Abstract
Mouse Aprt constructs that are highly susceptible to DNA methylation-associated inactivation in embryonal carcinoma cells were transfected into differentiated cells, where they were expressed. Construct silencing was induced by either whole-cell fusion of the expressing differentiated cells with embryonal carcinoma cells or by treatment of the differentiated cells with the DNA demethylating agent 5-aza-2′-deoxycytidine. Induction of silencing was enhanced significantly by the presence of a methylation center fragment positioned upstream of a truncated promoter comprised of two functional Sp1 binding sites. Initial silencing of the Aprt constructs was unstable, as evidenced by high spontaneous reversion frequencies (≈10−2). Stably silenced subclones with spontaneous reversion frequencies of <10−5 were isolated readily from the unstably silenced clones. These reversion frequencies were enhanced significantly by treatment of the cells with 5-aza-2′-deoxycytidine. A bisulfite sequence analysis demonstrated that CpG methylation initiated within the methylation center region on expressing alleles and that the induction of silencing allowed methylation to spread towards and eventually into the promoter region. Combined with the induction of revertants by 5-aza-2′-deoxycytidine, this result suggested that stabilization of silencing was due to an increased density of CpG methylation. All allelic methylation patterns were variegated, which is consistent with a gradual and evolving process. In total, our results demonstrate that silencing of mouse Aprt is a gradual process in the differentiated cells.
Aberrant silencing of tumor suppressor genes is now recognized to be as important as mutational events in human cancers (24). One of the hallmarks of silencing events is extensive CpG methylation of the affected gene promoters (3, 23). However, because malignant progression occurs over long periods of time in the body, it has been difficult to use tumor samples to determine whether the conversion of an actively expressed gene to one that has become heritably inactivated can occur in a single step, similar to the rapid fixation of mutagenic events (41) or, alternatively, if stable loss of expression is the result of a gradual process. It is also difficult to use tumors samples to determine when and how silencing of a particular gene is initiated, the relation between silencing and promoter region methylation (2, 9), and the source of promoter region methylation.
Because of limitations involved in using tumor specimens to model the interplay between loss of expression of tumor suppressors and DNA methylation, several groups have developed cellular systems to examine silencing of endogenous alleles or integrated transgenes. In one study, human mammary epithelial cells that underwent spontaneous or E6 papilloma gene-induced immortalization were examined for methylation and silencing of the p16 tumor suppressor gene (48). In this study, progressive methylation and loss of transcription were observed as a function of passage number, but a temporal relationship between these two events was not established. Another study examined the kinetics of silencing of a cDNA for green fluorescent protein (GFP) within a Moloney murine leukemia virus stably transfected into murine erthyroleukemia (MEL) cells (26). Cells that did not express GFP, suggesting integration in heterochromatic regions, were treated with the histone deacetylase inhibitor trichostatin A to activate GFP expression. In activated clones, GFP expression was then found to be lost gradually as a function of passage number, and this loss was correlated with progressive methylation of the GFP insert and the retroviral long terminal repeat. While both studies suggested a relation between loss of expression and increased methylation levels of the target genes, neither study identified the source of promoter region methylation.
In previous work, our investigators showed that removal or mutation of one of three consensus Sp1 binding sites from the mouse Aprt promoter sensitized it to stable, methylation-associated inactivation in embryonal carcinoma cells (31, 32). In these experiments the source of promoter region methylation was a DNA fragment, termed a methylation center, that attracts de novo DNA methylation (30). This fragment was placed upstream of the mutant promoters. Although these studies suggested that the spread of DNA methylation caused stable inactivation of the Aprt constructs, the integrated constructs were not shown to be expressed prior to their promoters being methylated. Therefore, the relation between silencing and DNA methylation was not revealed in these studies.
The work presented here examines the same Aprt constructs in differentiated cells, where they are readily expressed. Significantly, it was possible to induce silencing of these expressed constructs in the differentiated cells and to then follow a gradual process at the cellular and molecular levels. This process included the spread of DNA methylation from the methylation center fragment and the conversion of cells with unstably silenced phenotypes to those with stably silenced phenotypes. Taken as a whole, our work provides data to support a model (42) in which a severe drop in transcription removes a boundary that prevents the spread of DNA methylation to the promoter from a preexisting focus of methylation. According to this model, the spread of DNA methylation, in turn, leads to stabilization of the silencing process.
MATERIALS AND METHODS
Aprt constructs.
The creation of the constructs shown in Fig. 1 has been described elsewhere (31, 32).
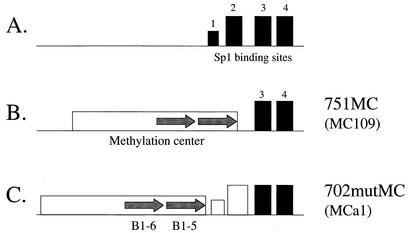
FIG. 1.
Constructs used to examine silencing in the differentiated cells. (A) The wild-type Aprt promoter is comprised of one nonconsensus Sp1 binding site (site 1) and three consensus binding sites (sites 2 to 4). (B) Sp1 binding sites 1 and 2 were deleted to create the 751MC construct (31), which has the methylation center fragment place upstream. The MC109 cell line used in this study contains three copies of this construct. (C) Sp1 binding sites 1 and 2 were mutated to eliminate any potential binding with Sp1 protein to create the 702mutMC construct (32). The MCa1 cell line used in this study contains three copies of this construct.
Cell culture and transfection.
All cell lines used in this study were derived from Aprt null P19 embryonal carcinoma cells (45) that lack both endogenous Aprt alleles. The isolation of the DIF-6 (differentiated morphology) (43) and 116U (embryonal carcinoma morphology) (40) cell lines were described elsewhere. The DIF-6 cells are also deficient for Hprt expression. All cells were grown routinely in Dulbecco's modified Eagle's medium (DMEM) (GibcoBRL) supplemented with 5% fetal bovine serum (Sigma) and 5% Serum Plus (JRH Biosciences).
All stable transfections were performed using cotransfection with the bacterial neo gene as described elsewhere (30, 31, 43). Briefly, 4 × 106 DIF-6 cells were transfected with 2 μg of neo plasmid and 10 μg of an Aprt construct (Fig. 1B and C), and the cells were selected for neo expression with 500 μg of G418 (GibcoBRL)/ml. A Southern blot analysis was then performed on G418-resistant clones to identify those that had also integrated the Aprt construct and, if so, to determine copy number. An APRT enzyme analysis (44) was used to identify expressing clones, and those with high-level expression (Table 1) were chosen for further analysis as discussed immediately below to identify transfectants suitable for this study.
TABLE 1.
Spontaneous and induced silencing frequencies for Aprt-expressing cell lines
| Cell line | Constructa | w/MCb | Sp actc | Inductiond | DAPr frequencye |
|---|---|---|---|---|---|
| MC109 | 751MC | Yes | 2.28 | None | 3.5 × 10−6 (3) |
| Fusion to EC | 1.5 × 10−2 (3) | ||||
| 5-aza-dC | 2.7 × 10−3 (5) | ||||
| MCa1 | 702mutMC | Yes | 1.51 | None | 9.5 × 10−6 (3) |
| Fusion to EC | 9.7 × 10−3 (8) | ||||
| 5-aza-dC | 1.2 × 10−3 (2) | ||||
| 702-5 | 702mut | No | 1.55 | None | <4.0 × 10−6 (2) |
| Fusion to EC | 8.5 × 10−5 (4)* | ||||
| 5-aza-dC | <5.9 × 10−6 (2) | ||||
| 751-3f | 751 | No | 2.25 | None | <1.4 × 10−6 (2) |
| Fusion to EC | <1.2 × 10−4 (5) | ||||
| 751-4f | 751 | No | 0.67 | None | <4.1 × 10−6 (5) |
| 5-aza-dC | <8.0 × 10−6 (2) |
See Fig. 1 for diagrams of constructs transfected into the cells. 702mut and 751 are versions of the 702mutMC and 751MC constructs, respectively, that lack the methylation center (MC) fragment.
