Bacteria produce a wide variety of both proteinaceous and nonproteinaceous molecules for defense, mediation of microbial competitions, and signaling purposes. Of the protein-based molecules, there are examples of large folded polypeptides that are translated conventionally from genes as well as smaller peptides that are assembled nonribosomally by peptide synthetases (41). The latter often contain modified amino acids, including those with altered chirality, N-methylation, and nonamide backbone bonds. Such modifications may give the producing organism an advantage, as the modified molecules are less susceptible to the normal proteolytic cleavage reactions of proteins. Cyclization is another strategy that has been used, exemplified by well-known natural products such as cyclosporin A and gramicidin S. The advantages of such a strategy for stabilizing peptides may be seen by the fact that a significant number of macrocyclic natural products have found pharmaceutical applications, including, for example, the widespread use of cyclosporin A as an immunosuppressive agent. Synthetic cyclic peptides are also widely used as lead molecules in the pharmaceutical industry.
The biosynthesis of cyclic nonribosomal peptides such as cyclosporin A and polyketides such as the antibiotic erythromycin, as well as hybrid peptide/polyketide drugs such as rapamycin, has recently been reviewed (41). Briefly, it involves the ordered condensation of monomer building blocks by an enzyme-driven process to produce a linear acyl chain that is cyclized by a thioester domain at the C-terminal end of the biosynthetic assembly line (41).
Over recent years, several examples of naturally occurring circular proteins fundamentally different from the nonribosomal cyclic peptides have been discovered (58). These molecules are true proteins in that they have a well-folded three-dimensional structure and are produced via translation of genes. Their only difference from conventional proteins is that their gene-coded precursor proteins are posttranslationally modified to join the N and C termini to produce a seamless circle of peptide bonds. Such circular proteins occur in a diverse range of organisms, from bacteria to plants and animals, but the focus here is on circular proteins produced by bacteria. In this review we describe the sequences and structures of these proteins and examine what is known about their biosynthesis. We compare them to other recently discovered circular proteins from higher organisms and speculate on the possible roles of backbone cyclization.
Circular proteins were unknown a decade ago, and the field is still in its infancy, but there are now enough examples known to make it timely to examine the structures and properties of bacterially produced circular proteins. Bacterial protein expression has also been used to facilitate the production of synthetic circular variants of noncyclic proteins, including β-lactamase (31) and green fluorescent protein (30). These studies have adapted intein-based methods to enable protein ligations that result in circular proteins. While the focus of this review is on naturally occurring circular proteins, the studies on artificially produced circular proteins highlight the importance and interest in this area. We note at the outset that we generally use the term circular rather than cyclic to emphasize the fact that the molecules that we are focusing on have a head-to-tail cyclized backbone rather than other cross-links, such as disulfide bonds, that might make just part of the structure cyclic. While the molecules that we examine are thus topologically circular, as we shall see, they fold into complex three-dimensional shapes.
SEQUENCES AND STRUCTURES
The currently known circular proteins from bacteria range in size from 21 to 78 amino acids. From the sequences summarized in Table 1, it is evident that while they vary widely in size and primary structure, a common theme among these proteins is a high proportion of hydrophobic residues. The structural data available for cyclic proteins from both microorganisms and higher organisms have been derived almost exclusively from nuclear magnetic resonance (NMR) analysis. In general, the structures are well defined and contain elements of regular secondary structure. Thus, apart from the fact that no termini are present, the structures are not fundamentally different from those of conventional linear proteins.
TABLE 1.
Sources, sequences, and activities of cyclic bacterial proteins
| Peptide | Source | No. of amino acids | Sequence | Charge | Structure | Activity |
|---|---|---|---|---|---|---|
| Microcin J25 | E. coli AY25 | 21 | GGAGHVPEYFVGIGTPISFYG | −1 | Compact fold containing β-strands | Antimicrobial (gram-negative, narrow spectrum) |
| Gassericin A (reutericin 6) | L. gasseri LA39, L. reuteri LA6 | 58 | IYWIADQFGIHLATGTARKLLDAMASGASLGTAFAAILGVTLPAWALAAAGALGATAA | 0 | Helical (predicted) | Antimicrobial (gram-positive, broad spectrum) |
| Bacteriocin AS-48 | E. faecalis | 70 | MAKEFGIPAAVAGTVLNVVEAGGWVTLTAVGSGGLSLLAAAGRESIKAYLKKEIKKKGKRAVTAW | +6 | Helical | Antimicrobial (gram-positive, broad spectrum) |
| TrbC | E. coli | 78 | SEGTGGSLPYESWLTNLRNSVTGPVAFALSIIGIVVAGGVLIFGGELNAFFRTLIFLVLVMALLVGAQNVMSTFFGRG | 0 | Helical (predicted) | Conjugative pili |
| T pilin | A. tumefaciens | 74 | QSAGGGTDPATMVNNICTFILGPFGQSLAVLGIVAIGISWMFGRASLGLVAGVVGGIVIMFGASFLGKTLTGGG | +1 | Helical (predicted) | Conjugative pili |
The smallest circular protein of bacterial origin, microcin J25 (MccJ25), was first isolated from Escherichia coli AY25 (54). Microcins are a group of antimicrobial peptides produced by members of the family Enterobacteriaceae under conditions of nutrient depletion that target microbes phylogenetically related to the producer strain (19). MccJ25 induces filamentation in an SOS-independent way (54). In attempts to identify the mode of action, a resistant strain of E. coli carrying a mutation in the rpoC gene coding for the β′ subunit of RNA polymerase was isolated (17). Subsequent experiments in which the wild-type rpoC gene was introduced into MccJ25-sensitive strains resulted in complete resistance, identifying RNA polymerase as the target of MccJ25 and possibly explaining the observed filamentation, which may result from impaired transcription of genes involved in cell division (17). Further mutational analysis has provided a more detailed understanding of the mode of interaction of MccJ25 with RNA polymerase (62). Other studies have shown that MccJ25 has the ability to disrupt the membrane of Salmonella enterica serovar Newport but not E. coli, suggesting that the mechanism of action might be different against different bacterial strains (52). Interestingly, the bioactivity of a thermolysin-linearized form of MccJ25 against E. coli strains is significantly reduced compared to the native form, although it retains significant activity against S. enterica serovar Newport (6). These findings suggest that the circular structure is probably more crucial for a specific protein-protein interaction than a nonspecific interaction with the bacterial membrane.
