Abstract
Neural basic helix-loop-helix (bHLH) transcription factors regulate neurogenesis in vertebrates. Signaling by peptide growth factors also plays critical roles in regulating neuronal differentiation and survival. Many peptide growth factors activate phosphatidylinositol 3-kinase (PI3K) and subsequently the Akt kinases, raising the possibility that Akt may impact bHLH protein function during neurogenesis. Here we demonstrate that reducing expression of endogenous Akt1 and Akt2 by RNA interference (RNAi) reduces neuron generation in P19 cells transfected with a neural bHLH expression vector. The reduction in neuron generation from decreased Akt expression is not solely due to decreased cell survival, since addition of the caspase inhibitor z-VAD-FMK rescues cell death associated with loss of Akt function but does not restore neuron formation. This result indicates that Akt1 and Akt2 have additional functions during neuronal differentiation that are separable from neuronal survival. We show that activated Akt1 enhances complex formation between bHLH proteins and the transcriptional coactivator p300. Activated Akt1 also significantly augments the transcriptional activity of the bHLH protein neurogenin 3 in complex with the coactivators p300 or CBP. In addition, inhibition of endogenous Akt activity by the PI3K/Akt inhibitor LY294002 abolishes transcriptional cooperativity between the bHLH proteins and p300. We propose that Akt regulates the assembly and activity of bHLH-coactivator complexes to promote neuronal differentiation.
Neural basic helix-loop-helix (bHLH) transcription factors, including neurogenin 1 to 3 (ngn1 to -3), NeuroD1 and -2, and MASH1, regulate neurogenesis in vertebrates (24). These proteins regulate the transcriptional events required for neural cell fate commitment, neuronal cell cycle withdrawal, and neuronal differentiation. The neural bHLH proteins can promote neuron formation from nonneural cells when expressed ectopically in the ectoderm of Xenopus laevis or zebra fish embryos, and forced expression of any of these proteins can promote neuronal differentiation of uncommitted mouse embryonal carcinoma cells (17, 34, 38, 41, 70). Targeted disruption of the genes encoding neural bHLH proteins in the mouse has demonstrated that these genes are required for the formation of subsets of neurons (5, 6, 19, 25, 40, 47, 51).
The neural bHLH proteins are transcriptional activators and function as heterodimers with E proteins such as E12 and E47 (37). In addition, bHLH proteins require the coactivators CREB-binding protein (CBP) and p300 to function as activators of transcription (49, 62, 67). The neural bHLH protein neuroD and the myogenic bHLH protein MyoD interact with the third Cys-His-rich zinc finger of CBP/p300 (16, 49, 61, 62, 75). CBP and p300 mediate interactions between the DNA-binding transcription factors and the RNA polymerase II transcriptional machinery to facilitate gene transcription (20, 21, 23). In addition, CBP and p300 possess intrinsic acetyltransferase (AT) activity and associate with proteins that possess AT activity, such as PCAF. Acetylation of histones and other proteins contributes to transcriptional activation and is involved in the nucleosomal remodeling that accompanies gene activation. Acetylation of MyoD increases its activity on muscle-specific promoters by increasing its affinity for DNA and its association with CBP/p300 (59, 60, 64). In addition to acetylation, signal-dependent phosphorylation of CBP and p300 is emerging as an important mode of regulation. Phosphorylation of the coactivators has been shown to regulate their recruitment to transcription factor complexes (31, 76).
Peptide growth factors, such as insulin-like growth factor and transforming growth factor alpha (TGF-α) and TGF-β, play critical roles in regulating neuronal cell differentiation and survival (2, 9, 18). Insulin-like growth factor, TGF-α, and TGF-β bind to their cognate receptors and activate intracellular signaling cascades. Phosphatidylinositol 3-kinase (PI3K) is activated by each of these peptide growth factors. Activation of PI3K leads to the production of the lipid second messengers phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate, which in turn activate the serine/threonine kinases Akt1 and -2 (also known as protein kinase B [PKB] α and β). Activated Akt phosphorylates a plethora of different targets and regulates glycogen synthesis, cell size, and cell survival (8). Akt1 and Akt2 are expressed during early neural development in the mouse (54), suggesting that these kinases may have a role in neurogenesis in addition to their role in promoting survival of mature neurons.
Regulation of bHLH transcriptional activity in neurogenesis by signal transduction pathways is largely unexplored. In the present study, we show that inhibition of expression of endogenous Akt by hairpin short interfering RNAs (siRNAs) reduces neuron generation in P19 cells transfected with a neural bHLH expression vector. The reduction in neuron generation by loss of Akt function is not solely due to decreased cell survival, because addition of a caspase inhibitor to hairpin-transfected cells promotes cell survival but does not restore neuron generation. One possible mechanism for Akt to regulate neuron formation is to modulate the activity of neurogenic transcription factor complexes. In support of this model, we demonstrate that activated Akt1 enhances the interaction between bHLH proteins and the transcriptional coactivator p300. Also, activated Akt significantly increases the transcriptional activity of a complex comprised of neurogenic bHLH proteins and the p300 coactivator. Furthermore, the bHLH-p300 transcriptional cooperativity is abolished when endogenous Akt activity is inhibited with the PI3K/Akt inhibitor LY294002. We propose that Akt regulates the assembly and activity of bHLH transcription factor complexes to promote neuronal differentiation.
MATERIALS AND METHODS
Plasmid expression vectors.
pGal-p300 and pGal-CBP vectors express fusion proteins of the Gal4 DNA binding domain (amino acids 1 to 147) fused to full-length p300 or CBP in pRc/RSV (14). pGal-CBP T1871A and pGal-p300 S1834A contain mutations of T to A at position 1871 in CBP and S to A at position 1834 in p300. The mutations were introduced into fragments of the CBP and p300 cDNAs using the QuikChange mutagenesis protocol with Pfu polymerase (Stratagene). These fragments were then inserted into the full-length expression vectors, and the final vectors were sequenced to confirm the presence of the expected mutation and the absence of extraneous mutations. pCS2+ΔPHDDAkt expresses an activated Akt1, amino acids 131 to 480, in which S473 and T308 were altered to D by PCR mutagenesis. Constitutively active (C/A) human SGK1 was inserted into pCS2+ (63, 70). pCS2+MASH1 and pCS2+ngn3 have been described previously (17). Myc epitope-tagged versions of these bHLH proteins were created by inserting the coding regions into pCS2+MT (63) or a derivative, pCS2+MTbgl2. Nuclear-localized green fluorescent protein (nlsGFP) was expressed from pCS2p+nls-eGFPbgl2, a derivative of pCS2+eGFP (17).
Vectors that express Akt1 and Akt2 hairpin siRNAs under the control of the mouse U6 promoter were constructed by inserting pairs of annealed DNA oligonucleotides into the mU6pro vector between the Bbs1 and XbaI sites as previously described (73). The first nucleotide of each predicted hairpin siRNA is G, which corresponds to the first nucleotide of the U6 snRNA; all templates include five T residues for RNA polymerase III termination. Each hairpin has a 28-nucleotide duplex (74) that includes two mismatched nucleotides in the sense strand near the center of the duplex region to facilitate sequence analysis of the DNA constructs. The mismatches have no effect on RNA interference (RNAi) (73). The XASH3 hairpin siRNA vector used as a control targets a Xenopus gene with no mammalian homolog and does not alter neuronal differentiation or expression vector function in P19 cells (74).
Luciferase assays.
P19 cells in 35-mm dishes were transfected with FuGENE (Roche) and appropriate combinations of expression vectors in the following amounts: pGal-p300 (250 ng), pGal-CBP (250 ng), pGal-ngn3 COOH (50 ng), pCS2+ ngn3 (500 ng; 100 ng in Fig. 3, below), pCS2+ΔPHDDAkt1 (150 ng), pCS2+C/A SGK (150 ng), and either 5XGal4-luciferase reporter plasmid pFR-luciferase (1 μg) or the E1X3 reporter (1 μg). Total DNA was kept constant by the addition of the appropriate amount of pCS2+ for all transfections. Luciferase was assayed 24 h (see Fig. 5) or 48 h (see Fig. 3) after transfection, using the Dual-Light luciferase and β-galactosidase reporter gene assay system (Tropix). Where noted, LY294002 (12.5 μM) was added to cells 5 h after transfection. All assays were normalized for transfection efficiency by using a cotransfected β-galactosidase expression vector (pCS2+cβGal; 75 ng) (73). Assays were performed in duplicate and/or in triplicate and repeated multiple times with similar results; representative experiments are shown below in Fig. 5 and 7.
