Abstract
The 1.8-Å resolution structure of the ATP-Mg2+-Ca2+-pyruvate quinary complex of Escherichia coli phosphoenolpyruvate carboxykinase (PCK) is isomorphous to the published complex ATP-Mg2+-Mn2+-pyruvate-PCK, except for the Ca2+ and Mn2+ binding sites. Ca2+ was formerly implicated as a possible allosteric regulator of PCK, binding at the active site and at a surface activating site (Glu508 and Glu511). This report found that Ca2+ bound only at the active site, indicating that there is likely no surface allosteric site. 45Ca2+ bound to PCK with a Kd of 85 μM and n of 0.92. Glu508Gln Glu511Gln mutant PCK had normal activation by Ca2+. Separate roles of Mg2+, which binds the nucleotide, and Ca2+, which bridges the nucleotide and the anionic substrate, are implied, and the catalytic mechanism of PCK is better explained by studies of the Ca2+-bound structure. Partial trypsin digestion abolishes Ca2+ activation (desensitizes PCK). N-terminal sequencing identified sensitive sites, i.e., Arg2 and Arg396. Arg2Ser, Arg396Ser, and Arg2Ser Arg396Ser (double mutant) PCKs altered the kinetics of desensitization. C-terminal residues 397 to 540 were removed by trypsin when wild-type PCK was completely desensitized. Phe409 and Phe413 interact with residues in the Ca2+ binding site, probably stabilizing the C terminus. Phe409Ala, ΔPhe409, Phe413Ala, Δ397-521 (deletion of residues 397 to 521), Arg396(TAA) (stop codon), and Asp269Glu (Ca2+ site) mutations failed to desensitize PCK and, with the exception of Phe409Ala, appeared to have defects in the synthesis or assembly of PCK, suggesting that the structure of the C-terminal domain is important in these processes.
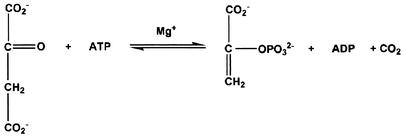
Escherichia coli phosphoenolpyruvate carboxykinase [ATP] (EC 4.1.1.49, PCK) plays a central role in gluconeogenesis by catalyzing the conversion of oxaloacetate (OAA) to phosphoenolpyruvate (PEP) (Fig. 1). This step is the first committed step in gluconeogenesis in E. coli, and PCK functions to divert tricarboxylic acid cycle intermediates toward gluconeogenesis (46). In fact, PCK is part of the gluconeogenic pathway in virtually all organisms, and PCK from mammalian and bacterial sources has been extensively studied in terms of kinetics of substrate binding (9, 28), possible active site residues (21, 27), the catalytic roles of metals (24, 30), and the reaction mechanism (1).
FIG.1.
PEP carboxykinase reaction.
Figure 1 shows that, in the presence of saturating concentrations of MgATP, PCK from E. coli is activated synergistically by Ca2+ or by Mn2+ (20). In the absence of Mg2+, there is no synergistic effect of adding Ca2+ and Mn2+. Furthermore, synergistic activation by Ca2+ but not activation by Mn2+ can be abolished by partial digestion of PCK by trypsin. Fluorescence binding with the Ca2+ analogue, Tb3+, indicated the presence of two Tb3+ sites on PCK with Kd values of 14 μM and 110 μM (20). Also, an X-ray crystallographic structure of a (Tb3+)2 complex of PCK (33) suggested the possibility of Ca2+ as an allosteric regulator of PCK via binding at both an internal active site and at a surface activating site (Glu508 and Glu511).
Activation of PCK by Mn2+ has been rationalized previously by the structure of a (ATP)-Mg2+-Mn2+-pyruvate complex of PCK with distinct binding sites for Mg2+ and for Mn2+ in the active site region (44). In order to truly identify the role of Ca2+ in the PCK kinetic mechanism, we have solved the crystal structure of the ATP-Mg2+-Ca2+-pyruvate quinary complex of PCK. This crystal complex contains Ca2+ at the active site and not at the putative allosteric site. This finding is corroborated by a binding study with 45Ca2+ and by mutagenesis of the surface site (Glu508 and Glu511), which was thought to be a potential allosteric site for Ca2+.
We have also investigated how partial digestion of PCK can abolish activation by Ca2+ (desensitize PCK) without affecting activation by Mn2+. N termini of tryptic peptides have been sequenced, and trypsin-sensitive sites have mutated. These mutated PCKs have been digested with trypsin and have been analyzed so that one can learn more about the mechanism of desensitization. Residues in the C-terminal domain removed by trypsin have undergone mutation or been deleted, and these PCKs have been analyzed so that one can learn more about the mechanisms of activation by Ca2+ and Mn2+.
MATERIALS AND METHODS
Media, bacterial strains, and plasmids.
Media and antibiotic concentrations have been described previously (19, 35). All strains were E. coli K12. MM294A (endA1 hsdR17 supE44 thi-1) was the original source of the pckA gene, cloned into plasmid pBR322 in plasmid HG26 (19). Strain HG89 (pckA4 pps-3 recA pyrD34 his-61 tyrA2 rpsL125 thi-1 lac argG::Tn5 srl::Tn10) was used for expression and purification of mutant PCKs (19).
Plasmid pHG41, with a unique ClaI site (at bp 1294 of pHG26), was prepared by cutting plasmid pHG26, which contains the pckA gene of E. coli (19, 35), with EcoRI (4359 bp of pBR322), filling in with DNA polymerase I (Klenow fragment), and ligating it to a fragment containing the other ClaI site (bp 0 of pHG26), which had similarly been filled in. PCR-mediated site-specific mutagenesis (see below) of pHG41was then used to create pHG50 with a unique AccI site at bp 1746 of pHG26. Plasmid pHG50 was used to construct pHG51 with an additional unique BssHII site at bp 2139 of pHG26 by the latter method. Plasmids pHG41, pHG50, and pHG51 all encoded the same wild-type PCK sequence and expressed high levels of PCK (up to 50% of soluble proteins) and were used for PCR mutagenesis.
Site-specific mutagenesis.
Two-step PCR mutagenesis (2) was used to construct substitution and deletion mutations in the pckA gene on plasmids. Codon usage tables for E. coli (36) were used to select relatively highly expressed codons for mutations. Silent mutations were used to introduce restriction sites to facilitate identification of mutant plasmids. Mutagenic primers and primers containing restriction endonuclease sites were synthesized by Life Technologies, Inc. EcoRI and XhoI were used to construct Arg2Ser. ClaI and AccI were used for Arg396Ser, Arg396(TAA) (termination codon at position 396), Phe409Ala, ΔPhe409 (deletion of Phe409), and Phe413Ala. SphI and ClaI were used for Asp269Glu. AccI and BssHII were used for the Glu508Gln Glu511Gln double mutant. ClaI and BssHII were used for Δ397-521 (deletion of residues 397 to 521). Mutations were sequenced between the two restriction sites, on both strands of DNA, by the Plant Biotechnology Institute, Saskatoon, Canada.
Protein purifications.
Cells were grown in 12-liter fermenter cultures grown on Luria-Bertani (LB) medium plus ticarcillin (35). Wild type and mutant PCKs were purified to homogeneity by the method of Goldie and Sanwal (20), except that CaHPO4 column chromatography was omitted and that no column steps were repeated. Small amounts of mutants Arg2Ser, Arg396Ser, and Arg2Ser Arg396Ser double mutant were partially purified from 300-ml cultures of LB medium plus ticarcillin by using 2-ml DEAE cellulose columns (37).
PCK enzyme assays.
PCK activity was assayed by the 14CO2 exchange assay (20), and protein concentration was determined by a modification of the method of Lowry (18) for crude extracts and a microbiuret method for purified proteins (4). Concentrations of metal ions and complexes were calculated as for binding assays by using the COMICS program, modified to accept bound or free ligands as input (20). Curves of Va-Vo/Eo versus [Ca2+] were fitted to hyperbolae by using the MENTON program (13).
Calcium binding.
Binding of 45Ca2+ to PCK was estimated by using the nitrocellulose filter method of Kawasaki et al. (26). Standard assay conditions for PCK were used: 100 mM Tris, pH 7.5, 20 μM EDTA, 1 mM MgATP, and 2.4 mM Mg2+ with 104 to 105 counts of 45CaCl2 per min per reaction (20). The assay volume was changed to 100 μl for filter binding. Concentrations of metal ions and complexes were calculated as for enzyme assays.