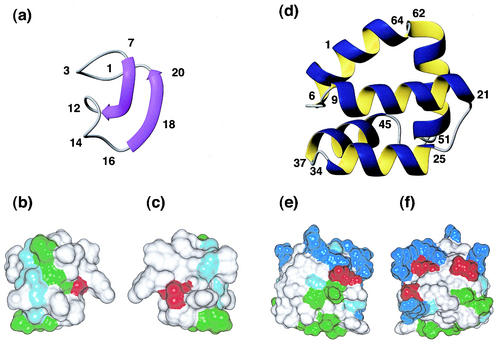
MccJ25 has been reported to contain a head-to-tail cyclized backbone based on enzyme cleavage data, sequencing, mass spectrometry, and NMR studies (6). It has been structurally characterized in methanol by NMR and proposed to adopt a highly compact globular structure, as shown in Fig. 1 (5). The structure has been described as a distorted antiparallel β-sheet that is twisted and folded back onto itself. Despite the highly hydrophobic nature of most of the residues in MccJ25, no real hydrophobic core is present due to its small size. Instead, most side chains are oriented towards the surface of the structure, forming hydrophobic patches, as indicated in Fig. 1 (panels b and c). The protection of the peptide backbone provided by these side chains may be responsible for the proteolytic stability of MccJ25 (5).
FIG. 1.
Solution structures of the circular bacterial proteins for which three-dimensional structures have been determined. (a) Ribbon representation of MccJ25 (PDB code 1HG6), with the β-strands shown as arrows. (b and c) Surface diagrams of MccJ25, with b in the same orientation as a and c rotated 180° about the y axis. White, green, and red represent hydrophobic, hydrophilic, and negatively charged residues, respectively. (d) Ribbon representation of AS-48 (PDB code 1E68), showing the five-helix bundle. (e and f) Surface diagrams of AS-48, with e in the same orientation as d and f rotated 180° about the y axis. White, green, blue, and red represent hydrophobic, hydrophilic, positively charged, and negatively charged residues, respectively. Glycine residues are shown in light blue.
It is interesting that a synthetic linear analogue did not fold correctly and did not have antibacterial activity even though a thermolysin-linearized derivative of the native peptide retained some structure and activity. Blond et al. suggested that folding into the native conformation may be assisted by a helper molecule in vivo (4). It is unusual for a small peptide lacking disulfide bonds to adopt such a well-defined structure as has been suggested for the native peptide, and it seems surprising that the additional constraint of a circular backbone would alone be sufficient to produce the observed fold. However, in our view there remain some inconsistencies in the spectroscopic data presented for MccJ25 and its thermolysin-linearized derivative that lead to questions about the exact structure of the peptide. At the time of writing, it remains unclear whether the peptide is in fact backbone cyclized or whether there are some other unusual chemical linkages stabilizing the structure. There may well be some revision of the primary structure as further investigations on this peptide are carried out.
Microcins produced by gram-negative bacteria have a counterpart in gram-positive bacteria, namely bacteriocins. The bacteriocins from lactic acid bacteria have been divided into four major classes based on size and structural features (40, 50, 60). Of interest here is class II, comprising small heat-stable peptides without lanthionine linkages, and more specifically subclass IIf, comprising atypical class II bacteriocins (60). At present this subclass has five members, of which two have been confirmed to be cyclic and one has been suggested to be cyclic based on sequence homology (60). These three members are discussed in further detail.
Gassericin A (GasA) has been isolated from two different strains of lactic acid bacteria, Lactobacillus gasseri LA39 (39) and Lactobacillus reuteri LA6 (57). When first described in 1991, then under the name reutericin 6, the peptide was thought to be significantly smaller, with an apparent molecular size of ≈3 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). However, subsequent work has shown that it carries a head-to-tail cyclization, comprises 58 residues, and has a mass of 5,652 Da (36, 38). No structural data are available, but its behavior on SDS-PAGE suggests a compact structure that also appears to be very stable, since heating for 60 min at 100°C does not destroy its inhibitory activity (57). More than 74% of the residues of GasA are hydrophobic and most probably exposed on the surface of the peptide, as evident from the fact that it cannot be eluted from a C18 column by methanol, acetonitrile, or 2-propanol (34). The secondary structure has been predicted to be helical, at least to some extent (38).
In addition to its antimicrobial activity against several Lactobacillus species, GasA is also active against several food-borne pathogenic bacteria, including Listeria monocytogenes, Bacillus cereus, and Staphylococcus aureus (36). GasA has been shown to be 98% identical to acidocin B, another bacteriocin isolated from Lactobacillus acidophilus M46 (46). While the chemical properties of acidocin B have not been fully characterized, the facts that this peptide differs from GasA at only a few positions and has a significantly lower apparent molecular weight on SDS-PAGE, consistent with what is observed for GasA, strongly suggest a macrocyclic structure.
Like GasA, bacteriocin AS-48 (AS-48), the second member of group IIf with a confirmed circular backbone, is also active against both related and unrelated gram-positive bacteria as well as several pathogenic organisms, including Staphylococcus, Enterococcus, and Salmonella species (1, 2, 24). According to Gálvez et al., AS-48 can interact directly with the cytoplasmic membrane of certain microorganisms without the mediation of surface receptors (24). AS-48 elicits its effects by inserting itself into the cytoplasmic membrane and forming pores. This renders the membranes permeable to ions and small molecules, leading to the release of cytoplasmic material and ultimately causing the lysis of sensitive cells (25). Because of its broad spectrum of antimicrobial activity and its temperature and pH stability, AS-48 has been proposed as a promising candidate for food biopreservation (1).
The three-dimensional structure of AS-48 as solved by NMR (26) is shown in Fig. 1. The fold is characterized by a globular arrangement of five α-helices connected by five short turn regions and enclosing a compact hydrophobic core. A very similar fold and membranolytic activity were earlier described for the linear mammalian protein NK-lysin from natural killer cells, suggesting a similar mechanism of action (26). Both NK-lysin and AS-48 have exceptional stability and high resistance to temperature denaturation (3, 9). Both contain a significant hydrophobic core and, in the case of NK-lysin, the fold is further stabilized by three disulfide bonds. While AS-48 lacks disulfide bonds, the additional stability is likely introduced from the circular backbone.
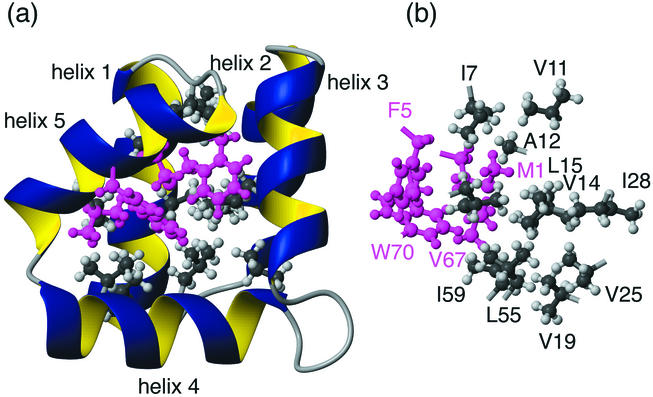
It is interesting that the backbone cyclization of the AS-48 precursor occurs at a point in the sequence corresponding to the middle of one of the α-helices (α5, spanning residues 64 to 5), which suggests that cyclization is absolutely necessary for the correct folding and function of AS-48 (8). This is supported by preliminary studies indicating that overexpressed linear (i.e., noncyclic) AS-48 does not adopt a native fold. In particular, the interactions of three of the internal hydrophobic residues of α5, Val67, Met1 and Phe5, with the hydrophobic core are thought to be essential for the stability of the five-helix globule (26). Figure 2 illustrates the hydrophobic interactions in the core and highlights the importance of these residues for the overall stability of AS-48. One would expect that in a linearized version of AS-48, the crucial helix, α5, would be unfolded and the important interactions with the core would be lost. This would likely disrupt the integrity of the core and have not only local structural effects but also major implications for the global fold.
