in vitro studies suggest that H1 subtypes may not be absolutely required for assembly of chromosomes or nuclei. By sequentially inactivating the genes for three mouse H1 subtypes (H1c, H1d, and H1e), we showed that linker histones are essential for mammalian development. Embryos lacking the three H1 subtypes die by mid-gestation with a broad range of defects. Triple-H1-null embryos have about 50% of the normal ratio of H1 to nucleosomes. Mice null for five of these six H1 alleles are viable but are underrepresented in litters and are much smaller than their littermates. Marked reductions in H1 content were found in certain tissues of these mice and in another compound H1 mutant. These results demonstrate that the total amount of H1 is crucial for proper embryonic development. Extensive reduction of H1 in certain tissues did not lead to changes in nuclear size, but it did result in global shortening of the spacing between nucleosomes.
DNA in the eukaryotic nucleus is organized into a highly compact nucleoprotein complex referred to as chromatin (48, 53). The histones constitute a family of proteins that are intimately involved in organizing chromatin structure. The nucleosome core particle, the highly conserved unit of chromatin organization in all eukaryotes, consists of an octamer of four core histones (H2A, H2B, H3, and H4) around which about 145 bp of DNA is wrapped. The chromatin fiber also contains a fifth histone, the linker histone (usually referred to as H1), which can bind to core particles and protect an additional ∼20 bp of DNA (linker DNA) from nuclease digestion. The precise location and stoichiometry of H1 within the chromatin fiber are uncertain (45, 46, 49), but in higher eukaryotes, there is, on average, nearly one H1 molecule for each core particle (48). Most of our knowledge about the role of H1 in chromatin structure is based on in vitro experiments. These studies indicate that two principal functions of linker histones are to organize and stabilize the DNA as it enters and exits the core particle and to facilitate the folding of nucleosome arrays into more compact structures (19, 38). Despite the presumed fundamental role of linker histones in chromatin structure, elimination of H1 in Tetrahymena, Saccharomyces cerevisiae, and Aspergillus nidulans and silencing of H1 in Ascobolus immersus showed that H1 is not essential in these unicellular eukaryotes (3, 34, 39, 41, 47).
In higher organisms, additional levels of control on chromatin organization and function are available because of the existence of multiple nonallelic linker histone variants or subtypes (6, 33). In mice, there are at least eight H1 subtypes, including the widely expressed subtypes H1a through H1e, the testis-specific subtype H1t, the oocyte-specific subtype H1oo, and the replacement linker histone H10 (26, 44). The genes for six of these subtypes, those that encode H1a to H1e and H1t, are present in a cluster on MMU13A2-3 (50), while the genes encoding H10 and H1oo are located on mouse chromosomes 15 and 6, respectively. The amino acid sequences of these H1 subtypes differ significantly; for example, H10 is only 30 to 38% identical to H1a to H1e, and even among H1a to H1e, the sequence divergence between certain subtypes approaches 40% (51). The different subtypes are also expressed differentially during development (27, 51). Certain H1 subtypes represent a major fraction of the linker histone in specific cell types; for example, H10 and H1e constitute 28 and 42%, respectively, of the H1 in adult mouse hepatocytes (42) and H1t constitutes 40 to 50% of the H1 in pachytene spermatocytes (25, 30).
Despite the extensive sequence divergence within the H1 family and despite the abundance of certain subtypes in specific tissues, we (17, 29, 42) and others (15, 18, 37) have found that elimination of any one of several different H1 subtypes by gene inactivation does not noticeably perturb mouse development. When considered along with the findings in yeast and Tetrahymena, these results may suggest that H1 linker histones also are not essential for mammalian development. However, we reported previously that the chromatin of animals lacking any one H1 subtype has a normal total H1-to-core histone ratio (17, 29, 42), suggesting that upregulation of the remaining subtypes is sufficient to maintain normal linker-to-core histone stoichiometry. Therefore, we sought to generate compound H1-null mice. We report here that elimination of three H1 subtypes (H1c, H1d, and H1e) together results in embryonic lethality, showing that linker histones are essential for proper mammalian development. Nevertheless, we found that marked reductions of linker histone content are tolerated in certain compound H1 mutant mice and lead to global reductions in nucleosome spacing.
MATERIALS AND METHODS
Disruption of H1 genes in ES cells and generation of chimeric mice.
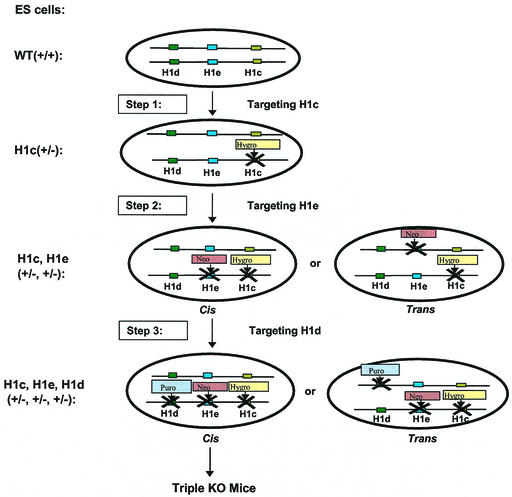
The procedures used for disruption of H1 genes in WW6 ES cells (23) and clones derived from these cells followed very closely the methods described previously (17). The H1c-Hygro targeting vector was described previously (17), except that the PGK-Hygro cassette was substituted for the PGK-Neo cassette. Two ES cell clones (WC1.4 and WC1.16) containing the modified H1c locus generated chimeras; chimeras from WC1.16 ranged from 85 to 99% on the basis of coat color, and all of these chimeras transmitted the modified H1c allele. For the second round of targeting to inactivate the H1e gene, clone WC1.16 was transfected with a linearized H1e targeting vector (17) containing the PGK-Neo cassette and a clone, WEC 9.6, that has a cis configuration of the H1c and H1e modified loci (see Results and reference 16) was used for the third round of targeting to inactivate the H1d gene. The H1d targeting vector (17) was linearized with NotI and transfected into WEC9.6 cells. Colonies resistant to 2 μg of puromycin (Sigma) per ml and 2 μM ganciclovir (Syntex) were isolated. Genomic DNA from 678 puromycin- and ganciclovir-resistant colonies was digested with XbaI and screened for homologous recombination events by Southern blot hybridization with a 0.55-kb 3′ H1d flanking region probe (17). Nine clones gave a 3.4-kb XbaI hybridizing fragment expected from the modified H1d allele. Seven clones were injected into C57BL/6 recipient blastocysts, and chimeric mice were derived. All cell lines generated chimeras ranging in chimerism from 80 to 99% on the basis of coat color. Male and female chimeras from each line were then mated with C57BL/6 mice. All seven lines transmitted the modified H1d allele. Three lines derived from ES cell clones (CDE7.3, CDE16.11, and CDE24.4) have the modified H1d allele in trans to the modified H1c and H1e alleles. Four lines derived from ES cell clones (CDE2.2, CDE3.19, CDE16.21, and CDE18.3) have the modified H1d allele in cis to the modified H1c and H1e alleles.
Mouse genotyping.
Mouse tail DNA and DNA from whole embryos, portions of embryos, or dissected yolk sacs were prepared as previously described (22). Genotyping of founder mice was carried out by Southern blot hybridization (17) and PCR assays. For compound H1 knockout lines, PCRs for both alleles of two or three H1 genes were performed to ensure the correct genotype. PCR was performed on mouse tail or embryo DNA with the following primers: H1c wild-type allele (H1c-WT), H1c gene-specific primer (Pcf; 5′ CTGCCACACCCAAAAAGG 3′) and H1c 3′ sequence-specific primer (Pcr; 5′ GAGCATAGAAGCCACCTACAAG 3′) (predicted size, 600 bp); H1c modified allele (H1c-Hygro), PGK-Hygro promoter PGK-specific primer-specific primer (pGK2; 5′ GCTGCTAAAGCGCATGCTCCA 3′) and Pcr (predicted band size, 400 bp); H1d wild-type allele (H1d-WT), H1d gene-specific primer (Pdf5t; 5′ AAGCCTAAAGCTTCTAAGCCG 3′) and H1d 3′ sequence-specific primer (Pdr8t; 5′ CTAGAGAACCCCCCTAATGC 3′) (predicted size, 410 bp); H1d modified allele (H1d-Puro), PGK-specific primers PGK2 and Pdr8t (predicted size, 305 bp); H1e wild-type allele (H1e-WT), H1e gene-specific primer (Pef; 5′ AACCAAAAACCTCCAAGCCT 3′) and H1e 3′ sequence-specific primer (Per; 5′ CCCTTAAAAGTTATCGACGTGG 3′) (predicted band size, 388 bp); H1e modified allele (H1e-Neo), PGK-Neo gene-specific primer (Pnf; 5′ TTTGAATGGAAGGATTGGAG 3′) and Per (predicted band size, 650 bp); H10 wild-type allele (H10-WT), H10 gene-specific primer (Pof; 5′AGAGCAGATCGCGAGTCAG 3′) and the H10 3′ sequence-specific primer (Por; 5′ GTTTGCTGTCCTTGCACAATAG 3′) (predicted band size, 860 bp); H10 modified allele (H10-Neo), PMCI-Neo gene-specific primer (Pmf; 5′ GATCGGCCATTGAACAAGAT 3′) and Por (predicted size, 1,075 bp).