Digestion of mutant PCKs with trypsin.
PCK (7 g) was incubated for 30 min at 30°C with limiting amounts of bovine trypsin (Sigma Scientific) in 40 μl of 10 mM Tris, pH 7.5, and 20 μM EDTA. Reactions were stopped by adding equal amounts of soya bean trypsin inhibitor (Sigma Scientific). (The trypsin inhibitor was estimated by Sigma to inhibit 2.5 times its weight of trypsin.) Wild-type PCK was estimated by using an E1cm1% of 13.2, and partially purified mutant PCKs were estimated by scanning sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels stained with Coomassie blue, with wild-type PCK as a standard. Reactions were then added to sample buffer, boiled for 2 min, and electrophoresed on SDS gels with 8 to 18% gradients of polyacrylamide. SDS gels of wild-type PCK were blotted on to Immobilon-P (polyvinylidine difluoride) membranes, stained with Coomassie blue, dried, and frozen at −20°C (32) for sequencing. N-terminal sequences of tryptic peptides were determined by the Protein Microsequencing Laboratory, Department of Biochemistry and Microbiology, University of Victoria, with an Applied Biosystems model 470A gas-phase sequencer.
X-ray crystallography.
PCK-ATP-Mg2+-Ca2+-pyruvate crystals were grown at 21°C by hanging drop vapor diffusion from 10-μl drops containing 6 mg of PCK ml−1, 5 mM CaCl2, 5 mM MgCl2, 2 mM ADP, 2 mM PEP, 1 mM EDTA, 200 mM ammonium acetate, 100 mM sodium acetate buffer (pH 4.8), 0.1 mM dithiothreitol, and 10% (wt/vol) polyethylene glycol 4000 as a precipitating agent. Under the crystallization conditions ADP and PEP reacted to produce ATP and pyruvate as occurred previously in the ATP-Mg2+-Mn2+-pyruvate complex of PCK (44). The drops were equilibrated against a 1-ml reservoir containing 100 mM sodium acetate (pH 4.8), 200 mM ammonium acetate, and 24% (wt/vol) polyethylene glycol 4000.
The long tabular crystal used for data collection took 5 days to grow to a size of 0.15 by 0.3 by 0.7 mm. Data were collected at the Photon Factory synchrotron radiation source (Tsukuba, Japan) on beamline BL18B by using a screenless Weissenberg camera with a crystal-to-film distance of 429.7 mm and a wavelength of 1.0 Å. Diffraction intensities were recorded on Fuji imaging plates and were digitized by using a Fuji BA-100 scanning system. The crystal was aligned with c parallel to the spindle axis. By using a single crystal, a total range of 105° of data were collected with a 7° oscillation range used for each imaging plate and an overlap of 0.5° between successive plates. Crystal alignment was done with Polaroid photography on the Weissenberg camera. Measurements from two oscillation photographs (one at the zero setting of the spindle dial and the other at 90 from the corrected zero setting) provided the missetting angles required for preliminary processing of the diffraction data with the DENZO program (38). All subsequent data reduction was performed by using the Collaborative Computational Project, Number 4 program suite (10). The complex crystallized in the monoclinic space group C2, with unit cell parameters as follows: a = 126.2, b = 95.2, c = 46.8, d = 95.2; with one monomer in the asymmetric unit. The estimated solvent content of the crystals is 47.4%, with a volume/mass ratio of 2.40 Å3 Da−1 (34). A total of 41,488 unique reflections were measured, comprising a data set that is 78.3% complete to 1.8 Å. Rsymm for the diffraction intensities, Io, to 1.8 Å is 3.7%, with an average redundancy factor of 2.84 and average I/sigma(I) of 16.6; the last shell (1.86 to 1.80 Å) is 69.4% complete, with an Rsymm of 45% and an average I/sigma(I) of 1.13.
The present structure is isomorphous, with the structure of PCK-ATP-Mg2+-Mn2+-pyruvate. The coordinates from this structure, minus substrates and water molecules, were used as a starting model for refinement. Rigid body refinement, followed by refinements by restrained conjugate gradient minimization and simulated annealing, were carried out by using the crystallography and nuclear magnetic resonance system (8) until convergence was reached. Substrates and water molecules were then added by visual inspection of calculated electron density maps by using TURBO-FRODO (A. Roussel, C. Fontecilla-Camps, and C. Cambillau, Abstr. Int. Union Crystallogr. 15th Int. Congr., abstr. MS-02.07.06, 1990) and were followed by iterative cycles of refinement and model adjustment. The progress of the refinement was monitored throughout by calculating the Rfree on a random test data set comprising 5% of the data with a final Rfree of 0.225. For the last refinement cycle, the working and test sets of data were combined by using all 38,351 reflections in the resolution range of 10 to 1.8 Å, with a final R of 0.1820. Refinement statistics are recorded in Table 1. Unless otherwise stated, all protein structure diagrams in this paper were drawn by using SETOR (14).
TABLE 1.
Final refinement statistics for Ca2+-Mg2+-ATP-pyruvate E. coli PCK complex
| Resolution limits (Å) (38,351 reflections) | 10.0-1.80 |
| [Last shell (Å) (2,917 reflections)] | [1.86-1.80] |
| Final R (all 10.0-1.80 Å data) | 0.18 |
| [Last shell] | [0.30] |
| Rfree∗ (5% of 10-1.80 Å data) | 0.22 |
| [Last shell] | [0.30] |
| No. of nonhydrogen protein atoms included in refinement | 4,167 |
| No. of solvent molecules (H2O) | 261 |
| rms deviation from ideality in bond lengths (Å) | 0.005 |
| rms deviation from ideality in bond angles (°) | 1.26 |
| rms deviation from ideality in dihedral angles (°) | 23.3 |
| rms deviation from ideality in improper angles (°) | 0.74 |
| Mean B factor for PCK Å2 | 28.2 |
| Mean B factor for ATP Å2 | 37.0 |
| Mean B factor for pyruvate Å2 | 52.4 |
| Mean B factor for waters Å2 | 38.2 |
| B factor for Mg2+ Å2 | 52.2 |
| B factor for Ca2+ Å2 | 49.5 |
RESULTS
Purification of PCK.
Twenty-four liters of fermenter culture produced yields of 1.5 to 2 g of purified wild-type PCK (microbiuret method) from strain MM294A/pHG41. This strain probably has a high copy number of pckA (similar to pUC plasmids), because the rop gene is deleted in pHG41 (19). Yields from E. coli without such a plasmid are typically about 2 to 3 mg for this size of culture (20). Twelve-liter cultures produced 0.6 to 1.2 g of mutant PCKs. Only two column chromatographic steps (DEAE cellulose and gel filtration) were required compared to four steps for purification from an E. coli strain without a high copy number plasmid.
X-ray crystallography.
Molecular replacement techniques were used to solve the crystal structure of the ATP-Mg2+-Ca2+-pyruvate E. coli PCK complex, and the structure was subsequently refined against 1.8-Å resolution data (Table 1). Of the 540 residues in the PCK molecule, 537 have been modeled, representing 99% of the protein scattering matter in the asymmetric unit. The first 3 amino acids of the N terminus were not modeled because of a lack of interpretable electron density for these residues. In previous complexes, a surface loop containing residues 393 to 400 was unable to be modeled satisfactorily but was approximately modeled by using glycine residues in this structure. All substrates and metal ions are visible in electron density maps. There are 261 water molecules, one ATP molecule, one magnesium ion, one pyruvate molecule, and one calcium ion in the crystal structure. Under the crystallization conditions, ADP and PEP reacted to form ATP and pyruvate, as occurred for the ATP-Mg2+- Mn2+-pyruvate E. coli PCK complex (44). Of the 537 residues in the model, 90% are located within the most-favored regions of the Ramachandran plot, with 9.4% of residues located in the next most-favored regions (40). The stereochemistry of the model is good, with root mean square (rms) deviations in geometry and average temperature factors as shown in Table 1.
