Abstract
The biotin-binding tetrameric proteins, streptavidin from Streptomyces avidinii and chicken egg white avidin, are excellent models for the study of subunit-subunit interactions of a multimeric protein. Efforts are thus being made to prepare mutated forms of streptavidin and avidin, which would form monomers or dimers, in order to examine their effect on quaternary structure and assembly. In the present communication, we compared the crystal structures of binding site W→K mutations in streptavidin and avidin. In solution, both mutant proteins are known to form dimers, but upon crystallization, both formed tetramers with the same parameters as the native proteins. All of the intersubunit bonds were conserved, except for the hydrophobic interaction between biotin and the tryptophan that was replaced by lysine. In the crystal structure, the binding site of the mutated apo-avidin contains 3 molecules of structured water instead of the 5 contained in the native protein. The lysine side chain extends in a direction opposite that of the native tryptophan, the void being partially filled by an adjacent lysine residue. Nevertheless, the binding-site conformation observed for the mutant tetramer is an artificial consequence of crystal packing that would not be maintained in the solution-phase dimer. It appears that the dimer-tetramer transition may be concentration dependent, and the interaction among subunits obeys the law of mass action.
Streptavidin from Streptomyces avidinii and avidin from chicken egg white are remarkably similar biotin-binding proteins that exhibit a nearly identical tertiary fold and tetrameric structure (21, 26, 31). Although the primary sequences are only moderately conserved, the respective binding site residues of both proteins acquire analogous positions and interact with biotin in the same manner (21).
Surprisingly, the fibropellins from the sea urchin harbor a C-terminal domain that is similar in sequence to streptavidin and avidin (3, 14). Seven of the 10 biotin-binding site residues, conserved in both streptavidin and avidin, are also conserved in the fibropellins. The most significant of the three remaining modifications are the replacement of Trp120 in streptavidin (Trp110 in avidin) by a lysine and of Trp79 (Trp70 in avidin) by arginine. In order to evaluate the consequences of such modifications to the biotin-binding site, we recently expressed the relevant mutations of avidin and streptavidin in a baculovirus-infected insect cell system. It was found that the W120K in streptavidin (W110K mutation in avidin) served to convert the native tetramer to a dimer in solution with a reduced affinity to biotin, thereby rendering the binding reversible (20). On the other hand, the W70R mutation in avidin severely impaired the interaction with biotin, and the double mutation inactivated binding completely (20).
The critical tryptophans of streptavidin and avidin (Trp120 and Trp110, respectively) are donated to the biotin-binding site from a neighboring monomer. Thus, the Trp residue contributes to biotin binding by being an important component of the hydrophobic cage, which accommodates biotin. Alterations of Trp120 to Phe or even Ala resulted in a decrease in affinity towards biotin, yet the tetrameric structure remained intact (5, 9, 30). In addition, the same Trp contributes to the stability of the 1-2 monomer-monomer interaction and thus to the quaternary assembly (29).
In order to understand why the single nonconventional W→K mutation leads to such a dramatic change in the properties of avidin and streptavidin, we decided to crystallize the mutated proteins and to study their structures. It was found that proteins in the crystalline state behave differently than those in solution. Upon crystallization from concentrated preparations of the corresponding mutant, the tetrameric form is recovered and new intermonomer interactions are formed—notably, a hydrophobic interaction between the lysine aliphatic segment and biotin. This interaction compensates for the hydrophobic interaction between biotin and the respective tryptophan in the native protein (W120 in streptavidin, W110 in avidin).
The present study demonstrates that the dimer-tetramer equilibrium is concentration and structure dependent and that the law of mass action also applies to the interaction among subunits in multimeric proteins.
MATERIALS AND METHODS
Crystallization and data collection: Savm-W120K.