FIG. 2.
Hydrophobic interactions in the core of AS-48. The hydrophobic side chains in the core are shown in grey (helices 1 to 4) and pink (helix 5) and labeled with residue numbers and single-letter amino acid codes. The importance of the side chains of helix 5 is clear from the extensive interactions with most other regions of the core. The compactness of the fold is highlighted by the fact that all of the residues shown here have less than 20% of their side chain surfaces exposed to the solvent. View b is rotated 90° around the y axis in relation to a.
Not all the hydrophobic residues in AS-48 are buried in the core; a significant number are also exposed to the solvent, resulting in hydrophobic patches. While only three of the helices, α1, α2, and α4, show modest amphipathic character, the overall structure is highly amphipathic, as illustrated in Fig. 1 (panels e and f). The distribution of the positive charges in AS-48 is highly asymmetrical, with most of the positively charged residues clustered in helix α4 and in the adjacent turn region between helices α4 and α5 (26). It is believed that this cluster of positively charged residues is responsible for the antimicrobial activity of AS-48 (26). The combination of an overall positive charge and an amphipathic character is optimal for interacting with and disrupting the negatively charged bacterial membrane and has been observed in a number of membrane-active antimicrobial proteins (53).
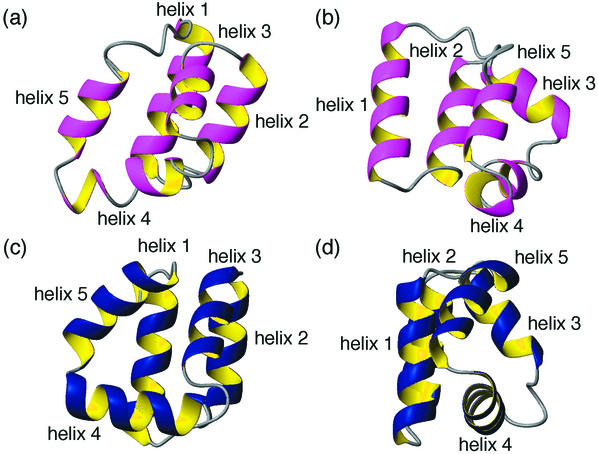
An indication that circular proteins are not unusual in terms of their structural features may be seen from the fact that the structure of AS-48 was predicted with high precision (to a root mean squares distance of 4.3 Å) in a recent protein structure prediction competition (51). In fact, AS-48 was the only circular protein included in the competition but was better predicted than all of the linear proteins. Figure 3 shows a comparison of the predicted structure and the one determined experimentally by NMR spectroscopy. It is interesting that the presence and extent of the five helical regions and their orientations in relation to each other were predicted very accurately. However, the packing of the side chains in the hydrophobic core is harder to predict, as illustrated by the fact that the predicted structure is significantly less compact. In particular, this is highlighted by helix 1, which is not closely associated with the molecular core in the predicted structure.
FIG. 3.
Comparison of the structure of AS-48 predicted in a recent blind test of protein structure prediction, CASP4 (51) (a and b) and the one determined by NMR spectroscopy (c and d). The helical regions are labeled 1 to 5, with helix 5 comprising the additional peptide bond linking the N and C termini. Views b and d are rotated 90° around the y axis in relation to a and c.
While all of the other circular proteins discussed so far are more or less involved in repelling other organisms from the producer, TrbC and T pilin have a very different role: they promote contact between cells. The pilins are the primary components of the bacterial conjugative system, a very efficient means of mediating horizontal gene transfer in a highly promiscuous manner (61). Kalkum et al. found that TrbC and T pilin, the subunits of the pili encoded by the IncP (RP4) and Ti plasmids, respectively, were proteins with their backbones cyclized by peptide bonds (35). Despite being very similar in function and size (78 versus 74 amino acids), TrbC and T pilin do not show a high degree of sequence similarity. However, there seem to be several conserved residues across various pilin homologues within the core region, including six absolutely conserved glycine residues (21, 43).
Although the solution structures of both TrbC and T pilin have not been resolved to date, there are some indications of what these proteins might look like. First, both contain a high proportion of hydrophobic amino acids (68% for TrbC and 70% for T pilin). A characteristic of IncP pili is their tendency to aggregate in bundles (21), which suggests that the surface of the pili is hydrophobic, and therefore it can be surmised that at least some extended hydrophobic areas exist on the surface of the TrbC subunits comprising the pili. On polyacrylamide gels, the linear form of T pilin has lower mobility than the mature circular protein, indicating that the three-dimensional structures of the two proteins most likely differ significantly (43). Finally, secondary-structure prediction programs have identified two putative transmembrane helices in both the circular pilins (21).
It is known that T pili are very durable and highly resistant to various chemical treatments that are known to destroy other pili (44, 45). Whether the circular character of T pilin confers this exceptional stability remains to be confirmed but seems likely.
BIOSYNTHESIS (CLOSING THE RING)
Cyclization of the protein backbone differs from other posttranslational modifications in that it is not possible to discern from the mature protein where in the sequence it has taken place, only that it has. The discovery of gene sequences encoding precursor proteins has provided insight into this “silent” event by allowing the identification of the amino acids involved in the head-to-tail linkage. Yet for a majority of circular proteins, relatively little is known about the mechanism that governs the joining of the termini. No apparent homology exists between the amino acids involved in the formation of the de novo peptide bond or the flanking residues across the different types of circular proteins. The cyclization of MccJ25, for instance, has been reported to involve a peptide bond between two glycine residues (55), while in AS-48 the link occurs between a methionine and a tryptophan residue (47). This covers the complete spectrum of amino acid types from the smallest, least sterically hindered to large, bulky ones. This disparity, in addition to the different functions and structures of the circular proteins, makes a ubiquitous mechanism of cyclization seem unlikely.
