Abstract
Bacillus subtilis PurR represses transcription of several genes involved in purine synthesis, metabolism, and transport and cofactor synthesis. PurR binds specifically to DNAs containing an inverted repeat of a 14-nucleotide “PurBox” located in the upstream control regions of genes in the PurR regulon. Further biochemical investigation of the interaction of PurR with a series of shortened upstream DNA fragments of the pur operon determined the minimum length and specificity elements of the operator. The relative affinities of the two PurBoxes differ significantly, such that upstream PurBox1 (−81 to −68 relative to the transcription start site) is designated “strong” and downstream PurBox2 (−49 to −36) is designated “weak.” Two PurBoxes are required for high-affinity PurR binding, and one of these must be strong. The shortest DNA construct with high affinity for PurR is a 74-bp perfect palindrome in which weak PurBox2 and its flanking sequences are replaced by strong PurBox1 and flanking sequences. Two PurR dimers bind to this symmetric construct. Phosphoribosylpyrophosphate (PRPP), the effector molecule that reduces affinity of PurR for DNA, requires one weak PurBox in the DNA construct to inhibit PurR binding. PRPP binds, as expected, to a PRPP-motif in PurR. A tracks outside the central conserved CGAA sequence of the PurBox may facilitate DNA bending, leading to a proposal for strong and weak designations of PurBoxes in the control regions of other genes regulated by PurR.
The Bacillus subtilis purR gene product is a negative regulator of expression of several genes relevant to purine biosynthesis. The purR regulon includes most genes encoding purine biosynthetic enzymes (the pur operon and purA), purR itself, and genes for purine transport (pbuG, pbuO, and pbuX) and metabolism (xpt and guaC) and cofactor biosynthesis (folD and glyA) (1, 13, 19). Cells grown in minimal medium supplemented with adenine produce pur operon-encoded proteins at 15- to 30-fold lower levels than when grown in medium without adenine (1, 13, 19). In all cases, expression of regulated genes is increased in purR strains and is unaffected by adenine supplementation.
The PurR protein is a 62-kDa homodimer with high affinity for large segments of DNA within the control regions of the pur, purA, purR, and pyr operons (2, 15, 19). Footprinting and deletion analyses have shown that PurR interacts with a region between nucleotides −147 and −21 relative to the transcription start site of the pur operon (15, 19, 20). DNase I protection experiments revealed a pattern of protected and hypersensitive regions with approximately 10-bp spacing, leading to the suggestion that DNA may wrap around PurR (2, 15, 19). Support for this hypothesis was obtained from experiments showing that PurR induces right-handed supercoils in DNA and that multiple PurR dimers bind control region DNA (15). The determinants of binding specificity within the large control region are unknown apart from a critical tetranucleotide between −75 and −72 with the sequence GAAC. Identical tetranucleotides occur as inverted repeats separated by ∼24 nucleotides in all four control regions known to bind PurR. However, the downstream tetranucleotide does not appear to be critical to PurR binding (15).
Transcription of the Lactococcus lactis pur operon (5) is activated by a regulatory protein, also named PurR due to its having 50% sequence identity with the B. subtilis PurR. Based on genetic data and comparison of sequences upstream of regulated genes, L. lactis PurR was proposed to activate pur transcription by binding a 13-nucleotide, cis-acting PurBox with the consensus sequence 5′-AWWWCCGAACWWT-3′, where “W” indicates an A or T nucleotide (4). In B. subtilis, the −75GAAC−72 sequence that is critical to PurR control of pur transcription is located within a consensus PurBox. The purR regulon was expanded beyond purine biosynthetic genes by sequence analysis of all promoter regions in the B. subtilis genome (13). Wherever inverted pairs of potential 14-nucleotide PurBoxes were found in promoter regions with 16- to 17-nucleotide spacing, the downstream genes were shown to be regulated by purR. In contrast, the PurBoxes in L. lactis occur as singles or as tandem repeats.
The affinity of B. subtilis PurR for control region DNA is influenced by an effector molecule, as expected for a bacterial repressor. Among all nucleotides and nucleotide derivatives tested, only 5-phosphoribosyl-1-pyrophosphate (PRPP) was found to affect the DNA affinity of PurR (19). PRPP is a central metabolite of nucleotide synthesis and salvage as well as being the starting material for purine biosynthesis. PRPP inhibits PurR-DNA binding in vitro and thus may be a transcription inducer in vivo. The addition of adenine to the B. subtilis culture medium diminishes the cellular pool of PRPP (10). This effect is thought to be responsible for the observed decrease in pur transcription by adenine in vivo. PurR contains a 12-residue, PRPP-binding motif that is found in members of the homologous PRT family (17). Most PRT proteins are phosphoribosyltransferases, enzymes of nucleotide synthesis or salvage that have PRPP as a substrate. Thus, PurR may have evolved from a nucleotide enzyme. The crystal structure of PurR (see reference 16) confirms that it is a member of the homologous PRT family. In addition to the PRT domain, PurR has an N-terminal winged-helix domain. Winged-helix domains are a subfamily of the helix-turn-helix (HTH) family, whose members are widely deployed for sequence-specific DNA binding. A recognition helix in the HTH motif is generally responsible for DNA sequence recognition.
Here we report the results of biochemical studies to investigate in more detail the interaction between PurR and the pur control region. Different affinities of the two PurBoxes for PurR led to construction of a minimal 74-nucleotide sequence with two strong-binding PurBoxes and with high affinity for the protein. One weak-binding PurBox is required for PRPP interference. The crystal structure of an analog complex shows that PRPP binds PurR at the expected site. The symmetric 74-nucleotide DNA sequence binds two PurR dimers.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains DH5α, SURE (Stratagene), and B834(DE3) were used for plasmid construction, for growth of plasmids containing multiple copies of DNA constructs, and for purR overexpression, respectively. DNAs containing different lengths of the pur operon upstream control region were generated by PCR from plasmid pDE264 (2). HindIII and EcoRI restriction sites were engineered at the ends of PCR primers (Table 1). PCR products were digested with HindIII and EcoRI and were used to replace the corresponding fragment of pET24a+.
