Abstract
The bacterial protein encoded by the gene ychF is 1 of 11 universally conserved GTPases and the only one whose function is unknown. The crystal structure determination of YchF was sought to help with the functional assignment of the protein. The YchF protein from Haemophilus influenzae was cloned and expressed, and the crystal structure was determined at 2.4 Å resolution. The polypeptide chain is folded into three domains. The N-terminal domain has a mononucleotide binding fold typical for the P-loop NTPases. An 80-residue domain next to it has a pronounced α-helical coiled coil. The C-terminal domain features a six-stranded half-barrel that curves around an α-helix. The crablike three-domain structure of YchF suggests the binding site for a double-stranded nucleic acid in the cleft between the domains. The structure of the putative GTP-binding site is consistent with the postulated guanine specificity of the protein. Fluorescence measurements have demonstrated the ability of YchF to bind a double-stranded nucleic acid and GTP. Taken together with other experimental data and genomic analysis, these results suggest that YchF may be part of a nucleoprotein complex and may function as a GTP-dependent translation factor.
The protein encoded by the gene ychF is 1 of 11 universally conserved GTPases and the only one whose function is unknown (7, 19, 24). GTPases are key regulators of protein synthesis, cell cycling and differentiation, and hormone signaling (4). Despite the functional diversity, all GTPases have the same core structure and act as molecular switches. The YchF family is characterized by a remarkably high sequence conservation. For instance, the Escherichia coli and human proteins are 47% identical. Forty-six residues of approximately 360 are strictly invariant throughout the family. The YchF sequence contains the Walker A (41) nucleotide-biding motif GXXXXGKT/S (where X denotes any amino acid), which defines the so-called P-loop involved in the binding of phosphates of nucleoside triphosphate (NTP) and the Walker B motif with a conserved aspartate that binds a water-bridged Mg ion. The common fold includes a parallel β-sheet flanked by α-helices, where the P-loop follows the N-terminal (central) β-strand, and the Walker B motif resides in the adjacent β-strand.
According to the most recent classification of P-loop NTPases (19), YchF belongs to the TRAFAC class that includes translation factors (hence the name of the class), Ras-related GTPases, heterotrimeric G proteins, septins, dynamins, and motor ATPases such as kinesin and myosin. The members of the class are characterized by a conserved threonine in the region between the Walker A and B motifs. This residue binds the Mg2+ required for NTP hydrolysis. Structurally, the distinctive feature of the TRAFAC class is an antiparallel β-strand next to the Walker B strand. From the functional point of view, these proteins are typical molecular switches that change the conformation upon NTP hydrolysis. Two regions are particularly sensitive to the presence of the γ-phosphate, the loop preceding the antiparallel β-strand (switch I) and the loop following the Walker B strand (switch II).
The presence of a glycine-rich motif in the switch II region as well as the conserved motif YXFXTXXXXXG in the switch I region are distinctive features of the Obg superfamily (19). Besides the YchF family, it includes four other protein families: Obg, DRG, Ygr210, and NOG1. The YchF and Ygr210 families form one branch of the superfamily with the YchF proteins present in bacteria and eukaryotes and the Ygr210 proteins present in archaea and fungi. The NTPase domains of all members of the superfamily share significant sequence similarity. For instance, YchF and YhbZ (the Obg homolog) from E. coli are 32% identical over a fragment of 210 residues. However, the rest of the structure is quite different, as are the regulatory pathways these proteins are involved in (6).
The specificity of G proteins to guanine is determined by the conserved NKXD motif, also known as G4 (5), located at the end of strand β6 (Fig. 1). A peculiar feature of YchF is the lack of the conserved lysine in this sequence and the replacement of the aspartate with glutamate. This is a unique exception among the TRAFAC GTPases which, in the absence of experimental data, raises questions regarding the nucleotide specificity of YchF.
FIG. 1.
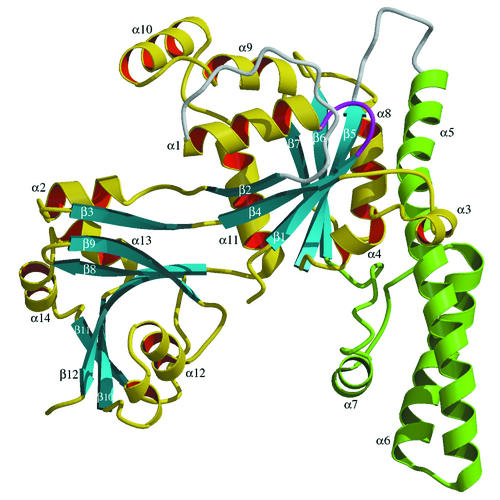
Ribbon presentation of the polypeptide fold of YchF. The α-helical domain is shown in green. Disordered loops are shown in white. The NTP-binding P-loop is shown in purple. The figure was produced with Molscript (18) and Raster3D (23).