“Yes” indicates construct contained methylation center fragment; “no” indicates methylation center fragment was not present.
Specific activities listed as nanomoles of adenine converted to AMP per minute per milligram of protein.
Induction conditions were fusion to embryonal carcinoma (EC) cells or treatment of cells with 3 μM 5-aza-dC. “None” indicates no treatment of cells and thus defines the spontaneous (background) level of DAP-resistant cells.
The frequency of cells resistant to 80 μg of DAP/ml. The number of independent experiments are shown in parentheses. For spontaneous and 5-aza-dC experimental determinations, the frequencies shown are averages. For fusion experimental determinations, the frequencies were determined by pooling all data obtained. The asterisk indicates a total of two DAP-resistant clones.
The 751-3 cells were unusually sensitive to 5-aza-dC. Therefore, a second cell line, 751-4, was tested with this agent instead.
There were two possible outcomes for the cotransfected plasmids. One was that the Aprt and neo plasmids would integrate on separate chromosomes and hence be unlinked. The second was that these plasmids would integrate on the same chromosome and hence be linked (i.e., syntenic). Linkage would allow us to use G418 selection for the neo construct to also maintain the Aprt constructs within the same cells regardless of their expression levels. To identify those transfectants in which the Aprt and neo constructs were linked, 105 cells from each Aprt-containing transfectant were plated in 100-mm dishes in the presence of 80 μg of 2′6-diaminopurine (DAP)/ml with or without 500 μg of G418/ml. DAP selects for cells that have lost significant Aprt expression. Those cotransfectants that yielded DAP-resistant clones at frequencies of approximately 10−4 when G418 was present were excluded from further analysis, as this constituted evidence that the neo and Aprt constructs were unlinked. Of a total of nine transfectant cell lines that were tested, four fell into this category. The remaining five Aprt-containing transfectants yielded a low frequency or undetectable numbers of DAP-resistant clones in the presence of G418, demonstrating linkage of the neo and Aprt constructs, and they were chosen for further experiments (Table 1). These cells lines were routinely maintained in G418-containing medium for all remaining experiments presented in this report.
APRT assay.
The assay for APRT enzymatic activity was performed as described elsewhere (44).
Induction of silencing by whole-cell fusion
For whole-cell fusions, 6.7 × 106 116U embryonal carcinoma cells and 2.3 × 106 differentiated transfectants (Table 1) were plated together in 10-cm tissue culture dishes and allowed to adhere for 6 h. The cells were then fused by removing the medium, rinsing twice with 1× phosphate-buffered saline, and exposing them to 1.5 ml of a 50% solution of polyethylene glycol (PEG 1000) dissolved in serum-free DMEM for 125 s at room temperature. PEG 1000 was a gift of Brett Spear. The PEG solution was removed by rinsing four times with 20 ml of phosphate-buffered saline, and fresh medium was added. The cells were allowed to recover overnight, trypsinized, and plated at a density of 1 × 105 to 2 × 105 cells per 15-cm dish in DMEM supplemented with 1 mg of G418/ml. The following day the medium was changed to DMEM supplemented with G418, 50 μM azaserine, and 60 μM hypoxanthine to select hybrid cells. Selection was maintained for 12 to 14 days until fusion clones were present.
Fusion-induced silencing of Aprt constructs was assessed by changing the medium on the fusion selection plates to DMEM supplemented with G418 and DAP. The number of DAP-resistant clones was determined following 7 to 10 days of selection. DAP-resistant fusion clones expanded for further analysis were maintained under DAP-G418 selection. Approximately 1,000 to 2,000 viable hybrid clones were obtained per fusion experiment.
Induction of silencing by exposure to 5-aza-dC.
To determine the ability of 5-aza-2-deoxycytidine (5-aza-dC) to induce silencing, 8 × 105 differentiated transfectants were plated in a T75 flask and exposed to 3 μM 5-aza-dC for 24 h. One week later the cells were plated at densities of 1 × 105 to 1.5 × 105 cells per 100-mm dish and exposed to DAP-G418 medium. DAP-resistant clones were identified after 10 to 14 days of selection, and some clones were isolated and expanded for further analysis
Determination of reversion frequencies.
To determine reversion frequencies, 105 cells were plated per 100-mm dish and exposed to medium containing 50 μM azaserine and 60 μM adenine (AzA medium). Growth in AzA medium requires significant Aprt expression.
Determination of Aprt expression by RT-PCR.
Total RNA was isolated with an RNeasy mini kit (Qiagen) with on-column DNase I digestion according to the manufacturer's instructions. Two micrograms of total RNA was used for first-strand cDNA synthesis, using a Superscript II cDNA synthesis kit with a poly(dT) primer according to the manufacturer's instructions (GibcoBRL). Following RNase H digestion, 1 μl of the first-strand cDNA reaction mixture for each sample was subjected to PCR with primers specific for Aprt gene transcripts (AP-4S, 5′-CAG AGA GTG GTC ATT GTG GAT G; AP-5A, 5′-CGG TAG CTC ACA AAG GTC ACT TAG) or Neo (Neo-S, 5′-GAC TGG GCA CAA CAG ACA ATC; neo-A, 5′-CAC AGT CGA TGA ATC CAG AAA AG). The number of PCR cycles for each primer set that yielded sufficient product within the linear phase of amplification was determined empirically. Because neo expression was identical in all derivatives from a given parental cell line, the neo-specific reverse transcription-PCR (RT-PCR) product served to demonstrate equivalent sample input amounts and efficiency of the RT reaction for the corresponding Aprt-specific RT-PCR products.
Bisulfite sequencing analysis.
BsrI-digested genomic DNA was modified with a 5.36 M urea, 3.44 M sodium bisulfite solution as described previously (49). PCR amplification of modified samples, cloning of PCR products, and sequence analysis were also as described elsewhere (49), except that primer S1 (upstream of B1 repetitive elements) was paired with primer ACA-29 (5′-AAA AAC AAA AAA AAA ATA AAT ATC AAC AC; downstream of Aprt promoter), and the resulting PCR products were TA-cloned directly without performing a seminested PCR.
FISH analysis.
The fluorescence in situ hybridization (FISH) analysis for Aprt integration sites was performed as described elsewhere (37) using the Aprt gene as the probe. No endogenous Aprt alleles were present in the transfected cells.
RESULTS
Mutant Aprt constructs are expressed in differentiated cells.
The mouse Aprt promoter is comprised of three consensus Sp1 binding sites (sites 2 to 4) and a nonconsensus site (site 1) that has little if any function (Fig. 1A) (32). Binding sites 3 and 4 are required for maximal levels of Aprt transcription, whereas binding site 2 is believed to be dispensable for transcription (16). However, this binding site is required to prevent silencing of Aprt in embryonal carcinoma cells, as its removal (31) or mutation (32) renders Aprt susceptible to methylation-associated silencing when the promoter is placed downstream of the cis-acting methylation center (Fig. 1B and C). This center contains two B1 repetitive elements, termed B1-5 and B1-6 (49).