TABLE 1.
Primers used in this studya
| Primer | Sequence (5′ to 3′) |
|---|---|
| Primers for upstream sites with a HindIII restriction site | |
| Pri−177F | TCTGCCTGTAAGCTTAATGGAAGAGCCTATTTTTATGG |
| Pri−143F | GAAAAAAAGCTTAATGGGTCAATTCAGATCG |
| Pri−123F | ATTCAGAAGCTTCCGTGCGGGAAAAAAATCG |
| Pri−91F | GTATTTGAAGCTTAATTGATCTAAAACACG |
| Pri−83F | ATTCAGAAGCTTAAAACACGAACATTAGTAG |
| Pri−75F | TGATCTAAAGCTTGAACATTAGTAGAATGAATTTTTG |
| Pri−69F | AAACACGAAGCTTAGTAGAATGAATTTTTG |
| Pri−54F | AGAATGAAGCTTTGTATCGTTCGATAATATCG |
| Primers for downstream sites with an EcoRI restriction site | |
| Pri+24R | TCCCAGAATTCTATGAGGTCGTGTTTTG |
| Pri−6R | TTCAGAATTCTCTTAACAACGGACATG |
| Pri−31R | CATGGATGAATTCAACGATATTATCGAACG |
| Pri−42R | CAACGGAATTCTCGAACGATACAAAAATTC |
| Pri−60R | ATACAAGAATTCATTCTACTAATGTTCGTG |
| Pri−75R | CTACTAGAATTCGTGTTTTAGATCAATTTC |
Restriction sites are underlined.
Isolation and labeling of DNA.
DNA fragments were purified from agarose gel (QIAGEN), dephosphorylated with alkaline phosphatase, labeled with [γ-32P]ATP by use of T4 polynucleotide kinase, and repurified. In some cases, nonspecific plasmid DNA was added to the DNA fragment by digestion with XhoI instead of HindIII, which added 15 extra bp to the 5′ end or with NdeI instead of EcoRI, which added 46 extra bp to the 3′ end.
Purification of PurR protein.
The NdeI-HindIII fragment of pR6H (15) was subcloned into pET24a+. Plasmid pET24PurR6H was transformed into E. coli B834(DE3). To increase the amount of protein in the soluble cell extract, the expression and lysis conditions were modified slightly from those described previously (15). Transformed cells were grown in Luria-Bertani medium with kanamycin for 4 h at 37°C and then for 16 h at 30°C. Cells from a 1-liter culture were harvested by centrifugation and resuspended in 20 ml of buffer A (50 mM sodium phosphate [pH 8.0], 500 mM NaCl, 15 mM imidazole) containing 1% phenylmethylsulfonyl fluoride. Cells were disrupted by three passages through a French press at 10,000 lb/in2. The resulting extract was clarified by centrifugation at 18,000 × g for 30 min, and 25 ml of a 50% suspension of Ni2+ nitrilotriacetic acid-agarose (QIAGEN) equilibrated with buffer A was added to the supernatant. The suspension was stirred gently for 30 min and then poured into a 1.5-cm-diameter column. The column was washed sequentially with 200 ml of buffer A and 200 ml of 50 mM imidazole in buffer B (50 mM sodium phosphate [pH 6.0], 500 mM NaCl, 10% glycerol). Bound PurR was eluted in a 200-ml gradient of 50 to 300 mM imidazole in buffer B. Fractions were analyzed by electrophoresis on sodium dodecyl sulfate-12% polyacrylamide gels. Fractions of the essentially pure protein were combined and dialyzed against 10 mM HEPES (pH 8.0)-50 mM (NH4)2SO4-300 mM NaCl. Purified PurR was concentrated by ultrafiltration to approximately 10 mg/ml and frozen at −20°C in 10- to 50-μl aliquots. The yield was ∼15 mg of 95% pure PurR from 1 liter of culture.
Protein-DNA binding.
Gel retardation assays were carried out as described previously (15). A typical 20-μl assay mixture contained 10 mM HEPES (pH 7.6), 50 mM KCl, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 5 mM dithiothreitol, 5 μg of double-stranded dI-dC per ml, approximately 10 fmol of 32P-end-labeled DNA probe, and PurR protein. After incubation for 20 min at room temperature, 10 μl of the mixture was loaded onto a 5% polyacrylamide gel in 90 mM Tris borate (pH 8.3)-2 mM EDTA and electrophoresed for about 1 h at 10 V/cm. Dried gels were visualized by autoradiography and quantitated with an InstantImager (Packard). Apparent Kd values, which represent the PurR concentrations required for 50% saturation, were calculated as described previously (15). The PurR molar concentration was based on the 32-kDa subunit. PRPP inhibition of PurR-DNA binding was measured by gel retardation, using 47 nM or 150 nM PurR and a series of increasing PRPP concentrations. In a control sample, the radioactivity of bound DNA in the absence of PRPP (bound PRPP(0)) was used to establish the 0% inhibition point. In samples containing PRPP, total DNA was the sum of counts in shifted and unshifted bands. Inhibition (%) = 100 × [unbound (%) − unbound PRPP(0) (%)]/bound PRPP(0) (%).
Construction and large-scale purification of the 74-bp DNA palindrome.