The crystal structure of YchF was determined in order to assist with the functional assignment of the protein. The YchF protein from Haemophilus influenzae (HI0393 in The Institute for Genomic Research numbering) was cloned and expressed, and the crystal structure was determined at a 2.4-Å resolution. Analysis of the structure and binding experiments suggest that YchF may be a GTP-dependent nucleic acid-binding protein.
MATERIALS AND METHODS
Expression and purification of YchF.
The ychF gene was cloned by PCR, with the genomic H. influenzae DNA as a template, into the pET15b vector (Novagen) and expressed in the E. coli strain B834(DE3). The protein was purified on a Poros HQ50 anion-exchange column and then on an HS20 cation-exchange column; dialyzed against 50 mM Tris-HCl (pH 7.5), 0.1 mM dithiothreitol, and 0.1 mM EDTA; and concentrated to 15 mg/ml for crystallization. The molecular mass of 39.6 kDa measured by matrix-assisted laser desorption ionization-mass spectrometry corresponded to the value calculated from the amino acid sequence. The equilibrium sedimentation indicated that the protein is a monomer in solution (data not shown).
Crystallization and structure determination.
YchF crystals were obtained at room temperature in hanging drops from 0.1 M Bicine (pH 7.5), 0.3 M magnesium acetate, and 13% polyethylene glycol 8000. They belong to the space group P21 with the following cell parameters: a, 61.0; b, 93.6; c, 85.2 Å; and β, 100.1°. There are two molecules in the asymmetric unit.
X-ray diffraction data from one native crystal (Table 1) were collected at 100°K on the IMCA-CAT beamline 17-ID at the Advanced Photon Source (Argonne, Ill.) with a Mar charge-coupled device detector. Heavy atom derivatives were measured on a Bruker rotating anode generator with a Mar345 image plate detector. All data were processed with HKL2000 (30). The structure was solved by multiple isomorphous replacement with uranium (5 mM UNO3 soaked for 1 h) and gold [9 mM KAu(CN)4 soaked for 1 h] derivatives. A single uranium site and two gold sites were located by SOLVE (39) and were used in phase refinement with SHARP (10). The atomic model was built by using O (15) and refined with REFMAC (26) to an R factor of 0.208 for all data in the resolution range of 10 to 2.4 Å (Rfree = 0.247 for 5% data). Crystallographic calculations were performed with the CCP4 program suite (8). No restraints on the noncrystallographic symmetry were imposed during refinement. Refinement statistics are given in Table 1. One residue, Val287 in chain B, has unusual main-chain torsion angles (φ = 28.1°; ψ = −110.2°) that correspond to the epsilon region of the Ramachandran plot. The electron density for this residue is clear and unambiguous. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (2) under the accession code 1JAL.
TABLE 1.
X-ray data and refinement statisticsa
| Parameter (unit) | Result |
|---|---|
| Resolution (Å) | 10-2.4 (2.46-2.40) |
| No. of unique reflections | 36,325 (2,379) |
| Completeness (%) | 99.4 (98.8) |
| Redundancy | 5.4 (5.2) |
| Rsym (Σ|I − 〈I〉|)/ΣI | 0.044 (0.242) |
| <I/σI> | 34.3 (4.4) |
| Rcryst (Σ||Fo| − |Fc||)/Σ|Fo|) | 0.208 (0.222) |
| Rfree (5% data) | 0.247 (0.264) |
| No. of protein atoms | 5,243 |
| No. of water molecules | 484 |
| RMSDb in bonds (Å) | 0.016 |
| RMSD in angles (°) | 1.5 |
| Mean B factors (Å2) from: | |
| Chain A | 52.4 |
| Chain B | 67.9 |
| Wilson plot | 55.1 |
Values for the highest-resolution shell are given in parentheses.
RMSD, RMS deviation.
Fluorescence measurements.