FIG. 3.
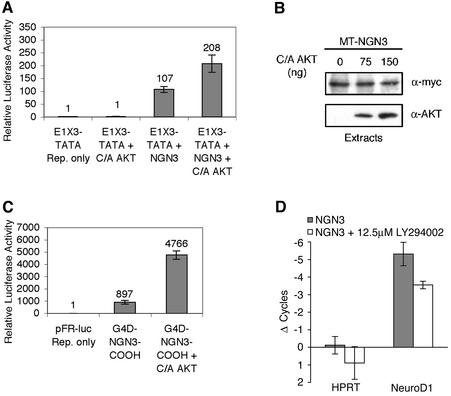
Activated Akt cooperates with ngn3 to increase transcription. (A) P19 cells were transfected with expression vectors for ngn3, activated Akt1, and a multimerized E-box driving a luciferase reporter (E1X3-TATA). (B) Activated Akt does not alter the level of the ngn3 protein. P19 cells were transfected with expression vectors for Myc epitope-tagged (MT) ngn3 and increasing concentrations of activated Akt1. Extracts were prepared, and MT-Ngn3 levels were assessed by Western blot analysis with anti-Myc antibody. (C) The carboxyl-terminal transactivation domain of ngn3 mediates Akt-regulated transcriptional cooperativity. P19 cells were transfected with expression vectors for Gal4-DBD fused to the ngn3 carboxyl-terminal transactivation domain (G4D-ngn3-COOH), constitutively active Akt1, or vector control, and the pFR Gal4-responsive luciferase reporter. (D) ngn3-induced expression of NeuroD1 is reduced in the presence of the PI3K/Akt inhibitor LY294002. NeuroD1 and hypoxanthine phosphoribosyltransferase (control) mRNAs were quantitated by real-time RT-PCR from P19 cells transfected with an ngn3 vector, with or without LY294002, as indicated. The change in threshold-crossing cycle is shown for each mRNA relative to that for transfection of a control vector (a decrease in threshold crossing corresponds to an increase in the mRNA level). The reduction in NeuroD1 expression in the presence of LY294002 is significant (P < 0.02 by Student's t test). Standard deviations are indicated in panels A, C, and D.
FIG. 5.
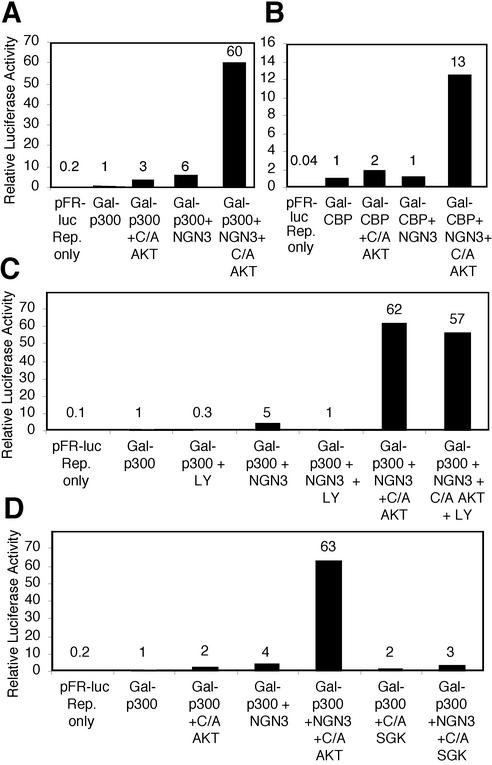
Activated Akt1 enhances the transcriptional activity of ngn3 in complex with the transcriptional coactivators p300 and CBP. P19 cells were transfected with expression vectors for the Gal4-DBD p300 fusion protein (Gal-p300) or Gal4-DBP-CBP fusion protein (Gal-CBP), ngn3, the pFR Gal4-responsive luciferase reporter, and constitutively active Akt1 (C/A Akt) or constitutively active Sgk1 (C/A SGK). The PI3K/Akt inhibitor LY294002 (12.5 μM) was added at the time of transfection, as indicated. (A) ngn3 and C/A Akt1 cooperatively enhance the transcriptional activity of Gal-p300. (B) ngn3 and C/A Akt1 cooperatively enhance the transcriptional activity of Gal-CBP. (C) Inhibition of the endogenous PI3K/Akt signaling pathway with LY294002 decreases the transcriptional activity of Gal-p300 in combination with ngn3, but not in the presence of C/A Akt1. (D) C/A Akt1, but not C/A SGK, enhances the transcriptional activity of ngn3 and Gal-p300.
FIG. 7.
Mutation of the Akt consensus phosphorylation site in p300 does not alter the cooperativity between ngn3, p300, and Akt. P19 cells were transfected with expression vectors for Gal4-p300 or Gal4-p300 S1834A, ngn3, constitutively active (C/A) Akt1 or vector control, and the pFR Gal4-responsive luciferase reporter.
Real-time reverse transcription-PCR (RT-PCR).
P19 cells in 35-mm dishes (1.6 × 105 cells per dish) were transfected with 0.5 μg of CS2p+eGFPBgl2 and 3 μg of CS2+MT or CS2+mNgn3 using FuGENE. Six hours after transfection, cells were changed to Opti-MEM (Invitrogen) plus 1% fetal bovine serum. A 12.5 μM concentration of LY294002 in dimethyl sulfoxide, or dimethyl sulfoxide alone (control), was added 22 h after transfection. RNA was isolated 30 h after transfection. RNA was DNase treated (per the manufacturer's instructions; RQ1 DNase; Promega), extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1), ethanol precipitated, and resuspended in RNase-free water. RNA yield was determined using the Ribogreen assay (Molecular Probes). Two micrograms of each RNA was reverse transcribed in a 10-μl reaction volume with random 12-mer primers (NEB) and Superscript II (Invitrogen). For each PCR, 1/40 of the cDNA was amplified using a Bio-Rad iCycler in a 25-μl reaction volume with 1:100,000 SYBR Green I (Molecular Probes), using a thermal profile of 15 s at 95°C, 30 s at 59°C, and 15 s at 72°C, followed by 15 s at 82°C for real-time fluorescence measurement for each cycle. The threshold-crossing cycle for each sample was determined using iCycler analysis software. The induction of NeuroD1 transcription (i.e., the decrease in threshold-crossing cycle) was determined for CS2+ngn3 with or without LY294002 relative to that for the CS2+MT control transfection. Data presented are an average from three transfections (with triplicate PCRs for each cDNA sample). PCR primers were as follows: HPRT forward, CAAACTTTGCTTTCCCTGGT; HPRT reverse, CAAGGGCATATCCAACAACA (HPRT primer sequences were kindly provided by Mohammad Othman and Anand Swaroop, University of Michigan); NeuroD1 forward, TCCAGGGTTATGAGATCGTC; NeuroD1 reverse, ACACTCATCTGTCCAGCTTGGG (17).
Coassociation assays.