Crystals of the streptavidin W120K mutant (Savm-W120K) were obtained by the vapor diffusion hanging drop method, with a Hampton Research crystal screen I (condition no. 28) (16, 28), at 20°C. A 3-μl drop contained 1.5 μl of the protein (10 mg/ml) and 1.5 μl of the reservoir solution. The 1-ml reservoir solution contained 15% polyethylene glycol (PEG) 8000, 0.2 M sodium acetate, and 0.1 M sodium cacodylate (pH 6.45). Crystallization conditions were further refined by various crystallization parameters. Improved crystals were obtained at 20°C, with reservoir solutions consisting of either 20% PEG 8000, 0.2 M sodium acetate, and 0.1 M cacodylate buffer (pH 7.05) or 25% PEG 6000, 0.3 M sodium acetate, and 0.1 M cacodylate buffer (pH 6.45). The crystals belonged to the monoclinic space group P21, with four Savm-W120K monomers in the asymmetric unit, with cell dimensions of a = 50.43 Å, b = 100.41 Å, c = 52.51 Å, and β = 112.12°. Diffraction data were collected from a single crystal at 100 K on an MAR 165-mm-diameter charge-coupled device detector, by using synchrotron radiation at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, at beam line ID14-1 (λ = 0.93 Å). Data were integrated and scaled by using the HKL suite (24).
Crystallization and data collection: Avm-W110K.
The avidin W120K mutant (Avm-W110K) crystals were obtained by the vapor diffusion hanging drop method at 20°C, with the Hampton Research crystal screen I (16). A 3-μl drop contained 1.5 μl of the protein (4 mg/ml) and 1.5 μl of the reservoir solution. The initial diamond-shaped crystals grew after 24 h, and the 1-ml reservoir solution contained 2.5 M sodium formate and 0.1 M acetate buffer (pH 4.6). The diamond-shaped crystals diffracted poorly to a resolution of 7.5 Å and could not be indexed. However, rectangular crystals were obtained from a reservoir solution containing 32% polyethylene glycol monoethylether (MPEG) 2000 and 0.1 M Tris-HCl (pH 8.5). Crystallographic data were collected at 100 K with an Oxford Cryosystem Cryostream cooling device from a single crystal on an MAR 165-mm-diameter charge-coupled device detector at beam line 14-1 at ESRF (λ = 0.93 Å). The crystal belonged to the orthorhombic P21212 space group with cell parameters of a = 67.70 Å, b = 77.45 Å, and c = 42.89 Å and two Avm-W110K monomers in the asymmetric unit. Data were integrated, reduced, and scaled by using the HKL suite (24).
RESULTS
Structure determination of Savm-W120K and Avm-W110K.
The structure of Savm-W120K was solved via molecular replacement methods, by using AMoRe (23) implemented in the CCP4 suite (6), with both monomeric and dimeric streptavidin (Protein Data Bank [PDB] code 2IZF) as search models (19). The solution for the tetrameric model in AMoRe had an R value of 33.7% and a correlation coefficient of 70.2% at a resolution range of 10.0 to 4.5 Å. The structure was initially refined by using the rigid body protocol in CNS (4) at a resolution range of 30.0 to 4.4 Å with a final R value of 31.0% (Rfree = 29.1%). During the preparation and purification of Savm-W120K, biotin was used to release the molecule from the iminobiotin column (12). The initial electron density maps (Fo-Fc and 3Fo-2Fc) calculated after rigid body refinement thus indicate the presence of biotin molecules in the four binding sites (Fig. 1A). The structure was further refined by simulated annealing with the slow cooling protocol in CNS (4) with data from 50.0 to 1.7 Å (Table 1). The Fobs values were scaled anisotropically (B11 = 3.17 Å2, B12 = 0.0 Å2, B22 = 0.43 Å2, B13 = 1.91 Å2, B23 = 0.0 Å2, and B33 = −3.60 Å2), and a bulk solvent correction was applied (17). The structure was built into electron density maps by using the graphics program O (18). The model of the Savm-W120K consists of residues 16 to 133 for monomer 1, residues 16 to 134 for monomer 2, residues 16 to 134 for monomer 3, and residues 16 to 135 for monomer 4, with 4 biotin molecules and 123 solvent molecules. The model was refined to a resolution range of 50 to 1.7 Å with a crystallographic R value of 21.2% and an Rfree value of 24.0% (Table 1).