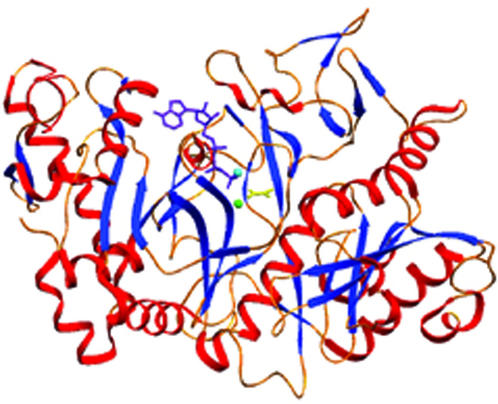
E. coli PCK is a monomeric, globular protein that is part of the α/β protein class (Fig. 2). It contains two distinct domains, a 265-residue C-terminal domain, and a 275-residue N-terminal domain. Between the two domains, the active site is found at the bottom of a deep cleft. Upon binding of ATP-Mg2+ to this site, domain closure is induced via a 20° rotation of the N- and C-terminal domains (44, 45). This closure is extremely important for catalysis, as it traps substrates, positions catalytic side chains of active site residues, and excludes bulk solvent.
FIG. 2.
Ribbon diagram indicating the tertiary structure and secondary structure elements of PCK. Helices are shown in red, Â-strands in blue, and random coil regions and turns in gold. Mg2+ is the light blue sphere, Ca2+ is the light green sphere, ATP is purple, and pyruvate is shown in yellow.
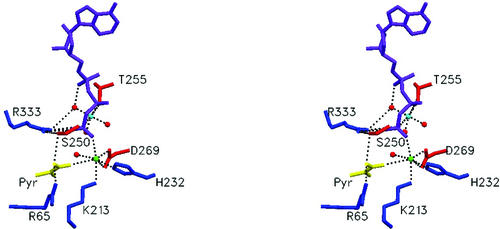
At the active site of PCK, the Ca2+-Mg2+ metal cluster is bound (Fig. 3). The distance between Ca2+ and Mg2+ is 5.18 Å. This observation again lends support to the concept of dual metal activation in enzymes, whereby Mg2+ serves to bind and activate ATP and whereby a second cation activates both the pyruvate enolate anion and ATP.
FIG. 3.
Stereo view of substrate/metal binding in the active site. Mg2+ is the light blue sphere, Ca2+ is the pink sphere, and the water molecules are red spheres, ATP is purple, and pyruvate is shown in yellow.
The Mg2+ ion possesses an octahedral coordination sphere and is coordinated by three ordered water molecules, O of Thr255 and two oxygen atoms from the β- and γ-phosphoryl groups of ATP (Fig. 3). This is identical to the coordination of Mg2+ in the previously published ATP-Mg2+-Mn2+-pyruvate PCK complex structure. The average distance between magnesium and the six coordinating atoms in its inner sphere is 2.2 Å (Table 2), which again is identical to the PCK-ATP-Mg2+-Mn2+-pyruvate structure (44).
TABLE 2.
Summary of active site electrostatic interactions and hydrogen bonding in PCK-Ca2+-Mg2+-ATP-pyruvate complex
| Substrate | Coordinating group | Distance (Å) |
|---|---|---|
| Ca2+ | Nɛ2 H232 | 2.6 |
| Oδ1 D269 | 2.5 | |
| Oδ2 D269 | 3.0 | |
| O1γ ATP | 2.4 | |
| H2O 719 | 2.6 | |
| O1α PVT | 2.8 | |
| Nζ K213 | 2.7 | |
| Avg | 2.7 | |
| Mg2+ | O2β ATP | 2.1 |
| O3γ ATP | 2.2 | |
| Oγ T255 | 2.1 | |
| H2O 674 | 2.3 | |
| H2O 716 | 2.5 | |
| H2O 738 | 2.3 | |
| Avg | 2.2 | |
| PVT | ||
| O1α | Ca2+ | 2.8 |
| H2O 719 | 3.2 | |
| O1β | 65 NH2 | 2.8 |
| O2 | H2O 719 | 3.4 |
Comparison of PCK-ATP-Mg2+-Ca2+-pyruvate and PCK-ATP-Mg2+-Mn2+-pyruvate complexes.
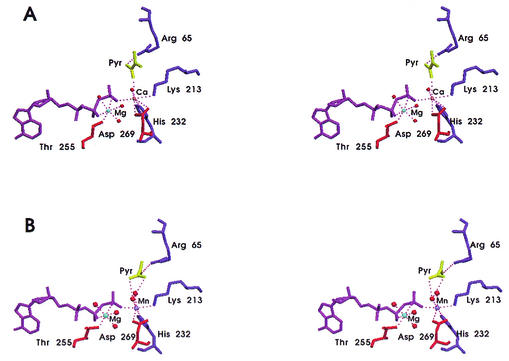
The coordination sphere of Ca2+ includes N2 from His232, two oxygen atoms from the side chain of Asp269, one oxygen atom from the γ-phosphoryl group of ATP, Nɛ from Lys213, the enolate carbonyl oxygen of pyruvate, and one ordered water molecule (Fig. 4A). The ATP/Mg2+ moiety is essentially identical in the two complexes (Fig. 4A and B). Ca2+ is in its often-observed seven-coordinate state, whereas Mn2+ is in its usual six-coordinate state. The calcium ion is coordinated by one oxygen atom of the γ-phosphoryl group of ATP, Nɛ from Lys213 and the N-2 from His232, similar to that of manganese in its complex. The coordination of calcium differs from that of manganese in these two complexes, with Ca2+ being coordinated by both oxygen atoms of Asp269, while Mn2+ is coordinated by one oxygen atom of Asp269; Ca2+ is coordinated to the carbonyl oxygen of pyruvate and to one water molecule, while Mn2+ is coordinated to two water molecules. The average group coordinating distance is 2.7 Å for Ca2+ and 2.3 Å for Mn2+. This coordination differs from that of Mn2+ in that Mn2+ is indirectly bridged via a second ordered water molecule to the enolate oxygen of pyruvate (44). The coordination of the Ca2+ by a lysine is unusual, yet coordination of this lysine to Mn2+ has been observed in the ATP-Mg2+-Mn2+-pyruvate quinary complex of E. coli PCK (44) and is analogous to the Tb3+ binary complex of E. coli PCK (33). This has been attributed to the neutrality of the side chain of Lys213, due to the high proportion of basic residues in the active site and the adjacent hydrophobic side chain of Phe413, thus lowering the pKa of the Lys213 side chain. The ATP remains bound in the syn conformation, which has been observed in all previously published E. coli PCK complexes (44, 45), as well as the structure of N-ethylmaleimide sensitive factor (49). The syn conformation is sterically strained yet promotes favorable interaction between ATP and PCK. As well, pyruvate is observed in the active site as the result of addition of a proton to the pyruvate enolate anion by PCK at pH 4.8 (Fig. 4 and Table 2). Pyruvate is bound to the positively charged pocket of the bottom of the active site, yet the global orientation of pyruvate differs from the previously solved ATP-Mg2+-Mn2+-pyruvate E. coli PCK quinary complex (44) (Fig. 4). The carboxyl oxygen atom O-1 of pyruvate binds directly to Ca2+ (2.8 Å) and a water molecule (2.6 Å), whereas the carbonyl oxygen atom of pyruvate was bound to Mn2+ indirectly through an ordered water molecule in the PCK-ATP-Mg2+-Mn2+-pyruvate quinary complex. The carbonyl oxygen atom, in this case, is hydrogen bonded to a water molecule (3.0 Å). Also, in the Ca2+-containing complex, Arg333 does not interact with pyruvate, whereas this side chain served to bridge ATP and pyruvate in the former, the ATP-Mg2+-Mn2+-pyruvate structure (44). In the Mn2+-bound structure, Arg65 neutralized the negative charges of both pyruvate carboxylate oxygen atoms, whereas Arg65 formed a hydrogen bond with the carboxylate oxygen O-1 (2.8 Å) in the ATP-Mg2+-Ca2+-pyruvate E. coli PCK quinary complex. In the Ca2+-containing complex the O atom of Ser250 is positioned in such a way that it is hydrogen bonded to both the pyruvate carboxylate oxygen O-1 and to the side chain of Arg333 (Fig. 3).
FIG. 4.
(A) Stereo view of the active site of PCK-ATP-Mg2+-Ca2+-pyruvate complex. Diagram of the global orientation of pyruvate in the active site of the ATP-Ca2+-Mg2+-pyruvate E. coli PCK quinary complex. Pyruvate is directly bridged to the γ-phosphoryl group of ATP via Ca2+. Ca2+ is the pink sphere, the water molecules are the red spheres, ATP is purple, and pyruvate is shown in yellow. (B) Stereo view of the active site of the PCK-ATP-Mg2+-Mn2+-pyruvate complex. Mn2+ is the purple sphere, the water molecules are the red spheres, ATP is purple, and pyruvate is shown in yellow.