A 37-bp DNA fragment was designed to encompass one-half of the desired palindromic DNA sequence—positions −93 to −60 of the pur control region with modifications at the ends to generate EcoRV and PstI restriction sites (Figure 1). Plasmid p1/2pur37 was constructed by inserting the 37-bp fragment into the EcoRV and PstI sites of a pUC18 derivative containing an EcoRI-BamHI-EcoRV-PstI-BglII-HindIII polylinker region. Tandem copies of the insert were created by the method of Tan et al. (18). Two features of the experimental design were critical to its success. First, duplication of inserts was ensured by the use of flanking restriction sites (BamHI and BglII) whose sticky ends can be ligated to produce a noncleavable product. Second, the half-palindrome had a sticky-end restriction site (PstI) on the end that became the center of the palindrome and a blunt-end site (EcoRV) on the other end. Plasmids containing 32 tandem repeats of the half-palindrome (i.e., p1/2pur37 × 32) were stable in the E. coli SURE strain, but those with 64 copies of the insert were unstable. Plasmid p1/2pur37 × 32 was digested with EcoRV to release the 37-bp half-palindrome with a 12-nucleotide spacer DNA, which was removed by digestion with PstI. The 74-bp palindrome was formed by self-ligation of the 37-bp fragment that was digested again with EcoRV to eliminate any multimers. The 37-bp (EcoRV- and PstI-digested insert), 49-bp (EcoRV-digested insert) and 74-bp (37-bp ligated insert) DNA fragments were purified on a 5-ml HiTrap Q HP ion-exchange column (Pharmacia) by using about 100 ml of a 300 to 1,500 mM sodium chloride gradient.
FIG. 1.
Construct for generating tandem repeats of pur half-palindrome. The 37-bp control region is shown in boldface; PurBox1 is shaded. Restriction sites are boxed and labeled; nucleotides changed to create restriction sites are underlined.
PurR-DNA binding stoichiometry.
PurR (300 μg) was incubated at room temperature for 15 min with or without the 74-bp DNA palindrome. A 10 to 30% sucrose gradient in binding buffer (10 mM HEPES [pH 7.6], 50 mM KCl, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 5 mM dithiothreitol) was made in 12-ml tubes by using a two-chamber linear gradient maker. Samples were loaded on top of the gradient and centrifuged for 15 h at 280,000 × g at 25°C by using an SW-41 Ti rotor in an L-7 55 ultracentrifuge (Beckman). The gradient was fractionated and analyzed by UV absorbance at 260 nm (ÅKTA; Pharmacia). Fractionated samples were analyzed for protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and for free and bound DNA by 5% native polyacrylamide gel electrophoresis.
Crystallography.
PurR and 1-α-pyrophosphoryl-2,3-α-dihydroxy-4-β-cyclopentane-methanol-5-phosphate (cPRPP) were cocrystallized at 20°C by vapor diffusion of a 1:1 mixture of protein solution (10 mg of PurR per ml, 10 mM HEPES [pH 8.0], 300 mM NaCl, 50 mM (NH4)2SO4, 10 mM MgCl2, 5 mM cPRPP) and well solution (100 mM HEPES [pH 7.5], 200 mM CaCl2, 28% polyethylene glycol 400). A crystal (dimensions, 0.1 by 0.1 by 0.1 mm) was removed from the growth solution and frozen directly in an N2 stream at 100 K. Diffraction data were collected at beamline ID-14 at the Advanced Photon Source and processed with HKL2000 (11). The space group was P21 (a, 58.4 Å; b, 135.7 Å; c, 82.1 Å; β, 95.1°) with two PurR dimers per asymmetric unit. The crystal form and crystallization conditions are unrelated to those of free PurR (16). The 2.22-Å data set was 98.3% complete with an overall Rsym of 5.5% (31.8% in the outermost shell). The structure was solved by molecular replacement using AMoRe (8) from the structure of free PurR (16). Following refinement with REFMAC5 (7), the final model includes two dimers of PurR, four molecules of cPRPP, and 574 waters. The cPRPP sites were not fully occupied in any of the four subunits. Electron density was especially weak for the cyclopentane ring of cPRPP, corresponding to ∼50% occupancy. Incomplete occupancy by cPRPP is not due to peculiarities of crystallization solutions; the conditions for crystal growth and for the PRPP interference experiments were similar with respect to buffer, pH, and divalent cation. Growth of this crystal form required cPRPP or PRPP in the protein solution, and there was no evidence that cPRPP bound elsewhere to the protein. cPRPP is an excellent PRPP analog and binds like PRPP to other PRT proteins (6). The protein model is complete except for residues 162 to 168 in a flexible loop and 8 to 10 residues at the C terminus, for which no density was visible. The final Rwork was 18.0%, Rfree was 23.4%, covalent bonds deviated from ideality by 0.009 Å (root mean square [RMS]), and 92% of the protein residues were in the most favorable regions of the Ramachandran plot. The refined model was deposited in the Protein Data Bank as entry 1p4a.
RESULTS
Mapping the boundaries of the PurR binding site.
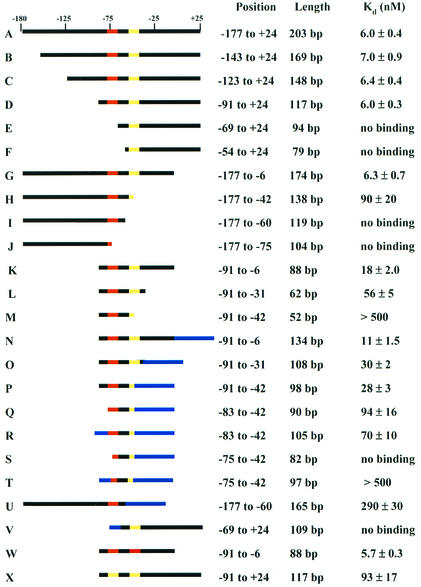
Previously, DNA recognition elements for PurR binding were localized to a region 20 to 150 bp upstream of the pur operon (15). Pseudopalindromic consensus PurBoxes are located between positions −81 and −38 relative to the transcription start site (−81PurBox1-N18-PurBox2−38 [5, 13]). To characterize the length and specificity elements of the pur operon control site, the affinity of PurR for the full-length pur control region and a series of shortened upstream DNA fragments was determined. The full-length wild-type pur operon control region (−177 to +24; Fig. 2) was amplified from the template pDE264 by using primers Pri-177F and Pri+24R (Table 1) and subcloned into pET24a+ to produce plasmid pETpur-177/24. Truncated constructs were produced in a similar fashion. PurR binding to purified DNAs was evaluated by gel retardation assay (Fig. 3A). The apparent Kd value, representing the PurR concentration required for 50% saturation of control site DNA, was 6.0 ± 0.4 nM for the full-length construct (Fig. 4, line A). This is similar to the 7.0 nM value reported for a control region from −147 to −21 (15).