Mouse embryo manipulation.
Mice of the appropriate genotype were set up for breeding in the afternoon, and each morning thereafter, females were checked for vaginal plugs. Embryos were staged as embryonic day 0.5 (E0.5) postcoitus at noon if a plug was found. Embryos for phenotyping were fixed with 4% paraformaldehyde overnight at 4°C, dehydrated with a series of 25, 50, 75, and 100% methanol at 4°C, and stored at −20°C. Embryos were passed through a decreasing methanol series to phosphate-buffered saline (PBS; 138 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate buffer, pH 7.4)-0.1% Tween 20 for phenotype characterization.
Preparation and analysis of histones.
Histone proteins from mouse tissues and embryos were prepared by extraction of chromatin with 0.2 N sulfuric acid as described previously (29, 42). The effluent from the high-performance liquid chromatography (HPLC) column was monitored at 214 nm, and the peaks were recorded with a Hewlett-Packard 1090 system. Peak areas were determined with a Hewlett-Packard peak integrator program.
Isolation of nuclei and chromatin and micrococcal nuclease digestion.
Spleen and thymus tissues were placed in Dulbecco modified Eagle medium with 10% fetal calf serum and gently minced between frosted glass slides, and the released lymphocytes were centrifuged at 200 × g for 10 min and then washed once in PBS. Splenocyte and thymocyte suspensions in PBS were centrifuged for 3 min at 800 × g, resuspended in 0.5% NP-40 in RSB (10 mM NaCl, 3 mM MgCl2, 10 mM Tris-HCl, pH 7.5) with 1 mM phenylmethylsulfonyl fluoride (PMSF) at 4°C, and homogenized by 10 to 15 strokes of pestle A in a Dounce homogenizer over 20 min. Released nuclei were centrifuged for 6 min at 1,000 × g, and the nuclear pellets were resuspended in RSB containing 0.5 mM PMSF. Liver tissue was cut into small pieces, rinsed for 3 min in PBS containing 1% sodium citrate, transferred to RSB containing 0.5 mM PMSF and 0.3 M sucrose, homogenized as described above, layered onto a 2 M sucrose cushion in RSB, and centrifuged for 30 min at 38,000 × g. The pellet was resuspended in RSB with 0.5 mM PMSF and 0.5% NP-40 added, and nuclei were prepared as for lymphocytes.
Nuclei were digested with 7 to 9 U of micrococcal nuclease (Boehringer Mannheim) per 100 μg of DNA for 5 min at 37°C in RSB containing 1 mM CaCl2 or with 15 to 30 U of enzyme per 100 μg of DNA for 5 min at 15°C. Reactions were terminated by addition of 5 mM Na-EDTA, and nuclei were pelleted for 5 min at 10,000 × g and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5).
DNA electrophoresis.
Chromatin released into TE buffer was treated with 1% sodium dodecyl sulfate-0.2% proteinase K for 1 h at 37°C, extracted with phenol-chloroform, and precipitated with alcohol in the presence of 0.3 M Na-acetate, and the pellet was resuspended in TE buffer. DNA electrophoresis was conducted with 1% agarose (type I; Sigma) in Tris acetate buffer with a 1-kb DNA ladder (Gibco BRL) as size markers. After ethidium bromide staining, gels were recorded digitally and band detection and DNA length assignment (on the basis of the positions of marker bands) were performed with Quantity One software (Bio-Rad). PsiPlot (Polysoftware International) was used for data analysis and graphing.
RESULTS
Sequential inactivation of three H1 histone genes shows that linker histones are essential for mammalian development.
As already mentioned, previous work from our laboratory showed that the chromatin of mice lacking any one of several H1 subtypes has a normal ratio of total H1 linker histones to core histones, indicating that synthesis of the remaining H1 subtypes can compensate for the inactivated subtype to maintain normal linker histone chromatin stoichiometry. To investigate the effect of disruption of linker-to-core histone stoichiometry, we sought to combine H1 histone-null mutations in a single strain. We chose first to combine mutations in the genes for H1c and H1e because, of the five somatic H1 subtypes, these two have very closely related amino acid sequences and their expression is often coordinately regulated (12, 13, 26, 51). We previously described mice null for either H1c or H1e (17). We attempted to obtain mice with a recombinant chromosome containing the mutant alleles for both genes by interbreeding H1c and H1e doubly heterozygous mutants. However, we failed to observe recombination in more than 1,000 meioses analyzed, probably because of the close proximity of the two genes on chromosome 13 (50). Therefore, we adopted a strategy of sequential gene inactivation in ES cells (Fig. 1). None of the H1 histone genes on chromosome 13 contain introns, and the coding region of each gene is about 600 nucleotides long. We designed each targeting vector to produce a null allele by deleting all of the H1 coding region and replacing it with a positive selectable marker gene. The first step in the sequential strategy was the generation of ES cells in which one allele of the gene for H1c is replaced with a PGK-Hygro cassette. The second step in the sequential gene inactivation strategy was inactivation of the H1e gene in H1c+/− ES cells. It also was necessary to identify ES cells in which the two modifications occurred in cis since only such ES cells are useful to generate homozygous double-mutant mice. Two H1c+/− ES cell lines were electroporated with an H1e targeting vector in which the PGK-Neo gene replaced the H1e coding region. Several H1c+/− H1e+/− clones were identified and injected into blastocysts. Chimeric mice were obtained, and several chimeras in which the modified H1c and H1e alleles cosegregated in agouti progeny were identified by PCR analysis of tail DNA and confirmed by Southern blot analysis (data not shown). H1cH1e++/−− double-heterozygous mice were interbred to generate homozygous H1c H1e double mutants. Figure 2A shows an example of PCR genotype analysis of F2 mice. Genotypes of founder mice were also confirmed by Southern blot analysis (data not shown). Of 179 F2 animals genotyped, 46 (26%) carried only the wild-type alleles, 93 (52%) were H1cH1e++/−− double heterozygotes, and 40 (22%) were H1c H1e double mutants. The ratio of the three classes of animals is not significantly different from the expected values for Mendelian transmission of the unmodified and modified chromosomes. Homozygous H1c H1e double-mutant mice are fertile, and male and female double mutants can mate to produce litters of normal size. Male double homozygotes were found to be smaller, on average, than wild-type littermates at postnatal day 7 (wild type, 5.52 g [standard error of the mean, 0.2 g; n = 5]; double mutant, 4.40 g [standard error of the mean, 0.1 g; n = 6]), as well as throughout adult life (data not shown). However, a size difference was not observed in females. Examination of sections from 35 different tissues of six mutants and six controls did not reveal any pathological or abnormal histological features.
FIG. 1.
Strategy for sequential inactivation of three H1 histone genes in mouse ES cells. Depicted are the chromosome homologues containing the three linked H1 genes (those encoding H1c, H1d, and H1e) to be targeted and the three steps used to target the genes, each with a different selectable marker gene. Also depicted are the cis and trans configurations of gene targetings that can occur in steps 2 and 3. WT, wild type; KO, knockout.
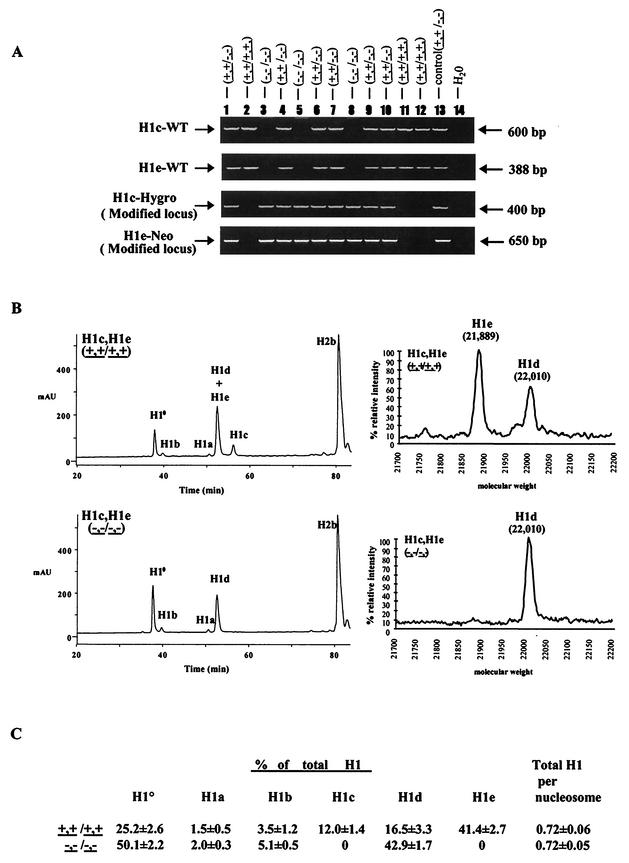
FIG.2.