Fluorescence was measured at 25°C with a Fluoromax-2 spectrofluorometer (Jobin Yvon). Binding of double-stranded DNA (dsDNA) was monitored by changes in fluorescence of tryptophan, which was measured with an excitation wavelength of 280 nm and a maximum emission spectrum of 346 nm. The reaction mixture contained 100 nM protein, 20 mM Tris-HCl (pH 7.5), and 50 mM NaCl. The DNA sample used in the experiment was digested plasmid DNA of about 6,000 bp. DNA was titrated up to the final concentration of 100 nM (per base pair). As a negative control, fluorescence was measured at the same concentrations of DNA in the absence of the protein (no significant fluorescence was detected).
The fluorescence of 2′(3′)-O-(2,4,6-trinitrophenyl)GTP (TNP-GTP) (Molecular Probes) was measured with an excitation wavelength of 408 nm and a maximum emission spectrum of 555 nm. The reaction mixture contained 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, and other components as indicated in the text. The titration curve was plotted as an increase in fluorescence with respect to the protein-free experiment versus the nucleotide concentration by using Kaleidagraph (Synergy Software).
RESULTS
Overview of the structure.
The YchF polypeptide is folded into three distinct domains (Fig. 1). The N-terminal (G) domain has a mononucleotide binding fold typical for the P-loop NTPases (33). A six-stranded mostly parallel β-sheet is flanked by α-helices on both sides. The strand order is 2-4-1-5-6-7, with strand β2 antiparallel to the other strands. A 23-residue loop following β2 forms part of the C-terminal (C) domain, which also includes residues 279 to 363. Domain C is dominated by the mixed β-sheet curved as a half-barrel around helix α12. An 80-residue polypeptide following strand β5 folds into the α-helical (A) domain consisting of two long α-helices (α5 and α6) and one short α-helix (α7). The loops connecting the three domains (Fig. 1) are not visible in the electron density, suggesting not only the flexibility of the loops but also the mobility of the corresponding domains.
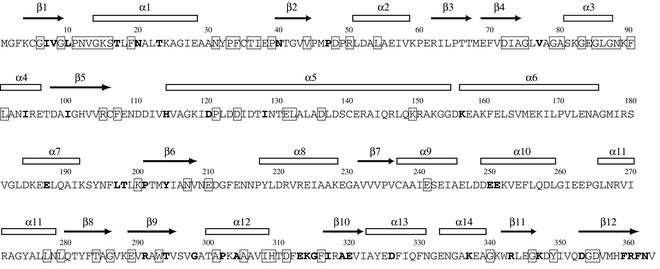
As a P-loop NTPase, YchF is characterized by the presence of two conserved sequence motifs involved in the binding of the phosphates of NTP (Fig. 2). The Walker A sequence G9XXXXGKS16 forms the P-loop between strand β1 and the following α-helix. The Walker B sequence D72XXG75 resides in the adjacent β-strand, β4, and contains an aspartic acid (Asp72) usually involved in the coordination of a Mg ion. Neither NTP nor Mg is observed in the present structure, although Mg2+ was in the crystallization solution.
FIG. 2.
Amino acid sequence of the H. influenzae YchF in one-letter code. Residues conserved in all family members with no more than one exception are enclosed in boxes. Residues with conservative replacements in two or three family members are in bold letters. Secondary structure elements calculated with DSSP (16) are indicated above the sequence.
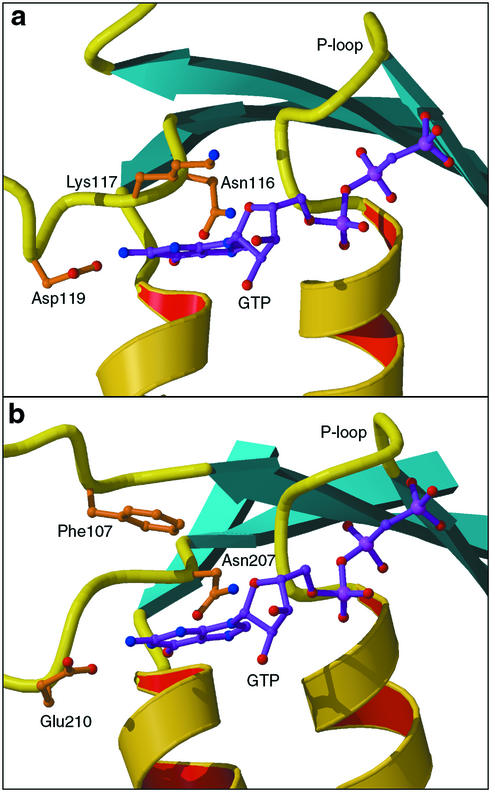
The specificity of G proteins to GTP is determined by the conserved NKXD motif (5) located at the end of strand β6. This sequence forms part of the GTP recognition site, where the lysine side chain stacks with the base, the aspartic acid residue forms a bidentate H bond to the N1 and N2 atoms of guanine, and the asparagine residue binds to the N7 atom (Fig. 3). The corresponding sequence in YchF is N207VNE210 with invariant Asn207 and Glu210. YchF has an unusual architecture of the guanine-binding site compared to other GTPases. Glu210 replaces the canonical aspartate and, together with Asn207, is positioned to bind the amino groups of guanine. The invariant Phe107 from another loop (β5) takes the role of the consensus lysine by providing the stacking interaction with the base. Thus, although somewhat unusual, the structure of the recognition site is consistent with the postulated guanine specificity of YchF.