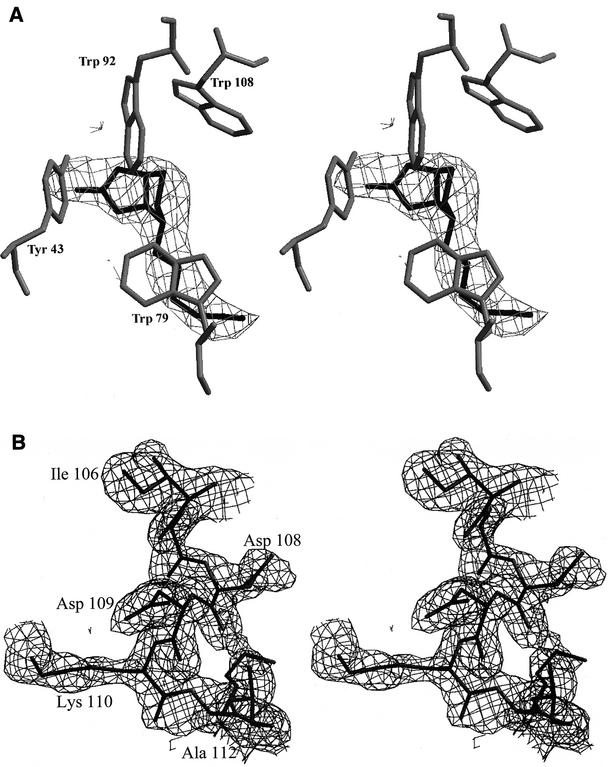
FIG. 1.
Electron density maps. (A) Stereo view of an Fobs-Fcalc difference map, calculated at 40.0 to 3.0 Å resolution after the initial stage of rigid body refinement and constructed at 2.0 σ with the superimposed coordinates of the final Savm-W120K model. Biotin was not included during this stage of refinement, and the map clearly indicates the position of the molecule in the binding site. (B) Stereo view of a 3Fobs-2Fcalc map of the L7,8 loop region after the initial cycle of refinement, calculated at 20 to 1.8 Å resolution and constructed at 1.0 σ with the superimposed coordinates of the final Avm-W110K model. During the first cycle of refinement, the residue at position 110 was altered to alanine and the electron density map clearly indicates the position of the extended side chain of lysine. The figures were constructed with Bobscript (8).
TABLE 1.
Data collection and refinement statistics
| Parameter (unit) | Result for:
|
|
|---|---|---|
| Savm-W120K | Avm-W110K | |
| Space group | P21 | P21212 |
| Resolution range (Å) | 40.0-1.7 | 20.0-1.8 |
| No. of unique reflections | 51,529 | 21,282 |
| Redundancy | 2 | 2.5 |
| Rsym(I)a | 4.1 (29.0)b | 5.6 (26.1)b |
| % Completeness | 97.3 (98.6)b | 98.2 (98)b |
| I/σ | 21.6 (3.9)b | 11.1 (2.9)b |
| No. of protein atoms | 3,534 | 1,800 |
| No. of ligand atoms | 64 | None |
| No. of solvent atoms | 123 | 94 |
| R factor (F > 1σ) | 21.2 | 20.5 |
| Rfreec | 24.0 | 22.7 |
| Average B factor (Å2) | ||
| Protein | 28.1 | 28.8 |
| Ligand | 21.9 | 31.8d |
| Solvent | 36.13 | 35.5 |
| RMSD from ideality | ||
| Bond length (Å) | 0.010 | 0.013 |
| Bond angle (°) | 1.6 | 1.7 |
| Ramachandran plot (PROCHECK) (%) | ||
| Favored | 89.90 | 93.30 |
| Allowed | 9.20 | 6.70 |
| Generously allowed | 0.90 | 0.00 |
| Disallowed | 0.00 | 0.00 |
Rsym (I) = Σ|I − 〈I〉|/ΣI.
The outer shell resolution range for Savm-W120K is 1.76 to 1.7 Å, Avm-W110K is 1.86 to 1.8 Å.
The test set is 5% of data at both complexes.
Structured water molecules in the biotin-binding site.