Analysis of H1c H1e double-knockout (KO) mice. (A) Genotype analysis of F2 mice from intercrosses of H1cH1e++/−− mice. Mouse tail DNA was analyzed by PCR (as described in Materials and Methods) for the wild-type H1c allele (H1c-WT), the wild-type H1e allele (H1e-WT), the modified H1c allele (H1c-Hygro), and the modified H1e allele (H1e-Neo). The deduced genotype of each embryo is indicated above each lane. The positions of the PCR products from the wild-type and modified alleles are indicated. Control reaction mixtures contained tail DNA from an H1c H1e double-heterozygous mutant mouse. (B) Analysis of histones extracted from livers of wild-type and H1c H1e double-homozygous mutant mice. The graphs on the left show results of reverse-phase HPLC analyses of approximately 100 μg of total liver histone extracts from a 20-week-old wild-type mouse (top) and an H1c H1e double-homozygous mutant (bottom). The abscissa represents elution time, and the ordinate represents absorbency at 214 Å. The identity of the histone subtype(s) in each peak is indicated. mAU, milli-absorbency units. The graphs on the right show results of time-of-flight mass spectrometry analysis of a fraction eluting between 52 and 54 min (corresponding to the peak marked H1d + H1e). The identities of the H1d and H1e subtypes detected in this analysis were shown previously (51). (C) H1 subtype composition of liver chromatin from wild-type and H1c H1e double-mutant mice. Data were calculated from HPLC analyses of wild-type and H1c H1e double-mutant strains like that shown in panel B. Values are means ± standard deviations of individual determinations made on three 5-month-old mice of each of the indicated genotypes. The percentage of total H1 was determined by the ratio of the A214 of the indicated H1 peak to the total A214 of all of the H1 peaks. Total H1 per nucleosome was determined by the ratio of the total A214 of all of the H1 peaks to half of the A214 of the H2b peak. The A214 values of the individual H1 peaks and the H2b peak were adjusted to account for the differences in the number of peptide bonds in each H1 subtype and H2b.
The foregoing results indicate that loss of H1c and H1e does not impair normal development, suggesting that other H1 subtypes compensate even in double-linker histone mutants. To investigate this possibility and to prove that the mutant alleles are nulls, total histone extracts of liver chromatin from H1c H1e homozygous mutants were analyzed by reverse-phase HPLC and time-of-flight mass spectrometry. Figure 2B shows that H1c and H1e are absent in the chromatin of homozygous double mutants, proving that the mutant alleles are nulls. Reverse-phase HPLC also separates all of the H1 linker histones from the nucleosomal core histones, allowing measurement of the linker histone-to-core histone stoichiometry in chromatin by determination of the ratio of the total amount of all of the H1 subtypes present to that of one of the core histones, e.g., H2b. The observed ratio of 0.7 H1 molecule per nucleosome in wild-type liver chromatin (Fig. 2C) is in good agreement with previous measurements of mouse liver chromatin by other methods (4) and with our previous determinations (17, 42). Although H1c and H1e normally constitute more than 50% of the linker histone in adult liver, the total H1-to-core histone ratio in H1c H1e double-null mutants is not different from that of wild-type littermates. Thus, even in the double mutants, synthesis of other H1 subtypes, primarily H10 and H1d, is sufficient to maintain a normal ratio of linker histones with respect to nucleosome core particles in chromatin. The existence of this compensatory mechanism may account for the normal development of the homozygous double-mutant mice. Nevertheless, as described below, the double-mutant animals proved to be very useful for generation of additional compound strains of mice with H1-deficient chromatin.
To disrupt the compensation observed in H1c H1e homozygous mutants, we next eliminated the H1d subtype because it is most closely related in sequence and expression pattern to H1c and H1e (27, 51), and it constituted a large fraction of the H1 remaining in the double mutant. Accordingly, we carried out a third step of gene inactivation in cis doubly targeted H1c+/− H1e+/− ES cells with an H1d-targeting vector in which a PGK-Puro gene replaced the H1d coding region. We identified four clones in which the modified H1d allele cosegregated with the mutant H1c and H1e alleles in agouti progeny of chimeric mice. Mice heterozygous for the three linked mutant alleles, derived from three independent ES cell clones, were intercrossed, and the progeny were genotyped by PCR assays for wild-type and modified alleles of two or three of the genes. Among a total of 638 F2 progeny analyzed, no triple-homozygous mutant animals were found (Table 1). The absence of triple-homozygous mutants in litters is not due solely to H1d deficiency because H1d-null mice obtained from three H1d-trans-targeted ES cell lines develop normally (17). Among the 638 F2 progeny, 239 were wild type and 399 were H1cH1d+++/−−− H1e+/−. The observed ratio of wild-type and H1cH1d+++/−−− H1e+/− mice is 1:1.67. This value deviates from the expected 1:2 Mendelian ratio (P = 0.0003), indicating that triple-heterozygous mutants are underrepresented in litters (17% of the heterozygous mutants died before birth). The timing and cause of their death are under investigation. A histopathology survey of tissues from six 4-month-old triple-heterozygous mutant mice did not show any consistent abnormalities.
TABLE 1.
Genotypes of embryos from H1cH1dH1e+++/−−− triple heterozygous mutant mice
| Stage | Total no. of embryos | No. of embryos
|
|||
|---|---|---|---|---|---|
| +++/+++ | +++/−−− | −−−/−−−a | Resorbedb | ||
| E7.5 | 55 | 14 | 32 | 7 | 2 |
| E8.5 | 62 | 22 | 29 | 10 | 1 |
| E9.5 | 230 | 56 | 123 | 32 (+4)* | 15 |
| E10.5 | 81 | 23 | 40 | 5 (+2)* | 11 |
| E11.5 | 50 | 11 | 18 | 1 (+4)* | 16 |
| E12.5 | 26 | 8 | 12 | 0 | 6 |
| E13.5 | 26 | 7 | 15 | 0 | 4 |
| Postnatal | 638 | 239 | 399 | 0 | NAc |
Numbers in parentheses are the numbers of homozygous mutant embryos that already had undergone autolysis.
Resorption indicates the number of instances in which embryos were not recovered and the decidua were very small.
NA, not available.
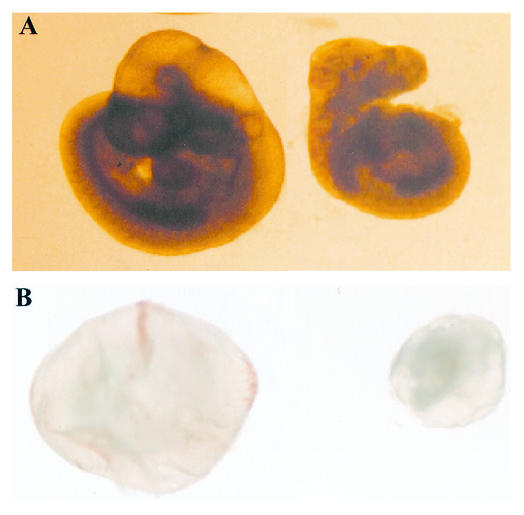
To determine when triple-homozygous mutant embryos die, embryos were obtained at various stages of gestation and genotyped (Fig. 3A). No homozygous mutants were recovered after E11.5, and they were increasingly underrepresented in litters recovered from E7.5 to E11.5 (Table 1). Homozygous mutant embryos analyzed from E8.5 to E10.5 exhibited a wide spectrum of abnormalities. Mutant embryos were most often considerably smaller than wild-type and heterozygous littermates, and even when they appeared grossly normal, their development was retarded, as judged by somite number. Only one-fourth of the expected number of triple-homozygous mutants were recovered at E10.5. The recovered mutant embryos at this stage were all smaller and less developed than wild-type littermates (Fig. 4A). Whereas wild-type embryos had 26 to 30 somites at this stage, the best-developed H1c H1d H1e triple-homozygous mutant embryos had only about 16 to 18 somites (Fig. 4A, right). In addition, the intact yolk sacs of the majority of E10.5 homozygous mutant embryos were smaller and paler, lacking visible blood or blood vessels, than their wild-type littermates (Fig. 4B). More detailed morphological observations were made on 21 triple-homozygous mutant embryos recovered at E9.5 (Fig. 5). Of these embryos, ∼50% (11 of 21) showed mild growth retardation relative to their wild-type siblings (Fig. 5A) and a variety of mild overt defects in development, each of low penetrance (Fig. 5B and C). Developmental aberrations included shortened tails, regions of excess tissue, mild neural tube defects, and failed chorioallantoic fusion. A second class of more severely affected embryos (8 of 21) showed significant growth retardation and additional variably penetrant disruptions in development (Fig. 5D and E). Among embryos of this class, we observed splayed anterior neural tubes, expanded pericardia, regions of tissue excess, failed chorioallantoic fusion, and developmental disproportion in which the posterior region of the embryo appeared to arrest in development and growth while the anterior regions continued to progress through proper heart looping and branchial arch development. Most embryos of this class had not undergone turning. In addition, among these 21 E9.5 embryos, morphologic analysis showed partial and complete axis duplication in two early somite stage embryos (Fig. 5F and G). Triple-homozygous mutant embryos were also underrepresented at both E8.5 and E7.5 (Table 1); the nature of the defects contributing to this early lethality is not known; however, analyses of embryos recovered at E8.5 support a phenotype distribution similar to that observed at E9.5.