For the CBP-bHLH coassociation assays, P19 cells in 60-mm dishes were transfected using FuGENE and 1.5 μg of MT-bHLH expression vector, 7 μg of pCS5+puro/GFP, and 450 ng of pCS2+ΔPHDD Akt1. Seven to 8 h after transfection, the cells were washed and medium with puromycin (10 μg/ml) was added to select for transfected cells. Approximately 14 to 16 h after transfection, the medium was removed and replaced with medium without puromycin. Cells were harvested 24 h after transfection and lysed in extract buffer (10 mM HEPES [pH 7.4], 1% Triton X-100, 2 mM EDTA, 0.1% beta-mercaptoethanol, 1% aprotinin, 50 mM NaF) with 50 mM NaCl. Endogenous CBP was immunoprecipitated using anti-CBP antibody (Santa Cruz), and the immunoprecipitate was captured on a 1:1 mixture of protein A-agarose (Sigma)-protein G-Sepharose (Calbiochem) beads. The immunoprecipitates were washed three times with extract buffer containing 100 mM NaCl and then analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 4 to 15% gradient gel; Bio-Rad) followed by Western blot analysis. CBP was detected using anti-CBP antibody (Santa Cruz), and the Myc epitope-tagged bHLH proteins were detected with anti-myc 9E10 antibody (Developmental Studies Hybridoma Bank).
Neuronal differentiation assays.
For RNAi experiments, P19 cells plated on murine laminin (Invitrogen)-coated dishes were transfected using FuGENE and 0.8 μg of U6 siRNA vector targeting Akt1, Akt2, Akt1 and Akt2, or XASH3 (control). The total amount of U6 siRNA vector per transfection was kept constant to 1.6 μg by addition of control XASH hairpin as needed. Each transfection also included 1.9 μg of ngn3 expression vector and 0.5 μg of NLS-GFP expression vector. As noted, the caspase inhibitor z-VAD-FMK (Calbiochem or Promega) at 20 μM was added at the time of transfection and in each subsequent medium change. Cells were fixed 4 days after transfection. Fixed cells were stained with the TuJ1 antibody to detect a neuron-specific tubulin, essentially as described previously (17). Cells were photographed with a video camera on a Zeiss Axiovert inverted microscope. NIH IMAGE software was used for cell counts for GFP; TuJ1 cells were counted manually.
RESULTS
Inhibition of endogenous Akt expression with hairpin siRNAs reduces neuron generation by ngn3.
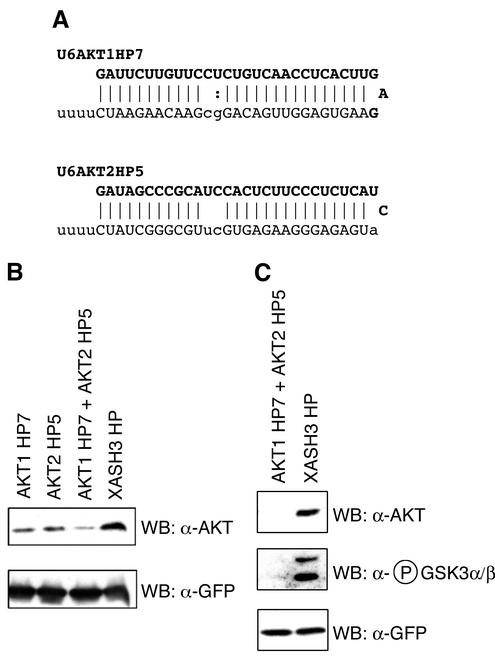
We and others have previously demonstrated that siRNAs or hairpin siRNAs expressed from an RNA polymerase III promoter such as the U6 snRNA promoter can effectively inhibit gene expression in mammalian cells by RNAi (7, 39, 45, 46, 55, 57, 66, 73, 74). We constructed U6 hairpin siRNA expression vectors directed against murine Akt1 and Akt2 (Fig. 1A). To test the effectiveness of the hairpin siRNAs at reducing the endogenous levels of the Akt kinases, we performed Western blot analysis on extracts from mouse P19 cells transiently transfected with combinations of siRNA hairpin expression vectors and biCS5puro/GFP. The inclusion of the biCS5puro/GFP vector in the transfection enabled the use of puromycin to rapidly select the transfected cell population (74). Hairpin siRNAs against Akt1 and Akt2 were effective at reducing the endogenous levels of Akt 1 and 2 compared with those of a control hairpin siRNA vector directed against the Xenopus XASH3 gene (74) (Fig. 1B). To determine whether the Akt hairpins reduced Akt levels sufficiently to decrease phosphorylation of Akt substrates, the phosphorylation state of serine 21 of glycogen synthase kinase 3α (GSK3α) and serine 9 of GSK3β, two well-characterized Akt substrates (71), was examined. Hairpin siRNAs against Akt1 and Akt2 were effective at reducing GSK3α/β Ser21/9 phosphorylation compared with that of the control hairpin (Fig. 1C); the reduction in GSK3α/β Ser21/9 phosphorylation closely followed the reduction of endogenous Akt protein levels.
FIG. 1.
Hairpin siRNA expression vectors against Akt1 and Akt2 reduce the endogenous levels of the Akt kinases and reduce GSK3α/β phosphorylation at Ser21/9. (A) Sequences and expected structures of hairpin siRNAs for Akt1 and Akt2 expressed from the mouse U6 promoter. Watson-Crick base pairs (|) and G:U base pairs (:) are shown. (B and C) P19 cells were transfected with expression vectors for biCS5puro/GFP and U6-driven hairpin siRNAs against Akt1 and/or Akt2, or XASH3 (a control hairpin siRNA). The caspase inhibitor z-VAD-FMK was added to cells at the time of transfection. Akt1 and Akt2 (B, upper panel) and GFP (transfection control) (B, lower panel) were detected by Western blot analysis of extracts prepared from transiently transfected puromycin-selected cells. The Akt antisera recognizes both Akt1 and Akt2, which migrate as a single band. Phospho-GSK3α/β is reduced when Akt1 and Akt2 expression is inhibited by RNAi (C, middle panel; upper band is GSK3α and lower band is GSK3β; other panels are as described for panel B).
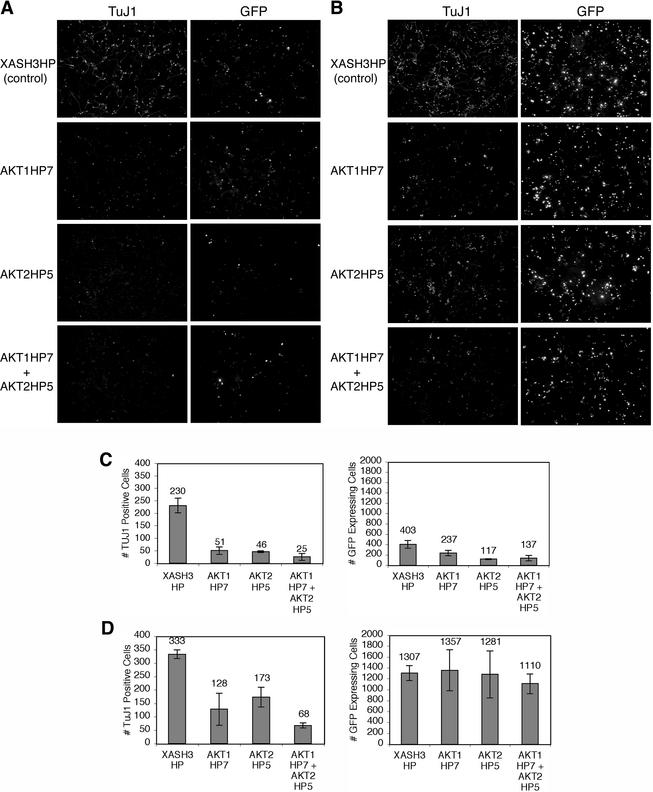
To investigate the role of endogenous Akt1 and Akt2 in bHLH-driven neuronal differentiation, we introduced the neural bHLH protein ngn3 into uncommitted P19 cells in combination with the U6-driven hairpin siRNAs directed against Akt1, Akt2, both Akt1 and Akt2, or the XASH3 control hairpin siRNA. In addition, the cells were cotransfected with a vector that expresses GFP, allowing the transfected cell population to be readily identified. By 4 days after transfection, many of the GFP-positive transfected cells adopted a neuronal morphology and expressed a neuron-specific tubulin protein, detected by indirect immunofluorescence with the TuJ1 antibody, as previously observed (17) (Fig. 2A). Coexpression of hairpin siRNAs against Akt1 or Akt2, each alone or in combination, with ngn3 reduced the number of TuJ1-positive neurons compared to the control XASH3 hairpin siRNA (Fig. 2A). Expression of the XASH3 hairpin siRNA does not reduce neuron formation in bHLH-transfected P19 cells (data not shown). Consistent with previous reports of the role of Akt in cell survival, introduction of hairpins targeting Akt1 and Akt2 also resulted in a decrease in the number of GFP-positive cells (Fig. 2A).