Differentiated cells derived from the embryonic carcinoma cells have a reduced capacity for de novo DNA methylation (43). To determine if the mutant constructs would inactivate in these cells, the constructs shown in Fig. 1B and C were transfected stably into the DIF-6 cell line (43) by cotransfection with the bacterial neo construct. For the 751MC construct (Fig. 1B) 10 of 13 transfectants expressed detectable levels of APRT enzymatic activity, and for the 702mutMC construct (Fig. 1C) 3 of 3 transfectants expressed APRT activity, for a total of 13 of 16 (81%) expressing transfectants (data not shown). In contrast, when these constructs were transfected stably into embryonal carcinoma cells, only 3 of 14 (21%) transfectants expressed detectable levels of APRT enzymatic activity (31, 32).
Induction of Aprt silencing in the differentiated cells.
Having demonstrated that the Aprt constructs containing the methylation center fragment could be expressed in the differentiated cell lines, we chose two transfectants (MC109 and MCa1) with high APRT specific activities (Table 1) for further testing to determine if it would be possible to induce silencing and then follow postinduction events. The high specific activities, which were comparable to those of highly expressing transfectants containing Aprt constructs lacking the methylation center (702-5, 751-3, and 751-4) (Table 1), suggested strongly that the constructs had integrated in expressed (i.e., euchromatic) regions of the genome. In both cases the expressing cells contained three copies of the Aprt constructs, thus preventing mutational inactivation from playing a major role in loss of Aprt expression. G418 selection was used to ensure retention of the constructs (see Materials and Methods for more details). A FISH analysis with an Aprt probe demonstrated that for the MC109 transfectant the three constructs had integrated into distinct sites on a single chromosome. For most MCa1 cells it appeared that the Aprt constructs had integrated into two discrete sites of a single chromosome, though some cells exhibited a second chromosome with a single hybridization signal. Low-frequency spontaneous silencing (10−5 to 10−6), as defined by the number of clones spontaneously resistant to DAP (Fig. 2), was observed for both cell lines. A Southern blot analysis demonstrated retention of the Aprt constructs in the silenced cells (data not shown). No spontaneous silencing events were observed for transfectants containing Aprt constructs lacking the methylation center fragment (Table 1).
FIG. 2.
Selection strategy for cells with silenced alleles. Parental cells (e.g., MC109) containing three copies of expressed Aprt constructs were either fused to embryonal carcinoma cells or treated with 5-aza-dC, and silenced clones were selected initially with DAP (e.g., 109AzcD4). These silenced clones were expanded and either selected in FA (e.g., 109AzcD4F12), subcloned in DAP (e.g., 109AzcD4S2), or grown in DAP as a mass culture for additional weeks. The different nomenclature strategies shown in this figure are used in Table 2 and in the text.
The comparison of Aprt construct expression in transfected embryonal carcinoma versus differentiated cells suggested that one or more factors present in the embryonal carcinoma cells could cause inactivation of the transfected Aprt constructs and that these putative factors were missing from the differentiated cells. To test this hypothesis, whole-cell fusions were performed between embryonal cells lacking Aprt and differentiated cells containing expressed Aprt constructs (MC109 and MCa1) (Fig. 1 and Table 1). The frequencies of DAP-resistant clones (Fig. 2) were found to be approximately 10−2 in proliferating hybrid cells (Table 1), demonstrating that the fusion process was a potent inducer of silencing. To determine if the methylation center fragment played a role in the cell fusion-induced silencing events, differentiated transfectants lacking the methylation center fragment (751-3 and 702-5) were also fused to the embryonal carcinoma cells. For one cell line (702-5), a total of two viable DAP-resistant clones were observed after fusion to the embryonal carcinoma cells, for an induction frequency of 8.5 × 10−5, compared with induced frequencies of approximately 1 × 10−2 for the transfectants containing constructs with the methylation center fragment. The other cell line tested (751-3) did not yield viable DAP-resistant clones (<1.2 × 10−4) when fused to the embryonal carcinoma cells (Table 1). These results suggest that the methylation center fragment contributed significantly to the silencing process induced by fusion of the differentiated cells with the embryonal carcinoma cells.
It has been reported by others that the demethylating agent 5-aza-dC can induce silencing of integrated bacterial gpt transgene in Chinese hamster cells (7). Therefore, we also tested the expressing cell lines to determine if 5-aza-dC exposure could induce silencing in the differentiated cells (Fig. 2). Induced silencing was observed for expressed constructs containing the methylation center fragment at frequencies of approximately 10−3 (Table 1). Silencing was not induced by 5-aza-dC in differentiated transfectants containing constructs that lacked the methylation center fragment.
Silencing of constructs with the methylation center fragment was not observed when the MC109 and MCa1 cells were exposed to ionizing radiation or benzo[α]pyrene (data not shown), demonstrating that these classical genotoxins could not induce silencing and, by extension, that DAP-resistant clones were not induced by the mutagenic effect of 5-aza-dC (22).
Silencing in the differentiated cells is initially unstable.
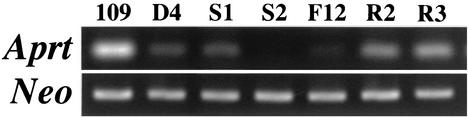
Inactivation of the mutant Aprt constructs in embryonal carcinoma cells was quite stable, as established by their inability to reactivate spontaneously (31). To establish if silencing in the differentiated cells was also stable, we determined spontaneous reversion frequencies for some DAP-resistant clones induced by whole-cell fusion and by 5-aza-dC treatment (109AzcD3, 109AzcD4, 109FusD2, 109FusD4, MCa1FusD2, and MCa1FusD8). Reversion frequencies of approximately 10−2 (Table 2) were observed for all of the tested clones, demonstrating that the silenced alleles could reactivate at relatively high frequencies. Therefore, silencing in these DAP-resistant clones was considered to be unstable (10, 11). The spontaneous reversion frequencies were not altered significantly after continued growth of the cells as mass cultures in DAP medium for up to 2 months (Table 2). We also examined APRT enzyme levels (data not shown) and mRNA levels (Fig. 3) in some of the DAP-resistant clones and found that in both cases the levels were reduced significantly, yet not completely, when compared with the expressing parental cells. The presence of residual APRT activity suggested that the unstable silencing events were incomplete (i.e., leaky). Spontaneous revertant cells regained significant amounts of Aprt mRNA (Fig. 3), though the regained levels were not as high as the parental expressing cells.
TABLE 2.