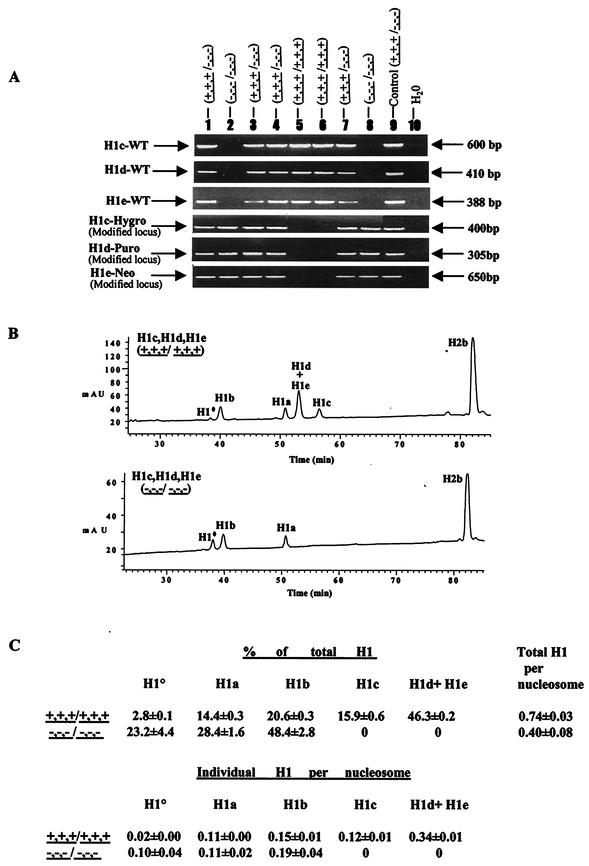
FIG.3.
Analysis of chromatin from H1c H1d H1e triple-null mouse embryos. (A) PCR genotype analysis of E7.5 embryos from intercrosses of H1c+/− H1d+/− H1e+/− mice. Embryo DNA was prepared and analyzed by PCR assays for H1c, H1d, and H1e wild-type (WT) and modified loci as described in Materials and Methods. The deduced genotype of each embryo is indicated above each lane. The positions of the PCR products from the wild-type and modified alleles are indicated. Control reaction mixtures contained tail DNA from an H1c H1d H1e triple-heterozygous mutant mouse. (B) Reverse-phase HPLC analysis of histones in extracts from E10.5 wild-type and homozygous H1c H1d H1e mutant embryos. Approximately 20 μg of total histone extract of chromatin from wild-type (top) and homozygous triple H1c H1d H1e mutant (bottom) E10.5 embryos were fractionated by reverse-phase HPLC. Other details are as in the legend to Fig. 2B. mAU, milli-absorbency units. (C) H1 subtype composition of chromatin from wild-type and H1c−/− H1d−/− H1e−/− E10.5 embryos. Data were calculated from HPLC analyses of wild-type and H1c H1d H1e triple-mutant embryos like that shown in panel B. Other details are as described in the legend to Fig. 2C.
FIG. 4.
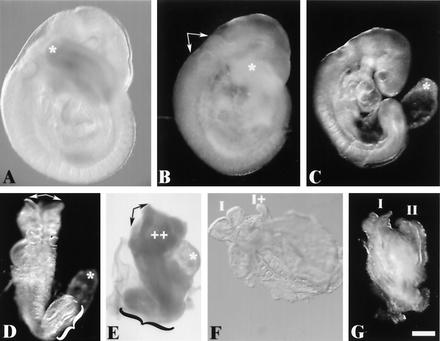
Gross morphology of wild-type and H1c H1d H1e triple-mutant embryos at E10.5. Approximately one-fourth of the expected number of homozygous mutant embryos are recovered at E10.5. (A) Morphology of an E10.5 wild-type embryo (left) and an H1c H1dH1e−−−/−− littermate (right). (B) Whole view of E10.5 conceptuses with yolk sacs, i.e., the wild type (left) and an H1cH1d H1e−−−/−−− littermate (right).
FIG. 5.
Phenotype of H1cH1dH1e−−−/−−− triple-homozygous mutant embryos recovered on E9.5. Approximately 60% of the expected number of homozygous mutant embryos are recovered at E9.5. The broad spectrum of abnormalities observed in recovered homozygous mutant embryos fall into three general classes. In the most predominant category (∼50%), representatives of which are shown in panels B and C, are embryos that were slightly developmentally delayed relative to their littermates (a representative wild-type embryo is shown in panel A) and presented a variety of incompletely penetrant abnormalities. Abnormalities observed in this class of mutants included occlusion of the rhombencephalic ventricle (arrows in panel B), a short tail (compare asterisks at the tail tips in panels A and B), and failure of the allantois to fuse with the chorion (asterisk in panel C). Mutant embryos falling into a second class, representatives of which are shown in panels D and E, are very developmentally delayed compared to their littermates and again variably presented developmental perturbations, including splayed anterior neural tubes (arrows in panels D and E), regions of excess tissue (++ in panel E), pericardial expansion (asterisk in panel E), failure of chorioallantoic fusion (asterisk in panel D), and caudal dysgenesis (braces in panels D and E). Mutant embryos in a third class are severely abnormal (F and G). They had not progressed past an early somite stage and in each case showed morphological evidence of partial (I and I+ in panel F) or complete (I and II in panel G) axis duplication. Embryos are viewed from the right side in panels A to C and E, from nearly dorsal in panel D, from ventral in panel F, and from a midline between the axes in panel G. Scale bar = 300 μm.
HPLC analysis of three mildly growth-retarded E10.5 triple-mutant embryos showed that, as expected, all three subtypes were absent from homozygous mutants (Fig. 3B), proving that the targeting of the H1d gene in cis doubly targeted H1c+/− H1e+/− ES cells created a null allele. The H1-to-core histone ratio in these embryos was found to be nearly 50% of that in wild-type embryos (Fig. 3C), showing that inactivation of the three H1 genes had indeed disrupted the compensation observed in single and double mutants. We conclude from these studies that, unlike the H1 subtypes of Tetrahymena and yeast, a minimum amount (∼50% the normal amount) of linker histones is required for proper mouse development (see Discussion).
We noted that the H10 subtype, which is correlated with cell cycle inhibition and terminal differentiation (7, 55), is specifically increased in triple-null mutant embryos (Fig. 3B and C). To investigate whether increased levels of H10 may contribute to embryonic lethality, we used the H1cH1dH1e+++/−−− strain described above and H10−/− mice (42) to generate H10−/−; H1cH1dH1e+++/−−− mice. Intercrosses of these animals did not produce viable progeny lacking all four subtypes (0 of 284 versus 1 of 4 expected), and E9.5 embryos null for all four subtypes (H10−/−; H1cH1dH1e−−−/−−−) were more severely retarded and abnormal than most E9.5 triple-null embryos. Therefore, an increased amount of H10 is unlikely to be the cause of the lethality of H1c H1d H1e triple-null embryos.
Marked reductions in linker histone content are tolerated in vivo.