FIG. 2.
Nucleotide sequence of the pur operon control region from B. subtilis. The wild-type nucleotide sequence is shown from position −177 to position +24 with respect to the site of transcription initiation (+1). The PurBoxes are shaded.
FIG. 3.
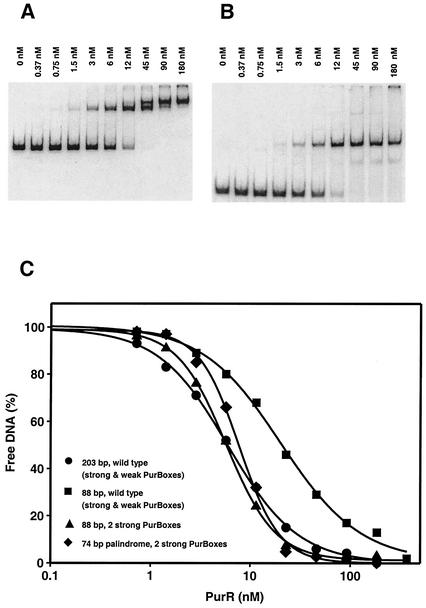
PurR-DNA binding. Gel mobility shift assays were run with 10 fmol of 32P-labeled DNA from the pur control region and various PurR concentrations. Gels are shown for the pur operon from position −177 to position +24 (A) and a 74-bp DNA palindrome with two strong PurBoxes (B). The calculated binding curves (C) are shown for the pur operon from position −177 to position +24 (Fig. 4, line A; •); the pur operon from position −91 to position −6 (▪; Table 2, A; Fig. 4, line K); and the pur operon from position −91 to position −6 with complementary PurBoxes (▴; Table 2, B; Fig. 4, line W) and a 74-bp DNA palindrome (⧫; Table 2, D).
FIG. 4.
Boundaries of the PurR DNA binding site. The top line represents the wild-type sequence of the B. subtilis pur operon from −177 to +24 relative to the transcription start site (from Fig. 2). PurBox1 (−81 to −68) is red, PurBox2 (−51 to −38) is yellow, and other wild-type regions are black. Nonspecific plasmid DNA generated by using alternative restriction sites is shown in blue. The stated nucleotide range is the position of the wild-type sequence relative to the transcription start site for each construct. The stated length is the total length, including any nonspecific DNA. Apparent Kd values for each fragment were determined by gel retardation assay. The actual DNA length is 1 to 2 unpaired nucleotides longer due to restriction digestion.
To map the 5′ boundary of the control region, we constructed and subcloned a series of truncations at positions −143, −123, −91, −69, and −54 relative to the transcription start site. PurR affinity was unchanged for fragments with 5′ ends at −143, −123, and −91 (Fig. 4, lines B through D). Essentially no binding was observed for DNA fragments with 5′ ends at either −69 or −54 (Fig. 4, lines E and F). For mapping of the 3′ boundary, truncations at positions −6, −42, −60, and −75 were constructed and subcloned. No change in the apparent Kd value was observed for the DNA fragment with a 3′ truncation at −6 (Fig. 4, compare lines A and G). A 15-fold increase in apparent Kd was observed when the 3′ position was at −42 (Fig. 4, line H), and no binding was detected when the 3′ boundary was at either −60 or −75 (Fig. 4, lines I and J). These data indicate that two PurBoxes are required for high-affinity PurR binding. Constructs having only one PurBox (Fig. 4, lines E, F, and I) had no detectable binding to PurR.
Effect of DNA length on PurR affinity.
The sequence between positions −91 and −6 relative to the transcription start site appears to include all PurR specificity elements. To define the minimal DNA required for high-affinity PurR binding, three double-end truncations were constructed (−91 to −6, −91 to −31, and −91 to −42). The −91 to −6 construct had threefold lower affinity (apparent Kd, 18 nM) than the full-length construct (Fig. 4, compare lines A and K). The −91 to −31 construct, which includes two PurBoxes, had an apparent Kd of 56 nM (Fig. 4, line L). The −91 to −42 construct, which includes only 10 of 14 nucleotides in downstream PurBox2, had barely detectable affinity (Kd >500 nM; Fig. 4, line M). The double-end truncations have lower affinity than predicted from the corresponding single-end truncations, indicating that overall DNA length contributes to PurR affinity (Fig. 4, compare line K to lines D and G; compare line M to lines D and H). To investigate the effect of DNA length on PurR affinity, plasmids bearing the double-end truncations were digested at alternative restriction sites in order to increase the size of the purified DNA by 46 nucleotides of nonspecific plasmid sequence at the downstream end or by 15 nucleotides at the upstream end. PurR affinity was increased for all three constructs with nonspecific DNA at the downstream end (Fig. 4, compare lines K through M with lines N through P). However, the same effect was not seen at the upstream end of the control region. PurR affinity was reduced threefold by the truncation of nine nucleotides beyond upstream PurBox1 (positions −91 to −83) and was not improved by the addition of nonspecific DNA (Fig. 4, lines P through R). A further truncation of seven nucleotides into upstream PurBox1 (−82 to −76) was devastating to PurR binding. The effect was not reversed by the addition of nonspecific DNA (Fig. 4, lines P, S, and T). This is in striking contrast to downstream PurBox2, where four nucleotides inside the PurBox can be substituted with nonspecific DNA with no effect on PurR affinity (Fig. 4, lines O and P).