All circular proteins for which the gene sequence has been determined originate from a precursor protein with an N-terminal signal peptide, implicating both cleavage and cyclization events in the maturation process. Whereas the AS-48, MccJ25, GasA, and T pilin precursors comprise a signal peptide followed by the mature peptide domain (33, 37, 47, 55), the TrbC precursor and the precursors of some circular proteins from plants and mammals also include N-terminal proregions and C-terminal domains (21, 32, 56). These extra domains, often conserved across a class of circular proteins, may play a role in organizing the residues of the mature protein in an orientation conducive to peptide bond formation. However, it appears that a suitable geometric arrangement is by itself not sufficient to result in cyclization; many proteins whose termini are situated in close proximity remain linear (49), including acyclic analogues of circular proteins themselves (14), and it is therefore likely that enzymatic processes also play a role. Proteolytic enzymes are obvious candidates for the necessary cleavage (and ligation) steps involved in the biosynthesis of macrocyclic proteins, but very few with the necessary specificity have been characterized.
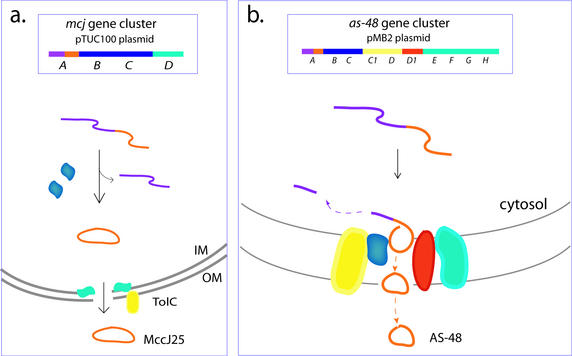
The biosynthesis of AS-48, MccJ25, and the pilins (little is known about GasA) involves auxiliary proteins, many of which are encoded on the same plasmid as the respective structural genes. As a result, these bacterial systems present excellent opportunities for dissecting the various components involved in protein cyclization. Three genes residing on the pTUC100 plasmid, mcjBCD, are necessary for expression of the MccJ25 phenotype in addition to the precursor gene mcjA (Fig. 4a) (55). The gene products McjB and McjC, both of which are required for the production of active protein, are thought to be involved in the maturation of MccJ25, but their exact function is not well understood. McjD, a putative ABC transporter protein involved in secretion, confers immunity to MccJ25. AS-48 production and immunity involve the coordinated expression of 10 genes, as-48ABCC1DD1EFGH, within the as-48 gene cluster on the pMB2 plasmid (Fig. 4b) (20). Of these, as-48ABCC1DD1 were initially identified as being necessary for the expression of the AS-48 phenotype and, except for the precursor protein As-48A, are all predicted to contain transmembrane helices, localizing them to the membrane (48). As-48B and As-48C have been implicated in the processing of As-48A to the mature cyclic protein. The presence of membrane-spanning domains in these proteins as well as the absence of linear AS-48 analogues led Martinez-Bueno et al. (48) to postulate that cleavage of the leader sequence and cyclization may be coupled to secretion. As-48C1DD1 and the recently discovered As-48EFGH proteins all play a role in AS-48 secretion and are required for the full expression of AS-48-mediated immunity (20).
FIG. 4.
Biosynthesis of the cyclic bacterial proteins MccJ25 and AS-48. (a) Four genes located within the mcj cluster, mcjABCD, are necessary for the production of and immunity to MccJ25. Maturation from the precursor McjA to MccJ25 requires both McjB and McjC (blue) and, due to the predicted absence of membrane-spanning domains in these proteins, probably occurs in the cytosol. McjD (aqua), which confers immunity to MccJ25, shows homology to bacterial ABC exporters and has been implicated in secretion of MccJ25 from the cell. A chromosomally encoded outer membrane protein, TolC (yellow), has since also been found to play a role in MccJ25 secretion (18). (b) Ten genes implicated in AS-48 production and immunity are located within the as-48 gene cluster, as-48ABCC1DD1EFGH, on the pMB2 plasmid. The gene products As-48ABCC1DD1 are necessary for AS-48 production and immunity and, except for the precursor protein As-48A, are all predicted to be located in the membrane. As-48B and As-48C (blue) are predominantly involved in AS-48 production. Their putative locations in the membrane may indicate that AS-48 processing and cyclization are coupled to secretion, as shown here. Alone, As-48D1 (red) is able to confer immunity on the cyclic protein AS-48, but As-48C1 and As-48D (yellow), the latter showing homology to ABC transporter proteins, enhance this resistance, and thus all three proteins have been implicated in AS-48 secretion. The as-49EFGH operon encodes another putative ABC transporter (aqua) which was recently shown to be required for full expression of AS-48 immunity. The precursor proteins are represented by purple strands, with the cyclic protein domains colored orange. IM, inner membrane; OM, outer membrane.
Similar to their bactericidal counterparts, the TrbC and T pilin precursors are proteolytically processed and cyclized, although rather than being secreted, the circular proteins are assembled into pilin filaments. Most of the proteins necessary for pilin biogenesis are encoded on the same plasmid as the structural gene. Eleven plasmid-encoded proteins, in addition to the precursor proteins, are essential to both TrbC and T pilin maturation (7, 27). Interestingly, these auxiliary proteins are predominantly involved in making up a membrane-spanning translocation complex that mediates both pilin assembly and function (Fig. 5). The only protein identified that has been implicated in the cyclization process is TraF, encoded on the RP4 plasmid along with TrbC. In contrast, processing of the T pilin is entirely independent of the Ti plasmid and is therefore assumed to be mediated by chromosomally derived proteins (21).
FIG. 5.