FIG. 2.
Inhibition of endogenous Akt by hairpin siRNAs reduces neuron generation by ngn3. (A) P19 cells were transfected with expression vectors for ngn3, nlsGFP, and U6-driven hairpin siRNAs against Akt1 (Akt1 HP7), Akt2 (Akt2 HP5), both Akt1 and Akt2, or a control, XASH3 (XASH3 HP). (B) Cells were transfected as described for panel A, but the caspase inhibitor z-VAD-FMK was added to cells at the time of transfection. Cells were fixed 4 days after transfection, and a neuronal β-tubulin was detected by indirect immunofluorescence with the antibody TuJ1. GFP was detected by epifluorescence. (C and D) The average number of GFP-positive cells and TuJ1-positive cells per field of view in hairpin-transfected cells without (C) or with (D) caspase inhibitor was determined. Averages are derived from counting the number of cells from three fields of view per transfection, from three independent transfections (standard deviations are indicated).
In order to determine whether there might be a requirement for Akt in differentiation in addition to a role for Akt in promoting cell survival, we assayed neuronal differentiation in the presence of the cell-permeable general caspase inhibitor z-VAD-FMK. z-VAD-FMK prevents cell death by inhibiting cysteine proteases that execute the cell death pathway (52, 53, 68, 69). z-VAD-FMK was added at the time of transfection of cells with hairpin siRNA vectors and the ngn3 and GFP expression vectors, and the cells were maintained in the presence of the inhibitor for the duration of the experiment. In the presence of the caspase inhibitor, the number of GFP-expressing cells with Akt hairpins significantly increased (Fig. 2B). Thus, the addition of the caspase inhibitor prevents cell death induced by the loss of Akt expression. However, the addition of the caspase inhibitor did not restore the number of TuJ1-positive neurons in cells transfected with Akt hairpins, suggesting a role for Akt1 and Akt2 in neuronal differentiation in addition to a role for Akt1 and Akt2 in promoting cell survival.
To quantify these observations, we counted the number of TuJ1-positive neurons and GFP-expressing cells. In the absence of caspase inhibitor, introduction of hairpins targeting Akt1 and Akt2 resulted in a threefold decrease in the number of GFP-labeled cells (Fig. 2C). However, the decrease in neuron formation (TuJ1-positive cells) when cells received hairpin siRNA vectors targeting Akt1 and Akt2 was ninefold compared to that with the control XASH3 hairpin siRNA vector (Fig. 2C). This result suggests that there may be a requirement for Akt during differentiation, as well as a requirement for Akt in promoting cell survival. In the presence of the caspase inhibitor, the number of GFP-expressing cells increased approximately threefold in cells transfected with the XASH3 control hairpin (Fig. 2D). Importantly, the number of GFP-expressing cells with Akt hairpins was equivalent to the number of GFP-expressing cells with the control hairpin (Fig. 2D). Thus, the addition of the caspase inhibitor prevents cell death induced by the loss of Akt expression. However, the addition of the caspase inhibitor does not restore the number of TuJ1-positive neurons to the control level. Neuron formation (TuJ1-positive cells) in the presence of the caspase inhibitor was reduced approximately fivefold by the combined action of the Akt1 and Akt2 hairpin siRNAs (Fig. 2D). These observations suggest that Akt1 and Akt2 have multiple roles, with functions in both cell survival and cell differentiation.
Akt regulates the transcriptional activity of ngn3.
Akt could regulate neuron formation by modulating bHLH transcriptional activity. Therefore, we investigated whether constitutively active Akt1 could enhance the ability of ngn3 to activate transcription of a multimerized E-box reporter, E1X3-TATA. The E1X3-TATA reporter contains three copies of the E1 E-box from the NeuroD1/β2 promoter, cloned into a TATA box-containing luciferase reporter (30). The constitutively activated Akt1 protein used in these experiments contains a deletion within the autoinhibitory plekstrin homology domain and substitutions of aspartic acids for Ser473 and Thr308, the regulatory activating phosphorylation sites within Akt1 (1, 3). Expression of ngn3 alone resulted in a significant increase in luciferase activity from the E1X3-TATA-driven reporter. Cotransfection of ngn3 with the constitutively activated Akt1 expression vector enhanced ngn3-dependent reporter activity by twofold; the activity of the E1X3 reporter was not increased by constitutively active Akt1 in the absence of ngn3 (Fig. 3A). To exclude the possibility that Akt1 activation increases the level of ngn3 protein in the transfected cells, we examined the effect of constitutively active Akt1 on a tagged ngn3 protein. The level of the tagged protein was not affected by increasing concentrations of constitutively active Akt1 (Fig. 3B). These results suggest that Akt1 can increase the ability of ngn3 to activate target gene expression.
Activated Akt potentially could increase the activity of the activation domain of ngn3 (e.g., by phosphorylation), or it could alter activation indirectly by modulating other aspects of ngn3 function, such as subcellular localization, DNA binding, or dimerization. Therefore, we tested whether the isolated transactivation domain of ngn3 was sufficient to cooperate with activated Akt1 in a reporter assay. The ngn3 carboxyl-terminal activation domain (C. Hart and D. L. Turner, unpublished data) was fused to the Gal4 DNA binding domain (Gal4D-ngn3 COOH). This construct does not contain the ngn3 bHLH domain that mediates DNA binding and dimerization. As shown in Fig. 3C, Gal4D-ngn3 COOH activates a Gal4-responsive luciferase reporter, and this activation is enhanced approximately fivefold by activated Akt. Thus, Akt strongly increases transcriptional activation by the isolated ngn3 activation domain, suggesting that modulation of transactivation is likely to be the major mechanism by which Akt modulates ngn3 function.
Members of the neurogenin family of bHLH transcription factors directly activate expression of NeuroD1 mRNA (10, 30, 42, 58). Real-time RT-PCR was used to quantify the expression of transcripts for endogenous NeuroD1 and a control gene for hypoxanthine phosphoribosyltransferase in response to expression of ngn3. P19 cells were transfected with expression vectors for ngn3 or vector control and then treated with the PI3K/Akt inhibitor LY294002 (12.5 μM) or vehicle control for the final 8 h prior to RNA harvest at 30 h after transfection. ngn3 increased expression of NeuroD1 (threshold crossing reduced by more than five cycles relative to vector control); however, in the presence of LY294002, the ngn3 activation of NeuroD1 expression was significantly attenuated (an increase in threshold crossing of 1.8 cycles relative to that of ngn3, which would be an approximately threefold reduction in the NeuroD1 mRNA level, for an amplification efficiency of 1.8 copies per cycle) (Fig. 3D). We conclude that the endogenous PI3K/Akt signaling pathway modulates the expression of NeuroD1 mRNA, an endogenous target gene of ngn3. Since Akt modulates the ability of ngn3 to activate transcription (Fig. 3A and C), it is likely that the decreased expression of NeuroD1 mRNA in the presence of LY294002 reflects reduced transcriptional activation of the NeuroD1 gene by ngn3.
Akt regulates the interaction between bHLH proteins and the transcriptional coactivators CBP and p300.