Requirement for two PurBoxes.
In the initial experiments, no PurR binding was detected to truncations having a single PurBox (Fig. 4, lines E and I). However, in both cases, the single PurBox was near the end of the DNA construct. DNA was added to the PurBox-proximal ends of these constructs by digesting the respective plasmids at alternative restriction sites. The extended construct containing downstream PurBox2 did not bind PurR in our assay, and the extended construct with upstream PurBox1 bound only very weakly (Fig. 4, lines U and V). Thus, two PurBoxes are required for high-affinity PurR binding. It also appears that upstream PurBox1 (−81 to −68) contributes more to high-affinity PurR binding than does downstream PurBox2 (−51 to −38). These results are consistent with the effects of mutations within the PurBox central tetranucleotide −75GAAC−72 (−47GTTC−44). Mutations in the upstream site diminished PurR binding, but those in the downstream site had little effect (15).
Strong and weak PurBoxes.
We next investigated the relative PurR affinities of upstream PurBox1 and downstream PurBox2. The sequences of PurBox1 and PurBox2 differ at 6 of 14 nucleotides (Table 2). DNAs bearing two identical PurBoxes in an inverted orientation were constructed. PurR affinity was increased threefold by replacement of downstream PurBox2 with upstream PurBox1 (Fig. 4, lines K and W). Conversely, affinity was decreased 15-fold by replacement of upstream PurBox1 with downstream PurBox2 (Fig. 4, lines D and X). A DNA containing a palindrome of upstream PurBox1 bound PurR with 19-fold greater affinity than a DNA with a palindrome of downstream PurBox2 (Fig. 4, lines W and X). Thus, we designate upstream PurBox1 a strong PurBox and downstream PurBox2 a weak PurBox.
TABLE 2.
PurR binding to symmetric constructs of the pur control region
| Construct | Sequence | Length (position) | Apparent Kd (nM) |
|---|---|---|---|
| A: Wild-type pur operon sequence encompassing two PurBoxesa | −81AAACACGAACATTAGTAGAATGAATTTTTGTATCGTTCGATAATA−37TTTGTGCTTGTAATCATCTTACTTAAAAACATAGCAAGCTATTAT | 88 bp (−91 to −6) | 18 ± 2.0 |
| B: pur operon nucleotide sequence with symmetric PurBoxes (7 mutations)a | −81AAACACGAACATTAGTAGAATGAATTTTTGTAATGTTCGTGTTTT−37TTTGTGCTTGTAATCATCTTACTTAAAAACATTACAAGCACAAAA | 88 bp (−91 to −6) | 5.7 ± 0.3 |
| C: pur operon nucleotide sequence with symmetric PurBox region (11 mutations)a | −81AAACACGAACATTAGTAGAATGCATTCTACTAATGTTCGTGTTTT−37TTTGTGCTTGTAATCATCTTACGTAAGATGATTACAAGCACAAAA | 88 bp (−91 to − 6) | 6.0 ± 0.4 |
| D: Half-palindrome sequenceb | −96(ATCGAAATTGATCTAAAACACGAACATTAGTAGACTGCA−58)2 | 74 bp | 7.8 ± 0.4 |
| (TAGCTTTAACTAGATTTTGTGCTTGTAATCATCTG)2 |
PurBoxes are in italics. Nucleotides mutated to introduce symmetry are underlined.
The half-palindrome between the EcoRV and PstI sites of p1/2pur37 self-ligates to form a 74-bp perfect palindrome. PurBox1 is in italics. Nucleotides matching the wild-type sequence are bolded; those changed to create restriction sites are underlined.
PurR bound to a double-end truncated control region (−91 to −6) having palindromic strong PurBoxes with an apparent Kd value of 5.7 nM and high cooperativity (Table 2, line B; Fig. 3C). Thus, the affinity of the full-length control region (−177 to +24; Kd = 6.0 nM) was recovered in a shorter DNA fragment with palindromic copies of strong PurBox1. No further increase in affinity occurred when four mutations were introduced in the intervening region (−67 to −52) to make the region encompassing both PurBoxes (−82 to −37) perfectly palindromic (Table 2, line C). The intervening region had no detectable sequence specificity, as we introduced other mutations to place a restriction site in this region, also without effect on PurR affinity (Table 2, line D).
Minimal length of the control region.
The −91 to −6 construct includes 10 nucleotides upstream of PurBox1 and 32 nucleotides downstream of PurBox2. We wished to define the shortest DNA sequence with high affinity for PurR, and we presume that this construct has equal amounts of DNA outside the PurBoxes. Results described above indicated that the region between −91 and −83 contains elements essential to PurR binding. Therefore, we designed a fully palindromic construct with flanking regions identical to the wild-type sequence beginning at position −93. With the addition of sites for two restriction enzymes (EcoRV and PstI), this was a 74-bp perfect palindrome (Table 2, line D). PurR bound this construct with an apparent Kd value of 7.8 nM and also exhibited high cooperativity (Fig. 3B and C). This is the shortest DNA fragment we identified with high affinity for PurR.
Inhibition by PRPP.