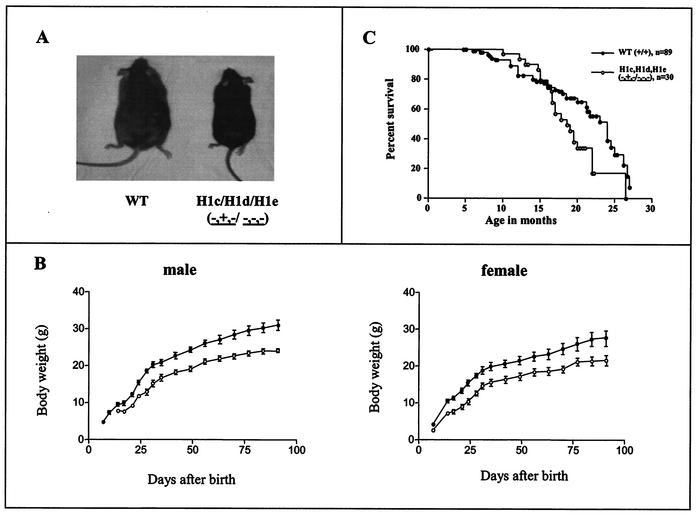
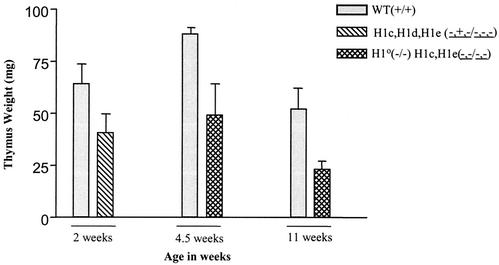
The foregoing results indicate that the compensatory mechanism present in the mouse H1 histone gene family is sufficient to maintain normal chromatin stoichiometry in the face of four null H1 alleles but not in the face of six null alleles. To determine whether a reduction in total H1 content is tolerated in mice, we generated two different strains of compound H1-null mice. Mice null for H1c and H1e but with one wild-type allele of H1d (H1cH1dH1e−+−/−−−) were produced by breeding H1c H1e double-null animals with H1c H1d H1e triple-heterozygous mutants. Mice null for H10, H1c, and H1e (H10−/−; H1cH1e−−/−−) were produced by intercrossing H10+/−; H1cH1e++/−− animals. Both types of mutants were very significantly underrepresented in litters (18% versus 50% expected [n = 269] and 2.32% versus 6.25% expected [n = 732], respectively), and in both cases, surviving mutants were much smaller shortly after birth and as adults (Fig. 6A and B and data not shown). We also studied the life span of H1c H1dH1e−+−/−−− mice. The Kaplan-Meier survival curve of the mutant appeared to differ slightly after about 17 months from that of wild-type controls (Fig. 6C), showing a very mildly shortened life span for H1cH1dH1e−+−/−−− mutant mice (P = 0.03). HPLC analyses of chromatin extracts from both these mutants and H10−/−; H1cH1e−−/−− mice showed that some tissues had 20 to 50% reductions in the total H1-to-nucleosome core ratio (Table 2). However, histopathology surveys of more than 35 tissue types did not reveal pathological or anatomical abnormalities, and most organs of the mutants were proportional to body size. In both strains, we found that total H1 stoichiometry was most severely depressed in the thymus, in which the H1-to-nucleosome ratio approached 50% of the wild-type level (Table 2). The H1-to-nucleosome ratios in the spleen and liver also were significantly reduced in the two strains, but not as much as in the thymus (Table2). Both strains also had significantly smaller thymi than their wild-type littermates (Fig. 7). Nevertheless, the histology of the thymus in both strains was normal.
FIG. 6.
H1cH1dH1e−+−/−−− mice are growth retarded. (A) Photograph of a representative H1cH1dH1e−+−/−−− mouse and a representative wild-type (WT) littermate at 4 months of age. (B) Growth curves of H1cH1dH1e−+−/−−− mice and littermate control mice. The left graph shows the mean body weight in grams ± the standard error of the mean for male mice (n = 7 for H1cH1e++/−− littermate controls, and n = 6 for H1cH1dH1e−+−/−−− mutant mice). The right graph shows the body weights of female mice (n = 7 for H1cH1e++/−− littermate controls, and n = 9 for H1cH1d−+−/−−− mutant mice). Symbols: •, controls; ○, mutants. (C) Kaplan-Meier survival curve of H1cH1dH1e−+−/−−− mutant mice and age-matched wild-type mice (P = 0.03).
TABLE 2.
H1 subtype composition and total H1 stoichiometry per nucleosome in wild-type and H1 compound-knockout micea
| Tissue and genotype | % of total H1b
|
Total H1/nucleosomec (no. of determinations) | |||||
|---|---|---|---|---|---|---|---|
| H1° | H1a | H1b | H1c | H1d | H1e | ||
| Thymus | |||||||
| Wild type (+/+) | 1.9 ± 1.6 | 10.7 ± 1.3 | 16.5 ± 0.9 | 26.7 ± 3.5 | 35.4 ± 2.7 | 9.6 ± 1.5 | 0.83 ± 0.10 (12) |
| H1°−/−; H1cH1e−−/−− | 0 | 17.0 ± 3.5 | 28.3 ± 3.0 | 0 | 54.7 ± 5.5 | 0 | 0.47 ± 0.26 (8) |
| H1cH1dH1e−+−/−−− | 5.9 ± 4.6 | 25.0 ± 3.0 | 35.4 ± 3.1 | 0 | 31.7 ± 2.6 | 0 | 0.41 ± 0.16 (6) |
| Liver | |||||||
| Adult | |||||||
| Wild type (+/+) | 29.0 ± 0.8 | 1.4 ± 0.6 | 15.8 ± 3.5 | 12.5 ± 1.0 | 14.3 ± 0.6 | 39.9 ± 2.5 | 0.79 ± 0.07 (5) |
| H1°−/−; H1cH1e−−/−− | 0 | 2.2 ± 0.3 | 6.3 ± 1.3 | 0 | 91.5 ± 1.6 | 0 | 0.64 ± 0.01 (4) |
| Neonatal | |||||||
| Wild type (+/+) | 9.5 ± 4.9 | 6.1 ± 2.4 | 14.2 ± 1.7 | 20.9 ± 4.5 | 30.3 ± 2.3 | 19.0 ± 1.5 | 0.76 ± 0.07 (5) |
| H1cH1dH1c−+−/−−− | 26.7 ± 6.5 | 9.9 ± 2.2 | 29.3 ± 4.6 | 0 | 32.7 ± 5.2 | 0 | 0.50 ± 0.15 (6) |
Data are from HPLC analyses of wild-type and H1 compound-knockout mice. Values are means ± standard deviations of individual determinations made on each of the indicated genotypes.
Determined by ratio of A214 of the indicated H1 peak to the total A214 of all H1 peaks. The A214 values of the individual H1 peaks were adjusted to account for the differences in the number of peptide bonds in each H1 subtype.
Determined by the ratio of the total A214 of all H1 peaks to half of the A214 of the H2b peak. The A214 values of the H1 and H2b peaks were adjusted to account for the differences in the number of peptide bonds in each H1 subtype and H2b.
FIG. 7.
Thymus weight is reduced in compound H1 knockout mice. Thymi from mice with the indicated genotypes were dissected and weighed at 2 weeks (wild type [WT] or littermate control, n = 9; H1cH1dH1e−+−/−−−, n = 10), 4.5 weeks (wild type, n = 2; H1o−/−; H1cH1e−−/−−−, n = 4), and 11 weeks (wild type, n = 7; H1o−/−; H1cH1e−−/−−, n = 2). Weights are presented as averages ± standard deviations.
Male and female mutants of both strains were fertile in crosses with wild-type animals, but crosses between mutants of the same strain often produced no litters or very small litters, which often contained primarily dead pups. These observations suggest that a combination of low maternal H1 mRNA abundance and low zygotic synthesis can lead to failure of embryogenesis. Taken together, the results indicate that deficits in total H1 content affect the probability of undergoing proper embryonic and postnatal development but that some embryos can tolerate a reduction in the level of H1 and survive to term and even grow to adulthood with some tissues exhibiting very substantial deficits in H1 content. Importantly, the birth of H1cH1dH1e−+−/−−− mice shows that a single wild-type allele of H1d is sufficient to partially rescue the embryonic lethality associated with nullizygosity for the H1c, H1d, and H1e subtypes.
Reduced linker histone content does not change nuclear size but leads to a global reduction in nucleosome spacing.
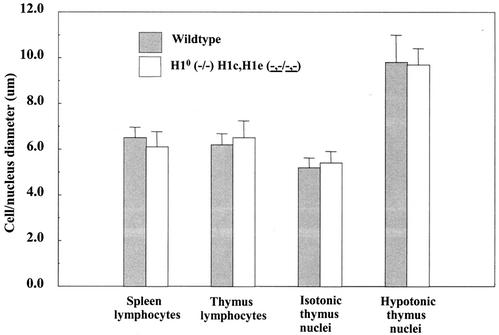
There are many potential effects of histone H1 depletion, ranging from large-scale changes at the level of the nucleus to local changes at the level of the nucleosome. To identify such effects, we made careful comparisons of features known or likely to be influenced by H1 levels. Since the net positive charge on H1 contributes to its ability to compact chromatin in vitro (8, 52), a significant loss of H1 may lead to an increase in nuclear size. Indeed, in Tetrahymena, macronuclei lacking H1 have a twofold increase in volume (41). Thymocytes and thymocyte nuclei are ideal for comparative measurements, being almost perfectly spherical and having a very narrow size distribution; in contrast, the pleiomorphy and large size variation of hepatocyte nuclei make such comparisons impracticable. Thymocytes from mice null for H10, H1c, and H1e and from wild-type animals were prepared gently in isotonic medium and allowed to attach to glass slides, and their diameters were recorded. Similarly, thymocyte nuclei were prepared and measured in isotonic medium. No significant differences were found in the diameter of either the cells or the nuclei (Fig. 8). To determine whether nuclei carry diffusible components, such as divalent cations or polyamines, that can compensate for the loss of H1, nuclei were allowed to equilibrate and swell in hypotonic medium prior to deposition on slides. Again, there were no significant differences in size (Fig. 8). This somewhat surprising result suggests that other fundamental changes in chromatin organization must compensate for the depletion of H1.