Activation of Glu508Gln Glu511Gln PCK by calcium.
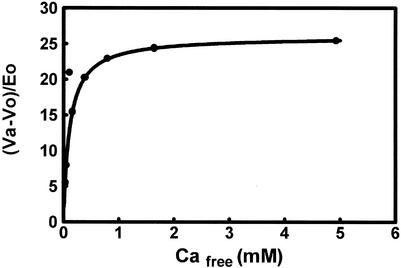
A fluorescence binding study with Tb3+ (20) and the (Tb3+)2 complex of PCK (33) suggested that there were probably two binding sites for Ca2+ on PCK. The surface Tb3+ binding site (Glu508 and Glu509) is colored red in Fig. 5A. Ca2+ and Mg2+ at the active site are colored green and cyan, respectively. We determined activation by Ca2+ for a mutant PCK with Glu508 and Glu509 both mutated to Gln. Plots of Va-Vo/Eo for the Glu508Gln Glu511Gln double mutant were very similar to those published for the wild type. Maximum activated velocity (Va) was 24 U/mg, Ka was 124 μM wild type with 1 mM MgATP and 2.4 mM Mg2+ (20), and Va was 26 U/mg. Ka was 110 μM for Glu508Gln Glu511Gln (Fig. 6). From these results, it appears to be unlikely that the surface Tb3+ site in the (Tb3+)2 complex of PCK (Glu508 and Glu511) is an allosteric site for Ca2+ activation.
FIG. 5.
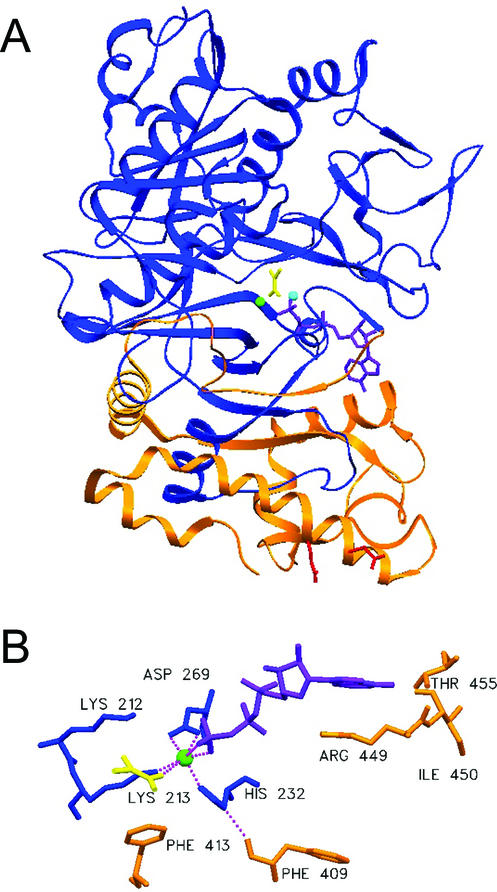
(A) PCK molecule showing C-terminal domain cleaved by trypsin (orange). The rest of PCK is colored blue. ATP and pyruvate are purple, Ca2+ is green, Mg2+ is cyan, and residues Glu508 and Glu511 are red. (B) View of residues interacting with Ca2+ in blue and C-terminal Phe413 and Phe409 (both orange) interacting with Lys213 and with His232, respectively.
FIG. 6.
Activation of Glu508Gln Glu511Gln PCK by Ca2+. PCK activity assayed by the 14CO2 exchange assay, with 20 mM OAA, 20 mM NaH14CO3, [Mg2+] of 2.4 mM, and [MgATP] of 1 mM.
Calcium binding.
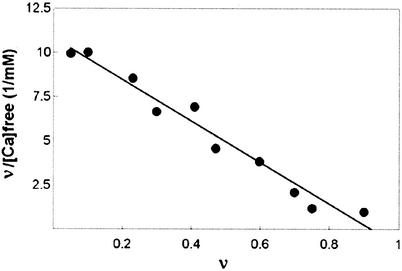
It was desirable to determine the nature of Ca2+ binding under conditions supporting enzyme activity and activation of PCK by Ca2+, since the ATP-Mg2+-Ca2+-pyruvate complex of PCK was crystallized under nonphysiological conditions at pH 4.8. 45Ca2+ bound to wild-type PCK with n of 0.92 and Kd of 85 μM in assay buffer at pH 7.5 (Fig. 7). The Glu508Gln Glu511Gln double mutant had similar parameters (n = 0.95 and Kd = 94 μM) (data not shown). This is further evidence that the surface Tb3+ site is probably not a Ca2+ binding site.
FIG. 7.
Scatchard plot of 45Ca2+ binding to wild-type PCK.
Digestion of mutant PCKs with trypsin.
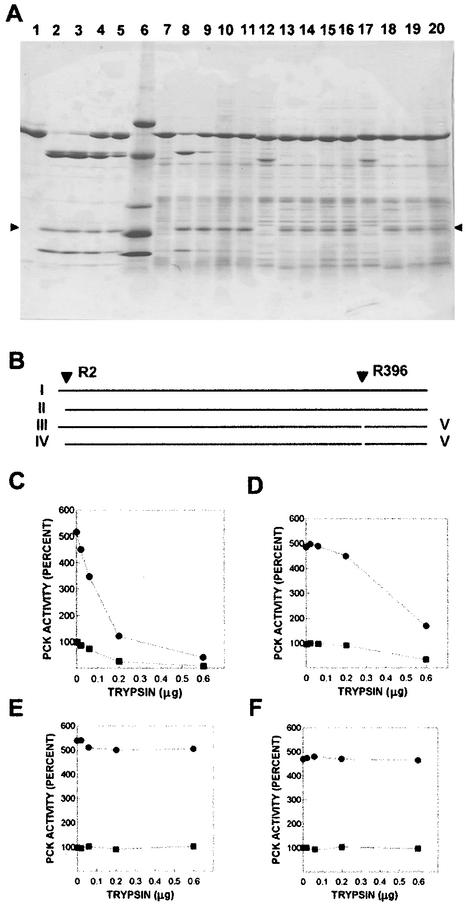
Previously, time-limited digestion of PCK with high concentrations of trypsin and PCK led to the formation of a lower-molecular-weight core enzyme, which retained normal activity assayed in the absence of Ca2+ but lost the ability to be activated by Ca2+ (20). The molecular weight change appeared to be complete in 15 min, but desensitization to activation by Ca2+ required 120 min. We completed concentration-limited digests of PCK with trypsin, to try to identify the trypsin-sensitive sites and to learn more about the mechanism of desensitization. Figure 8A, lanes 2 to 5, shows the results of digesting wild-type PCK with different amounts of trypsin. Fragments from similar experiments were subjected to N-terminal protein sequencing as described above. The 60-kDa band in lanes 4 and 5 appears as a doublet. The N-terminal sequence of the larger band (not shown) was the same as for native PCK (structure I in Fig. 8B). The smaller band had a N-terminal sequence that was similar but started with Val3, indicating that trypsin hydrolysis occurred after Arg2 (fragment II [Fig. 8B]). The same sequences were obtained for the larger and smaller bands in the doublet at 45 kDa, seen in lanes 2 to 5 (fragments III and IV, respectively). The N-terminal sequence of the 20-kDa band was identical to that of soya bean trypsin inhibitor, used in the experiment, and the N-terminal sequence of the 15-kDa band (fragment V) was identical to that of PCK, starting after Arg396. All N-terminal sequences were determined for at least six residues, and that of native PCK was determined for 44 residues. These results indicate that trypsin hydrolyzed PCK after residues Arg2 and Arg396. Wild-type PCK lost both Ca2+-activated and basal activities in parallel (Fig. 8C), indicating that PCK was not desensitized to Ca2+ activation in these concentration-limited digests.
FIG. 8.