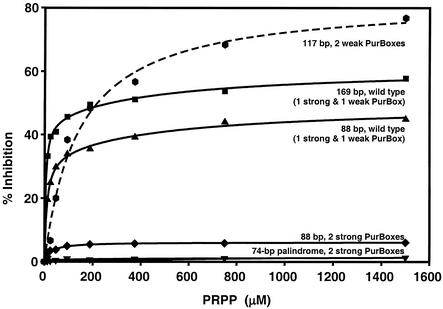
PRPP inhibits specific binding of PurR to the pur operon control site (19). We measured the effect of PRPP on binding of purified PurR to various DNA fragments (Fig. 5). In these experiments, PurR concentrations were near the end of the transition in the DNA binding curve (Fig. 3C). PurR binding to a 169-nucleotide construct containing the wild-type pur control region from −143 to +24 (one strong and one weak PurBox) was inhibited up to 60% by PRPP, while binding to a shorter 88-bp DNA (−91 to −6) was inhibited only up to 40%. When the DNA construct included two weak PurBoxes and no strong PurBox, PRPP inhibited binding by up to 80%. In contrast, PRPP had virtually no inhibitory effect on constructs with two strong PurBoxes, independent of the order of addition of components to the binding assay. Thus, at least one weak PurBox was required for PRPP inhibition of PurR-DNA binding in the gel retardation assay. Additionally, PRPP had a greater effect on binding longer DNA constructs. The observed incomplete inhibition of DNA binding by purified PurR is consistent with previous work, in which complete inhibition was observed only when the source of PurR was soluble cell extract (15, 19, 20). This raises the possibility that an unidentified molecule in the cell extract enhances the PRPP effect.
FIG. 5.
Effect of PRPP on PurR-DNA binding. Gel retardation assays were carried out on 32P-end-labeled DNA probes (10 fmol), PurR (47 nM, solid lines; 150 nM, dashed line), and various PRPPs. Relative to the transcription start site, the curves represent wild-type nucleotides −143 to +24 (▪; Fig. 4, line B), wild type −91 to −6 (▴; Table 2, A; Fig. 4, line K), −91 to −6 with complementary strong PurBoxes (⧫; Table 2, B; Fig. 4, line W), a 74-bp full palindrome with strong PurBoxes (▾; Table 2, D), and −91 to +24 with complementary weak PurBoxes ( ; Fig. 4, line X).
PRPP binding to PurR.
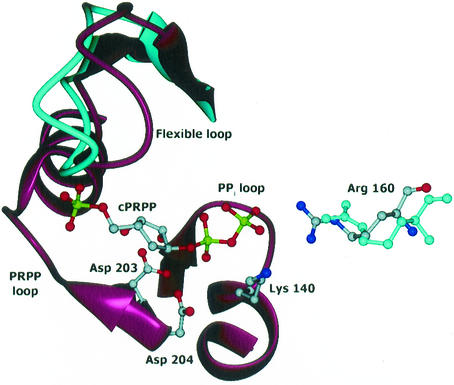
The site of PRPP binding to PurR was determined in the crystal structure of PurR complexed with the stable analog cPRPP. cPRPP binds to the anticipated PRPP binding site in the PRT domain of PurR (Fig. 6) (17). No divalent cations were apparent in any of the cPRPP sites. The structure of the PurR subunit in the cPRPP complex is similar to the free PurR subunit structure (the root mean square deviation [RMSD] is 0.32 Å for 160 Cα atoms). The largest conformational change within the subunit is a closing of the PRT flexible loop by 7 to 9° over bound cPRPP (Fig. 6). Within the subunit, cPRPP binding did not induce hinging between the winged-helix and PRT domains, which are oriented identically in the bound and free forms of PurR. Within the dimer, the angle between monomers differs by about 3° in the cPRPP complex and free PurR. This modest conformational change does not have repercussions that appear capable of changing DNA affinity significantly. However, the motion allows invariant Arg160 from the flexible loop of one subunit to contact the pyrophosphate group of cPRPP in the other subunit (Fig. 6). The PurR flexible loop behaves similarly to the flexible loops of other PRT family members with respect to both closure over bound PRPP and participation in PRPP binding to the partner subunit (17)
FIG. 6.
PRPP binding site in the crystal structure of PurR-cPRPP. The phosphate and pyrophosphate groups of cPRPP bind as expected. The flexible loop is above cPRPP in this view. Structural elements that change in response to cPRPP binding, the flexible loop and Arg160, are drawn in cyan for the free PurR structure. Side chains are also shown for invariant residues Asp203, Asp204, and Lys140, which contact cPRPP. The Arg160 side chain is from the second subunit of the dimer. This figure was prepared with DINO (DINO: Visualizing Structural Biology [2002] http://www.dino3d.org).
Large-scale purification of palindromic DNA.
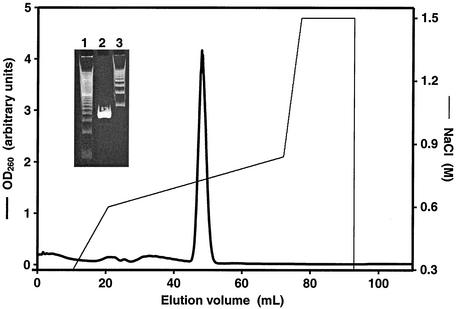
Purification of large DNA oligomers with palindromic or near-palindromic symmetry was complicated by the formation of DNA hairpins, which did not bind PurR. This problem was avoided in the small-scale preparations required for gel retardation assays by avoiding heat and chaotropic agents during purification. However, we also wished to characterize the minimal PurR-DNA complex in detail, including determination of a crystal structure. This requires milligram quantities of purified DNA. An additional complication is the instability in E. coli of plasmids bearing long palindromes, such as the minimal 74 bp described here. We employed a method, which was developed for structural studies of the nucleosome, to produce large quantities of palindromic DNA by isolation of tandem half-palindromes from plasmids grown in E. coli (18). The purified palindrome eluted as a single peak and was more than 99% pure by electrophoresis (Fig. 7). An 8-liter E. coli culture yielded 4 mg of purified 74-bp DNA palindrome.
FIG. 7.