Spontaneous reversion frequencies distinguish unstable and stable silencing eventsa
| Cell lineb | Selectionc | Reversion frequencyd
|
Phenotypea | |
|---|---|---|---|---|
| Spontaneous | 5-azc-dC exposuree | |||
| 109AzcD3 (3) | DAP | 1.6 × 10−2 | Unstable | |
| 109AzcD3 (8) | DAP | 2.4 × 10−2 | Unstable | |
| 109AzcD3 (12) | DAP | 7.9 × 10−3 | Unstable | |
| 109AzcD3S2 | DAP/DAP | <8.3 × 10−6 | Stable | |
| 109AzcD4 (3) | DAP | 2.6 × 10−2 | Unstable | |
| 109AzcD4 (8) | DAP | 7.8 × 10−3 | Unstable | |
| 109AzcD4 (12) | DAP | 6.4 × 10−3 | Unstable | |
| 109AzcD4F11 | DAP/FA | 4.7 × 10−6 | Stable | |
| 109AzcD4F12 | DAP/FA | 6.2 × 10−6 | 6.3 × 10−3 | Stable |
| 109AzcD4S1 | DAP/DAP | 2.3 × 10−2 | Unstable | |
| 109AzcD4S2 | DAP/DAP | 5.0 × 10−6 | 1.8 × 10−3 | Stable |
| 109AzcD4S3 | DAP/DAP | 2.3 × 10−3 | Unstable | |
| 109AzcD4S6 | DAP/DAP | 4.9 × 10−3 | Unstable | |
| 109FusD3 | DAP | 1.0 × 10−2 | Unstable | |
| 109FusD2 | DAP | 5.7 × 10−3 | Unstable | |
| 109FusD2S2 | DAP/DAP | <7.9 × 10−6 | 3.2 × 10−3 | Stable |
| 109FusD2S3 | DAP/DAP | 4.4 × 10−4 | NC | |
| 109FusD2S4 | DAP/DAP | 9.0 × 10−3 | Unstable | |
| 109FusD2S5 | DAP/DAP | 5.9 × 10−4 | NC | |
| 109FusD2S6 | DAP/DAP | <7.0 × 10−6 | Stable | |
| MCa1FusD2 | DAP | 4.8 × 10−3 | Unstable | |
| MCa1FusD8 | DAP | 2.5 × 10−2 | Unstable | |
Unstable silencing events defined by spontaneous reversion frequencies of >10−3 and stable silenced events defined by spontaneous reversion frequencies of <10−5. NC, not classified.
Cell lines with silenced Aprt alleles. Silencing was induced by 5-aza-dC (Azc) or by whole-cell fusion (Fus). Numbers in parentheses indicate weeks in culture after silencing was induced.
“DAP” indicates initial DAP-selected clone; “DAP/FA” indicates FA-selected subclone isolated from an unstably silenced DAP-resistant clone; “DAP/DAP” indicates DAP-selected subclone isolated from an unstably silenced DAP-resistant clone. See Fig. 2 for more details.
Reversion frequencies determined in medium containing 60 μM adenine and 50 μM azaserine.
Revertants induced by treating with 3μM 5-aza-dC.
FIG. 3.
RT-PCR analysis for Aprt expression. RNA samples were isolated from the parental MC109 cell line (109), a 5-azc-dC-induced DAP-resistant clone, 109AzcD4 (D4), an unstable (S1) and a stable (S2) subclone, a stable FA-selected subclone (F12), and two spontaneous revertants (R2 and R3) isolated from the S1 subclone. Expression from the cotransfected bacterial neo gene was used to confirm that equivalent amounts of RNA were used. See Materials and Methods for more details on methods and Table 2 and text for more details on cell lines.
If Aprt silencing was leaky at the cellular level in the induced DAP-resistant clones, we would expect that each cell within the population would be expressing a small amount of Aprt mRNA and protein. Alternatively, a cell population would appear leaky for expression if it were comprised of a small pool of highly expressing revertant cells and a far larger population of nonexpressing cells. To distinguish between these possibilities, we tested the cells of a newly isolated DAP-resistant clone (109AzcD4) (Table 2) for their ability to clone in the presence of DAP versus their ability to clone in the presence of 2-fluoroadenine (FA). FA has a tighter binding affinity than DAP for the APRT enzyme (25) and, therefore, it can kill DAP-resistant cells that express low levels of normal protein (i.e., that are leaky for expression). The relative cloning efficiency for the 109AzcD4 cells was 58.0% in DAP, but only 4.6% in FA. This result supports the notion that unstable silencing is leaky at the cellular level.
Stably silenced clones can be isolated readily from unstably silenced clones.
Although the majority of the 109AzcD4 cells were sensitive to FA, the presence of an FA-resistant variant population suggested that the cellular phenotype of unstable silencing was not an end point. To test for this possibility, we isolated two FA-selected subclones (109AzcD4F11 and 109AzcD4F12) (Fig. 2) and then determined their spontaneous Aprt reversion frequencies. These frequencies were approximately 5 × 10−6, compared with spontaneous reversion frequencies of approximately 1 × 10−2 for the 109AzcD4 cell line from which the FA-selected clones were derived (Table 2). This 2,000-fold difference in reversion frequencies demonstrated that FA selected for cells with stably silenced Aprt alleles and, by extension, that unstable silencing could progress to a more stable form of silencing.
To determine if stably silenced clones could arise in the absence of FA selection (i.e., if they were present prior to exposure to FA), we isolated subclones (Fig. 2) of 5-aza-dC-induced (e.g., 109AzcD4S2) and cell fusion-induced (e.g., 109FusD2S2) silenced clones in DAP and determined their spontaneous reversion frequencies. As shown in Table 2, many of the subclones still yielded spontaneous revertants at high frequencies (10−2 to 10−3), but some subclones appeared to be stably silenced, as defined by reversion frequencies of <10−5. These results demonstrated that a phenotypically unstable cell population is actually a mixture of cells that are unstably and stably silenced. Stably silenced subclones (109AzcD4S2 and 109AzcD4F11) exhibited markedly reduced or absent Aprt mRNA compared with an unstably silenced sib clone (109AzcD4S1) and the original unstably silenced DAP-resistant clone (109AzcD4) from which they were all derived (Fig. 3).
To determine if DNA methylation was involved in stable silencing, three of the stably silenced subclones were treated with 5-aza-dC, and reversion frequencies were found to increase 100-fold or greater compared with the spontaneous frequencies (Table 2). This result is consistent with a role for DNA methylation in the silencing process, at least for the maintenance of stable silencing.
Finally, to determine if stable inactivation could occur in a single induction step, we treated the MC109 cells with 5-aza-dC to induce silencing and compared the frequencies of induced DAP- and FA-resistant clones. In a first experiment, the frequency of DAP-resistant clones was 1.1 × 10−3, whereas no FA-resistant clones were observed (<10−5). In a second experiment, we plated a larger number of 5-aza-dC-treated MC109 cells in FA and in this case observed several FA-resistant clones at a frequency of 5.0 × 10−6. This is 200-fold lower than the frequency of 5-aza-dC-induced DAP-resistant clones. These experiments, coupled with those presented above, suggested that the probability of an allele becoming stably silenced is enhanced significantly if it can progress through an unstably silenced intermediate.
The induction of silencing allows the spread of DNA methylation from the methylation center region.