FIG. 8.
Nuclear size is not changed in lymphocytes depleted of H1. Spleen and thymus cells and nuclei of wild-type and H10 H1c H1e triple-null mice were equilibrated and measured in isotonic and chelating hypotonic buffers. Bars show mean diameters and standard deviations (n = 70 and 62, respectively, for wild-type and mutant spleen lymphocytes; n = 123 and 75, respectively, for wild-type and mutant thymus lymphocytes; n = 76 and 117, respectively, for wild-type and mutant isotonic thymus nuclei; n = 152 and 38, respectively, for wild-type and mutant hypotonic thymus nuclei).
One possible change in the properties of chromatin that may compensate for decreased H1 is a reduction in nucleosome spacing. Previous work has suggested a relationship between linker histones and the spacing between nucleosome core particles or nucleosome repeat length (NRL) (reviewed in reference 53). For example, in vitro chromatin reconstitution in drosophila egg extracts demonstrates that H1 has a powerful influence on NRL (5). However, inorganic cations or polyamines can exert similar effects in vitro and they can substitute for H1 in these reactions (5). To determine the relationship between H1 content and NRL of chromatin assembled in vivo, NRLs of wild-type and H1-depleted mutant tissues were determined from nucleosome ladders generated by micrococcal nuclease digestion of isolated nuclei. Precautions were taken to minimize potential artifacts caused by H1 exchange and nucleosome sliding during the procedure (see Materials and Methods). In all cases, the rates of digestion of wild-type and H1-depleted nuclei, as determined by the amounts of soluble chromatin released and the ratios of nucleosome oligomers to monomers, did not vary appreciably between wild-type and knockout nuclei (data not shown). Digestion conditions were selected to maximize the number of bands visible in gels, and in most cases, it was possible to observe lengths of up to nine nucleosomes (Fig. 9A). Plots of nucleosome number versus DNA length showed an excellent linear relationship, with the slope yielding the mean NRL (Fig. 9B). A potential source of artifacts in digestion experiments is the loss of linker DNA from the ends of cleaved polynucleosomes, the rate of which may be strongly influenced by the presence or absence of H1. The magnitude of this effect is inversely related to polynucleosome size and would be detectable on plots of DNA length versus nucleosome number as a reduction in the divergence of wild-type and mutant plots at higher nucleosome numbers. However, the data points were an excellent fit to the linear regression lines for the higher nucleosome numbers (Fig. 6B), showing that this “end” effect did not influence the NRL calculations. We established a conservative sensitivity limit for changes in NRL (ΔNRL) of 5 bp (see Materials and Methods).
FIG. 9.
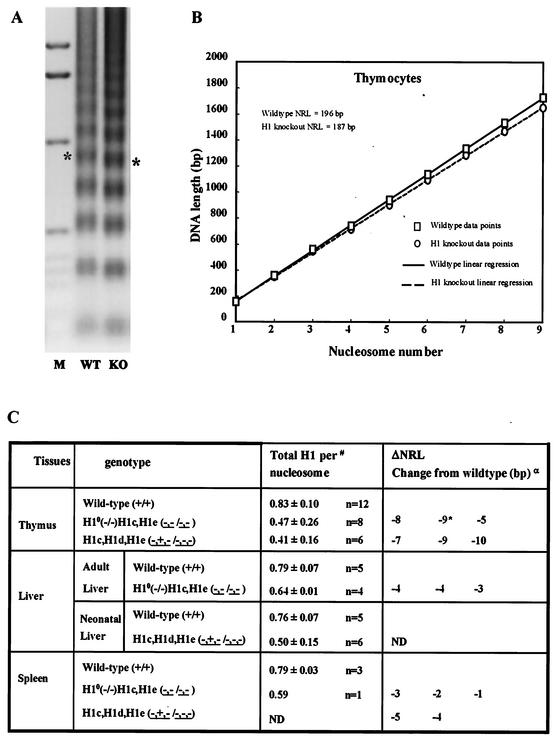
Nucleosome spacing is reduced in mutants with extensive reductions in total H1. (A and B) Thymus nuclei from wild-type (WT) and H10 H1c H1e triple-null (KO) mice. (A) Nucleosome repeat ladder obtained by micrococcal nuclease digestion of thymus nuclei as described in Materials and Methods. Lane 1 contains marker (M) DNA. Chromatin from the H1-deficient mutant nuclei migrates faster than that from wild-type nuclei, as illustrated by the asterisks at the pentanucleosome bands. (B) Plot of nucleosome number versus DNA length for thymus chromatin shown in panel A. Symbols show the data points, and lines are the linear regressions of the data points, which indicate a ΔNRL of 9 bp for these samples (see Materials and Methods for details). (C) Relationship between changes in H1 content and NRL. Superscripts: α, results of separate experiments (each comparison was done with different wild-type and knockout mice); #, from data in Table 2; *, data shown in panels A and B. ND, not determined.
Comparisons of ΔNRLs of wild-type and mutant thymus, spleen, and liver cells showed a clear correlation between the reduction in total H1 per nucleosome and ΔNRL (Fig. 9C). Thymus lymphocytes from two different compound-knockout strains, H10−/−; H1cH1e−−/−− and H1cH1dH1e−+−/−−−, with reductions in total H1 of 43 and 50% (about 0.45 molecule of H1 per nucleosome), showed very similar reductions in NRL, with a mean value of 8 bp. In contrast, the liver and spleen cells of mice with these genotypes had 75 to 80% of the wild-type H1 level and much smaller reductions in NRL. In every animal analyzed, spleen and liver NRLs for the knockouts were lower than for the wild type but the ΔNRLs were generally less than the 5 bp we established as significant. These results demonstrate that linker histones participate in determining the spacing between nucleosomes in vivo.
DISCUSSION
With the exception of yeast, which has a very low H1 content relative to that of nucleosomes, most of the eukaryotic cells examined have approximately equimolar amounts of H1 and nucleosome core particles (48). The abundance of H1 in chromatin and the existence of multiple specifically regulated subtypes in higher organisms suggest that H1 subtypes play an essential role in chromatin structure and function. However, until this report, studies of gene inactivation in a variety of organisms have failed to reveal an essential role for linker histones in vivo. Moreover, in vitro studies with cell extracts depleted of H1 also suggest that linker histones are not required for assembly of condensed chromosomes or nuclei (11, 32).
Linker histone genes have been completely eliminated only in Tetrahymena, yeast, and the filamentous fungus A. nidulans. Loss of H1 did not affect the growth or viability of these cells (34, 39, 41, 47). Elimination of H1 in Tetrahymena and yeast did affect the expression of specific genes, both positively and negatively (21, 40). Presumably, the affected genes were not essential for survival or the effects on them were not large enough to cause phenotypic consequences. Silencing of an H1 gene in another filamentous fungus, A. immersus, did lead to a shortened life span (3). Inactivation of individual H1 genes in several other organisms has generally not produced obvious phenotypic effects, although studies of Caenorhabditis elegans and tobacco have suggested a role for H1 subtypes in germ line development (24, 36). However, inactivation of the gene for the abundant, testis-specific histone H1t in mice had no apparent effect on spermatogenesis (15, 18, 29). Similarly, mice lacking the differentiation-associated linker histone H10, which is both the most divergent subtype and very abundant in certain tissues of adult mice, also did not exhibit obvious phenotypic defects (42). Loss of several other individual H1 subtypes also did not cause abnormalities in mice (17, 37).
The absence of a phenotype in strains of mice with a gene inactivated is often interpreted to mean that one or more other members of the gene family can compensate for the lost functions of the inactivated gene. This hypothesis may be difficult to prove, requiring the generation of compound mutants. This is an especially arduous task for members of a gene family that are tightly linked, such as the H1 genes. However, we were strongly motivated to carry out such studies because an analysis of chromatin from single H1-null mice showed that the stoichiometry of H1 to nucleosomes is normal in such mice, providing direct evidence for compensation. As shown here, the compensatory mechanism for linker histones in mice is sufficiently robust to permit loss of two abundant H1 subtypes (Fig. 2C). The mechanism seems to involve increased mRNA synthesis of specific H1 subtypes (Y.F. and A.I.S., unpublished observations), suggesting feedback or cross-regulation of transcription within the H1 gene family. Nevertheless, it is not sufficient to maintain a normal H1-to-nucleosome ratio in mice lacking three H1 subtypes. One explanation for the failure of compensation in the triple-null embryos is that the synthetic output of the remaining H1 genes is not sufficient. Alternatively, the compensation mechanism itself may depend on H1 stoichiometry or H1 subtype composition in the cells, which, if true, would also imply cross-regulation within the H1 gene family.