Purification of 74-bp DNA palindrome. The 74-bp DNA palindrome was purified by anion-exchange chromatography. The left axis and the thick line represent the UV absorbance at 260 nm (OD260). The thin line and right axis represent the NaCl gradient used for elution. Insert shows polyacrylamide gel electrophoresis of purified DNA palindrome. Lane 1, marker of 20-bp ladder; lane 2, 74-bp purified DNA; lane 3, marker of 100-bp ladder.
Stoichiometry of the DNA-PurR complex.
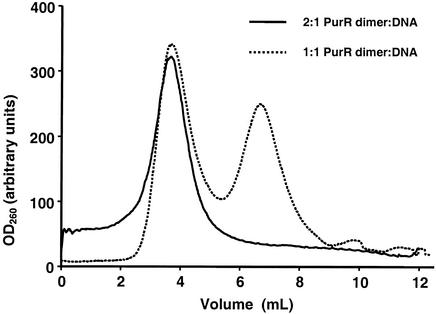
Repressor-operator complexes with PurR and fragments of pur operon con trol site DNA were observed by gel shift assay in stoichiometries of two to six PurR dimers per DNA fragment (15). However, these experiments were performed with a large molar excess of PurR. We used ultracentrifugation to measure the PurR-DNA stoichiometry of high-affinity complexes. In this experiment, PurR and the 74-bp DNA palindrome were incubated in various stoichiometries. The complexes, or free DNA, were analyzed by sedimentation in a 10 to 30% sucrose gradient. No free DNA was observed in a 2:1 mixture of PurR dimer to DNA (Fig. 8). As the proportion of DNA in the mixture was increased, free DNA was observed. About 50% free DNA was detected in the 1:1 mixture. We conclude from these results that two PurR dimers bind one 74-bp DNA palindrome.
FIG. 8.
PurR-DNA stoichiometry. Sedimentation analysis shows absorbance at 260 nm (OD260) for fractions in 10 to 30% sucrose gradients for 2:1 (solid line) and 1:1 (dashed line) PurR dimer-DNA mixtures.
DISCUSSION
PurR-DNA binding.
The PurBoxes in the pur control region are designated strong and weak because of greater PurR affinity for upstream, strong PurBox1 than for downstream, weak PurBox2 (Fig. 4, lines D and X, K and W). Two PurBoxes, at least one of which must be a strong PurBox, are required for PurR affinities similar to the levels observed with 100 or more bp of wild-type pur control region, as would be encountered by PurR in vivo. The flanking sequence upstream of strong PurBox1 also includes some specificity elements (Fig. 4, lines P through R). Maximum PurR affinity for a minimal DNA construct was achieved by the replacement of weak PurBox2 and its outer flanking sequence with the analogous sequences from strong PurBox1 (Table 2).
PurR binding to the minimal, 74-bp construct is highly cooperative (Fig. 3C), consistent with the stoichiometry of two PurR dimers per DNA construct (Fig. 8). We presume that in this complex, each PurR dimer interacts with specific sequences in one PurBox. Taken together, the positive cooperativity and stoichiometry are consistent with a binding mechanism in which one PurR dimer binds weakly to a strong PurBox and increases the affinity of a second dimer for the second PurBox. Positive cooperativity could be achieved either through protein-protein interactions or through deformation of DNA by the first PurR dimer. Up to six PurR dimers bind 127- to 196-bp segments from the pur control region, with the number of dimers correlated with DNA length (15). This is indicative of extensive, nonspecific PurR-DNA interactions as well as PurR dimer-dimer interactions.
We examined the crystal structures of PurR for potential dimer-dimer interactions. The PurR-cPRPP complex (this study) and free PurR (16) crystallize under different conditions and in different crystal forms. Thus, any crystal lattice contacts common to the two structures are candidates for a dimer-dimer contact that may form in solution. Among the dimer-dimer contacts in each crystal form, one—and only one—exists in both crystal lattices. The winged-helix domain in one subunit (β1: Leu23, and wing: Leu60, Val62, Pro63, Ala66, and Lys70) contacts the PRT domain in another dimer (flexible loop: Ile173, Asn174, Val176, and Ile183). This small contact (350 Å2 of buried surface per subunit) is predominantly hydrophobic and could facilitate cooperativity of PurR binding to DNA in solution and in vivo.
How then does DNA interact with PurR? DNA consistently binds to the most electropositive surface of proteins. The first of two strongly positive PurR surfaces is at the dimer interface of the winged-helix domain (16). Virtually all conserved residues in the winged-helix domain also map to this surface, which includes the N terminus, the start of the HTH recognition helix, and the wing. Because winged-helix and HTH domains have no known function apart from recognition of DNA sequences, we infer that the presence of a winged-helix domain in all PurR homologs and the conservation of its positively charged surface are due to a common DNA-binding function. Winged-helix domains of other proteins interact with DNA through contacts by the recognition helix (α3) and the wings (3). Thus, the conserved surface of the winged-helix dimer is the prime candidate for recognition of the conserved CGAA sequence in the center of the PurBox. The second strongly positive PurR surface surrounds the PRPP binding site at the opposite end of the molecule (16). This surface and the PRPP binding site are also conserved among PurR homologs. The structure and biochemical data are consistent with a model in which DNA contacts both electropositive ends of the PurR dimer by bending or kinking around the protein. The prevalence of repeated A and T nucleotides, or A tracks (9), in the PurBox and surrounding sequences could facilitate DNA bending.
Control of DNA binding by PRPP.
A major finding of our studies is that PRPP interference with DNA binding requires one weak PurBox in the DNA construct. Compared to wild-type constructs with one strong and one weak PurBox, PRPP has a greater effect on PurR binding to DNA constructs with two weak PurBoxes and has no effect on constructs with two strong PurBoxes (Fig. 5). This indicates that PRPP has a weak affinity for PurR and cannot disrupt the tightest PurR-DNA complexes. Even at high concentrations, PRPP did not completely abolish binding of purified PurR to wild-type pur control sequences or to shorter constructs (Fig. 5) (12, 20). It should be noted that PRPP completely abolished PurR binding to pur control sequences when the source of PurR was cell extract (12, 20), suggesting that other components of the extract may play a role.