FIG. 3.
Guanine recognition site in Ras (Protein Data Bank entry 121P), a typical G protein (a), and YchF (b). GTP is shown in purple, as observed in Ras and as modeled in YchF. The lysine residue of the consensus sequence NKXD (Lys117 in Ras) is spatially replaced with Phe107 in YchF.
Nucleotide binding.
Attempts to obtain crystal complexes with GTP or GDP by soaking native crystals and by cocrystallization failed. One possible explanation is that the conformational changes accompanying the nucleotide binding are inconsistent with the present crystal form.
Binding of GTP by YchF was assessed by measuring fluorescence of the nucleotide labeled with TNP. In the absence of Mg2+, the fivefold increase in fluorescence was observed upon the addition of 10 μM protein to 10 μM nucleotide. Titration of the nucleotide at a fixed protein concentration produced a hyperbolic curve that gave a binding constant for TNP-GTP of about 5 μM (Fig. 4).
FIG. 4.
Fluorescence of TNP-GTP in the absence and presence of GTP. The protein was present at a concentration of 2.5 μM. The increase in the fluorescence intensity of TNP-GTP with respect to that in the protein-free solution is plotted over the TNP-GTP concentration. The fluorescence intensity was recorded at 25°C at a wavelength of 555 nm, and the excitation wavelength was 408 nm.
To demonstrate the binding of the labeled nucleotide in the NTP site, competition experiments were performed with GTP and TNP-GTP. The protein concentration was 2.5 μM, GTP was added first, and TNP-GTP was titrated to the protein-GTP complex. In the control experiment, no GTP was added to the protein (Fig. 4). GTP at a concentration of 2.5 μM did not prevent binding of TNP-GTP. However, the excess of GTP (20 μM) inhibited binding of TNP-GTP. In the similar experiments with a reversed order of nucleotide binding (TNP-GTP first, then GTP), no nucleotide exchange was detected, even with a 100-fold excess of GTP (1 μM TNP-GTP and up to 100 μM GTP). These experiments demonstrate that TNP-GTP and GTP compete for the same binding site, although the labeled nucleotide has a much higher affinity.
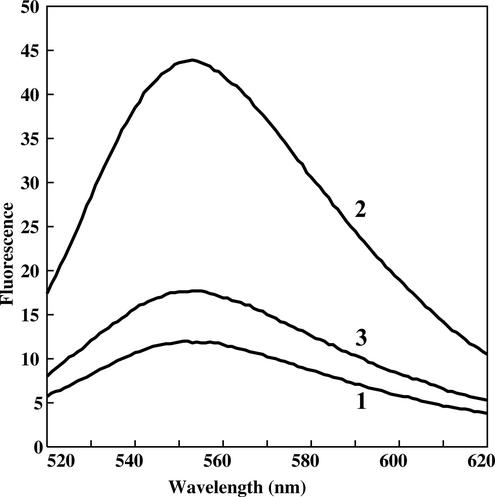
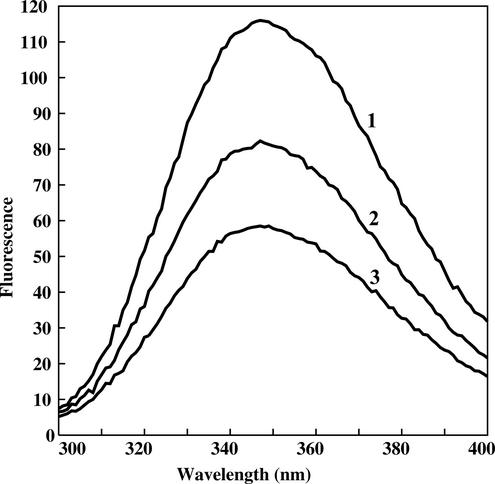
The effect of Mg2+ on the nucleotide binding was monitored in a separate experiment (Fig. 5). First, the complex with TNP-GTP was formed in the absence of Mg2+, which was visualized as an increase in fluorescence (Fig. 5, curve 2). Then Mg2+ was added to the reaction mixture, which caused an immediate quenching of fluorescence (Fig. 5, curve 3). We attribute the effect of the inhibition of nucleotide binding to the excess of Mg2+ rather than to the hydrolysis and release of the product by YchF. Inhibition of GTP binding at millimolar Mg2+ concentrations is typical for most of the G proteins (14, 22, 25, 36), though not for all of them. For instance, the Caulobacter crescentus CgtA protein, a member of the Obg family, shows a 60-fold increase in affinity to GTP (1.2 versus 65 μM) in the range of 5 to 15 mM Mg2+ and a decrease at higher concentrations of Mg2+ (20).