Digestion of wild-type and mutant PCKs with trypsin. (A) SDS-gradient PAGE (8 to 18% polyacrylamide) of purified wild-type and partially purified mutant PCKs treated with trypsin for 30 min as described in Materials and Methods. The arrows (▸, ◂) indicate the position of soya bean trypsin inhibitor. Lanes: 1, 7 μg of wild-type PCK (WT); 2, WT + 0.6 μg of trypsin; 3, WT + 0.2 μg of trypsin; 4, WT + 0.06 μg of trypsin; 5, WT + 0.02 μg of trypsin; 6, size standards; 7, Arg2Ser mutant; 8, Arg2Ser + 0.6 μg of trypsin; 9, Arg2Ser + 0.2 μg of trypsin; 10, Arg2Ser + 0.06 μg of trypsin; 11, Arg2Ser + 0.02 μg of trypsin; 12, Arg396Ser mutant; 13, Arg396Ser + 0.6 μg of trypsin; 14, Arg396Ser + 0.2 μg of trypsin; 15, Arg396Ser + 0.06 μg of trypsin; 16, Arg396Ser + 0.02 μg of trypsin; 17, Arg2Ser Arg396Ser double mutant; 18, Arg2Ser Arg396Ser + 0.6 μg of trypsin; 19, Arg2Ser Arg396Ser + 0.2 μg of trypsin; and 20, Arg2Ser Arg396Ser + 0.06 μg of trypsin. (B) Model of trypsin hydrolysis of wild-type PCK. I represents uncut PCK, with N terminus at the left; II, cut at Arg2; III, cut at Arg396; IV, larger product of cutting at Arg2 and at Arg396; and V, small C-terminal fragment. Size standards for lane 7: bovine serum albumin, 66 kDa; ovine serum albumin, 45 kDa; pepsin, 34.7 kDa; α-lactoglobulin, 18.4 kDa; and lysozyme, 14.3 kDa. Activity of PCK after digestion with different amounts of trypsin as described in Materials and Methods. (C) Wild-type PCK. (D) Arg2Ser PCK.(E) Arg396Ser PCK. (F) Arg2Ser Arg396Ser double mutant. All assays had 1 mM MgATP and 2.4 mM Mg2+. In panels C to F, • represents activity with 400 μM Ca2+ and ▪ represents activity with no Ca2+.
Mutant PCKs were constructed (Arg2Ser, Arg396Ser, and Arg2Ser Arg396Ser double mutant) to remove trypsin-sensitive sites. Digestion of these PCKs by trypsin and the effects on activation by Ca2+ are also shown in Fig. 8. Arg2Ser (lanes 7 to 11 in Fig. 8A and D) produced no doublet bands of 60 or 45 kDa. (Only fragments III and V appear in these digests.) The appearance of the 45- and 15-kDa bands (lane 8) coincided with about 70% loss of PCK activity, with or without Ca2+. Hydrolysis at Arg396 appeared to require higher trypsin concentrations in the Arg2Ser mutant than in the wild type; possibly hydrolysis at Arg2 facilitates the reaction at Arg396 in the wild type. The Arg396Ser mutant was cut only at Arg2, and the Arg2Ser Arg396Ser double mutant was not visibly affected by trypsin under these conditions. Digestion of the three mutant PCKs with higher concentrations of trypsin in time-limited digestions led to loss of detectable bands on gels and PCK activity (not shown). From these results, it seems unlikely that hydrolysis at either the Arg2 site or at the Arg396 site is sufficient for the desensitization of wild-type PCK in time-limited digests. The N-terminal sequence of PCK, desensitized in a time-limited digest, indicates that it has been cut at the Arg2 site. However, it is possible that hydrolysis at both sites could be required to facilitate hydrolysis at some other site when wild-type PCK is densensitized in a time-limited digest, since we are unable to desensitize either the Arg2Ser or the Arg396Ser PCKs in time-limited digests or to obtain fragments suitable for N-terminal sequencing from them under these conditions.
Attempts to obtain mutant PCKs desensitized to activation by Ca2+.
Figure 5A shows the C-terminal domain removed by hydrolysis at the Arg396 site in orange. Desensitized PCK is probably missing all of these residues, based on its size (20). We constructed mutations that remove all of this C-terminal domain, Arg396(TAA) (stop codon at residue 396), and since termination mutations often interfere with protein assembly and/or stability, Δ397-521(deletion of residues 397 to 521) was also constructed. It was hoped that the 19 C-terminal residues retained in Δ397-521 would stabilize the protein and protect it from cellular proteases. Phe409 and Phe413 of the C-terminal domain both have strong interactions with residues that coordinate Ca2+ in the active site. The carbonyl oxygen atom of Phe409 is hydrogen bonded with the N-1 hydrogen atom of His232, and Cɛ of Phe413 has a strong van der Waals interaction with Cγ of Lys213 (Fig. 5B). Phe409Ala, ΔPhe409 (deletion of Phe409), and Phe413Ala mutations were constructed to test the hypothesis that these residues are important for activation by Ca2+. A Lys213Ser mutation leads to loss of all activation by Ca2+ and by Mn2+ without significantly changing Vmax in the absence of Ca2+ or Mn2+, or Kms for substrates (37). We hypothesized that Phe409Ala, ΔPhe409, Phe413Ala, Δ397-521, or the Arg396(TAA) mutation might lead to loss of activation by Ca2+ but not to loss of activation by Mn2+, since partial digestion of PCK by trypsin has that affect. An Asp269Glu mutation was also constructed, since the larger Glu side chain might crowd the active site, preventing activation by Ca2+ but not by the smaller Mn2+ ion.
Isolates of E. coli strain HG89 (pckA4 pps recA1) containing plasmids with the Phe409, ΔPhe413Ala, Δ397-521, or the Arg396(TAA) mutant pckA gene all failed to synthesize detectable mutant PCKs, as demonstrated by lack of any enzyme activity and lack of an identifiable 60-kDa band on SDS gels of crude extracts. They were all unable to grow on minimal medium with succinate as the carbon source, indicating that these mutant pckA genes failed to complement the pckA4 mutation. It is likely that these mutations all interfere with the synthesis, assembly, or stability of PCK. Attempts to produce PCK protein from these mutants by using lower temperatures, protease-deficient strains, or strains with plasmids overexpressing groEL or dnaK chaperonin systems have failed. It is likely that the C-terminal domain and the contacts of Phe409 and Phe413 are important for the synthesis, assembly, or stability of PCK. However, the Phe409Ala mutant PCK was successfully expressed. Phe409Ala mutant PCK appeared to have normal kinetic parameters and positive complementation of pckA4 and was activated normally by Ca2+ or Mn2+ (not shown). Probably the carbonyl oxygen atom of Ala at position 409 is able to hydrogen bond with Lys213—the side chain of Phe409 points away from the active site and may not be required for PCK activity or activation by Ca2+.
DISCUSSION
The significance of the ATP-Mg2+-Ca2+-pyruvate E. coli PCK quinary structure lies in the observation of Ca2+ bound only at the active site and not at the proposed allosteric site Glu508 Glu511 (33). This is corroborated by observations of normal Ca2+ activation in the Glu508Gln Glu511Gln double mutant and of binding of 45Ca2+ to only one site on wild type and on Glu508Gln Glu511Gln double mutant PCKs. The presence of two different divalent cations binding at two separate regions of the active site has previously been observed in the ATP-Mg2+-Mn2+-pyruvate quinary complex (44). As well, many other enzymes that catalyze phosphoryl transfer between anionic substrates require two divalent cations for optimal catalytic activity, such as MutT dGTPase (17), pyruvate kinase (3, 22), and glutamine synthetase (29).
The role of intracellular free Ca2+ in E. coli has been assessed recently (25). Two distinct Ca2+ transport systems have been studied. As in higher organisms, Ca2+ may regulate the cell cycle, since some cell cycle mutants were defective in Ca2+ homeostasis. There is evidence for significant Ca2+ binding proteins in E. coli, such as FtsZ and Acp. There is also evidence for a role of intracellular Ca2+ in the regulation of chemotaxis and in the regulation of several genes.