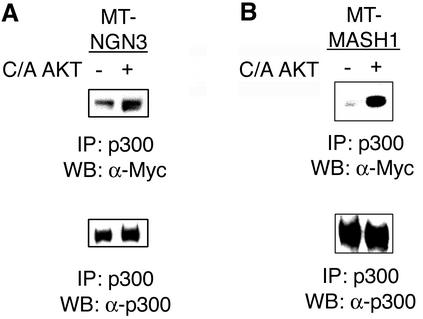
The neural bHLH proteins are known to function as transcriptional activators in part by recruiting the coactivators CBP and p300 to target genes. Activated Akt could modulate transactivation by regulating the recruitment of the coactivators by ngn3. We used coimmunoprecipitation to determine whether activated Akt1 enhanced the association between p300 and bHLH proteins. P19 cells were transiently transfected with an expression vector that encoded a Myc epitope-tagged ngn3, with and without constitutively active Akt1. Extracts were prepared, and endogenous p300 was immunoprecipitated. p300 and coassociated ngn3 were detected by Western blot analysis using antibodies directed against p300 or the Myc epitope tag of ngn3. An increase in the association of ngn3 with p300 was observed in the presence of constitutively active Akt1 (Fig. 4A). Quantitation of the Western blots by densitometric analysis revealed that the interaction between ngn3 and p300 increased by approximately 3.5-fold. A similar result was obtained with a second bHLH protein, MASH1 (Fig. 4B). The interaction of MASH1 and p300 increased by approximately fivefold when cells were transfected with constitutively active Akt1.
FIG. 4.
Activated Akt1 enhances complex formation between p300 and bHLH proteins. P19 cells were transfected with expression vectors for Myc epitope-tagged ngn3 (MT-ngn3) (A) or MT-MASH1 (B) with and without constitutively active Akt1. Endogenous p300 was immunoprecipitated from extracts 24 h after transfection and subjected to SDS-PAGE. p300 and coassociated Myc epitope-tagged bHLH proteins were detected by Western blot analysis with antibodies directed against p300 or the Myc epitope tag.
The coimmunoprecipitation assays provided evidence of increased complex formation by activated Akt. To determine whether increased complex formation is accompanied by increased transcriptional activity, we introduced combinations of expression vectors for p300 fused to the Gal4 DNA binding domain (Gal4 DBD-p300), ngn3, and constitutively active Akt1 into P19 cells, and transcriptional activation was determined with a Gal4-responsive luciferase reporter construct (14). Expression of either ngn3 or constitutively active Akt1 increased Gal4 DBD-p300-mediated reporter activity by three- to sixfold (Fig. 5A). Interestingly, coexpression of ngn3 and activated Akt1 synergistically increased Gal4 DBD-p300-mediated reporter activity by an additional 10-fold (60-fold activation). Thus, Akt-mediated enhanced complex formation (Fig. 4) is accompanied by enhanced complex activity (Fig. 5). Cooperation between ngn3 and Akt1 also extends to CBP: activated Akt1 in combination with ngn3 enhanced the transcriptional activity of a Gal4 DBD-CBP fusion protein 13-fold (Fig. 5B). We observed similar cooperation between activated Akt1 and other neural bHLH proteins, including ngn1 and NeuroD1, when expressed in combination with Gal4 DBD-CBP (data not shown).
To assess the involvement of the endogenous PI3K/Akt signaling pathway in the transcriptional cooperativity observed between ngn3 and p300, the activity of ngn3 together with Gal4 DBD-p300 in the presence and absence of the PI3K inhibitor LY294002 was determined. The fivefold activation observed when Gal4 DBD-p300 and ngn3 were cotransfected was abolished upon addition of LY294002 (Fig. 5C). However, LY294002 addition did not reduce activation by cotransfected Gal4 DBD-p300, ngn3, and the constitutively activated Akt1, which is not regulated by PI3K lipid products. The observations that LY294002 abolishes cooperative activation between Gal4 DBD-p300 and ngn3 and that the lipid-independent, activated Akt1 complements the LY effect on transcriptional cooperativity support a role for the endogenous PI3K/Akt signaling pathway in the regulation of neural bHLH protein-coactivator function.
Serum- and glucocorticoid-inducible kinase (SGK) is regulated by PI3K signaling and is highly related within its kinase domain to Akt family members (35, 56). Both Akt and SGK phosphorylate an RXRXX(S/T) consensus substrate recognition site. However, Akt preferentially phosphorylates consensus sites with a bulky hydrophobic residue adjacent to the carboxyl-terminal phosphoacceptor site (35, 56). Thus, although SGK and Akt have substrates in common (77), they are likely to have unique substrates as well. We investigated whether constitutively active SGK enhances the transcriptional activity of ngn3 in complex with p300. In contrast to constitutively active Akt1, constitutively active SGK1 did not cooperate with ngn3 to enhance the transcriptional activity of a Gal4 DBD-p300 fusion protein (Fig. 5D). Thus, the regulation of neural bHLH-coactivator function is likely to be the result of the action of Akt on a substrate(s) not shared with SGK.
Akt coassociates with CBP in vivo.
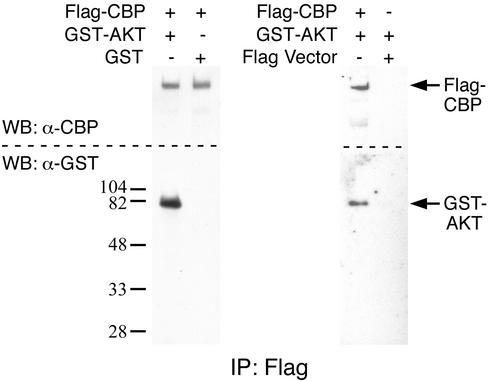
To determine if Akt can be found in complex with CBP in vivo, coprecipitation experiments were performed. HEK293 cells were transfected with expression constructs for Flag epitope-tagged CBP, Flag vector control, glutathione S-transferase (GST), and GST wild-type Akt1. Flag-CBP or Flag (control) was immunoprecipitated and subjected to SDS-PAGE followed by Western blot analysis with antibodies directed against the epitope tags to detect Flag-CBP and coassociated GST-Akt1. CBP coassociates with GST-Akt1 but not GST alone; also, Flag alone does not coassociate with GST-Akt1 (Fig. 6).
FIG. 6.
Akt and CBP coassociate in vivo. Flag-CBP or Flag (expressed from Flag vector [control]) was immunoprecipitated from lysates prepared from HEK293 cells transfected with the indicated constructs. The immunoprecipitates were subjected to SDS-PAGE followed by Western blotting with antibodies directed against CBP or the epitope tag on Akt (GST).
Mammalian CBP and p300 (human and mouse) share a single conserved canonical Akt consensus phosphorylation site, Ser1834 in p300 and Thr1871 in CBP. Phosphorylation of this consensus site by Akt has been reported to disrupt the interaction between CBP and C/EBPβ (26). The Akt consensus phosphorylation site is located in the region that binds the bHLH proteins MyoD, NeuroD1, and probably ngn1 and ngn3 (36, 62, 75), raising the possibility that phosphorylation of this site by Akt could enhance the interaction between neural bHLH proteins and CBP/p300, leading to enhanced complex activity. To assess whether phosphorylation of this site by Akt mediates the cooperativity in transcriptional activity between p300 and bHLH proteins, we generated a nonphosphorylatable p300 mutant protein by substituting alanine for serine at the phosphoacceptor site within the Akt consensus site. Expression vectors for the Gal4 DNA binding domain fused to wild-type or alanine mutant p300, ngn3, and constitutively active Akt1 were introduced into P19 cells, and transactivation was determined with a Gal4-responsive luciferase reporter construct. Together, ngn3 and Akt1 enhance the transcriptional activity of either the wild-type or alanine p300 mutant fusion protein (Fig. 7). A similar result was obtained with a CBP mutant at the Thr1871 Akt phosphoacceptor site (data not shown). Therefore, phosphorylation of the conserved Akt consensus site in p300 and CBP is unlikely to mediate cooperativity between ngn3, p300/CBP, and Akt1.