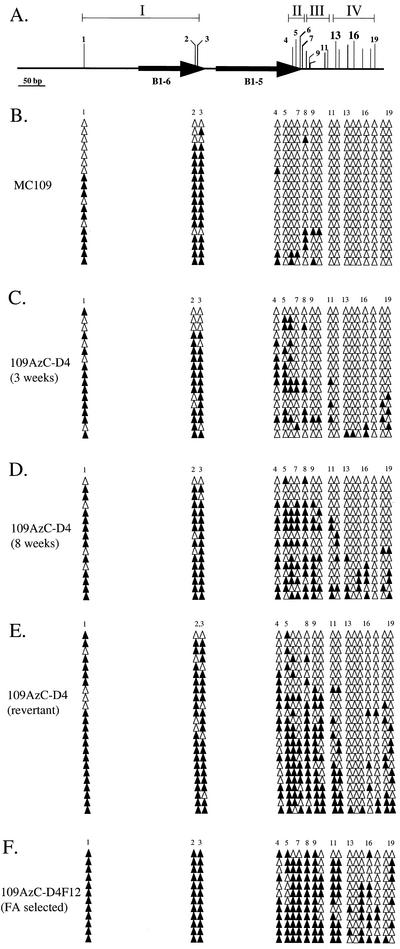
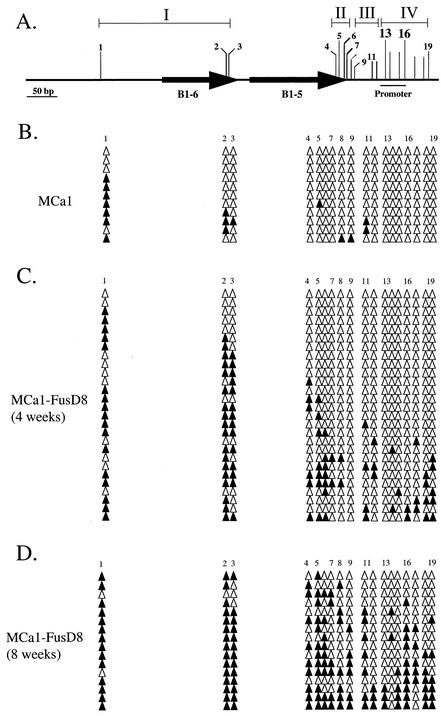
Four phenotypes were described with the above work: the parental expressing cells, unstably silenced cells (reversion frequencies of >10−3), stably silenced cells (reversion frequencies of <10−5), and revertant cells. Bisulfite sequencing was used to determine the relations between CpG methylation and these cellular phenotypes. For the purpose of this analysis, we grouped CpG sites on the 751MC construct (Fig. 1B) in the expressing MC109 cells into four regions (Table 3; Fig. 4A). Region I includes three CpG sites, two within the B1-6 repetitive element (sites 2 and 3) and an upstream CpG site (site 1). Region II includes four CpG sites (sites 4 to 7) within the B1-5 repetitive element. Region III includes five CpG sites (sites 8 to 12) located between the B1-5 element and Sp1 binding site 3. These CpG sites are remnants of the polylinker cloning site used to make the Aprt constructs shown in Fig. 1. Region IV contains seven CpG sites from the Aprt promoter region (sites 13 to 19), beginning with CpG site 13 in Sp1 binding site 3 and including CpG site 16 in Sp1 binding site 4. The four regions are essentially the same for the 702mutMC construct (Fig. 1C) in the expressing MCa1 cells, except there is no CpG site 10 (Fig. 5A).
TABLE 3.
| Cells | Relevant informationb | CpG methylation (%) in region:
|
|||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| MC109 | Expressing parental (4B) | 81 | 8 | 7 | 0 |
| 109AzcD4 | Silenced ∼3 weeks (4C) | 71 | 37 | 12 | 8 |
| 109AzcD4 | Silenced ∼8 weeks (4D) | 83 | 50 | 40 | 13 |
| 109AzcD4 | Silenced ∼12 weeks | 92 | 42 | 38 | 22 |
| 109AzcD4R | Revertant (4E) | 82 | 54 | 34 | 13 |
| 109AzcD4F12 | Stable subclone (4F) | 100 | 79 | 80 | 29 |
| 109AzcD4S2 | Stable subclone | 91 | 70 | 56 | 27 |
| MCa1 | Expressing parental (5B) | 33 | 2 | 9 | 0 |
| MCa1FusD8 | Silenced ∼4 weeks (5C) | 60 | 20 | 8 | 9 |
| MCa1FusD8 | Silenced ∼9 weeks (5D) | 88 | 56 | 34 | 32 |
Regions are shown in Fig. 1 and discussed in the text.
Cell phenotype and matching figure (in parentheses) showing allele-by-allele methylation patterns.
FIG. 4.
Methylation patterns for the expressing MC109 and derived silenced cells. (A) CpG sites analyzed were divided into four regions (see text for details). Arrows represent the B1-5 and B1-6 repetitive elements within the methylation center. CpG sites 13 and 16 are located within Sp1 binding sites 3 and 4, respectively. (B to F) Bisulfite sequencing analyses to identify methylated (closed triangles) and unmethylated (open triangles) CpG sites.
FIG. 5.
Methylation patterns for the expressing MCa1 and derived silenced cells. (A) CpG sites analyzed were divided into four regions (see text for details). Arrows represent the B1-5 and B1-6 repetitive elements within the methylation center. CpG sites 13 and 16 are located within Sp1 binding sites 3 and 4, respectively. There is no CpG site 10 on this construct. (B to D) Bisulfite sequencing analyses to identify methylated (closed triangles) and unmethylated (open triangles) CpG sites.
CpG methylation was observed in the expressing parental cell lines, but it was mostly limited to region 1, with 81 and 33% of available CpG sites methylated in the expressing MC109 and MCa1 cells, respectively (Table 3 and Fig. 4B and 5B). For both cell lines, less than 10% of CpG sites in regions II and III were methylated, and no methylation was observed in region IV. These results demonstrated that the methylation center could function at a reduced level in the differentiated cells and that the two remaining Sp1 binding sites in the promoter were sufficient to block the spread of DNA methylation in these cells.
Construct methylation was next examined in 5-azc-dC-induced (109AzcD4) and cell fusion-induced (MCa1-FusD8) Aprt-deficient clones that were unstably silenced, using DNA isolated within 3 to 4 weeks after the start of DAP selection. In both cases the overall levels of construct methylation increased for these silenced alleles when compared with those in the parental expressing cells, with the observed patterns showing that methylation had begun spreading from the methylation center towards the Sp1 binding sites (Table 3; Fig. 4C and 5C). Nonetheless, most alleles (63 to 65%) did not exhibit any CpG methylation in region IV, which includes the Sp1 bindings sites. CpG methylation for the Aprt constructs was examined after the silenced cell lines were maintained in culture under DAP selection for an additional 1 and 2 months (Table 3, Fig. 3D and 4D, and additional data not shown). Increased methylation levels were observed when compared with the initially silenced cells, with the patterns again revealing increased spread of DNA methylation from the methylation center fragment towards the Sp1 binding sites on many alleles. However, relatively little change was observed between the 8- and 12-week time points (Table 3), which was the longest time point that any clonally derived cultures was maintained. We also examined the methylation pattern for a revertant cell line that was selected and maintained in AzA medium. In this case, approximately one-third (30%) of the alleles were not methylated in region IV and the remaining alleles were methylated (70%) (Fig. 4E).
For a final analysis, we examined CpG methylation in an FA-selected subclone, 109AzcD4F12 (Fig. 4F; Table 3). This subclone was characterized as being stably silenced (Table 2). CpG methylation levels were found to be highest for all regions in this stably silenced subclone and continued spreading of methylation towards the promoter was again apparent (Fig. 3D; Table 3), though methylation of the Sp1 binding sites was clearly not required for silencing to occur. Similar results were obtained for a second stably silenced clone, 109AzcD4S2 (Table 3). In total, the methylation analysis demonstrated that the spread of DNA methylation from the methylation center region towards the promoter occurred almost entirely subsequent to the acquisition of DAP resistance (i.e., initial silencing of Aprt).