Ca2+ concentrations in E. coli are difficult to measure due to the smallness of the cells and to the presence of two cellular membranes. The best experiments involve expressing a recombinant bioluminescent Ca2+ binding protein, aequorin, in E. coli cells, although cells have to be shaken with the lipophilic cofactor, coelentrazine, for hours before measurements can be taken (25) and although it is difficult to estimate the concentration of aequorin bound to its cofactor in the cells. Ca2+ concentrations were reported to be 200 to 300 nM in exponentially growing cells, and slow fluctuations into the low-micromolar range were observed under some conditions, including the stationary phase of growth. These estimated concentrations are lower than the Ka for activation of PCK by Ca2+ (70 to 100 μM) but are close enough to be significant, given the technical difficulties inherent in the measurements. The kinetic and physical data clearly indicate that Ca2+ activates PCK in vitro. The evidence for a role of Ca2+ in cellular regulation in E. coli suggests that activation of PCK by Ca2+ may be important in vivo. Also, expression of the pckA gene increases 100-fold in the stationary phase after growth on enriched media, probably to facilitate synthesis of glycogen, since expression of glg genes and synthesis of glycogen increase markedly in the stationary phase (19). It is possible that activation of PCK by Ca2+ plays a role in stationary-phase glycogen synthesis. Transport of Ca2+ is known to be energy dependent, and the observed increases in Ca2+ during the stationary phase could be due to decreased rates of transport (25).
There are many ways to account for behavioral differences observed between Ca2+ and Mg2+ in biological systems, especially their selective specificity for different regions of the PCK active site (15, 31). In terms of ionic radii, Ca2+ and Mg2+ differ significantly. The ionic radius of Mg2+ is 0.65 Å, which is consistent with its perfect fit into the 1.40-Å hole provided by the octahedral all-oxygen coordination sphere (48). Ca2+, with an anionic radius of 1.0 Å, is too big for the 1.40-Å hole but fits easily into the “looser” mixed coordination sphere of nitrogen and oxygen ligands that constitute the bridging site (41). As proof, the average distance between Mg2+ and its coordinating oxyanions is 2.2 Å, whereas the average distance between Ca2+ and its ligands is 2.7 Å (Table 2).
Though both Ca2+ and Mg2+ are hard Lewis acids (23, 41), Ca2+ has a filled outer 3p shell, rendering its ligand interactions purely electrostatic. It thus exhibits a strong preference for oxygen ligands over nitrogen or sulfur ligands, but large variations are possible in its coordination number, geometry, and ligand type (29, 31). For example, ligand-to-donor atom distances vary more (2.3 ± 0.3 Å) for Ca2+ than for Mg2+ (2.0 ± 0.1 Å) (11, 12). As well, Ca2+-carboxylate interactions are very common in proteins, and in this structure, Ca2+ coordinates with O1 and O2 of Asp269 with a coordination number of seven (12). Again, this is characteristic of the tendency of Ca2+ to vary its number of coordinating ligands. The asymmetric coordination of Ca2+ to O-1 and O-2 of Asp269 (2.5 and 3.0 Å) results in a coordination sphere with five planar and two axial ligands. Mg2+, on the other hand, possesses an unfilled outermost 3s orbital, tends towards covalent bond formation, and is characteristically targeted toward a binding site of an octahedron of oxygen atoms with a fixed Mg2+-to-O distance of 2.05 ± 0.05 Å, which is clearly exhibited in this structure (29, 31). Thus, the phosphate-dominated binding site that PCK offers is perfectly suited for Mg2+ binding and is consistent with its major role as a cofactor for enzymes that catalyze the hydrolysis or cleavage of polyphosphates.
The coordination of Ca2+ by nitrogen ligands is unusual, yet coordination by two nitrogens is observed in this structure. Ca2+ binding to the Ca2+-sequestering aminocarboxylates EDTA and nitrilotriacetic acid (NTA) has been investigated in the crystal structures of Ca(CaEDTA) · 7H2O and Na(CaNTA) (5). The EDTA structure contained two independent Ca2+ ions, one of which was coordinated by eight ligands, including two nitrogens with distances of 2.71 and 2.62 Å. One Ca2+ ion was found in the NTA complex with sevenfold coordination; one nitrogen ligand was identified 2.63 Å away from Ca2+. The antibiotic ionophore A23187 is a monocarboxylic acid that binds and transports divalent cations across membranes. The crystallographic structure indicated that one Ca2+ ion (CN = 7) was coordinated by two molecules of A23187, including two nitrogen atoms of the benzoxazole ring system (2.69 and 2.58 Å) (42). Finally, the complex of calcium (IMP) contained five Ca2+ ions, one of which was coordinated to the purine nitrogen atom N-7 and through two water-mediated interactions to the carbonyl group O-6 and the phosphate group; the Ca2+-to-N7 distance was 2.73 Å, and all Ca-to-O distances lay in the range of 2.3 to 2.5 Å (7).
Ca2+, like Mn2+, appears to possess a much higher affinity for the bridging site between ATP and the anionic substrate. Previously, binding of Mg2+ was not observed in this position in the ATP-Mg2+-Mn2+-pyruvate (44), ATP-Mg2+-oxalate (45), and ATP-Mg2+-pyruvate complexes of E. coli PCK (L.W. Tari, H. Goldie, and L. T. J. Delbaere, unpublished data). The looser, more electrostatic bonding of Ca2+ with its coordinating ligands probably allows for faster diffusion of substrates in and out of the active site. Moreover, the direct coordination of pyruvate by Ca2+ allows for direct nucleophilic attack on the γ-phosphoryl group by the enolate of pyruvate, as well as much closer orientation of the pyruvate enolate anion with ATP, allowing for a shorter distance for phosphoryl transfer. Ca2+ is also kinetically superior to Mg2+, as it undergoes rapid replacement of waters in its inner coordination sphere approximately 103 times faster than Mg2+. Replacement of inner-sphere water occurs at a rate of 108.4 s−1 for Ca2+ and 105.2 s−1 for Mg2+ (16). It is probable that the lower water exchange rate for Mg2+ is due to its restricted ability to facilitate substitution by reducing or enlarging its coordination sphere from six donors. In this way, the greater radii of Ca2+ exclude interfering waters and allow for direct coordination of the enolate of pyruvate as well as increasing exchange of inner-sphere products for reactants. Furthermore, Ca2+ is 10 times more efficient than Mn2+ in terms of water exchange (16). Previous studies of PCK illustrated loss of the synergistic effects of Ca2+ in the presence of saturating amounts of Mg2+ upon digestion of the enzyme with trypsin and loss of C-terminal residues. Activation by Mn2+ in the presence of Mg2+, however, was retained, with a decreased Km of 0.009 mM (20). Hydrolysis at Arg2 and at Arg396, trypsin-sensitive sites identified in this work, is not sufficient for desensitization of PCK but may be necessary in addition to hydrolysis at some other site or sites by trypsin. Peptide mapping and sequencing of the desensitized PCK should be done to determine its primary structure. It would also be worthwhile to obtain a high-resolution structure of desensitized PCK, since the phenomena of retention of activity after loss of many residues interacting with the active site and the loss of activation by Ca2+ but not by Mn2+ are very interesting.
Possible effect of partial trypsin cleavage of E. coli PCK.
Loss of the C-terminal residues 397 to 540 would account for about 25% of the enzyme, resulting in exposure of hydrophobic residues to solution. Removal of such a large amount of the enzyme almost certainly results in global changes that are difficult to estimate. Residues 449 to 455 surround and interact with the adenine ring of ATP, and loss of this region may result in loss of specificity for ATP binding. This perturbation of the ATP binding site may result in loss of tight binding of ATP now that only the ribose ring and triphosphate chain can bind specifically. Movement of the γ-phosphate could result in loss of the Ca2+ binding oxygen ligand. Another small portion of the lost region consisting of residues 406 to 417 is found near the active site region. The Phe413 side chain points directly into the active site region, forming van der Waals interactions with Cγ of Lys213. Lysine side chains are generally flexible and are largely located on the surface of a protein in contact with the aqueous solvent. Lys213 and Lys212, however, are found in the active site and have specific chemical functions. It has been noted that Lys213 possesses rotation around its C—C bond, which is normally sterically unfavorable; thus, Phe413 must assist in constraining the Lys213 side chain to a single position in the active site. Free movement of the side chain of Lys213 in aqueous solution would take it out of proximity with the metal ion that it coordinates, as well as away from Lys212, Arg65, Arg333, and other positively charged amino acids. Thus, Lys213 likely could not remain deprotonated and would no longer act as a ligand for the Ca2+ ion. Phe413 may also be important in the assembly or stability of PCK, since the Phe413Ala mutation led to a loss of detectable PCK protein.
Hydrogen bonding also occurs between the Phe409 carbonyl oxygen atom and N-1 of His232. This residue helps orient the imidazole ring of His232 so that the N-2 atom is pointing directly towards the Ca2+ ion. Loss of Phe409 (ΔPhe409 mutant) also led to loss of detectable PCK, while loss of the side chain only (Phe409Ala) had no observed effect.