FIG. 5.
Fluorescence spectra of TNP-GTP in the absence and presence of YchF and Mg2+. Line 1, 2.5 μM TNP-GTP; line 2, 2.5 μM TNP-GTP plus 2.5 μM YchF; line 3, 2.5 μM TNP-GTP plus 2.5 μM YchF plus 2 mM Mg2+. The emission spectrum was recorded at 25°C, and the excitation wavelength was 408 nm.
Nucleic acid binding.
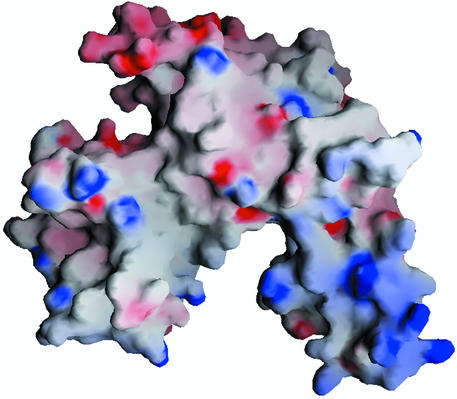
The crablike three-domain structure of YchF, with the A and C domains forming claws, suggests the binding site for a double-stranded nucleic acid in the cleft between the domains. The idea is supported by the clustering of positively charged residues at the inner surfaces of domains A and C (Fig. 6). Helix α7 of domain A and helix α12 of domain C, which face each other, are likely to be key elements of nucleic acid binding. The distance between them as measured between Cα atoms of Gln189 and Lys303 is 20.4 Å, which is about the size of the nucleic acid duplex.
FIG. 6.
Electrostatic surface potential calculated with GRASP (28). Positive charges are blue, and negative charges are red. The orientation of the molecule is as shown in Fig. 1.
The ability of YchF to bind nucleic acids was estimated by measuring changes in the fluorescence of tryptophan upon addition of dsDNA. There are two tryptophan residues in YchF, and one of them, Trp293, is located at the putative nucleic acid-binding site. Its environment is expected to change upon binding nucleic acid, which would result in the change in fluorescence. The experiment was performed with a digested plasmid DNA as a substrate. Upon addition of dsDNA to the protein sample, fluorescence decreased by about 50% (Fig. 7), indicating changes in the tryptophan environment.
FIG. 7.
Quenching of fluorescence of tryptophan upon addition of dsDNA to the YchF protein. The protein concentration was 100 nM. Line 1, no dsDNA; line 2, 50 nM (per base pair) dsDNA; line 3, 100 nM (per base pair) dsDNA.
DISCUSSION
Although the overall architecture of YchF has no analogues in the Protein Data Bank, each of the domains has topological matches in the protein universe. A helical coiled coil resembles that observed in the transcript cleavage factor GreA (38) and in seryl-tRNA synthetase (SerRS) (3). In both proteins, the coiled-coil domain is involved in binding RNA substrates. GreA contacts the 3′ end of the transcript attached to RNA polymerase with the tip of the coiled-coil finger. The helical arm of SerRS is in contact with the TΨC and D loops and the variable stem of tRNASer.
Domain C has a topology typical for the ubiquitinlike superfamily of α+β proteins (SCOP) (27). Besides ubiquitin-related proteins, the family includes Ras-binding domains of different effectors and, most notably, the N-terminal domain of threonyl-tRNA synthetase (ThrRS) (32). The similarity of the latter to YchF is striking: the root mean square (RMS) deviation of 62 common Cα atoms is only 2.0 Å. The domain is not involved in the binding of the cognate tRNAThr, and its function remains unknown. At the sequence level, the N-terminal domain of ThrRS represents the conservative TGS domain (PF02824 in the Pfam database) (1), which was also found in guanosine polyphosphatase SpoT and some GTPases (42).