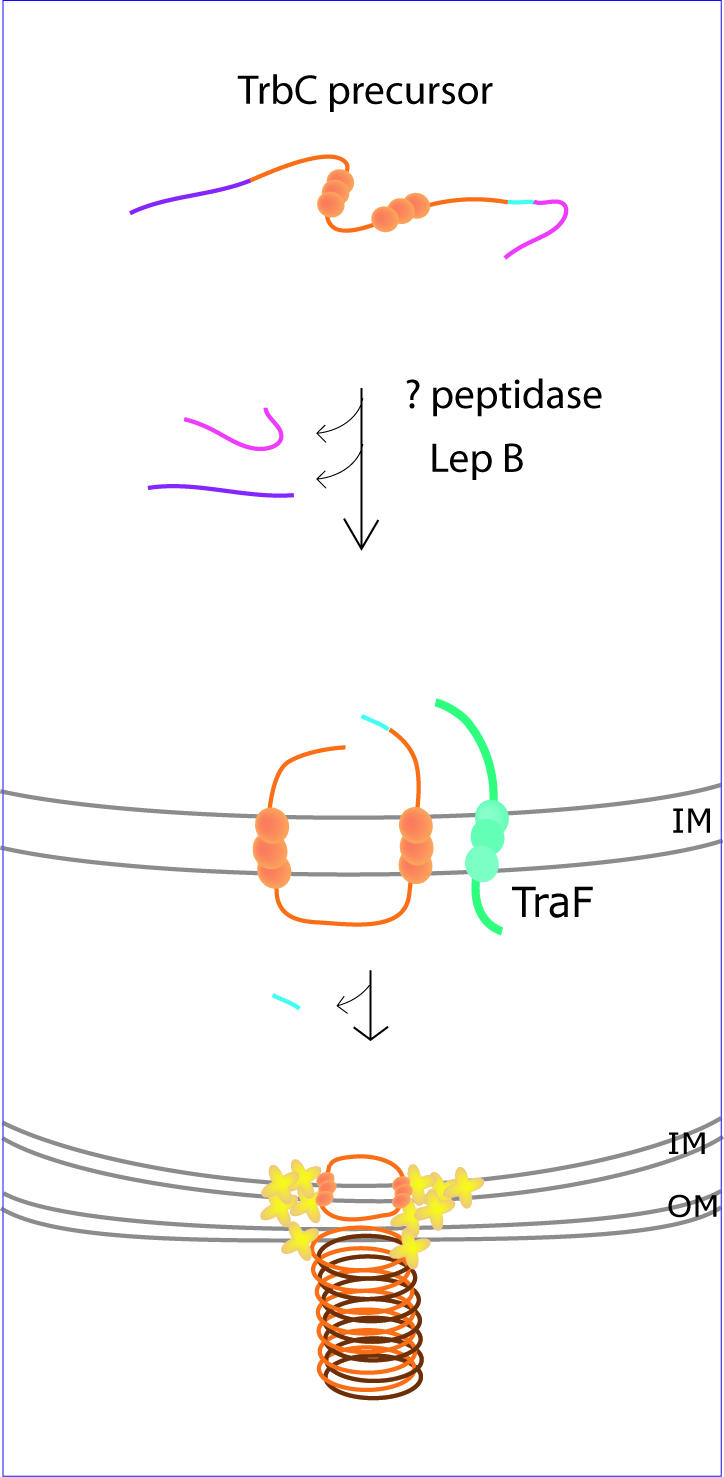
Biosynthesis of cyclic bacterial protein TrbC pilin. The TrbC precursor is processed at both termini prior to insertion of the protein into the inner membrane, where cyclization takes place. An unidentified peptidase is responsible for proteolytic cleavage at the C terminus (pink), while the chromosomally encoded peptidase LepB removes an N-terminal peptide (purple). The membrane-spanning TraF protein (aqua), encoded on the RP4 plasmid with the precursor, is thought to catalyze cyclization of TrbC in a concerted event that involves simultaneous removal of a C-terminal tetrapeptide (light blue) (refer to text for details). The cyclic TrbC units (orange) are then transferred to the cell surface and assembled into pilin filaments with the aid of a translocation complex comprised, at least in part, of components also encoded on the RP4 plasmid (yellow crosses). The balls on the proteins indicate transmembrane segments. IM, inner membrane; OM, outer membrane. Adapted in part from Kalkum et al. (35).
Maturation of TrbC from a 145-residue precursor to a 78-residue circular protein involves three proteolytic cleavages and a cyclization event (Fig. 5) (21). In the first instance, a 27-amino-acid peptide is removed from the C terminus of the precursor by an as yet unidentified enzyme. The subsequent removal of a 36-amino-acid N-terminal signal peptide is performed by LepB, a chromosomally encoded signal peptidase I, to generate a protein that corresponds to linear TrbC with a C-terminal tetrapeptide. The ultimate cleavage and cyclization are attributed to TraF, a plasmid-encoded protein homologous to the leader peptidases. Mutation studies of TraF at residues corresponding to those conserved in leader peptidases suggested that, like these serine proteases, TraF functions via a Ser-Lys catalytic dyad (22).
A putative cyclization mechanism has been proposed in which the final cleavage and peptide bond formation occur as a single concerted event via an acyl intermediate catalyzed by TraF, transferring the energy released from the cleavage reaction directly to the formation of a peptide bond. The absence of linear TrbC molecules and the presence of the tetrapeptide in cyclization-deficient mutants support this contention, but the involvement of an extraneous enzyme or alternative mechanism cannot be discounted. It is interesting that no linear analogues identical in sequence to any naturally occurring circular proteins have been isolated. Both TrbC and TraF span the cytoplasmic membrane during the course of their interaction, most probably to optimize contact between the relevant residues. In other circular proteins, disulfide bonds and a tight globular structure may supplant the need for membrane interaction.
VirB, the T pilin precursor, consists of a 47-residue N-terminal signal peptide followed by the sequence of the mature pilin protein (74 residues) (33). As in the TrbC system, proteolytic cleavage of the signal peptide is also carried out by a chromosomally encoded peptidase similar to LepB. However no TraF homolog is present on the Ti plasmid. Interestingly, cyclization of T pilin was found to occur in Agrobacterium tumefaciens but not in E. coli, indicating that the enzyme or mechanism responsible for cyclization is of chromosomal origin and species specific (43).
The diversity of both bacterial and nonbacterial precursor sequences suggests that a single mechanism of cyclization is unlikely to be common to all circular proteins. However, certain parallels exist between the biosynthetic pathways of different circular bacterial proteins. The similarity of precursor organization, the opportunity for coupled cleavage and cyclization events, and the common accessory components, such as membrane transporters, often coexpressed with the respective proteins may point towards a common approach to cyclization, albeit one which has become species specialized over evolutionary time. Alternatively, the substantial benefits to protein stability associated with forming that final peptide bond may have been realized independently numerous times and culminated in the existence of multiple mechanisms to achieve the same end.
The biosynthesis of circular proteins from plant and animal cells is not as well understood as it is in bacterial systems and is complicated by the lack of an efficient expression system. It will be interesting to follow developments in the characterization of components involved in the cyclization of bacterial proteins and to access the potential for any “cross-reactivity” between the novel processing proteins of circular proteins from different sources.
OTHER NATURALLY OCCURRING CIRCULAR PROTEINS
The currently known circular proteins from higher organisms all contain one or more disulfide bonds, which contrasts with the absence of such bonds in bacterial circular proteins. They range in size from 14 to approximately 30 amino acids, as indicated in Fig. 6. The largest family of circular proteins in higher organisms is the plant cyclotides (11). They are typically about 30 amino acids in size, contain an N- to C-cyclized backbone, and incorporate three disulfide bonds arranged in a cystine knot topology (12). In this motif, an embedded ring in the structure formed by two disulfide bonds and their connecting backbone segments is penetrated by the third disulfide bond. The combination of this knotted and strongly braced structure with a circular backbone renders the cyclotides impervious to enzymatic breakdown and makes them exceptionally stable. The cyclotides have a diverse range of biological activities, ranging from uterotonic action to anti-human immunodeficiency virus and neurotensin antagonism (13).