Even after the loss of the C-terminal domain, the carbonyl oxygen atom of Gly248, which has van der Waals interactions with His232, may cause rotation and movement of the imidazole ring in order to hydrogen bond with N-1 of His232. This could result in the loss of another ligand that coordinates Ca2+. Phe413 also has van der Waals interactions with the guanidium side chain of Arg65. Loss of these residues may result in movement of Arg65 and loss of the hydrogen bond between Arg65 and O-1 of pyruvate. Thus, pyruvate binding may be diminished or pyruvate may be forced to bind in a different orientation, resulting in a decrease of catalytic activity. It is probable, however, that elimination of Ca2+ activation is due to movement of its ligands, such as the γ-phosphate of ATP, Lys213, His232, and pyruvate, as a result of loss of surrounding “constraining residues.” Ca2+ interactions with its ligands are mainly electrostatic, and loss of up to four ligands would abolish its relatively weak binding. Mn2+, on the other hand, strongly interacts with its ligands and may still be able to bind to the active site even if some of its ligands are no longer restrained in conformation. Alternatively, even with the loss of constraining residues, Mn2+ may be able to force the side chains of Arg65, Lys213, and His232 and the γ-phosphate of ATP to remain in coordinating positions in order to continue activating PCK catalysis.
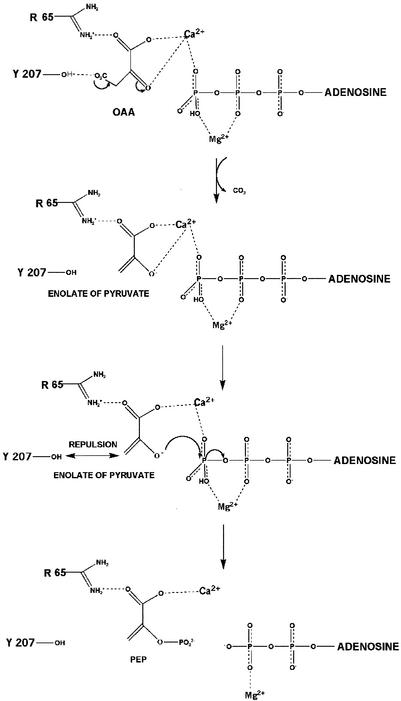
Catalytic mechanism based on the structure of the PCK-ATP-Mg2+-Ca2+-pyruvate complex.
The role of Ca2+ in PCK catalysis is to bind one of the oxygen atoms of the γ-phosphate of ATP, increasing the electrophilicity of the γ-phosphate by polarizing another P—O bond. The proposed mechanism of phosphoryl transfer (Fig. 9) is as follows: OAA binds in the first coordination sphere of Ca2+, which promotes decarboxylation of OAA to form the enolate of pyruvate. Ca2+ then serves to electrostatically stabilize the pyruvate enolate anion via the carboxylate oxygen atom O-1, and the carboxylate oxygen atom O-1 forms a hydrogen bond with Arg65. Furthermore, superposition of OAA over the pyruvate binding site indicates that the second carboxyl group can interact with Arg333, stabilizing the molecule further. Arg333 also could bridge OAA and an oxygen of the γ-phosphate of ATP. Since the pyruvate enolate anion is directly bound to Ca2+, displacement of water molecules is not required, allowing for faster substrate exchange than does Mn2+. Ca2+ also neutralizes electrostatic repulsion between the enolate of pyruvate and the γ-phosphate of ATP, moving the two substrates closer together and orienting the enolate oxygen atom so that it is correctly positioned to attack and displace the γ-phosphoryl group (Fig. 9). The hydroxyl group of Tyr207 may also sterically repel the CH2 group of the enolate of pyruvate, again moving the enolate oxygen closer to the γ-phosphate. Tyr207 may play a similar role in other ATP-dependent PCKs, as it is part of a PCK-specific domain made up of residues 204 to 213 (Gly-Thr-Trp-Tyr-Gly-Gly-Glu-Met-Lys-Lys) that is identical between species with the exception of residues at position 3. Arg333 and Arg65, as well, are identical among all ATP-dependent enzymes sequenced, indicating their importance in interacting with the pyruvate intermediate formed during PCK catalysis (33).
FIG. 9.
Proposed reaction mechanism for the synthesis of PEP by E. coli PCK. Arg65 forms a salt bridge interaction with the carboxylate oxygen atom of OAA. Arg333 further stabilizes the molecule by forming a salt bridge with another carboxylate oxygen atom, thus bridging ATP and OAA. OAA binds in the first coordination sphere of Ca2+, which promotes decarboxylation to form the pyruvate enolate intermediate. Ca2+ electrostatically stabilizes the enolate anion via the carboxylate oxygen and neutralizes the repulsions between the enolate and the γ-phosphoryl group of ATP. Ca2+ serves to orient the two substrates and position the enolate oxygen to attack and displace the γ-phosphoryl group. The hydroxyl group of Tyr207 may also move the substrates closer together by repulsion of the CH2 group of the enolate.
The role of Mg2+ in coordination of β- and γ-phosphoryl oxygen atoms has been observed many times in structures such as Ef-Tu (6), DNA gyrase B (47), and H-ras-p21 (39). As a Lewis acid, Mg2+ polarizes the P—O bond in the γ-phosphoryl group by rendering the γ-phosphate electrophilic, increasing the possibility of nucleophilic attack by the enolate of pyruvate (44). By neutralizing electrostatic repulsion between the negatively charged β- and γ-phosphate oxygen atoms, Mg2+ and Lys254 stabilize an extended form of the triphosphate chain with eclipsed β- and γ-phosphate oxygens, forming a sterically strained, high-energy transition state. By lowering the free energy of activation, Mg2+ moves the anionic substrates closer together and promotes the in-line transfer of the γ-phosphoryl group to the enolate oxygen of pyruvate (Fig. 9). The Mg2+-ATP substrate positions are nearly identical in the ATP-Mg2+-Mn2+-pyruvate quinary complex (44) and ATP-Mg2+-pyruvate quaternary complex (unpublished data), and all support the formation of an associative transition state with SN2-like displacement of the γ-phosphate by the enolate of pyruvate. The SN2-like displacement of the phosphoryl group has been illustrated by a recent structure of the E. coli PCK ADP-AlF3-Mg2+ quaternary complex, whereby AlF3 mimics the phosphoryl group being transferred (43).
It is observed from crystallographic studies of the ATP-Ca2+-Mg2+-pyruvate E. coli PCK quinary complex that Mg2+ and Ca2+ possess two distinct binding sites. Mg2+ correctly positions and activates ATP, whereas Ca2+ serves to activate the PCK catalytic reaction by directly bridging and activating ATP and the enolate anion of pyruvate. In this structure, the global orientation of pyruvate has changed completely to a more logical orientation by surrounding side chains and tighter interaction with Ca2+. The chemical properties of Ca2+, observed changes in active site residue and substrate orientation, and studies of mutant PCKs indicate that Ca2+ may serve as a superior but not allosteric activator in dual metal ion-facilitated phosphoryl transfer.
Acknowledgments
We thank N. Watanabe, N. Sakabe, and K. Suzuki from the Photon Factory for assistance with data collection.
A.S. and R.W. gratefully acknowledge financial support from the Colleges of Medicine and Graduate Studies, University of Saskatchewan. This work was funded by a Canadian Institutes of Health Research grant to L.T.J.D. (no. MT-10162) and by grants from NSERC and NSC Technologies to H.G.
REFERENCES
- 1.Ash, D. E., F. A. Emig, S. A. Chowdhury, Y. Satoh, and V. A. Schramm. 1990. Mammalian and avian liver phosphoenolpyruvate carboxykinase. Alternate substrates and inhibition by analogues of oxaloacetate. J. Biol. Chem. 265:7377-7384. [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl. 1992. Short protocols in molecular biology, vol. 2. John Wiley & Sons, New York, N.Y.
- 3.Baek, Y. H., and T. Nowak. 1982. Kinetic evidence for a dual cation role for muscle pyruvate kinase. Arch. Biochem. Biophys. 217:491-497. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, J. L. 1962. Techniques in protein chemistry, p. 294-295. Elsevier Publishing Co., Amsterdam, The Netherlands.