Despite the fact that the crystals of Avm-W110K belonged to the same space group with similar cell parameters to those of avidin in its glycosylated and deglycosylated forms (22, 25), we decided to solve the structure via molecular replacement techniques in order to examine the packing of the Avm-W110K molecule. The search model for molecular replacement was a monomer of avidin (the 2AVI model), and the translation search solution at the resolution range of 10.0 to 4.0 Å consisted of two Avm-W110K monomers with an R value of 37.7% and a correlation coefficient of 55.5%. For the molecular replacement process, position 110 was altered to alanine and all biotin, carbohydrate, and solvent molecules were removed. The structure was initially refined using rigid body protocols in CNS (4) at a resolution range of 20.0 to 4.0 Å, resulting in an R value of 36.7% (Rfree = 37.7%). The model was further refined by using simulated annealing protocols in CNS (4). The initial electron density maps after simulated annealing refinement clearly indicated the conformation of the Lys-110 residue in the structure of Avm-W110K (Fig. 1B). The Fobs values were scaled anisotropically (B11 = −1.01 Å2, B12 = 0.0 Å2, B22 = 0.57 Å2, B13 = 0.0 Å2, B23 = 0.0 Å2, and B33 = 0.44 Å2), and a bulk solvent correction was applied (17). The structure was built into electron density maps by using the graphics program O (18). The structure of the Avr-W110K consists of residues 2 to 37 and 42 to 123 for monomer 1, and residues 3 to 35, 45 to 86, and 88 to 123 for monomer 2. The structure contains 94 solvent molecules and was refined to a resolution of 1.8 Å with a crystallographic R value of 20.5% and an Rfree value of 22.7% (Table 1). The coordinates of Savm-W120K (1NQM) and Avm-W110K (1NQN) are available at the RCSB PDB (2).
The first remarkable observation from the crystal structures is that both Savm-W120K and Avm-W110K mutants retained their inherent tetrameric organization in the crystalline state, although they exist as dimers in solution (20). The overall folds of both the Savm-W120K and the Avm-W110K monomers were similar to those of the wild-type streptavidin and avidin, as described previously (21, 26, 31). Each monomer consists of 8 antiparallel β-strands, which form a classical β-barrel, wide at one end and narrow at the other, where the biotin-binding site is located at the wide end of the barrel.
Comparison of native and mutant streptavidin structures.
The structure of Savm-W120K contains biotin in each of its four binding sites. In the mutant, the previously described flexible L3,4 loop (10) was found to be in the closed and ordered conformation, forming two hydrogen bonds with the biotin ligand. One of the H bonds exists between Ser45 Oγ and the biotin uredio nitrogen, and the second exists between Asn49 N and one of the biotin carboxylate oxygens. More than 100 streptavidin structures have been reported that crystallize in space groups and cell parameter systems, containing one, two, or four monomers in the asymmetric unit. We therefore selected an appropriate standard for comparison of the mutant structure. In this context, the tetrameric streptavidin structures were categorized into several groups reflecting the different crystallographic symmetries. Comparison of the tertiary and quaternary structures of Savm-W120K was thus conducted by using a reference group of structures that crystallized in the same space group (P21) and nearly identical cell parameters as those observed for the mutant.
There are currently 3 reported structures in the PDB that are isomorphous and thus maintain full identity in space group and cell parameters with that of mutant Savm-W120K. One is the biotin-complexed streptavidin at pH 4.5 (PDB code 1SWE) (10), and the other two are the biotin complexes of the W120A and W120F mutations of streptavidin (PDB codes 1SWR and 1SWP, respectively) (9). The corresponding biotin-free streptavidin structures belong to the same P21 space group, although the cell parameters differ. By superimposing Savm-W120K monomer 1 with the corresponding wild type (116 Cα pairs) and the W120A (115 Cα pairs) and W120F (117 Cα pairs) structures, relatively low root mean square deviation (RMSD) values were obtained (0.40, 0.36, and 0.39 Å, respectively). The quaternary arrangements among the latter streptavidin structures were examined by superimposing the individual monomers and by assessing the subsequent fit of the resultant dimers and tetramers. The observed RMSD values were comparatively low (0.7 to 1.15 Å or 234 to 236 Cα pairs for Savm-W120K relative to the wild type and W120F and W120A mutants). The major deviations in the streptavidin structures are observed in the L7,8 loop, which contains the designated mutation (Fig. 2A). Lys120 in Savm-W120K results in a conformational change in the L7,8 loop, where the W120F and W120A mutations in the same site essentially maintain similar conformations as for the wild-type streptavidin. Thus, with reference to Savm-W120K, the RMSD values for the L7,8 loop (residues 110 to 124) of the designated structures are 0.42, 0.38, and 0.45 Å for the wild type, W120F, and W120A, respectively, whereas the corresponding Cα distances for residue 120 are 1.39, 1.11, and 1.25 Å. The Lys120 side chain in Savm-W120K adopts a conformation that, in the crystal, results in the interaction of the aliphatic segment of its side chain with the valeric acid moiety of biotin (Fig. 3A).