Thus, both domains A and C appear to be related to RNA-binding proteins, and their presence in YchF suggests a similar feature for this protein. The ability of YchF to bind dsDNA further supports the contention that some nucleic acid may be a natural substrate for YchF. The putative nucleic acid-binding surface of YchF may not be limited to helices α7 and α12, which are positioned to embrace the nucleic acid duplex. The extended binding surface of domain C may include His309 and Asp311 adjacent to helix α7 and a cluster of polar residues Arg318, Glu320, Lys337, Glu338, and Arg360 on the outer surface of the domain. Most of these residues are conserved. All together, they form a continuous patch on the domain surface, suggesting a possibility that nucleic acid winds around the C domain. Given the small size of the domain, it would require a sharp bending of the nucleic acid, which is unlikely for linear dsDNA but possible for the loopy structure of RNA.
Domain G has all the features of the GTP-dependent molecular switches (4, 34). These proteins exist in two major conformations, and the switch between them depends on the GTP-GDP exchange. Two regions adjacent to the nucleotide site transmit the signal between the active center and periphery of the protein. The switch I region includes strand β2 and the preceding loop. A threonine residue (Thr36 in YchF) at the tip of the loop binds the γ-phosphate of GTP and Mg2+ and moves away from the active site upon GTP hydrolysis (the switch I region is completely disordered in the present nucleotide-free structure). A glycine residue (Gly75 in YchF) of the Walker B motif is a key element of switch II. Located at the end of the β-strand, it binds the γ-phosphate and promotes a conformational rearrangement of the following loop upon GTP hydrolysis. Glycine is conserved in this position to maintain the restrained conformation of the chain (φ/ψ = 134/160° in YchF). The following glycine-rich loop forms a short α-helix (α3). The high level of sequence conservation of both switch regions in the YchF family (Fig. 2) indicates their functional importance, which is likely related to the regulatory function typical for G proteins.
Ras-like G proteins are characterized by a relatively low intrinsic rate of GTP hydrolysis (37). Structural and mutagenesis studies on the H-ras p21 protein have suggested a key role of Gln61 in activating a water molecule for a nucleophilic attack on the γ-phosphate of GTP (31). It was also noted that the Q61E mutant of H-ras p21 had a 20-fold-greater rate of GTP hydrolysis than the wild-type protein, whereas the substitution of Gln61 by other amino acids reduced the GTPase rate (11, 12). Gln61 immediately follows the Walker B motif in p21. While Gln61 is conserved in the ras-like G proteins, this position is occupied by a histidine residue in elongation factors (which is in accord with their lower GTPase activities) and by hydrophobic residues in the other members of the TRAFAC class (19), suggesting that they lack an efficient catalytic machinery for GTP hydrolysis. With the highly conserved leucine residue (Leu76) in this position, the YchF structure does not indicate any alternative residue that may act as a general base in catalysis. It seems likely that YchF and other members of the family are not GTPases but rather sensors of the relative levels of GTP and GDP.
The putative nucleic acid-binding domains A and C are in direct contact with the switch regions of domain G, so that the conformational changes upon GTP-GDP transition may be transmitted to the nucleic acid site and regulate the nucleic acid-binding ability of the protein. The 23-residue loop protruding from domain G (residues 45 to 67) is part of domain C and is also closely associated with the switch I region. The coiled coil of domain A is in contact with the glycine-rich helix α3, which is part of switch II. The long helical structure of domain A implies a lever-like mechanism of conformational changes associated with nucleic acid binding. The involvement of the coiled coil in the binding a nucleic acid is expected due to a number of conserved basic residues at the tip of the α-hairpin.
Based on the analysis of the sequences, YchF has been assigned to the Obg superfamily, which includes four other families of GTPases (19). Obg is an essential gene that is involved in DNA replication in C. crescentus (21) and Streptomyces griseus (29) and is associated with the ribosome (35). Several members of the superfamily, including YchF, possess the TGS domain related to the RNA-binding proteins (42). These observations, taken together with the universal phyletic distribution of the proteins, allowed Leipe and coauthors to suggest that these proteins may be uncharacterized translation factors (19).