- 5.Barnett, B. L., and V. A. Uchtman. 1979. Structural investigations of calcium-binding molecules. 4. Calcium binding to aminocarboxylates. Crystal structures of Ca(CaEDTA)Q7H2O and Na(CaNTA). Inorg. Chem. 18:2674-2678. [Google Scholar]
- 6.Berchtold, H., L. Reshetnikova, C. O. A. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365:126-132. [DOI] [PubMed] [Google Scholar]
- 7.Brown, E. A., and C. E. Bugg. 1980. Calcium-binding to nucleotides: structure of a hydrated calcium salt of inosine 5′-monophosphate. Acta Crystallogr. Sect. B 36:2597-2604. [Google Scholar]
- 8.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., Y. Sato, and V. L. Schramm. 1991. Isotope trapping and positional isotope exchange with rat and chicken liver phosphoenolpyruvate carboxykinase. Biochemistry 30:4143-4151. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 11.Da Silva, F. J. J. R., and R. J. P. Williams. 1991. Magnesium: an introduction to its biochemistry. In N. J. Birch (ed.), Magnesium and the cell. Academic Press, Boston, Mass.
- 12.Einspahr, H., and C. E. Bugg. 1984. Crystal structure studies in calcium complexes and implications for biological systems. Met. Ions Biol. Syst. 17:51-97. [Google Scholar]
- 13.Eisenthal, R., and A. Cornish-Bowden. 1974. A comparison of seven methods for fitting the Michaelis-Menten equation. Biochem. J. 139:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, S. V. 1993. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11:134-138. [DOI] [PubMed] [Google Scholar]
- 15.Forsén, S., and J. Kördel. 1994. Calcium in biological systems, p. 107-167. In I. Bertini, H. B. Gray, S. J. Kippard, and J. S. Valentine (ed.), Bioinorganic chemistry. University Science Books, Mill Valley, Calif.
- 16.Frey, C. M., and J. Stuehr. 1974. Simple complexes. Met. Ions Biol. Syst. 1:51.
- 17.Frick, D. N., D. J. Weber, J. R. Gillespie, M. J. Bessman, and A. S. Mildvan. 1994. Dual divalent cation requirement of the MutT dGTPase: kinetic and magnetic resonance studies of the metal and substrate complexes. J. Biol. Chem. 269:1794-1803. [PubMed] [Google Scholar]
- 18.Geiger, P. J., and S. P. Bessman. 1972. Protein determination by Lowry's method in the presence of sulfhydryl reagents. Anal. Biochem. 49:467-473. [DOI] [PubMed] [Google Scholar]
- 19.Goldie, H., and V. Medina. 1990. Physical and genetic analysis of the phosphoenolpyruvate carboxykinase (pckA) locus from Escherichia coli K12. Mol. Gen. Genet. 220:191-196. [DOI] [PubMed] [Google Scholar]
- 20.Goldie, H., and B. Sanwal. 1980. Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of Escherichia coli. J. Biol. Chem. 255:1399-1405. [PubMed] [Google Scholar]
- 21.Guidinger, P. F., and T. Nowak. 1991. An active-site lysine in avian liver phosphoenolpyruvate carboxykinase. Biochemistry 30:8851-8861. [DOI] [PubMed] [Google Scholar]
- 22.Gupta, R. K., M. Oesterling, and A. S. Mildvan. 1976. Dual divalent cation requirement for activation of pyruvate kinase: essential roles of both enzyme- and nucleotide-bound metal ions. Biochemistry 15:2881-2887. [DOI] [PubMed] [Google Scholar]
- 23.Hay, R. W. 1984. Bio-inorganic chemistry, vol. 22. John Wiley & Sons, Hoboken, N.J.
- 24.Hebda, C. A., and T. Nowak. 1982. Phosphoenolpyruvate carboxykinase: Mn2+ and Mn2+ substrate complexes. J. Biol. Chem. 257:5515-5522. [PubMed] [Google Scholar]
- 25.Holland, I. B., H. E. Jones, A. K. Campbell, and A. Jacq. 1999. An assessment of the role of intracellular free Ca2+ in E. coli. Biochimie 81:901-907. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki, H., H. Kasai, and T. Okuyama. 1985. Protein analyses and reagents: microscale assay of calcium-binding activity of proteins and peptides using a nitrocellulose membrane. Anal. Biochem. 148:297-302. [DOI] [PubMed] [Google Scholar]
- 27.Krautwurst, H., S. Bazaes, F. D. González, A. M. Jabalquinto, P. A. Frey, and E. Cardemil. 1998. The strongly conserved lysine 256 of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase is essential for phosphoryl transfer. Biochemistry 37:6295-6302. [DOI] [PubMed] [Google Scholar]
- 28.Krebs, A., and W. A. Bridger. 1980. The kinetic properties of phosphoenolpyruvate carboxykinase of Escherichia coli. Can. J. Biochem. 58:309-318. [DOI] [PubMed] [Google Scholar]
- 29.Liaw, S. H., and D. Eisenberg. 1994. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme-substrate complexes. Biochemistry 33:675-681. [DOI] [PubMed] [Google Scholar]
- 30.Maggini, S., F. B. Stoecklin-Tschan, S. Mörikofer-Zwez, and P. Walter. 1993. A physiological role of Mn2+ in the regulation of cytosolic phosphoenolpyruvate carboxykinase from rat liver is unlikely. Biochem. J. 292:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, R. 1984. Bioinorganic chemistry of calcium. Met. Ions Biol. Syst. 17:1-49. [Google Scholar]
- 32.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 33.Matte, A., H. Goldie, R. M. Sweet, and L. T. J. Delbaere. 1996. Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold. J. Mol. Biol. 256:126-143. [DOI] [PubMed] [Google Scholar]
- 34.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 35.Medina, V., R. Pontarollo, D. Glaeske, H. Tabel, and H. Goldie. 1990. Sequence of the pckA gene of Escherichia coli: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. J. Bacteriol. 172:7151-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novakovski, B. A. A. 2001. Mutations which alter substrate and metal ion kinetics of Escherichia coli phosphoenolpyruvate carboxykinase. M.Sc. dissertation. University of Saskatchewan, Saskatoon, Canada.
- 38.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In J. C.W. Carter and R. M. Sweet (ed.), Methods in enzymology: macromolecular crystallography. Academic Press, New York, N.Y. [DOI] [PubMed]
- 39.Pai, E., W. Kabsch, U. Krengel, K. C. Holmes, J. John, and A. Wittinghofer. 1989. Structure of the guanine-nucleotide-binding domain of the H-ras oncogene product p21 in the triphosphate conformation. Nature 341:209-214. [DOI] [PubMed] [Google Scholar]
- 40.Ramakrishnan, C., and G. N. Ramachandran. 1965. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys. J. 5:909-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson, W. G. 1998. Chemistry and biochemistry of calcium, p. 1-25. In B. E. C. Nordin (ed.), Calcium in human biology: human nutrition review. Springer-Verlag, New York, N.Y.
- 42.Smith, G. D., and W. L. Duax. 1976. Crystal and molecular structure of the calcium ion complex of A23187. J. Am. Chem. Soc. 98:1578-1580. [Google Scholar]
- 43.Sudom, A. M., L. Prasad, H. Goldie, and L. T. J. Delbaere. 2001. The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF3. J. Mol. Biol. 314:83-92. [DOI] [PubMed] [Google Scholar]
- 44.Tari, L. W., A. Matte, H. Goldie, and L. T. J. Delbaere. 1997. Mg2+-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions. Nat. Struct. Biol. 4:990-994. [DOI] [PubMed] [Google Scholar]
- 45.Tari, L. W., A. Matte, U. Pugazhenthi, H. Goldie, and L. T. J. Delbaere. 1996. Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase. Nat. Struct. Biol. 3:355-363. [DOI] [PubMed] [Google Scholar]
- 46.Utter, M. F., and H. M. Kolenbrander. 1972. Formation of oxaloacetate by CO2 fixation on phosphoenolpyruvate, p. 117-168. In P. Boyer (ed.), The enzymes. Academic Press, New York, N.Y.
- 47.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dodson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]
- 48.Williams, R. J. P. 1993. Magnesium: an introduction to its biochemistry, p. 15-30. In Nicholas J. Birch (ed.), Magnesium and the cell. Academic Press, Boston, Mass.
- 49.Yu, R. C., P. I. Hanson, R. Jahn, and A. T. Brunger. 1998. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat. Struct. Biol. 5:803-811. [DOI] [PubMed] [Google Scholar]