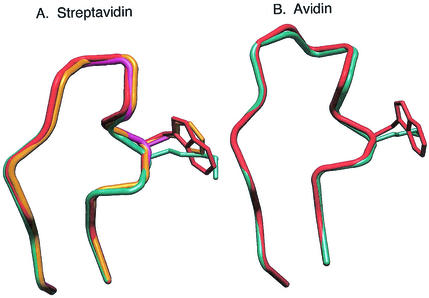
FIG. 2.
Superposition of the L7,8 loop in streptavidin and avidin. (A) For streptavidin, four biotin-complexed structures were compared: wild type (red), W120F (yellow), W110A (magenta) and Savm-W120K (cyan). The comparison clearly indicates that the L7,8 loops in the W120F and W120A mutants have conformations similar to that of wild-type streptavidin. However, the L7,8 loop in Savm-W120K shows a different conformation in positions 120 and 121 compared to the other structures. (B) In avidin, the L7,8 loop is one amino acid residue larger than that of streptavidin, and the comparison was conducted between apo-avidin (red) and Avm-W110K (cyan). The major conformational change in the avidin L7,8 loop occurs in residues 105 to 106, and the conformation in position 110 is relatively conserved.
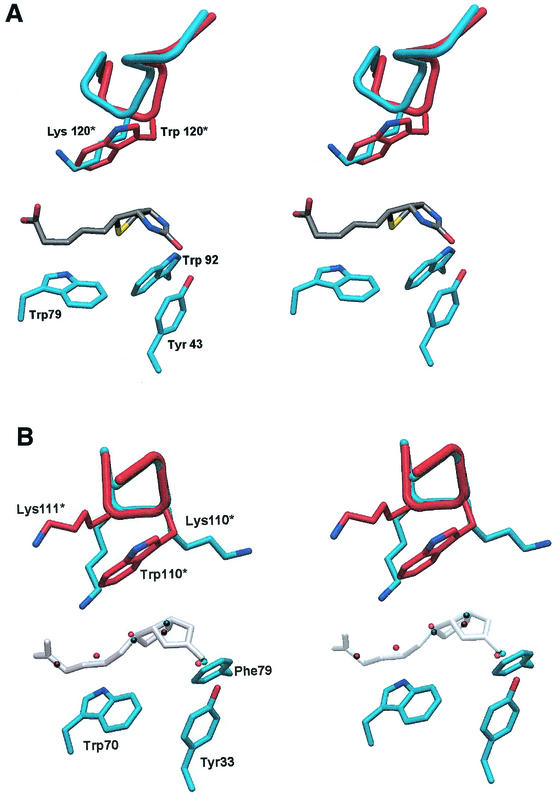
FIG. 3.
(A) Stereo view of a region of the biotin-binding site of Savm-W120K. Biotin is shown in dark gray, and side chains of Tyr43, Trp79, and Trp92 are shown in cyan. A segment of the L7,8 loop from wild-type streptavidin from an adjacent monomer is superimposed (shown in red). The figure clearly indicates that, although the main chain in this region exhibits minor changes, the conformation of Lys120 is consistent with that of Trp120 from wild-type streptavidin. The aliphatic component of Lys120 thus maintains the hydrophobic nature that substitutes for the aromatic ring system of Trp120. (B) Stereo view showing the superposition of Avm-W110K (cyan) and apo-avidin (red) from the aspect of the biotin-binding site shown in panel A. Biotin from the avidin-biotin complex is shown in shadowed white as a reference for the structured water molecules in the binding site. The positions of the five water molecules (red) in the apo-avidin binding site collectively anticipate that of the biotin molecule. Only three water molecules (cyan) are observed in Avm-W110K, and they assume positions consistent with those in apo-avidin that anticipate the bicyclic ring system of biotin. The L7,8 loop maintains the main chain conformation in this region; however, the side chain of Lys110 is positioned in a direction opposite that of Trp110. Moreover, the side chain of the neighboring residue, Lys111, partially accommodates the void in the biotin-binding site caused by the lack of the aromatic moiety.