DISCUSSION
Rodent Aprt has proven useful for examining various parameters of gene silencing and/or DNA methylation in mammalian cells (6, 28, 31, 32, 36). One advantage of this gene over other silencing models is that it is possible to use selective pressure to isolate either Aprt-deficient or Aprt-expressing cells, which allows selective processes that occur during cancer progression to be mimicked. In this study we examined silencing of expressed mouse Aprt constructs transfected into differentiated cells. It was possible to conduct this study because Aprt expression was maintained at high levels in these cells, as evidenced by very low spontaneous silencing frequencies (Table 1), presumably due to integration in actively expressed regions of the genome. Only G418 selection was used to retain the mouse Aprt constructs in the cells. Two methods were found to induce silencing of the Aprt constructs: fusion of the differentiated cells with embryonal carcinoma cells, and treatment of the differentiated cells with 5-aza-dC. Although we do not yet understand how either of these methods induced silencing (see below), they were extremely useful to allow us to mark the approximate time when silencing was initiated and then to follow postinduction events as they unfolded. This analysis led to a number of interesting observations that, in total, allow us to describe a new model for a relation between silencing and the spread of DNA methylation in mammalian cells.
The allele-by-allele analysis shown in Fig. 4 and 5 revealed that the spread of DNA methylation is progressive (i.e., methylation levels continue to increase with time) but not processive (i.e., methylation of CpG sites does not occur sequentially on a given allele). Because there are multiple Aprt alleles per cell, the changes in methylation patterns that are observed could be occurring both within cells and between cells. Progressive methylation has also been shown for silenced p16 alleles in transformed human mammary epithelial cells (48) and for GFP constructs in MEL cells (26). Moreover, our analysis revealed a dynamic process, because almost all alleles examined from a given subclone exhibited distinct methylation patterns. These observations can only be explained by assuming that methylation patterns in the silenced cells are in flux, which suggests that allele-specific pressures can convert nonmethylated sites to being methylated and, perhaps less frequently, change methylated sites to being nonmethylated. This shifting of methylation status for specific CpG sites could explain the variegated methylation patterns that were observed in this study and in other studies with cultured cells (26, 39, 48) and tumor specimens (8, 14, 15, 29, 33).
Our ability to select for reversion events allowed us to demonstrate a wide variation in the extent or “stability” of silencing, because spontaneous reversion frequencies for silenced clones ranged over almost 4 orders of magnitude, from 1.6 × 10−2 to 6.2 × 10−6. Significantly, all DAP-resistant clones when first isolated exhibited high spontaneous reversion frequencies of approximately 10−2, indicating that silencing was relatively unstable when first induced. These reversion frequencies could drop dramatically when the cells were either subcloned in DAP, which yielded clones with the full range of reversion frequencies, or selected in FA, which yielded clones with spontaneous reversion frequencies of <10−5. FA has a tighter binding affinity for the APRT enzyme than DAP (25) and, therefore, provides a second level of selection for further reductions in Aprt expression. In contrast to these methods, continued passage of the cells for up to 2 months had minimal impact on reversion frequencies, despite a gradual increase in methylation levels. This apparent contradiction can be explained if we assume that only one unstable allele is required in a cell to allow a revertant cell to arise, which is consistent with the methylation pattern observed in a revertant cell line (see below). Taken together, these results suggest that progression to stable silencing occurs stochastically at the cellular level.
The difference in APRT binding affinity between DAP and FA was further exploited by demonstrating that the induction of DAP-resistant clones by 5-aza-dC occurred approximately 200-fold more often than the induction of FA-resistant clones. Although FA-resistant clones were rarely isolated directly from the parent expressing cells, they could readily be isolated from DAP-resistant clones. This observation suggests that once silencing is induced it can progress, and that it is possible to use selection to rapidly convert a population that is heterogeneous for stable and unstable alleles to one that is homogenous for stably silenced alleles. Progression of silencing was also observed in a study of GFP silencing in MEL cells (see the introduction), where it was shown that early in the silencing process GFP expression could be could be reactivated by treating the cells with either a histone deacetylase inhibitor or with 5-aza-dC, but as the cells continued to grow in culture the constructs became refractory to the individual drug treatments (26).
At this time we can separate Aprt silencing into two cellular phenotypes: (i) unstable, as defined by spontaneous reversion frequencies of >10−3 and FA sensitivity; and (ii) stable, as defined by spontaneous reversion frequencies of <10−5 and FA resistance. The expressing cells can also be divided into two phenotypes: (i) the parental cells and (ii) the revertant cells. Straightforward correlations between the different phenotypes and the different DNA methylation patterns are difficult, because some overlap was observed when comparing methylation patterns for the different phenotypes. For example, none of the alleles in the expressing parental cells were methylated in region IV, which includes the Sp1 promoter elements, but alleles lacking promoter region methylation also represented the majority of alleles in early cultures of DAP-resistant cells. In addition, these cells, which were unstably silenced, contained a subset of alleles that were methylated in their promoter regions. Alleles that were methylated in their promoter regions represented the overwhelming majority of alleles observed in cells defined as stably silenced (>95%) (Fig. 4F, Table 3, and data not shown). A mixture of alleles with unmethylated and methylated promoter regions, at an approximate 1:2 ratio, was observed in a revertant cell line.
With regards to alleles that are unmethylated in their promoter regions, we speculate that they are either expressed or unstably repressed. This hypothesis predicts that alleles with unmethylated promoters in the parental and revertant cells are expressed and that similarly appearing alleles in unstably silenced cells are substantially repressed, yet revertible. It does not, however, explain the chromatin or other changes that account for the marked difference in expression between expressing and nonexpressing alleles that are unmethylated in their promoter regions. With regards to alleles that are methylated in their promoter regions, we speculate that they are stably silenced regardless of whether they are found in cells that can or cannot revert spontaneously. Because each cell examined contains three Aprt alleles, it is likely that many unstably silenced cells contain mixtures of revertible and nonrevertible alleles, which could explain why the revertant cell examined in detail (Fig. 4E) has a mixture of alleles, with one-third being unmethylated in their promoter regions. These alleles are presumably expressed in the revertant cell.
A role for DNA methylation in stable silencing is apparent from the observation that revertant cells could be induced by treating stably silenced cells with 5-aza-dC (Table 2). However, no specific CpG site or group of sites, including those within the Sp1 binding sites, could be implicated in stable silencing, suggesting that other explanations will be required to explain how methylation and stable silencing are linked, such as a density effect. Other investigators have suggested methylation density effects with both plasmid- and transgene-based systems (5, 13, 21, 27), which presumably involve attraction of methyl binding proteins that in turn recruit chromatin-modifying proteins (35, 47), but exact relations between methylation density and silencing have not been established with these systems either. Additional work will be necessary to better define the relations between methylation, chromatin changes, and silencing phenotypes in our system. Finally, the observation that essentially all alleles change from being unmethylated in their promoter regions in expressing cells to being methylated in stably silenced cells argues against one or two of the Aprt constructs in the parental cells being repressed via a methylation-independent mechanism. If this were the case, these alleles would not have become methylated in the stably silenced cells.
While we cannot explain fully how the variable methylation patterns account for different expressing and silenced phenotypes, based on the observations that were made in this study we can develop a model that portrays silencing of Aprt as a gradual and progressive process involving the spread of DNA methylation from an upstream focus within the methylation center fragment. Briefly, our model assumes that silencing initiates with a sharp, albeit incomplete, drop in transcription that removes a barrier to methylation spreading that normally protects the promoter. This barrier may result from binding of transcription factor to the Sp1 binding sites (32), and it would be compromised when transcription factor binding is significantly reduced or lost shortly after the induction of silencing. Silencing of Aprt is proposed to be initially incomplete, as demonstrated by residual APRT enzymatic activity and mRNA and by high spontaneous reversion frequencies, which in total indicate that restoration of transcription factor binding will occur sporadically during the silencing process. If so, the barrier to methylation spreading will occasionally reappear on silenced alleles, only to disappear when transcription factor binding is lost anew. An alternating pattern of transcription factor binding and loss is speculated to create an evolving scenario in which methylation will spread towards the promoter in a halting process, and average transcription levels will gradually drop. This dynamic scenario could explain variegated methylation patterns, and it could also explain why correlations between methylation and stability of silencing are difficult to make conclusively. Finally, the endgame of the evolving process is predicted to occur when methylation density increases to a point where it becomes sufficient to lock in the silenced state.