DISCUSSION
Neural bHLH transcription factors are required for formation of neurons in mammals (24), and forced expression of a neural bHLH protein is sufficient to drive neuronal differentiation of uncommitted cells (17, 38, 41, 70). Here we have shown that the serine-threonine kinases Akt1 and Akt2 are required for neuronal differentiation of mouse P19 cells in response to forced expression of the neural bHLH protein ngn3. While Akt1 and Akt2 are known to be involved in neuronal survival, our results indicate that Akt1 and Akt2 have additional functions during neuronal differentiation that are separable from neuronal survival. We observed that activated Akt increased transactivation of a reporter by ngn3 or by the isolated activation domain of ngn3. We also found that PI3K-Akt signaling modulates expression of an endogenous ngn3 target gene, NeuroD1. In addition, we found that activated Akt1 can promote the formation of complexes between neural bHLH proteins and the coactivators CBP and p300. Taken together, these results suggest a novel role for Akt in regulating neuronal differentiation at the level of transcription. We suggest that one function of the Akt proteins during neuronal differentiation is to regulate the interaction between neural bHLH proteins and the CBP/p300 coactivators.
We coexpressed ngn3 with U6-driven hairpin siRNAs directed against Akt1, Akt2, or both Akt1 and Akt2. The hairpin siRNAs effectively reduced the endogenous levels of the Akt kinases. Decreased Akt levels led to decreased cell survival and decreased neuron formation in the ngn3-transfected cells. By culturing the transfected cells in the presence of a cell-permeable general caspase inhibitor, we observed an increase in cell survival but not a concomitant increase in neuron formation, indicating that the effects of Akt on bHLH-driven neuron formation are distinct from its effects on cell survival. While both Akt1 and Akt2 are expressed at high levels during neurogenesis (54), they have not been previously shown to have a role in neuron formation or the early steps of neuronal differentiation, although Akt function has been implicated in later aspects of neuronal differentiation, such as axon branching (44). Targeted disruption of either Akt1 or Akt2 in mice has no apparent effect on neuron formation or differentiation (11-13). In contrast, inhibition of either kinase by RNAi led to a substantial reduction in neuron formation from ngn3-transfected P19 cells. This may reflect differences between P19 cells and neural progenitors in the level of Akt signaling, or the acute loss of Akt1 or Akt2 expression during the ngn3 transfections may not allow sufficient time for compensatory mechanisms to operate (e.g., upregulation of the remaining Akt1/2 kinase or another kinase such as Akt3). RNAi against Akt1 and Akt2 together reduced neuron formation in ngn3-transfected P19 cells more than RNAi against either kinase individually, suggesting that the Akt1 and Akt2 kinases have partially redundant functions. The effect of targeted disruption of both Akt1 and Akt2 in mice has not been reported yet.
The neural bHLH proteins NeuroD, ngn1 to -3, and MASH1, as well as the myogenic bHLH protein MyoD, interact with the coactivators CBP and p300 (references 16, 36, 49, 61, 65, and 67 and this work). CBP/p300 facilitate the assembly of active transcription factor complexes in part by bringing transcription factors and basal components of the transcriptional machinery into proximity, as well as by acetylation of histones and transcription factors through an intrinsic AT activity (20, 21, 23). Our data indicate that activated Akt can increase transcriptional activation by ngn3. Furthermore, activated Akt can increase the amount of ngn3 or MASH1 proteins that are associated with CBP/p300, providing a likely mechanism for the increased transcriptional activity. Activated SGK, which phosphorylates a consensus site similar but not identical to that of Akt, does not enhance transcriptional cooperativity between bHLH proteins and coactivators. This observation suggests that the regulation of neurogenic transactivation function is a specific effect of Akt, arising from the action of Akt on a substrate(s) not shared with SGK. Akt also augments the transcriptional activity of other neural bHLH proteins in complex with CBP and p300, including NeuroD1 and ngn1. In addition, activated Akt can enhance transcriptional cooperativity between the myogenic bHLH protein MyoD and p300 (unpublished observations). Thus, Akt is likely to be a general regulator of coactivator interactions with the bHLH family of transcription factors. LY294002, a pharmacological inhibitor of PI3K/Akt, abolishes ngn3-CBP transcriptional cooperativity, but this effect can be overcome by activated Akt1. This observation indicates that the endogenous PI3K/Akt signaling pathway is required for neural bHLH-coactivator interactions. In addition, we have found that Akt coassociates with CBP. Coupled with the requirement for Akt during ngn3-driven neuron formation, our results suggest that Akt is likely to control neuron formation and differentiation at least in part by modulating neural bHLH-coactivator interactions. One possible mechanism to explain our results would be that Akt, in complex with CBP/p300, regulates the interaction between bHLH proteins and coactivators by phosphorylating CBP/p300 and/or the bHLH proteins.
Phosphorylation of coactivators can regulate their recruitment to transcription factor complexes and thereby impact transcriptional events. For example, phosphorylation of CBP by protein kinase C recruits CBP to the AP-1 complex (76), while phosphorylation by calmodulin kinase IV contributes to CREB/CBP-dependent transcription events (31). Moreover, as CBP/p300 are thought to be rate-limiting components for transcriptional activation in the cell (29, 33), phosphorylation is one means by which cells can rapidly and reversibly modulate coactivator-transcription factor interactions to regulate transcription. Guo et al. reported that Akt phosphorylates CBP/p300 within the CH3 domain and that this phosphorylation disrupts the interaction between CBP and C/EBPβ (26). NeuroD, MyoD, and probably other bHLH proteins can bind to CBP/p300 via the CH3 domain. However, we found that alteration of the phosphoacceptor site within the Akt consensus site in CBP and p300 (Thr1871 in CBP and S1834 in p300) did not affect the ability of Akt to potentiate transcriptional cooperativity between the coactivators and ngn3. Therefore, it is unlikely that phosphorylation of CBP/p300 by Akt at T1871/S1834 contributes to the increase in transcriptional cooperativity. It remains possible that Akt enhances interactions between the bHLH proteins and CBP/p300 by phosphorylating CBP and p300 at a noncanonical phosphorylation site.
Another explanation for our observations is that Akt could phosphorylate the neural bHLH transcription factors and increase their affinity for CBP/p300, leading to transcriptional cooperativity. For example, phosphorylation of CREB by protein kinase A or Akt enhances its association with CBP/p300 (15, 22). Although ngn3 contains a possible consensus phosphorylation site for Akt in its N terminus, no conserved Akt consensus phosphorylation site is shared among the various neural bHLH proteins. Alteration of the potential phosphoacceptor serine to alanine in the potential ngn3 Akt consensus site does not alter ngn3 function in P19 cells or reduce the effect of Akt on ngn3-CBP cooperativity (unpublished observations). Furthermore, while function of the isolated activation domain of ngn3 is enhanced by activated Akt, this domain does not contain a consensus phosphorylation site for Akt. Therefore, it is unlikely that neural bHLH protein function is regulated through direct phosphorylation by Akt.
Akt is known to modulate the functions of other kinases, raising the possibility that Akt could regulate bHLH-coactivator interactions indirectly. Candidate kinases that could mediate the effect of Akt include GSK3α/β. Akt phosphorylates GSK3α and GSK3β and negatively regulates their enzymatic activity (71). Marcus et al. reported that GSK3β inhibits the function of XNeuroD1 and prevents neurogenesis in Xenopus (43). Vetter and colleagues demonstrated that the integrity of a GSK3 consensus phosphorylation site in Xenopus and mouse neuroD is critical for proper regulation of the timing of NeuroD function (48). Taken together, these observations suggest that inhibition of GSK3 activity may be a prerequisite for neurogenesis to proceed in the appropriate time and place. However, although GSK3α and -β were phosphorylated at Ser21/Ser9 in P19 cells under our experimental conditions, reduction of GSK3α and GSK3β function using hairpin siRNAs (74) does not increase neuron formation by ngn3-transfected P19 cells, nor does it complement the deficit in neuron formation generated by the Akt hairpin siRNAs (data not shown). In addition, expression of GSK3β Ser9Ala, a mutant kinase that cannot be inhibited by Akt phosphorylation, does not block the ability of activated Akt1 to increase ngn3-CBP transcriptional cooperativity (data not shown). Thus, negative regulation of GSK3 by Akt is unlikely to mediate either the requirement for Akt function during ngn3-driven neuronal differentiation or the ability of Akt to promote functional and physical cooperativity between the neural bHLH proteins and CBP/p300. It remains possible that modulation of GSK3 function by Akt regulates bHLH function and neuron formation in other contexts.