By sequentially inactivating three H1 genes in ES cells, preparing chimeric mice, and breeding heterozygous progeny, we have demonstrated that, unlike the H1 subtypes of Tetrahymena and yeast, linker histones are essential in mammals. Embryos lacking H1c, H1d, and H1e die by mid-gestation with a broad range of defects, extending from mild growth retardation and developmental delay to very severe abnormalities in various embryonic structures. The breadth of the phenotypes and time of lethality are consistent with the ubiquitous expression of these three H1 subtypes (1, 9, 27) and presumably also reflect the stochastic aspects of developmental processes.
We found that the chromatin of homozygous triple-null embryos has about 50% of the normal amount of H1 (Fig. 3C). This observation may suggest that, as in haploinsufficiency syndromes, having 50% of the normal amount of a gene product, in this case, H1 in chromatin, disrupts normal development. This hypothesis is also supported by the finding of severely affected thymus development in two types of compound H1-null mutants with H1-to-nucleosome ratios in the thymus approaching 50% of normal (see Results and Table 2). In contrast, the ratios in the livers of the same animals are reduced only 20 to 35% and liver development appears to be normal. How can having 50% of normal levels of linker histones disrupt development? We suggest that this level of reduction in H1-to-nucleosome stoichiometry leads to specific gene expression changes that perturb normal developmental processes. The relatively nonspecific abnormalities and variation in the time of death found for the triple-null embryos suggest that the expression of numerous genes in multiple cell types is affected. A principal defect in cell division is less likely because the embryos progress quite far. Another possible explanation for the embryonic lethality is that H1c, H1d, and H1e, which are the most closely related subtypes, constitute an essential subgroup of the mouse H1 subtypes, i.e., that, together, they function as a single unique subtype. This possibility seems unlikely because inactivation of the genes encoding the highly divergent H10 and H1t subtypes did not produce phenotypes in the corresponding tissues in which these subtypes are very abundant (29, 42). Furthermore, we have found that several different types of other single- and double-H1-null mutants are normal (this report and reference 17). It is noteworthy that in each of these single- and double-H1-null mutants, the H1-to-nucleosome core ratio is normal (Fig. 2 and references 17, 29, and 42). The fact that any single H1-null strain and several double-H1-null strains develop normally indicates that it is not any individual H1 or H1 subgroup, but rather the total amount of H1, i.e., the H1/nucleosome ratio, that is important for proper development. Furthermore, while H1c H1e double-null mutants are normal, the H10 H1c H1e triple-null mutants are very underrepresented in litters and the surviving mutants are retarded in growth. In addition, we also attempted to generate H10−/−; H1cH1dH1e−+−/−−− mice. Such mice were even more underrepresented in litters than either H1cH1d H1e−+−/−−− or H10−/−; H1cH1e−−/−− pups, and the few H10−/−; H1cH1dH1e−+−/−−− mice that were born all died within 3 days after birth (data not shown). These results further support the idea that the total amount of H1 is a key factor that influences mouse development. Another possible cause of the lethality may be that the marked deficiency of H1 allows for deposition of excess amounts of other molecules in chromatin that are deleterious to proper development. We tested one candidate, H10, that is increased about fivefold in the triple-null embryos and is normally expressed in large amounts only as cells become quiescent or undergo differentiation and terminal cell division (55). Inhibition of embryonic development by excess H10 was excluded by our finding that embryos lacking H10 and the three other subtypes (H10−/−; H1cH1dH1e−−−/−−−) also die by midgestation. Thus, proper chromatin function during development appears to require a minimum amount (∼50%) of the normal level of H1. Nevertheless, we found that substantial reductions in H1 content are tolerated in vivo. It may be that these reductions in H1 stoichiometry are withstood because H1 continuously exchanges among its binding sites in cellular chromatin (28, 31).
Linker histones are highly basic molecules, the typical mammalian H1 having 50 to 60 positively charged amino acids (48). As contributors to the electrostatic balance within chromatin and nuclei, they have a major impact on the extent of chromatin compaction in vitro (8, 52). Reduction of the H1 content of nuclei by ∼50% is therefore expected to be accompanied by either a compensatory mechanism to restore the electrostatic balance, a decreased level of chromatin compaction, or both.
In this context, our observations of a marked reduction in NRL in thymocytes and other cells with multiple H1 genes inactivated (Fig. 9) suggest that this phenomenon is, at least in part, a charge compensation mechanism, and indeed, the number of positive charges lost per nucleosome approximately balances the reduction in DNA negative charge due to the NRL changes. Correlations between H1 content and NRL have been observed in other systems: nuclei from ox cerebral cortex neurons have a short NRL (162 bp) compared with glial nuclei (201 bp) and a corresponding change in H1 content from 0.45 molecule to 1.04 molecules per nucleosome (35). Overexpression of H10 in cultured mouse fibroblasts resulted in a 15-bp increase in NRL (20), and Tetrahymena lacking macronuclear H1 has an NRL that is ∼10 bp shorter than that of the wild type (M. A. Gorovsky, personal communication). Inclusion of linker histones in in vitro reconstitution systems results in increased NRLs (5) and can also restore the native spacing of randomized nucleosomal arrays (43). The present work, in which we directly compared NRLs of normal and H1-deficient nuclei from the same mammalian cell type, significantly extends these earlier observations and suggests an underlying common mechanism. During chromatin replication, core histones are first bound to newly replicated DNA, at which time the NRL is short; only later, as H1 is added, is the mature NRL established (2). It is therefore possible that a deficit of new H1 molecules during replication in the compound H1-null cells results in failure of the normal NRL to be established.
Tetrahymena macronuclei lacking H1 are larger than wild-type macronuclei (41), presumably because of the absence of the normal level of chromatin condensation. This phenomenon was not seen in H1-depleted mouse cells. We carefully compared wild-type thymocyte nuclei, which are spherical and have a very narrow size distribution, with their counterparts from the H10-, H1c-, and H1e-null strains (mean ΔNRL, −8 bp), but we did not detect a size difference. Since we also found no differences in nuclear size after equilibration in hypotonic chelating buffer (Fig. 8), it seems unlikely that compensation for the loss of H1 positive charges involves increased levels of diffusible di- or polyvalent cations.
The studies reported here finally put to rest the possibility that H1 is nonessential in mammals. In recent years, the potential functions of linker histones have been expanded from chromatin condensation and the consequent general inhibition of transcription to include more specific gene regulatory activities (10, 14, 40, 54). The compound H1 knockout mice described here provide a unique approach to the identification of specific mammalian genes that are especially sensitive to H1 stoichiometry. Such genes are likely to have very important roles in mammalian embryonic development.
Acknowledgments
This work was supported by National Institutes of Health grants CA 79057 (A.I.S.) and GM43786 (C.L.W.).
We thank the AECOM Cancer Center Gene Targeting Facility (particularly Harry Hou for injection of ES cells), the AECOM Laboratory of Macromolecular Analysis, and the AECOM Comparative Pathology Facility. We thank Julianna Ayala, Changsheng Li, and Hui Xu for technical assistance and Robert Russell and Eric Bouhassira for helpful discussions.