FIG. 6.
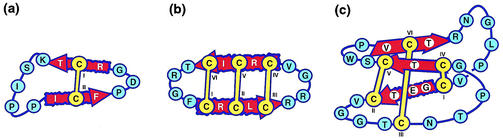
Schematic representations of sequences and structures of nonbacterial circular proteins, including (a) SFTI-1, (b) RTD-1, and (c) kalata B1 from the cyclotide family. Sequences are represented by the single-letter amino acid code, and the disulfide bonds are highlighted by yellow lines connecting the cysteine residues. β-Sheets are shown as red arrows.
Recently, another group of Cys-rich macrocyclic peptides, MCoTI-I and MCoTI-II, from a plant in the Cucurbitaceae family (29) were discovered. Unlike the previously reported cyclotides, these molecules have trypsin-inhibitory activity. The three-dimensional structure of MCoTI-II has recently been determined and contains a cyclic cystine knot motif (23, 28), suggesting that these Cucurbitaceae peptides are related to the cyclotide family. SFTI-1 is a much smaller cyclic peptide from plants that also has trypsin-inhibitory activity. It contains just a single disulfide bond, as illustrated in Fig. 6.
The only circular peptide so far directly discovered in animals is RTD-1 (56) and its homologues RTD-2 and RTD-3, found in rhesus monkey leucocytes. Very recently it was reported that human bone marrow also expresses a pseudogene that apparently encodes an antimicrobial peptide, retrocyclin, similar in sequence to RTD-1 (10). These molecules are like the cyclotides in that they have a circular backbone and three disulfide bonds, but differ in being about half the size of the cyclotides and having a laddered rather than a knotted arrangement of the disulfide bonds. A schematic illustration of the structures of representative circular proteins from higher organisms is shown in Fig. 6. As noted earlier, relatively little is known about biosynthesis of the disulfide-containing cyclic peptides from higher organisms, but the potential complexity is illustrated by the fact that the 18-amino-acid peptide RTD-1 is the product of two genes and two head-to-tail ligation reactions of the encoded 9-amino-acid peptides (56).
ROLE OF THE CIRCULAR BACKBONE
One obvious question that is raised by the discovery of macrocyclic peptides in various organisms is what the advantages are of such a modification. Intuitively, the answer involves improving the stability of peptides by removing possible sites for exoproteases and constraining the conformation of the termini, leading to an entropic advantage in binding interactions. Recent studies on the influences of linearization on naturally occurring circular proteins have suggested both a structural and functional role for the cyclic backbone.
Cleaving AS-48 with cyanogen bromide results in a linear form that is unable to maintain the native structure (8). Based on this result, it appears that the cyclic backbone is required to maintain the three-dimensional structure of AS-48. However, as discussed earlier, AS-48 was opened by hydrolyzing the Tyr70-Met1 bond, which is in the middle of α-helix 5. Cleaving in a different region, such as a loop, may result in retention of the overall structure. Further analogues are required before this can be properly assessed. However, the results do suggest that α-helix 5 itself is crucial for maintaining the structure and that cyclization appears to have a significant structural role, as it occurs specifically in an element of secondary structure and not in a loop region.
Studies on the effects of breaking the backbone of nonbacterial circular proteins have suggested that the cyclic backbone is not essential for maintaining the overall fold in these cases. Synthetic linear derivatives of RTD-1 (56, 59), SFTI-1 (42), and the cyclotides (14, 15) all retain some elements of native secondary structure. The additional restraints of the disulfide bonds in the nonbacterial circular proteins are likely to play a major role in maintaining the overall structure. Studies on the naturally occurring macrocyclic trypsin inhibitor MCoTI-II (29) also suggest that the circular backbone is not required to maintain the overall fold, as linear homologues with similar structures exist in nature (29). Furthermore, the loop that may be regarded as the linker region that joins the termini to form the cyclic structure is disordered in the NMR-derived structures of MCoTI-II (23, 28). This disorder suggests that the role of cyclization in this particular case is not to rigidify the structure, as might be imagined. Preventing attack by exoproteases has been suggested as a significant role for the circular backbone in this molecule (23, 28)
Analyses of structural stability have also provided information on the role of the circular backbone. AS-48 was found to be extremely resistant to heat- and denaturant-induced unfolding (9). It was shown to denature only when the temperature reached 102°C, and at low temperature it did not unfold even in 8 M urea. It appears that the circular backbone is responsible for this stability, as the other structural features are quite standard for a protein of this size (9). Because of the lack of structure in the linear form of AS-48, it was not possible to perform a rigorous analysis of the thermodynamic contributions of the circular backbone. However, this has been possible for an artificially generated cyclic PIN1 WW domain, which has been shown to posses an improved thermodynamic stability compared to the linear wild type (16). This protein, which comprises 34 amino acids, adopts a triple-stranded β-sheet structure in which the two termini are close together (≈10 Å apart) on one face of the molecule. In this study, it was concluded that the size of the linker used for cyclization must be optimal to prevent the introduction of strain, which would destabilize the native fold. For an optimal linker, a stabilization of up to 1.7 kcal/mol was reported.
In addition to influences on the overall fold and stability, the cyclic backbone also affects biological activity. Linear analogues of RTD-1, SFTI-1, and the cyclotides all show decreased biological activities relative to the native peptides. This indicates that the circular backbone is critical for maintaining the native level of activity. Overall, while the role of the circular backbone is by no means fully understood, it appears to be involved in improving stability and biological activity and in some cases may be involved in the structural integrity.
CONCLUDING REMARKS
Although circular proteins have been discovered only over the last decade, they are now found in a wide range of organisms, and it is likely that many more will be discovered in the next few years. Circular proteins of bacterial origin adopt well-defined three-dimensional structures and have a high degree of thermal stability and resistance to denaturation by chaotropes. It is clear from examples such as AS-48 that circular proteins behave very much like conventional proteins in terms of their three-dimensional structures, i.e., that the sequence contains all of the intrinsic information required for folding into a defined three-dimensional shape. This is illustrated by the high fidelity of protein structure prediction in the case of AS-48. However, there remain some fascinating questions on the structural biology of circular proteins, including how and why the mechanism of cyclization evolved.
Acknowledgments
This work was supported in part by a grant from the Australian Research Council (D.J.C.). D.J.C. is an Australian Research Council Senior Fellow.