Our data support a model in which Akt signaling regulates the assembly and/or stability of neural bHLH transcription factor-coactivator complexes. We favor the possibility that Akt directly, or acting via a downstream kinase, phosphorylates a protein(s) that acts in the complex with the coactivators and the bHLH proteins to promote complex formation or stability. Alternately, Akt could inhibit a negative regulator of bHLH-coactivator interactions. Recently it has been reported that Akt can negatively regulate the corepressor N-CoR and promote glial differentiation (28). In mice, targeted disruption of N-CoR leads to premature neuronal differentiation (32), raising the possibility that Akt phosphorylation could inhibit N-CoR function during neuronal differentiation as well as during glial differentiation. Although N-CoR has not been linked to neural bHLH function, it has been reported that N-CoR negatively regulates activation by the myogenic bHLH protein MyoD (4), and N-CoR can compete with CBP/p300 for binding to transcription factors (72).
While our data implicate Akt in the regulation of neural bHLH protein function and neuron formation, they do not exclude an additional role for Akt and bHLH proteins in neuronal survival. Akt has been implicated in cell survival in many systems, and a subset of the neural bHLH proteins has also been implicated in neuronal survival (47, 51). Akt may modulate the expression of bHLH target genes that are involved in neuronal survival, subsequent to neuron formation.
Akt may also influence cell fate decisions within the developing nervous system. In addition to promoting neurogenesis, the bHLH protein ngn1 titrates CBP/p300 to inhibit gliogenesis during early neural development, and other neural bHLH proteins are likely to function similarly (27, 50, 67). In the presence of neural bHLH proteins, Akt could promote neuronal cell fates and simultaneously suppress glial cell fates by enhancing the interaction between the bHLH proteins and CBP/p300.
Acknowledgments
We thank Dan Goldman and Robert Davis for discussions. We gratefully acknowledge David Wilson, Holly Sucic, and Chris Hart for assistance in constructing vectors, Robert Thompson and Paresh Patel for discussions on real time RT-PCR, and Robert Davis, Ming-Jer Tsai, and Kun-Liang Guan for kindly providing plasmids.
This work was supported by a Research Scholar Grant (RSG-01-177-01-MGO) from the American Cancer Society (A.B.V.), a grant from the Walther Cancer Institute (A.B.V.), NIH grant NS38698 (D.L.T.), and a grant from the University of Michigan Biomedical Research Council (D.L.T.).
REFERENCES
- 1.Alessi, D. R., M. Andjelkovic, B. Caudwell, P. Cron, N. Morrice, P. Cohen, and B. A. Hemmings. 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15:6541-6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. F., M. A. Aberg, M. Nilsson, and P. S. Eriksson. 2002. Insulin-like growth factor-I and neurogenesis in the adult mammalian brain. Brain Res. Dev. Brain Res. 134:115-122. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, M., O. Batista, A. Bellacosa, P. Tsichlis, and P. K. Vogt. 1998. The Akt kinase: molecular determinants of oncogenicity. Proc. Natl. Acad. Sci. USA 95:14950-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, P., M. Downes, P. Lau, J. Harris, S. L. Chen, Y. Hamamori, V. Sartorelli, and G. E. Muscat. 1999. The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Mol. Endocrinol. 13:1155-1168. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Arie, N., H. J. Bellen, D. L. Armstrong, A. E. McCall, P. R. Gordadze, Q. Guo, M. M. Matzuk, and H. Y. Zoghbi. 1997. Math1 is essential for genesis of cerebellar granule neurons. Nature 390:169-172. [DOI] [PubMed] [Google Scholar]
- 6.Brown, N. L., S. Kanekar, M. L. Vetter, P. K. Tucker, D. L. Gemza, and T. Glaser. 1998. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 125:4821-4833. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Brunet, A., S. R. Datta, and M. E. Greenberg. 2001. Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr. Opin. Neurobiol. 11:297-305. [DOI] [PubMed] [Google Scholar]
- 9.Cameron, H. A., T. G. Hazel, and R. D. McKay. 1998. Regulation of neurogenesis by growth factors and neurotransmitters. J. Neurobiol. 36:287-306. [PubMed] [Google Scholar]
- 10.Cau, E., G. Gradwohl, C. Fode, and F. Guillemot. 1997. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124:1611-1621. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W. S., P. Z. Xu, K. Gottlob, M. L. Chen, K. Sokol, T. Shiyanova, I. Roninson, W. Weng, R. Suzuki, K. Tobe, T. Kadowaki, and N. Hay. 2001. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, H., J. Mu, J. K. Kim, J. L. Thorvaldsen, Q. Chu, E. B. Crenshaw III, K. H. Kaestner, M. S. Bartolomei, G. I. Shulman, and M. J. Birnbaum. 2001. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 292:1728-1731. [DOI] [PubMed] [Google Scholar]
- 13.Cho, H., J. L. Thorvaldsen, Q. Chu, F. Feng, and M. J. Birnbaum. 2001. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J. Biol. Chem. 276:38349-38352. [DOI] [PubMed] [Google Scholar]
- 14.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 15.Du, K., and M. Montminy. 1998. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273:32377-32379. [DOI] [PubMed] [Google Scholar]
- 16.Eckner, R., T. P. Yao, E. Oldread, and D. M. Livingston. 1996. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10:2478-2490. [DOI] [PubMed] [Google Scholar]
- 17.Farah, M. H., J. M. Olson, H. B. Sucic, R. I. Hume, S. J. Tapscott, and D. L. Turner. 2000. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127:693-702. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, A. J., B. D. Dierks, and T. A. Reh. 2002. Exogenous growth factors induce the production of ganglion cells at the retinal margin. Development 129:2283-2291. [DOI] [PubMed] [Google Scholar]
- 19.Fode, C., G. Gradwohl, X. Morin, A. Dierich, M. LeMeur, C. Goridis, and F. Guillemot. 1998. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron 20:483-494. [DOI] [PubMed] [Google Scholar]
- 20.Giles, R. H., D. J. Peters, and M. H. Breuning. 1998. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 14:178-183. [DOI] [PubMed] [Google Scholar]
- 21.Giordano, A., and M. L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell Physiol. 181:218-230. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez, G. A., and M. R. Montminy. 1989. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675-680. [DOI] [PubMed] [Google Scholar]
- 23.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 24.Guillemot, F. 1999. Vertebrate bHLH genes and the determination of neuronal fates. Exp. Cell. Res. 253:357-364. [DOI] [PubMed] [Google Scholar]
- 25.Guillemot, F., L. C. Lo, J. E. Johnson, A. Auerbach, D. J. Anderson, and A. L. Joyner. 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75:463-476. [DOI] [PubMed] [Google Scholar]
- 26.Guo, S., S. B. Cichy, X. He, Q. Yang, M. Ragland, A. K. Ghosh, P. F. Johnson, and T. G. Unterman. 2001. Insulin suppresses transactivation by CAAT/enhancer-binding proteins beta (C/EBPβ). Signaling to p300/CREB-binding protein by protein kinase B disrupts interaction with the major activation domain of C/EBPβ. J. Biol. Chem. 276:8516-8523. [DOI] [PubMed] [Google Scholar]
- 27.Hatakeyama, J., K. Tomita, T. Inoue, and R. Kageyama. 2001. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development 128:1313-1322. [DOI] [PubMed] [Google Scholar]
- 28.Hermanson, O., K. Jepsen, and M. G. Rosenfeld. 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419:934-939. [DOI] [PubMed] [Google Scholar]
- 29.Horvai, A. E., L. Xu, E. Korzus, G. Brard, D. Kalafus, T. M. Mullen, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 1997. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc. Natl. Acad. Sci. USA 94:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, H. P., M. Liu, H. M. El-Hodiri, K. Chu, M. Jamrich, and M. J. Tsai. 2000. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol. Cell. Biol. 20:3292-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Impey, S., A. L. Fong, Y. Wang, J. R. Cardinaux, D. M. Fass, K. Obrietan, G. A. Wayman, D. R. Storm, T. R. Soderling, and R. H. Goodman. 2002. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 34:235-244. [DOI] [PubMed] [Google Scholar]