REFERENCES
- 1.Adenot, P. G., E. Campion, E. Legouy, C. D. Allis, S. Dimitrov, J. Renard, and E. M. Thompson. 2000. Somatic linker histone H1 is present throughout mouse embryogenesis and is not replaced by variant H1 degrees. J. Cell Sci.113:2897-2907. [DOI] [PubMed]
- 2.Annunziato, A. T., and R. L. Seale. 1983. Chromatin replication, reconstitution and assembly. Mol. Cell. Biochem. 55:99-112. [DOI] [PubMed] [Google Scholar]
- 3.Barra, J. L., L. Rhounim, J.-L. Rossignol, and G. Faugeron. 2000. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol. Cell. Biol. 20:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, D. L., and J. O. Thomas. 1981. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 9:5883-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank, T. A., and P. B. Becker. 1995. Electrostatic mechanism of nucleosome spacing. J. Mol. Biol. 252:305-313. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. T. 2001. Histone variants: are they functionally heterogeneous? Genome Biol. Rev. 2:0006.1-0006.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, D. T., B. T. Alexander, and D. B. Sittman. 1996. Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 24:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, D. J., and T. Kimura. 1990. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 211:883-896. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, H. J., C. Oblin, and M. Bustin. 1992. Developmental regulation of chromatin composition during mouse embryogenesis: somatic histone H1 is first detectable at the 4-cell stage. Development 115:791-799. [DOI] [PubMed] [Google Scholar]
- 10.Crane-Robinson, C. 1999. How do linker histones mediate differential gene expression? Bioessays 21:367-371. [DOI] [PubMed] [Google Scholar]
- 11.Dasso, M., S. Dimitrov, and A. P. Wolffe. 1994. Nuclear assembly is independent of linker histones. Proc. Natl. Acad. Sci. USA 91:12477-12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doenecke, D., W. Albig, H. Bouterfa, and B. Drabent. 1994. Organization and expression of H1 histone and H1 replacement histone genes. J. Cell. Biochem. 54:423-431. [DOI] [PubMed] [Google Scholar]
- 13.Dong, Y., A. M. Sirotkin, Y. S. Yang, D. T. Brown, D. B. Sittman, and A. I. Skoultchi. 1994. Isolation and characterization of two replication-dependent mouse H1 histone genes. Nucleic Acids Res. 22:1421-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou, Y., C. A. Mizzen, M. Abrams, C. D. Allis, and M. A. Gorovsky. 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell 4:641-647. [DOI] [PubMed] [Google Scholar]
- 15.Drabent, B., P. Saftig, C. Bode, and D. Doenecke. 2000. Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem. Cell Biol. 113:433-442. [DOI] [PubMed] [Google Scholar]
- 16.Fan, Y., S. A. Braut, Q. Lin, R. H. Singer, and A. I. Skoultchi. 2001. Determination of transgenic loci by expression FISH. Genomics 91:66-69. [DOI] [PubMed] [Google Scholar]
- 17.Fan, Y., A. Sirotkin, R. G. Russell, J. Ayala, and A. I. Skoultchi. 2001. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H10 replacement subtype. Mol. Cell. Biol. 21:7933-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fantz, D. A., W. R. Hatfield, G. Horvath, M. K. Kistler, and W. S. Kistler. 2001. Mice with a targeted disruption of the H1t gene are fertile and undergo normal changes in structural chromosomal proteins during spermiogenesis. Biol. Reprod. 64:425-431. [DOI] [PubMed] [Google Scholar]
- 19.Georgel, P. T., and J. C. Hansen. 2001. Linker histone function in chromatin: dual mechanisms of action. Biochem. Cell. Biol. 79:313-316. [PubMed] [Google Scholar]
- 20.Gunjan, A., B. T. Alexander, D. B. Sittman, and D. T. Brown. 1999. Effects of H1 histone variant overexpression on chromatin structure. J. Biol. Chem. 274:37950-37956. [DOI] [PubMed] [Google Scholar]
- 21.Hellauer, K., E. Sirard, and B. Turcotte. 2001. Decreased expression of specific genes in yeast cells lacking histone H1. J. Biol. Chem. 276:13587-13592. [DOI] [PubMed] [Google Scholar]
- 22.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo—a laboratory manual, second edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Ioffe, E., Y. Liu, M. Bhaumik, F. Poirier, S. M. Factor, and P. Stanley. 1995. WW6: an embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proc. Natl. Acad. Sci. USA 92:7357-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jedrusik, M. A., and E. Schulze. 2001. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development 128:1069-1080. [DOI] [PubMed] [Google Scholar]
- 25.Lennox, R. W., and L. H. Cohen. 1984. The alterations in H1 histone complement during mouse spermatogenesis and their significance for H1 subtype function. Dev. Biol. 103:80-84. [DOI] [PubMed] [Google Scholar]
- 26.Lennox, R. W., and L. H. Cohen. 1984. The H1 subtypes of mammals: metabolic characteristics and tissue distribution, p. 373-397. In G. S. Stein, J. L. Stein, and W. F. Marzluff (ed.), Histone genes: structure, organization and regulation. John Wiley & Sons, Inc., New York, N.Y.
- 27.Lennox, R. W., and L. H. Cohen. 1983. The histone H1 complements of dividing and nondividing cells of the mouse. J. Biol. Chem. 258:262-268. [PubMed] [Google Scholar]
- 28.Lever, M. A., J. P. Th'ng, X. Sun, and M. J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873-876. [DOI] [PubMed] [Google Scholar]
- 29.Lin, Q., A. Sirotkin, and A. I. Skoultchi. 2000. Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol. Cell. Biol. 20:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meistrich, M. L., L. R. Bucci, P. K. Trostle-Weige, and W. A. Brock. 1985. Histone variants in rat spermatogonia and primary spermatocytes. Dev. Biol. 112:230-240. [DOI] [PubMed] [Google Scholar]
- 31.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 32.Ohsumi, K., C. Katagiri, and T. Kishimoto. 1993. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science 262:2033-2035. [DOI] [PubMed] [Google Scholar]
- 33.Parseghian, M. H., and B. A. Hamkalo. 2001. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem. Cell. Biol. 79:289-304. [PubMed] [Google Scholar]
- 34.Patterton, H. G., C. C. Landel, D. Landsman, C. L. Peterson, and R. T. Simpson. 1998. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 273:7268-7276. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, E. C., D. L. Bates, T. D. Prospero, and J. O. Thomas. 1984. Neuronal nuclei and glial nuclei from mammalian cerebral cortex: nucleosome repeat lengths, DNA contents and H1 contents. Eur. J. Biochem. 144:353-360. [DOI] [PubMed] [Google Scholar]
- 36.Prymakowska-Bosak, M., M. R. Przewloka, J. Slusarczyk, M. Kuras, J. Lichota, B. Kilianczyk, and A. Jerzmanowski. 1999. Linker histones play a role in male meiosis and the development of pollen grains in tobacco. Plant Cell 11:2317-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabini, S., K. Franke, P. Saftig, C. Bode, D. Doenecke, and B. Drabent. 2000. Spermatogenesis in mice is not affected by histone H1.1 deficiency. Exp. Cell Res. 255:114-124. [DOI] [PubMed] [Google Scholar]
- 38.Ramakrishnan, V. 1997. Histone H1 and chromatin higher-order structure. Crit. Rev. Eukaryot. Gene Expr. 7:215-230. [DOI] [PubMed] [Google Scholar]
- 39.Ramon, A., M. I. Muro-Pastor, C. Scazzocchio, and R. Gonzalez. 2000. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol. Microbiol. 35:223-233. [DOI] [PubMed] [Google Scholar]
- 40.Shen, X., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86:475-483. [DOI] [PubMed] [Google Scholar]
- 41.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82:47-56. [DOI] [PubMed] [Google Scholar]
- 42.Sirotkin, A. M., W. Edelmann, G. Cheng, A. Klein-Szanto, R. Kucherlapati, and A. I. Skoultchi. 1995. Mice develop normally without the H10 linker histone. Proc. Natl. Acad. Sci. USA 92:6434-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein, A., and P. Kunzler. 1983. Histone H5 can correctly align randomly arranged nucleosomes in a defined in vitro system. Nature 302:548-550. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, M., J. D. Hennebold, J. Macfarlane, and E. Y. Adashi. 2001. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 128:655-664. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, J. O. 1999. Histone H1: location and role. Curr. Opin. Cell Biol. 11:312-317. [DOI] [PubMed] [Google Scholar]
- 46.Travers, A. 1999. The location of the linker histone on the nucleosome. Trends Biochem. Sci. 24:4-7. [DOI] [PubMed] [Google Scholar]
- 47.Ushinsky, S. C., H. Bussey, A. A. Ahmed, Y. Wang, J. Friesen, B. A. Williams, and R. K. Storms. 1997. Histone H1 in Saccharomyces cerevisiae. Yeast 13:151-161. [DOI] [PubMed] [Google Scholar]
- 48.van Holde, K. E. 1989. Chromatin. Springer-Verlag, New York, N.Y.
- 49.Vignali, M., and J. L. Workman. 1998. Location and function of linker histones. Nat. Struct. Biol. 5:1025-1028. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Z. F., T. Krasikov, M. R. Frey, J. Wang, A. G. Matera, and W. F. Marzluff. 1996. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1 Mb. Genome Res. 6:688-701. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z. F., A. M. Sirotkin, G. M. Buchold, A. I. Skoultchi, and W. F. Marzluff. 1997. The mouse histone H1 genes: gene organization and differential regulation. J. Mol. Biol. 271:124-138. [DOI] [PubMed] [Google Scholar]
- 52.Widom, J. 1989. Toward a unified model of chromatin folding. Annu. Rev. Biophys. Biophys. Chem. 18:365-395. [DOI] [PubMed] [Google Scholar]
- 53.Wolffe, A. P. 1998. Chromatin: structure and function. Academic Press, San Diego, Calif.
- 54.Wolffe, A. P., S. Khochbin, and S. Dimitrov. 1997. What do linker histones do in chromatin? Bioessays 19:249-255. [DOI] [PubMed] [Google Scholar]
- 55.Zlatanova, J., and D. Doenecke. 1994. Histone H1 zero: a major player in cell differentiation? FASEB J. 8:1260-1268. [DOI] [PubMed] [Google Scholar]