Comparison of native and mutant avidin structures.
The monomeric model of the apo Avm-W110K maintained features similar to those of the native apo-avidin structures. The L3,4 loop (the loop connecting strands β3 to β4) was partially disordered, thus exposing the biotin-binding site to solvent molecules. The biotin-binding site of Avm-W110K contained three ordered water molecules, which mimic the position of the bicyclic ring system of biotin (Fig. 3B). As described previously (21), there are five conserved solvent molecules in the biotin-binding site of native apo-avidin, and the positions of three of them are in agreement with the binding site water molecules observed in the Avm-W110K structure. The disorder in the L3,4 loop is consistent with previous observations of avidin complexes, where the L3,4 loop maintains a high propensity for disorder in the apo form and in complex with biotinylated molecules (13, 27). Superimposition of one of the Avm-W110K monomers with apo-avidin resulted in a relatively low RMSD (0.58 Å for 112 Cα pairs). Upon maintaining the latter superposition of the two monomers (i.e., Avm-W110K and apo-avidin), the quaternary structure remained similar as well and the RMSD between the second monomer in the asymmetric unit was 0.7 Å (for 111 Cα pairs). The superposition of the remaining two native avidin and Avm-W110K monomers (keeping monomer 1 superimposed) resulted in an RMSD of 0.76 Å (for 223 Cα pairs).
The W110K mutation is located in the L7,8 loop connecting strands β7 and β8. In Avm-W110K, the L7,8 loop remained in a conformation similar to that of the wild-type avidin (Fig. 2B). The main differences between the L7,8 loops of the native and mutant protein lies in residues Asn104 and Asp105, where the Cα distances between the two structures were 0.68 and 0.72 Å, respectively. The positions of the Cα of Trp110 of the wild type and of Lys110 of the mutant remained similar, with respective distance differentials of only 0.44 and 0.41 Å for both monomers. The side chain of Lys110 of the mutant, however, attains a conformation different from that of Trp110 of the wild-type protein, i.e., Lys110 is not directed into the biotin-binding site per se but juts out in the opposite direction (Fig. 3B). In one of the monomers, the Lys side chain (Nɛ) forms an H bond with the main chain Ile117(O) from the adjacent molecule. In addition, the adjacent residue, Lys111, undergoes a conformational change in the mutant relative to that of the wild-type protein. The void resulting from the lack of Trp110 is now partially filled by the side chain of Lys111 (Fig. 3B).
DISCUSSION
This study indicates that the structure of a protein in dilute aqueous solution is not always that seen in a crystal. Specifically, mutations of Trp 110 of avidin and streptavidin result in proteins that are dimers in solution and tetramers in the crystal. In this study, we show that upon crystallization of a protein, which has been demonstrated biochemically to be a dimer in solution, the protein associates into a tetramer.
An unconventional mutation (W→K) of the tetrameric biotin-binding proteins avidin and streptavidin has recently been described (20). This particular Trp of both native proteins is special in that it is inserted into the biotin-binding pocket of one monomer from the peripheral position of another. Hence, Trp-110 of avidin and Trp-120 of streptavidin play a critical role in both the binding of the biotin ligand and the stability of the tetrameric structure. It was previously demonstrated that this single W→K mutation caused both proteins to dimerize in solution, such that the dimers still bound biotin, albeit with reduced affinity (20). Surprisingly, both mutant proteins crystallized as tetramers with the same parameters as the native avidin and streptavidin.
The W→K modification serves to dramatically alter the character of the hydrophobic interaction contributed by the Trp residue to the biotin-binding pocket. As a rule, lysines are normally exposed to the environment, and it appears that the mutation of the hydrophobic residue to a charged residue generates sufficient disruptive force in solution to convert the tetrameric protein to a dimer, presumably due to the interaction of the Lys with the environmental water and salts.