Another indication that YchF is involved in translation comes from the genome-scale project aimed at the characterization of multiprotein complexes in Saccharomyces cerevisiae (13). By using tandem-affinity purification (TAP), it was shown that the yeast homolog of YchF, YBR025c, interacts with the components of the multisubunit translation elongation factor eEF1.
Furthermore, a relation of YchF to ribosomal proteins can be deduced from the analysis of the gene expression in E. coli under conditions of genotoxic stress. YchF appears to be among the genes downregulated in response to the DNA damage (17). Changes in gene expression after treatment of E. coli culture with mitomycin C were assessed with DNA array technology. The expression profile of ychF in these experiments correlates with the genes related to protein biosynthesis, including those coding for ribosomal proteins, which are also downregulated.
Finally, in many bacterial genomes including both gram-negative and gram-positive species, ychF is located next to the pth gene, which codes for peptidyl-tRNA hydrolase. This protein ensures the recycling of peptidyl-tRNAs produced through abortion of translation. The phylogenetic pattern of pth is similar to that of ychF, suggesting that the proteins may be functionally related. Northern blot experiments showed that both pth and ychF are cotranscribed in a bicistronic transcript (9).
The exceptionally high level of amino acid conservation typical for ribosomal proteins suggests a key role for YchF in the cell. Most of the conserved residues are implicated in binding GTP and nucleic acid as discussed above. On the other hand, the amino acid conservation in the YchF family extends far beyond the GTP-binding motifs and is not limited to the putative nucleic acid-binding site. Of particular interest is the cluster of acidic residues in the fragment containing residues 241 to 258. These residues form a negatively charged surface centered on helix α10 in the G domain. Although most of them are not strictly conserved, the overall negative charge of the surface is preserved. The surface may be involved in interactions with other components of the putative ribonucleoprotein complex. One possibility is that helix α10 mimics a nucleic acid duplex and thus competes for a binding site on an effector protein. An example of such a molecular mimicry is a C-terminal α-helix in DinI, a negative regulator of the SOS response in E. coli (40). By competing for the DNA-binding site of RecA, DinI interferes with the activation of RecA.
The crystal structure of the YchF homolog from yeast has been recently solved by Lima and coworkers (Protein Data Bank entry 1NI3). The two structures are very similar and can be superimposed with the RMS deviation of 2.3 Å over 306 common Cα atoms. The similarity is particularly strong in the TGS (C-terminal) domains, the RMS deviation for which is only 0.8 Å. The largest deviation between the structures, up to 15 Å for the equivalent Cα atoms, is in the switch II region. This glycine-rich fragment forms a helix (α3) in the E. coli protein, whereas in the yeast protein it lacks any secondary structure. Although the difference may be partly due to crystal contacts that involve this fragment in the yeast structure, it indicates the conformational flexibility of switch II, which is well documented for other G proteins. In spite of different crystallographic environments, both structures reveal the switch I region and the linker between domains G and A as the most mobile parts of the protein molecule, as can be judged from their high atomic B factors. Apparently, the flexibility of these functionally important parts of the molecule is an intrinsic feature of the protein rather than a crystallographic artifact.
The crystal structure of YchF is the first three-dimensional structure obtained for a protein family widely represented in all forms of life. Analysis of the structure and available experimental data support the contention that YchF may function as a GTP-dependent nucleic acid-binding protein. Its involvement in translation is a distinct possibility. These results provide a basis for further studies of this undoubtedly important protein family.
Acknowledgments
The work was supported by the National Institutes of Health grant P01-GM57890. Use of the Advanced Photon Source was supported by the Office of Science, Basic Energy Sciences, U.S. Department of Energy, under contract no. W-31-109-Eng-38.
Certain commercial materials, instruments, and equipment are identified in this manuscript in order to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials, instruments, or equipment identified is necessarily the best available for the purpose.