- 32.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 33.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 34.Kim, C. H., Y. K. Bae, Y. Yamanaka, S. Yamashita, T. Shimizu, R. Fujii, H. C. Park, S. Y. Yeo, T. L. Huh, M. Hibi, and T. Hirano. 1997. Overexpression of neurogenin induces ectopic expression of HuC in zebrafish. Neurosci. Lett. 239:113-116. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, T., M. Deak, N. Morrice, and P. Cohen. 1999. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem. J. 344(Pt. 1):189-197. [PMC free article] [PubMed] [Google Scholar]
- 36.Koyano-Nakagawa, N., D. Wettstein, and C. Kintner. 1999. Activation of Xenopus genes required for lateral inhibition and neuronal differentiation during primary neurogenesis. Mol. Cell. Neurosci. 14:327-339. [DOI] [PubMed] [Google Scholar]
- 37.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 38.Lee, J. E., S. M. Hollenberg, L. Snider, D. L. Turner, N. Lipnick, and H. Weintraub. 1995. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268:836-844. [DOI] [PubMed] [Google Scholar]
- 39.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 40.Ma, Q., Z. Chen, I. del Barco Barrantes, J. L. de la Pompa, and D. J. Anderson. 1998. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20:469-482. [DOI] [PubMed] [Google Scholar]
- 41.Ma, Q., C. Kintner, and D. J. Anderson. 1996. Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87:43-52. [DOI] [PubMed] [Google Scholar]
- 42.Ma, Q., L. Sommer, P. Cserjesi, and D. J. Anderson. 1997. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J. Neurosci. 17:3644-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcus, E. A., C. Kintner, and W. Harris. 1998. The role of GSK3β in regulating neuronal differentiation in Xenopus laevis. Mol. Cell. Neurosci. 12:269-280. [DOI] [PubMed] [Google Scholar]
- 44.Markus, A., J. Zhong, and W. D. Snider. 2002. Raf and akt mediate distinct aspects of sensory axon growth. Neuron 35:65-76. [DOI] [PubMed] [Google Scholar]
- 45.McManus, M. T., C. P. Petersen, B. B. Haines, J. Chen, and P. A. Sharp. 2002. Gene silencing using micro-RNA designed hairpins. RNA 8:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyagishi, M., and K. Taira. 2002. U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20:497-500. [DOI] [PubMed] [Google Scholar]
- 47.Miyata, T., T. Maeda, and J. E. Lee. 1999. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, K. B., M. L. Schneider, and M. L. Vetter. 2002. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron 34:183-195. [DOI] [PubMed] [Google Scholar]
- 49.Mutoh, H., F. J. Naya, M. J. Tsai, and A. B. Leiter. 1998. The basic helix-loop-helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells. Genes Dev. 12:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nieto, M., C. Schuurmans, O. Britz, and F. Guillemot. 2001. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29:401-413. [DOI] [PubMed] [Google Scholar]
- 51.Olson, J. M., A. Asakura, L. Snider, R. Hawkes, A. Strand, J. Stoeck, A. Hallahan, J. Pritchard, and S. J. Tapscott. 2001. NeuroD2 is necessary for development and survival of central nervous system neurons. Dev. Biol. 234:174-187. [DOI] [PubMed] [Google Scholar]
- 52.O'Neill, G. M., and E. A. Golemis. 2001. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol. Cell. Biol. 21:5094-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orike, N., G. Middleton, E. Borthwick, V. Buchman, T. Cowen, and A. M. Davies. 2001. Role of PI 3-kinase, Akt and Bcl-2-related proteins in sustaining the survival of neurotrophic factor-independent adult sympathetic neurons. J. Cell Biol. 154:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owada, Y., A. Utsunomiya, T. Yoshimoto, and H. Kondo. 1997. Expression of mRNA for Akt, serine-threonine protein kinase, in the brain during development and its transient enhancement following axotomy of hypoglossal nerve. J. Mol. Neurosci. 9:27-33. [DOI] [PubMed] [Google Scholar]
- 55.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park, J., M. L. Leong, P. Buse, A. C. Maiyar, G. L. Firestone, and B. A. Hemmings. 1999. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 18:3024-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paul, C. P., P. D. Good, I. Winer, and D. R. Engelke. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20:505-508. [DOI] [PubMed] [Google Scholar]
- 58.Perron, M., K. Opdecamp, K. Butler, W. A. Harris, and E. J. Bellefroid. 1999. X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl. Acad. Sci. USA 96:14996-15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polesskaya, A., and A. Harel-Bellan. 2001. Acetylation of MyoD by p300 requires more than its histone acetyltransferase domain. J. Biol. Chem. 276:44502-44503. [DOI] [PubMed] [Google Scholar]
- 60.Polesskaya, A., I. Naguibneva, A. Duquet, E. Bengal, P. Robin, and A. Harel-Bellan. 2001. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 21:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puri, P. L., M. L. Avantaggiati, C. Balsano, N. Sang, A. Graessmann, A. Giordano, and M. Levrero. 1997. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu, Y., A. Sharma, and R. Stein. 1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 18:2957-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rupp, R. A., L. Snider, and H. Weintraub. 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8:1311-1323. [DOI] [PubMed] [Google Scholar]
- 64.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 65.Sharma, A., M. Moore, E. Marcora, J. E. Lee, Y. Qiu, S. Samaras, and R. Stein. 1999. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol. Cell. Biol. 19:704-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun, Y., M. Nadal-Vicens, S. Misono, M. Z. Lin, A. Zubiaga, X. Hua, G. Fan, and M. E. Greenberg. 2001. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104:365-376. [DOI] [PubMed] [Google Scholar]
- 68.Tang, G., J. Yang, Y. Minemoto, and A. Lin. 2001. Blocking caspase-3-mediated proteolysis of IKKβ suppresses TNF-alpha-induced apoptosis. Mol. Cell 8:1005-1016. [DOI] [PubMed] [Google Scholar]
- 69.Tapon, N., K. Nagata, N. Lamarche, and A. Hall. 1998. A new rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-κB signalling pathways. EMBO J. 17:1395-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner, D. L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8:1434-1447. [DOI] [PubMed] [Google Scholar]
- 71.Woodgett, J. R. 2001. Judging a protein by more than its name: GSK-3. Sci. STKE 2001:RE12. [DOI] [PubMed]
- 72.Xu, L., R. M. Lavinsky, J. S. Dasen, S. E. Flynn, E. M. McInerney, T. M. Mullen, T. Heinzel, D. Szeto, E. Korzus, R. Kurokawa, A. K. Aggarwal, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1998. Signal-specific co-activator domain requirements for Pit-1 activation. Nature 395:301-306. [DOI] [PubMed] [Google Scholar]
- 73.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, J. Y., J. Taylor, S. L. DeRuiter, A. B. Vojtek, and D. L. Turner. 2003. Simultaneous inhibition of GSK3α and GSK3β using hairpin siRNA expression vectors. Mol. Ther. 7:228-236. [DOI] [PubMed] [Google Scholar]
- 75.Yuan, W., G. Condorelli, M. Caruso, A. Felsani, and A. Giordano. 1996. Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem. 271:9009-9013. [DOI] [PubMed] [Google Scholar]
- 76.Zanger, K., S. Radovick, and F. E. Wondisford. 2001. CREB binding protein recruitment to the transcription complex requires growth factor-dependent phosphorylation of its GF box. Mol. Cell 7:551-558. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, B. H., E. D. Tang, T. Zhu, M. E. Greenberg, A. B. Vojtek, and K. L. Guan. 2001. Serum- and glucocorticoid-inducible kinase SGK phosphorylates and negatively regulates B-Raf. J. Biol. Chem. 276:31620-31626. [DOI] [PubMed] [Google Scholar]