Interestingly, the tetrameric avidin contains 5 molecules of structured water in the biotin-binding site, which sits in the apo protein in a structure similar to that of biotin. Upon binding biotin, the structured water vacates the site. In the mutated avidin, however, only 3 molecules of water were found in the crystal structure, indicating a different binding site architecture, resulting from replacement of the native hydrophobic Trp residue with a charged Lys. Indeed, the contribution to the binding site from the adjacent monomer, reflected in the two tandem lysines (residues 110 and 111), is completely different than that observed in the original protein. In fact, the conformation of the tandem lysines is dictated by the crystal packing contacts, since in solution this interaction would not exist for the mutated avidin dimer. In this context, the mutated lysine cannot emulate the critical role played by Trp110 in stabilizing the tetrameric structure in solution.
Despite the dramatic effects and consequences of the W→K modification, all the other interface residues and their potential interactions are retained in the mutant proteins. During the crystallization process, the latter interactions overcome the effects of the mutated residue and the protein crystallizes as a tetramer. Interestingly, concanavalin A is known to undergo pH- and temperature-dependent tetramer-dimer exchange in solution (1). Nevertheless, the molecule almost invariably crystallizes in the tetrameric form (11, 15). Likewise, dimer-tetramer equilibrium would exist in solution for the avidin and streptavidin W→K mutants. Concentration of the protein solution would favor tetramer formation, culminating in their crystallization. Consequently, the dimer-tetramer exchange appears to depend on protein concentration, and the process of crystallization can be viewed as a manifestation of macromolecular crowding (7). The law of mass action would thus apply to the interaction between subunits and, hence, to protein-protein interactions in general. From the present study, it follows that care must be taken in interpreting results determined at concentrations and in environments differing from those in the cell. Otherwise, anomalous interactions may be observed.
Acknowledgments
We thank the ESRF staff for assistance during data collection.
This research was supported in part by the Israeli Science Foundation.
Y.P. and Y.E.-D. contributed equally to the scientific research.
REFERENCES
- 1.Agrawal, B. B., and I. J. Goldstein. 1968. Protein-carbohydrate interaction. VII. Physical and chemical studies on concanavalin A, the hemagglutinin of the jack bean. Arch. Biochem. Biophys. 124:218-229. [DOI] [PubMed] [Google Scholar]
- 2.Berman, H. M., T. Battistuz, T. N. Bhat, W. F. Bluhm, P. E. Bourne, K. Burkhardt, Z. Feng, G. L. Gilliland, L. Iype, S. Jain, P. Fagan, J. Marvin, D. Padilla, V. Ravichandran, B. Schneider, N. Thanki, H. Weissig, J. D. Westbrook, and C. Zardecki. 2002. The protein data bank. Acta Crystallogr. D 58:899-907. [DOI] [PubMed] [Google Scholar]
- 3.Bisgrove, B. W., and R. A. Raff. 1993. The SpEGF III gene encodes a member of the fibropellins: EGF repeat-containing proteins that form the apical lamina of the sea urchin embryo. Dev. Biol. 157:526-538. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., et al. 1998. Crystallographic and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Chilkoti, A., T. Boland, B. D. Ratner, and P. S. Stayton. 1995. The relationship between ligand-binding thermodynamics and protein-ligand interaction forces measured by atomic force microscopy. Biophys. J. 69:2125-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodson, E. J., M. Winn, and A. Ralph. 1997. Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 277:620-633. [DOI] [PubMed] [Google Scholar]
- 7.Ellis, R. J. 2001. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26:597-604. [DOI] [PubMed] [Google Scholar]
- 8.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 9.Freitag, S., I. Le Trong, A. Chilkoti, L. A. Klumb, P. S. Stayton, and R. E. Stenkamp. 1998. Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex. J. Mol. Biol. 279:211-221. [DOI] [PubMed] [Google Scholar]