An important aspect of our model is that it does not require global cellular changes in the methylation machinery. Instead, it only requires changes in expression at the level of individual alleles. A similar model of silencing preceding promoter methylation has been proposed by others, but in this model it was speculated that the observed methylation spreads from random seeding of de novo methylation (38). It is also possible that in some cases allele specific DNA methylation change could occur due to perturbations at the cellular level (17, 18), including altered expression for DNA methyltransferases. Consistent with these possibilities, overexpression of DNA methyltransferase has been shown to cause the spread of DNA methylation towards active promoters in cultured cells (20, 46). Methylation spreading at a number of loci also occurs as a function of age (1). Therefore, there are possible scenarios that can explain our observations other than the model we have presented here and in more detail elsewhere (42). Most likely, however, there are a variety of pathways, ranging from allele specific to genome-wide, that can convert an actively expressed allele to one that is methylated and stably inactivated in malignant cells.
Our model is useful for envisioning silencing as a process, instead of a single event, but it does not address the mechanisms by which the initial drop in transcription occurs. Studies by Costa and colleagues have shown that nickel compounds have the potential to induce silencing in mammalian and yeast cells (12), and they have suggested that this is due to the prevention of histone acetylation (50). This possibility would explain why nickel is effective both in a system that has DNA methylation (mammalian cells) and in a system that lacks DNA methylation (yeast cells), because the initial change will be at the level of chromatin. Costa and colleagues have also shown that 5-aza-dC can induce silencing of integrated bacterial gpt genes in mammalian cells (7), and we have confirmed that this will occur for mouse Aprt. We have shown further that fusion of the differentiated cells with embryonal cells is also a potent inducer of silencing. In both cases, the presence of the methylation center dramatically increased the probability that silencing could be induced. While we do not know at this time whether these methods of silencing induction are working via similar or different pathways, several possible mechanisms can be proposed. One is that a repressive factor present in the embryonal carcinoma cells, but absent in the differentiated cells, can interact with the methylation center fragment. This factor would either be introduced to the differentiated cells by fusion with embryonal carcinoma cells or expressed when the gene is reactivated by 5-aza-dC treatment. A second possibility is that both cell fusion and 5-aza-dC exposure cause global changes in chromatin that can include the methylation center region, resulting in silencing of the nearby Aprt promoter. Further work will be required to distinguish between these and other possibilities.
In summary, the data obtained demonstrate that silencing of mouse Aprt in differentiated cells is a gradual process that initiates with a dramatic drop in transcription and then involves the spread of DNA methylation towards the promoter. At least two phenotypes were associated with the silencing process, one characterized by very high spontaneous reversion frequencies (≈10−2) and sensitivity to FA, and the second characterized by low or undetectable spontaneous reversion frequencies (<10−5) and resistance to FA. This latter phenotype could be reversed by treating the cells with 5-aza-dC, suggesting a role for DNA methylation in locking in the silencing process. While the presence of the methylation center fragment contributed to the initiation of silencing, it is less clear if methylation within this center played a role. However, the data demonstrate that once silencing is initiated, as defined by cellular resistance to DAP, the methylation center acts as a seed region from which DNA methylation spreads. Although unstable and stable silencing represent two definable features of this model, we do not believe that the transition from one to the other occurs as a discrete event. Instead, the variegated patterns of methylation that are observed are more consistent with an evolving and prolonged process by which an allele that has retained a low-level potential for transcription becomes one that loses this potential irrevocably. Finally, although we note that this model applies directly only to mouse Aprt, the presence of unstable phenotypes coupled with variegated methylation patterns suggests that such instability could be observed in malignant cells, as demonstrated for contrasting E-cadherin phenotypes within cultures of human breast cancers (19). Moreover, the presence of methylated repetitive elements near a variety of tumor suppressor genes in human cells (3, 4, 20) suggests that additional aspects of this model will be applicable for understanding at least a subset of silencing processes that occur in human cancers.
Acknowledgments
This work was supported by NIH grants CA92114 and CA76528 (M.S.T.) and CA97021 (M.J.T.), an NRSA postdoctoral award (CA90038) (P.A.Y.), and an NIH training grant (T32 GM08617) (R.W.B.).
We thank Brett Spear for helpful advice on the whole-cell fusions.
REFERENCES
- 1.Ahuja, N., and J. P. Issa. 2000. Aging, methylation and cancer. Histol. Histopathol. 15:835-842. [DOI] [PubMed] [Google Scholar]
- 2.Baylin, S., and T. H. Bestor. 2002. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell 1:299-305. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S. B. 2002. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin. Cancer Biol. 12:331-337. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., J. G. Herman, J. R. Graff, P. M. Vertino, and J.-P. Issa. 1998. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 72:141-196. [PubMed] [Google Scholar]
- 5.Boyes, J., and A. Bird. 1992. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 11:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Raxin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 7.Broday, L., Y.-W. Lee, and M. Costa. 1999. 5-Azacytidine induces transgene silencing by DNA methylation in Chinese hamster cells. Mol. Cell. Biol. 19:3198-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron, E. E., S. B. Baylin, and J. G. Herman. 1999. p15(INK4B) CpG island methylation in primary acute leukemia is heterogeneous and suggests density as a critical factor for transcriptional silencing. Blood 94:2445-2451. [PubMed] [Google Scholar]
- 9.Clark, S. J., and J. Melki. 2002. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene 21:5380-5387. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, G. E., P. L. Bishop, and M. S. Turker. 1993. Hemidemethylation is sufficient for chromatin relaxation and transcriptional activation of a methylated APRT gene in mouse P19 embryonal carcinoma cell line. Somat. Cell. Mol. Genet. 19:221-229. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, G. E., N. H. Khattar, P. L. Bishop, and M. S. Turker. 1992. At least two distinct epigenetic mechanisms are correlated with high frequency “switching” for APRT phenotypic expression in mouse embryonal carcinoma stem cells. Somat. Cell. Mol. Genet. 18:215-226. [DOI] [PubMed] [Google Scholar]
- 12.Costa, M., J. E. Sutherland, W. Peng, K. Salnikow, L. Broday, and T. Kluz. 2001. Molecular biology of nickel carcinogenesis. Mol. Cell. Biochem. 222:205-211. [PubMed] [Google Scholar]