REFERENCES
- 1.Abriouel, H., M. Maqueda, A. Gálvez, M. Martínez-Bueno, and E. Valdívia. 2002. Inhibition of bacterial growth, enterotoxin production, and spore outgrowth in strains of Bacillus cereus by bacteriocin AS-48. Appl. Environ. Microbiol. 68:1473-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abriouel, H., E. Valdívia, A. Gálvez, and M. Maqueda. 1998. Response of Salmonella choleraesuis LT2 spheroplasts and permeabilized cells to the bacteriocin AS-48. Appl. Environ. Microbiol. 64:4623-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, M., T. Curstedt, H. Jornvall, and J. Johansson. 1995. An amphipathic helical motif common to tumourolytic polypeptide NK-lysin and pulmonary surfactant polypeptide SP-B. FEBS Lett. 362:328-332. [DOI] [PubMed] [Google Scholar]
- 4.Blond, A., M. Cheminant, D. Destoumieux-Garzón, I. Ségalas-Milazzo, J. Péduzzi, C. Goulard, and S. Rebuffat. 2002. Thermolysin-linearized microcin J25 retains the structured core of the native macrocyclic peptide and displays antimicrobial activity. Eur. J. Biochem. 269:6212-6222. [DOI] [PubMed] [Google Scholar]
- 5.Blond, A., M. Cheminant, I. Ségalas-Milazzo, J. Péduzzi, M. Barthélémy, C. Goulard, R. Salomón, F. Moreno, R. Farías, and S. Rebuffat. 2001. Solution structure of microcin J25, the single macrocyclic antimicrobial peptide from Escherichia coli. Eur. J. Biochem. 268:2124-2133. [DOI] [PubMed] [Google Scholar]
- 6.Blond, A., J. Péduzzi, C. Goulard, M. J. Chiuchiolo, M. Barthélémy, Y. Prigent, R. A. Salomón, R. N. Farías, F. Moreno, and S. Rebuffat. 1999. The cyclic structure of microcin J25, a 21-residue peptide antibiotic from Escherichia coli. Eur. J. Biochem. 259:747-755. [DOI] [PubMed] [Google Scholar]
- 7.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobos, E. S., V. V. Filimonov, A. Gálvez, M. Maqueda, E. Valdívia, J. C. Martínez, and P. L. Mateo. 2001. AS-48: a circular protein with an extremely stable globular structure. FEBS Lett. 505:379-382. [DOI] [PubMed] [Google Scholar]
- 9.Cobos, E. S., V. V. Filimonov, A. Gálvez, E. Valdívia, M. Maqueda, J. C. Martínez, and P. L. Mateo. 2002. The denaturation of circular enterocin AS-48 by urea and guanidinium hydrochloride. Biochim. Biophys. Acta 1598:98-107. [DOI] [PubMed] [Google Scholar]
- 10.Cole, A. M., T. Hong, L. M. Boo, T. Nguyen, C. Zhao, G. Bristol, J. A. Zack, A. J. Waring, O. O. Yang, and R. I. Lehrer. 2002. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA 99:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craik, D. J., N. L. Daly, T. Bond, and C. Waine. 1999. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294:1327-1336. [DOI] [PubMed] [Google Scholar]
- 12.Craik, D. J., N. L. Daly, and C. Waine. 2001. The cystine knot motif in toxins and implications for drug design. Toxicon 39:43-60. [DOI] [PubMed] [Google Scholar]
- 13.Craik, D. J., S. Simonsen, and N. L. Daly. 2002. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 5:251-260. [PubMed] [Google Scholar]
- 14.Daly, N. L., and D. J. Craik. 2000. Acyclic permutants of naturally occurring cyclic proteins. Characterization of cystine knot and beta-sheet formation in the macrocyclic polypeptide kalata B1. J. Biol. Chem. 275:19068-19075. [DOI] [PubMed] [Google Scholar]
- 15.Daly, N. L., S. Love, P. F. Alewood, and D. J. Craik. 1999. Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry 38:10606-10614. [DOI] [PubMed] [Google Scholar]
- 16.Deechongkit, S., and J. W. Kelly. 2002. The effect of backbone cyclization on the thermodynamics of beta-sheet unfolding: stability optimization of the PIN WW domain. J. Am. Chem. Soc. 124:4980-4986. [DOI] [PubMed] [Google Scholar]
- 17.Delgado, M. A., M. R. Rintoul, R. N. Farías, and R. A. Salomón. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado, M. A., J. O. Solbiati, M. J. Chiuchiolo, R. N. Farías, and R. A. Salomón. 1999. Escherichia coli outer membrane protein TolC is involved in production of the peptide antibiotic microcin J25. J. Bacteriol. 181:1968-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Destoumieux-Garzón, D., J. Péduzzi, and S. Rebuffat. 2002. Focus on modified microcins: structural features and mechanisms of action. Biochimie 84:511-519. [DOI] [PubMed] [Google Scholar]
- 20.Diaz, M., E. Valdívia, M. Martínez-Bueno, M. Fernandez, A. S. Soler-González, H. Ramirez-Rodrigo, and M. Maqueda. 2003. Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 69:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eizenbrandt, R., M. Kalkum, E. M. Lai, R. Lurz, C. I. Kado, and E. Lanka. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274:22548-22555. [DOI] [PubMed] [Google Scholar]
- 22.Eizenbrandt, R., M. Kalkum, R. Lurz, and E. Lanka. 2000. Maturation of IncP pilin precursors resembles the catalytic Dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felizmenio-Quimio, M. E., N. L. Daly, and D. J. Craik. 2001. Circular proteins in plants: solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J. Biol. Chem. 276:22875-22882. [DOI] [PubMed] [Google Scholar]
- 24.Gálvez, A., M. Maqueda, M. Martínez-Bueno, and E. Valdívia. 1989. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against gram-positive and gram-negative bacteria and other organisms. Res. Microbiol. 140:57-68. [DOI] [PubMed] [Google Scholar]
- 25.Gálvez, A., M. Maqueda, M. Martínez-Bueno, and E. Valdívia. 1991. Permeation of bacterial cells, permeation of cytoplasmic and artificial membrane vesicles, and channel formation on lipid bilayers by peptide antibiotic AS-48. J. Bacteriol. 173:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González, C., G. M. Langdon, M. Bruix, A. Gálvez, E. Valdívia, M. Maqueda, and M. Rico. 2000. Bacteriocin AS-48, a microbial cyclic polypeptide structurally and functionally related to mammalian NK-lysin. Proc. Natl. Acad. Sci. USA 97:11221-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heitz, A., J. F. Hernandez, J. Gagnon, T. T. Hong, T. T. Pham, T. M. Nguyen, D. Le-Nguyen, and L. Chiche. 2001. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry 40:7973-7983. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez, J. F., J. Gagnon, L. Chiche, T. M. Nguyen, J. P. Andrieu, A. Heitz, T. Trinh Hong, T. T. Pham, and D. Le Nguyen. 2000. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39:5722-5730. [DOI] [PubMed] [Google Scholar]