- 10.Freitag, S., I. Le Trong, L. Klumb, P. S. Stayton, and R. E. Stenkamp. 1997. Structural studies of the streptavidin binding loop. Protein Sci. 6:1157-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamodrakas, S. J., P. N. Kanellopoulos, K. Pavlou, and P. A. Tucker. 1997. The crystal structure of the complex of concanavalin A with 4′-methylumbelliferyl-alpha-D-glucopyranoside. J. Struct. Biol. 118:23-30. [DOI] [PubMed] [Google Scholar]
- 12.Heney, G., and G. A. Orr. 1981. The purification of avidin and its derivatives on 2-iminobiotin-6-aminohexyl-Sepharose 4B. Anal. Biochem. 114:92-96. [DOI] [PubMed] [Google Scholar]
- 13.Huberman, T., Y. Eisenberg-Domovich, G. Gitlin, T. Kulik, E. A. Bayer, M. Wilchek, and O. Livnah. 2001. Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences. J. Biol. Chem. 276:32031-32039. [DOI] [PubMed] [Google Scholar]
- 14.Hunt, L. T., and W. C. Barker. 1989. Avidin-like domain in an epidermal growth factor homolog from a sea urchin. FASEB J. 3:1760-1764. [DOI] [PubMed] [Google Scholar]
- 15.Jain, D., K. J. Kaur, and D. M. Salunke. 2001. Plasticity in protein-peptide recognition: crystal structures of two different peptides bound to concanavalin A. Biophys. J. 80:2912-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jancarik, J., and S.-H. Kim. 1991. Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 24:409-411. [Google Scholar]
- 17.Jiang, J. S., and A. T. Brunger. 1994. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J. Mol. Biol. 243:100-115. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., and M. Kjeldgaard. 1997. Electron density map interpretation. Methods Enzymol. 277:173-208. [DOI] [PubMed] [Google Scholar]
- 19.Katz, B. A. 1997. Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH. J. Mol. Biol. 274:776-800. [DOI] [PubMed] [Google Scholar]
- 20.Laitinen, O. H., K. J. Airenne, A. T. Marttila, T. Kulik, E. Porkka, E. A. Bayer, M. Wilchek, and M. S. Kulomaa. 1999. Mutation of a critical tryptophan to lysine in avidin or streptavidin may explain why sea urchin fibropellin adopts an avidin-like domain. FEBS Lett. 461:52-58. [DOI] [PubMed] [Google Scholar]
- 21.Livnah, O., E. A. Bayer, M. Wilchek, and J. L. Sussman. 1993. Three-dimensional structures of avidin and the avidin-biotin complex Proc. Natl. Acad. Sci. USA 90:5076-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livnah, O., and J. L. Sussman. 1990. Crystal forms of avidin. Methods Enzymol. 184:90-93. [DOI] [PubMed] [Google Scholar]
- 23.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 24.Otwinowski, Z., and W. Minor. 1997. Processing X-ray data collected in oscillation mode. Methods Enzymol. 276:307-325. [DOI] [PubMed] [Google Scholar]
- 25.Pähler, A., W. A. Hendrickson, M. A. Gawinowicz Kolks, C. E. Argarana, and C. R. Cantor. 1987. Characterization and crystallization of core streptavidin. J. Biol. Chem. 262:13933-13937. [PubMed] [Google Scholar]
- 26.Pähler, A., J. L. Smith, and W. A. Hendrickson. 1990. A probability representation for phase information from multiwavelength anomalous dispersion. Acta Crystallogr. A 46:537-540. [DOI] [PubMed] [Google Scholar]
- 27.Pazy, Y., T. Kulik, E. A. Bayer, M. Wilchek, and O. Livnah. 2002. Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin. J. Biol. Chem. 277:30892-30900. [DOI] [PubMed] [Google Scholar]
- 28.Pazy, Y., O. H. Laitinen, B. Ravoy, M. S. Kulomaa, M. Wilchek, E. A. Bayer, and O. Livnah. 2001. Crystallization and preliminary X-ray analysis of W120K mutant of streptavidin. Acta Crystallogr. D 57:1885-1886. [DOI] [PubMed] [Google Scholar]
- 29.Sano, T., and C. R. Cantor. 1995. Intersubunit contacts made by tryptophan 120 with biotin are essential for both strong biotin binding and biotin-induced tighter subunit association of streptavidin. Proc. Natl. Acad. Sci. USA 92:3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sano, T., M. W. Pandori, X. Chen, C. L. Smith, and C. R. Cantor. 1995. Recombinant core streptavidins. A minimum-sized core streptavidin has enhanced structural stability and higher accessibility to biotinylated macromolecules. J. Biol. Chem. 270:28204-28209. [DOI] [PubMed] [Google Scholar]
- 31.Weber, P. C., D. H. Ohlendorf, J. J. Wendoloski, and F. R. Salemme. 1989. Structural origins of high-affinity biotin binding to streptavidin. Science 243:85-88. [DOI] [PubMed] [Google Scholar]