REFERENCES
- 1.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biou, V., A. Yaremchuk, M. Tukalo, and S. Cusack. 1994. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263:1404-1410. [DOI] [PubMed] [Google Scholar]
- 4.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125-132. [DOI] [PubMed] [Google Scholar]
- 5.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 6.Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 10:1581-1592. [DOI] [PubMed] [Google Scholar]
- 7.Caldon, C. E., P. Yoong, and P. E. March. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41:289-297. [DOI] [PubMed] [Google Scholar]
- 8.Collaborative Computational Project, number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Vera, L. R., J. M. Galindo, and G. Guarneros. 2002. Transcriptional analysis of the gene encoding peptidyl-tRNA hydrolase in Escherichia coli. Microbiology 148:3457-3466. [DOI] [PubMed] [Google Scholar]
- 10.De La Fortelle, E., and G. Bricogne. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276:472-494. [DOI] [PubMed] [Google Scholar]
- 11.Der, C. J., T. Finkel, and G. M. Cooper. 1986. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 44:167-176. [DOI] [PubMed] [Google Scholar]
- 12.Frech, M., T. A. Darden, L. G. Pedersen, C. K. Foley, P. S. Charifson, M. W. Anderson, and A. Wittinghofer. 1994. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry 33:3237-3244. [DOI] [PubMed] [Google Scholar]
- 13.Gavin, A. C., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 14.Hall, A., and A. J. Self. 1986. The effect of Mg2+ on the guanine nucleotide exchange rate of p21N-ras. J. Biol. Chem. 261:10963-10965. [PubMed] [Google Scholar]
- 15.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 17.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 19.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 20.Lin, B., K. L. Covalle, and J. R. Maddock. 1999. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J. Bacteriol. 181:5825-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maddock, J., Bhatt, A., Koch, M., and Skidmore, J. 1997. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J. Bacteriol. 179:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menard, L., E. Tomhave, P. J. Casey, R. J. Uhing, R. Snyderman, and J. R. Didsbury. 1992. Rac1, a low-molecular-mass GTP-binding-protein with high intrinsic GTPase activity and distinct biochemical properties. Eur. J. Biochem. 206:537-546. [DOI] [PubMed] [Google Scholar]
- 23.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 24.Mittenhuber, G. 2001. Comparative genomics of prokaryotic GTP-binding proteins (the Era, Obg, EngA, ThdF (TrmE), YchF and YihA families) and their relationship to eukaryotic GTP-binding proteins (the DRG, ARF, RAB, RAN, RAS and RHO families). J. Mol. Microbiol. Biotechnol. 3:21-35. [PubMed] [Google Scholar]
- 25.Molina y Vedia, L., C. A. Ohmstede, and E. G. Lapetina. 1990. Properties of the exchange rate of guanine nucleotides to the novel rap-2B protein. Biochem. Biophys. Res. Commun. 171:319-324. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 27.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, S., M. Itoh, and K. Ochi. 1997. Molecular cloning and characterization of the obg gene of Streptomyces griseus in relation to the onset of morphological differentiation. J. Bacteriol. 179:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 31.Pai, E. F., U. Krengel, G. A. Petsko, R. S. Goody, W. Kabsch, and A. Wittinghofer. 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sankaranarayanan, R., A. C. Dock-Bregeon, P. Romby, J. Caillet, M. Springer, B. Rees, C. Ehresmann, B. Ehresmann, and D. Moras. 1999. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 97:371-381. [DOI] [PubMed] [Google Scholar]
- 33.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci 15:430-434. [DOI] [PubMed] [Google Scholar]
- 34.Schweins, T., and A. Wittinghofer. 1994. GTP-binding proteins. Structures, interactions and relationships. Curr. Biol. 4:547-550. [DOI] [PubMed] [Google Scholar]
- 35.Scott, J. M., J. Ju, T. Mitchell, and W. G. Haldenwang. 2000. The Bacillus subtilis GTP binding protein obg and regulators of the σB stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 182:2771-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo, H. S., C. H. Choi, H. Y. Kim, J. Y. Jeong, S. Y. Lee, M. J. Cho, and J. D. Bahk. 1997. Guanine-nucleotide binding and hydrolyzing kinetics of ORrab2, a rice small GTP-binding protein expressed in Escherichia coli. Eur. J. Biochem. 249:293-300. [DOI] [PubMed] [Google Scholar]
- 37.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66:639-678. [DOI] [PubMed] [Google Scholar]
- 38.Stebbins, C. E., S. Borukhov, M. Orlova, A. Polyakov, A. Goldfarb, and S. A. Darst. 1995. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature 373:636-640. [DOI] [PubMed] [Google Scholar]
- 39.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voloshin, O. N., B. E. Ramirez, A. Bax, and R. D. Camerini-Otero. 2001. A model for the abrogation of the SOS response by an SOS protein: a negatively charged helix in DinI mimics DNA in its interaction with RecA. Genes Dev. 15:415-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf, Y. I., L. Aravind, N. V. Grishin, and E. V. Koonin. 1999. Evolution of aminoacyl-tRNA synthetases-analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9:689-710. [PubMed] [Google Scholar]