- 30.Iwai, H., A. Lingel, and A. Pluckthun. 2001. Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J. Biol. Chem. 276:16548-16554. [DOI] [PubMed] [Google Scholar]
- 31.Iwai, H., and A. Pluckthun. 1999. Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 459:166-172. [DOI] [PubMed] [Google Scholar]
- 32.Jennings, C., J. West, C. Waine, D. J. Craik, and M. A. Anderson. 2001. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 98:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, A. L., E. M. Lai, K. Shirasu, and C. I. Kado. 1996. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 178:5706-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabuki, T., T. Saito, Y. Kawai, J. Uemura, and T. Itoh. 1997. Production, purification and characterization of reutericin 6, a bacteriocin with lytic activity produced by Lactobacillus reuteri LA6. Int. J. Food Microbiol. 34:145-156. [DOI] [PubMed] [Google Scholar]
- 35.Kalkum, M., R. Eizenbrandt, R. Lurz, and E. Lanka. 2002. Tying rings for sex. Trends Microbiol. 10:382-387. [DOI] [PubMed] [Google Scholar]
- 36.Kawai, Y., Y. Ishii, J. Uemura, H. Kitazawa, T. Saito, and T. Itoh. 2001. Lactobacillus reuteri LA6 and Lactobacillus gasseri LA39 isolated from faeces of the same human infant produce identical cyclic bacteriocin. Food Microbiol. 18:407-415. [Google Scholar]
- 37.Kawai, Y., T. Saito, H. Kitazawa, and T. Itoh. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 62:2438-2440. [DOI] [PubMed] [Google Scholar]
- 38.Kawai, Y., T. Saito, M. Suzuki, and T. Itoh. 1998. Sequence analysis by cloning of the structural gene of gassericin A, a hydrophobic bacteriocin produced by Lactobacillus gasseri LA39. Biosci. Biotechnol. Biochem. 62:887-892. [DOI] [PubMed] [Google Scholar]
- 39.Kawai, Y., T. Saito, T. Toba, S. K. Samant, and T. Itoh. 1994. Isolation and characterization of a highly hydrophobic new bacteriocin (gassericin A) from Lactobacillus gasseri LA39. Biosci. Biotechnol. Biochem. 58:1218-1221. [DOI] [PubMed] [Google Scholar]
- 40.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 41.Kohli, R. M., and C. T. Walsh. 2003. Enzymology of acyl chain macrocyclization in natural product biosynthesis. Chem. Commun. (Cambridge) 2003:297-307. [DOI] [PubMed]
- 42.Korsinczky, M. L., H. J. Schirra, K. J. Rosengren, J. West, B. A. Condie, L. Otvos, M. A. Anderson, and D. J. Craik. 2001. Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J. Mol. Biol. 311:579-591. [DOI] [PubMed] [Google Scholar]
- 43.Lai, E. M., R. Eizenbrandt, M. Kalkum, E. Lanka, and C. I. Kado. 2002. Biogenesis of T pili in Agrobacterium tumefaciens requires precize VirB2 propilin cleavage and cyclization. J. Bacteriol. 184:327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai, E. M., and C. I. Kado. 2002. The Agrobacterium tumefaciens T pilus composed of cyclic T pilin is highly resilient to extreme environments. FEMS Microbiol. Lett. 210:111-114. [DOI] [PubMed] [Google Scholar]
- 45.Lai, E. M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 46.Leer, R. J., J. M. van der Vossen, M. van Giezen, J. M. van Noort, and P. H. Pouwels. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629-1635. [DOI] [PubMed] [Google Scholar]
- 47.Martínez-Bueno, M., M. Maqueda, A. Gálvez, B. Samyn, J. Van Beeumen, J. Coyette, and E. Valdívia. 1994. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez-Bueno, M., E. Valdívia, A. Gálvez, J. Coyette, and M. Maqueda. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27:347-358. [DOI] [PubMed] [Google Scholar]
- 49.McManus, A. M., N. F. Dawson, J. D. Wade, L. E. Carrington, D. J. Winzor, and D. J. Craik. 2000. Three-dimensional structure of RK-1: a novel alpha-defensin peptide. Biochemistry 39:15757-15764. [DOI] [PubMed] [Google Scholar]
- 50.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 51.Pillardy, J., C. Czaplewski, A. Liwo, J. Lee, D. R. Ripoll, R. Kazmierkiewicz, S. Oldziej, W. J. Wedemeyer, K. D. Gibson, Y. A. Arnautova, J. Saunders, Y. J. Ye, and H. A. Scheraga. 2001. Recent improvements in prediction of protein structure by global optimization of a potential energy function. Proc. Natl. Acad. Sci. USA 98:2329-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rintoul, M. R., B. F. de Arcuri, R. A. Salomón, R. N. Farías, and R. D. Morero. 2001. The antibacterial action of microcin J25: evidence for disruption of cytoplasmic membrane energization in Salmonella newport. FEMS Microbiol. Lett. 204:265-270. [DOI] [PubMed] [Google Scholar]
- 53.Rosengren, K. J., A. M. McManus, and D. J. Craik. 2002. The structural and functional diversity of naturally occurring antimicrobial peptides. Curr. Med. Chem. Anti-infect. Agents 1:319-341. [Google Scholar]
- 54.Salomón, R. A., and R. N. Farías. 1992. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solbiati, J. O., M. Ciaccio, R. N. Farías, J. E. González-Pastor, F. Moreno, and R. A. Salomón. 1999. Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J. Bacteriol. 181:2659-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang, Y. Q., J. Yuan, G. Osapay, K. Osapay, D. Tran, C. J. Miller, A. J. Ouellette, and M. E. Selsted. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 286:498-502. [DOI] [PubMed] [Google Scholar]
- 57.Toba, T., S. K. Samant, E. Yoshioka, and T. Itoh. 1991. Reutericin 6, a new bacteriocin produced by Lactobacillus reuteri LA6. Lett. Appl. Microbiol. 13:281-286. [Google Scholar]
- 58.Trabi, M., and D. J. Craik. 2002. Circular proteins-no end in sight. Trends Biochem. Sci. 27:132-138. [DOI] [PubMed] [Google Scholar]
- 59.Trabi, M., H. J. Schirra, and D. J. Craik. 2001. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from Rhesus macaque leukocytes. Biochemistry 40:4211-4221. [DOI] [PubMed] [Google Scholar]
- 60.van Belkum, M. J., and M. E. Stiles. 2000. Nonlantibiotic antibacterial peptides from lactic acid bacteria. Nat. Prod. Rep. 17:323-335. [DOI] [PubMed] [Google Scholar]
- 61.Waters, V. L. 2001. Conjugation between bacterial and mammalian cells. Nat. Genet. 29:375-376. [DOI] [PubMed] [Google Scholar]
- 62.Yuzenkova, J., M. Delgado, S. Nechaev, D. Savalia, V. Epshtein, I. Artsimovitch, R. A. Mooney, R. Landick, R. N. Farias, R. Salomon, and K. Severinov. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin j25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]