- 13.Curradi, M., A. Izzo, G. Badaracco, and N. Landsberger. 2002. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell. Biol. 22:3157-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodge, J. E., A. F. List, and B. W. Futscher. 1998. Selective variegated methylation of the p15 CpG island in acute myeloid leukemia. Int. J. Cancer 78:561-567. [DOI] [PubMed] [Google Scholar]
- 15.Domann, F. E., J. C. Rice, M. J. Hendrix, and B. W. Futscher. 2000. Epigenetic silencing of maspin gene expression in human breast cancers. Int. J. Cancer 85:805-810. [DOI] [PubMed] [Google Scholar]
- 16.Dush, M. K., M. R. Briggs, M. E. Royce, D. A. Schaff, J. A. Tischfield, and P. J. Stambrook. 1988. Identification of DNA sequences required for mouse APRT gene expression. Nucleic Acids Res. 16:8509-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteller, M., and J. G. Herman. 2002. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J. Pathol. 196:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Fruhwald, M. C., and C. Plass. 2002. Global and gene-specific methylation patterns in cancer: aspects of tumor biology and clinical potential. Mol. Genet. Metab. 75:1-16. [DOI] [PubMed] [Google Scholar]
- 19.Graff, J. R., E. Gabrielson, H. Fujii, S. B. Baylin, and J. G. Herman. 2000. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J. Biol. Chem. 275:2727-2732. [DOI] [PubMed] [Google Scholar]
- 20.Graff, J. R., J. G. Herman, S. Myöhänen, S. B. Baylin, and P. M. Vertino. 1997. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem. 272:22322-22329. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh, C. L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson-Grusby, L., P. W. Laird, S. N. Magge, B. J. Moeller, and R. Jaenisch. 1997. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl. Acad. Sci. USA 94:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 24.Klein, G. 2002. Introduction: genetic and epigenetic contributions to tumor evolution. Semin. Cancer Biol. 12:327-330. [DOI] [PubMed] [Google Scholar]
- 25.Krenitsky, T. A., S. M. Neil, G. B. Elion, and G. H. Hitchings. 1969. Adenine phosphoribosyltransferase from monkey liver. Specificity and properties. J. Biol. Chem. 244:4779-4784. [PubMed] [Google Scholar]
- 26.Lorincz, M. C., D. Schubeler, S. C. Goeke, M. Walters, M. Groudine, and D. I. Martin. 2000. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 20:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorincz, M. C., D. Schubeler, S. R. Hutchinson, D. R. Dickerson, and M. Groudine. 2002. DNA methylation density influences the stability of an epigenetic imprint and Dnmt3a/b-independent de novo methylation. Mol. Cell. Biol. 22:7572-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macleod, D., J. Charleton, J. Mullins, and A. Bird. 1994. Sp1 binding sites in the mouse aprt gene promoter are required to prevent methylation of CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 29.Melki, J. R., P. C. Vincent, R. D. Brown, and S. J. Clark. 2000. Hypermethylation of E-cadherin in leukemia. Blood 95:3208-3213. [PubMed] [Google Scholar]
- 30.Mummaneni, P., P. L. Bishop, and M. S. Turker. 1993. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J. Biol. Chem. 268:552-558. [PubMed] [Google Scholar]
- 31.Mummaneni, P., K. A. Walker, P. L. Bishop, and M. S. Turker. 1995. Epigenetic gene inactivation induced by a cis-acting methylation center. J. Biol. Chem. 270:788-792. [DOI] [PubMed] [Google Scholar]
- 32.Mummaneni, P., P. Yates, J. Simpson, J. Rose, and M. S. Turker. 1998. The primary function of a redundant Sp1 binding site in the mouse aprt gene promoter is to block epigenetic gene inactivation. Nucleic Acids Res. 26:5163-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nosaka, K., M. Maeda, S. Tamiya, T. Sakai, H. Mitsuya, and M. Matsuoka. 2000. Increasing methylation of the CDKN2A gene is associated with the progression of adult T-cell leukemia. Cancer Res. 60:1043-1048. [PubMed] [Google Scholar]
- 34.Rose, J. A., P. A. Yates, J. Simpson, J. A. Tischfield, P. J. Stambrook, and M. S. Turker. 2000. Biallelic methylation and silencing of mouse Aprt in normal kidney cells. Cancer Res. 60:3404-3408. [PubMed] [Google Scholar]
- 35.Rountree, M. R., K. E. Bachman, J. G. Herman, and S. B. Baylin. 2001. DNA methylation, chromatin inheritance, and cancer. Oncogene 20:3156-3165. [DOI] [PubMed] [Google Scholar]
- 36.Siegfried, Z., S. Eden, M. Mendelsohn, X. Feng, B. Z. Tsuberi, and H. Cedar. 1999. DNA methylation represses transcription in vivo. Nat. Genet. 22:203-206. [DOI] [PubMed] [Google Scholar]
- 37.Smith, L., A. Plug, and M. Thayer. 2001. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci. USA 98:13300-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, J. Z., C. Stirzaker, J. Harrison, J. R. Melki, and S. J. Clark. 2002. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene 21:1048-1061. [DOI] [PubMed] [Google Scholar]
- 39.Stimson, K. M., and P. M. Vertino. 2002. Methylation-mediated silencing of TMS1/ASC is accompanied by histone hypoacetylation and CpG island-localized changes in chromatin architecture. J. Biol. Chem. 277:4951-4958. [DOI] [PubMed] [Google Scholar]
- 40.Turker, M., K. A. Walker, C. D. Jennings, I. Mellon, A. Yusufji, and M. Urano. 1995. Spontaneous and ionizing radiation induced mutations involve large events when selecting for loss of an autosomal locus. Mutat. Res. 329:97-105. [DOI] [PubMed] [Google Scholar]
- 41.Turker, M. S. 1998. Estimation of mutation frequencies in normal mammalian tissues and the development of cancer. Semin. Cancer Biol. 8:407-419. [DOI] [PubMed] [Google Scholar]
- 42.Turker, M. S. 2002. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene 21:5388-5393. [DOI] [PubMed] [Google Scholar]
- 43.Turker, M. S., P. Mummaneni, and P. L. Bishop. 1991. Region and cell type-specific de novo DNA methylation in cultured mammalian cells. Somat. Cell. Mol. Genet. 17:151-157. [DOI] [PubMed] [Google Scholar]
- 44.Turker, M. S., A. C. Smith, and G. M. Martin. 1984. High frequency “switching” at the adenine phosphoribosyltransferase locus in multipotent mouse teratocarcinoma stem cells. Somat. Cell. Mol. Genet. 10:55-69. [DOI] [PubMed] [Google Scholar]
- 45.Turker, M. S., P. J. Stambrook, J. A. Tischfield, A. C. Smith, and G. M. Martin. 1989. Allelic variation linked to the adenine phosphoribosyltransferase locus in a mouse teratocarcinoma cell line and feral-derived mouse strains. Somat. Cell. Mol. Genet. 15:159-166. [DOI] [PubMed] [Google Scholar]
- 46.Vertino, P. M., R. W. Yen, J. Gao, and S. B. Baylin. 1996. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol. 16:4555-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wade, P. A. 2001. Methyl CpG-binding proteins and transcriptional repression. Bioessays 23:1131-1137. [DOI] [PubMed] [Google Scholar]
- 48.Wong, D. J., S. A. Foster, D. A. Galloway, and B. J. Reid. 1999. Progressive region-specific de novo methylation of the p16 CpG island in primary human mammary epithelial cell strains during escape from M0 growth arrest. Mol. Cell. Biol. 19:5642-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates, P. A., R. W. Burman, P. Mummaneni, S. Krussel, and M. S. Turker. 1999. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 274:36357-36561. [DOI] [PubMed] [Google Scholar]
- 50.Zoroddu, M. A., L. Schinocca, T. Kowalik-Jankowska, H. Kozlowski, K. Salnikow, and M. Costa. 2002. Molecular mechanisms in nickel carcinogenesis: modeling Ni(II) binding site in histone H4. Environ. Health Perspect. 110(Suppl. 5):719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]