PRPP binds in the same manner to the same site in PurR as in other PRT proteins (Fig. 6). However, weak affinity of PurR for PRPP results in incomplete occupancy of the PRPP site by the PRPP analog in the crystal structure of the cPRPP complex. This is consistent with a model in which PRPP blocks one part of an extensive DNA binding site. The manner in which cPRPP binds PurR also may explain the super-repressed phenotype observed for substitutions of Asp203 and Asp204 with Ala (20). Dual carboxyl side chains are a hallmark of the PRPP sites of PRT proteins. In most PRT proteins, bound Mg2+ is coordinated by the ribose hydroxyls of PRPP, which are hydrogen bonded to the dual carboxylates. However, no Mg2+ was bound in the crystal structure of cPRPP-PurR, despite its inclusion in crystallization solutions, and the ribose analog was poorly ordered. If the PRPP site in PurR is designed not to bind Mg2+, then elimination of a carboxyl side chain should enhance PurR affinity for both PRPP and DNA. Under normal conditions, the enhanced affinities of PRPP and DNA would balance one another to produce little or no net effect on repression levels in the mutants relative to the wild type. However, growth with adenine supplements would reduce intracellular PRPP levels so that the enhanced affinity of DNA would predominate, thus creating the super-repressed phenotypes of D203A and D204A. An Mg2+-excluded binding site would also account for the poor affinity of PurR for PRPP.
Strong and weak PurBoxes.
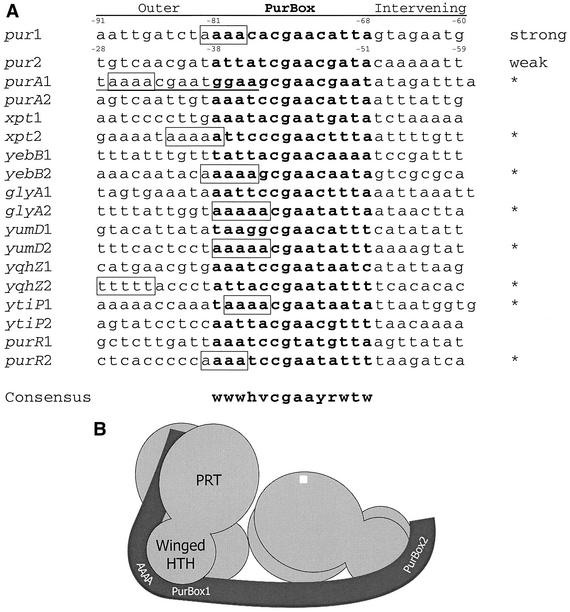
The PRPP disruption results demonstrate the physiological significance of nonidentical PurBoxes. PRPP is thought to induce pur transcription in B. subtilis because it is the only molecule known to interfere with PurR-DNA binding (19). PRPP levels are reduced in cells grown in medium supplemented with adenine (10), which also represses transcription of B. subtilis genes regulated by PurR (1, 13, 19). One weak PurBox is required for PurR to be removed from DNA under the control of PRPP (Fig. 5). Thus, the control region of each of these genes should include one strong PurBox for PurR binding and one weak PurBox for PRPP control. We compared the upstream and downstream PurBox sequences in the control region of each gene (Fig. 9A). The central CGAA sequence is virtually invariant in B. subtilis PurBoxes. The flanking sequences within and outside the PurBox are notable for the high concentration of A-T base pairs. We expect DNA to curve or kink as it binds PurR, and A tracks (A4-5 or T4-5) can facilitate DNA bending. Thus, we propose strong and weak designations for the PurBoxes of each gene based on the presence of A tracks. For all genes except purA, the choice was clear. For purA, we note that an alternative PurBox1 with a strong A track exists exactly 10 bp upstream of putative PurBox1. The reexamination of PurBox sequences also led to a refinement of the PurBox consensus for B. subtilis relative to that proposed earlier (13) (Fig. 9A).
FIG. 9.
Strong and weak PurBoxes. (A) Alignment of B. subtilis PurBox sequences. For each control region regulated by strong and weak PurBoxes (13), sequences are shown for the upstream PurBox (labeled “1”), for the complement of the downstream PurBox (labeled “2”), for 10 nucleotides outside the PurBox region, and for the inter-PurBox region. PurBox sequences are in boldface. Potential A track sequences within the PurBox or in the outer region are boxed. Potential A tracks in the intervening region were not considered because pur operon mutations that eliminated an A track in this region had no impact on PurR affinity (Table 2). Based on the presence of an A track, proposed strong boxes are marked with *. Alternative PurBox1 for purA is underlined. The B. subtilis-specific consensus sequence is shown at the bottom; W indicates either A or T; H is A, C or T; V is A, C, or G; Y is C or T; and R is A or G. (B) Model for PurR-DNA binding. Two PurR dimers (light gray) bind a DNA (dark gray) containing two PurBoxes. According to this model, each PurBox is recognized by the conserved, positively charged surface of a winged-helix dimer. PurR dimer-dimer contacts are like the crystal lattice contact seen in both PurR crystal forms. An A track at the outer edge of the strong PurBox kinks the DNA, thereby allowing contact with the positively charged surface of the PRT domain and with the conserved, positively charged surface surrounding the PRPP site (top of the labeled dimer).
Model for PurR-DNA binding.
A model that takes into account the biochemical and structural data on PurR is shown in Fig. 9B. The model has several key features. The central CGAA sequence of each PurBox is recognized by the conserved surface of the winged-helix domain of the PurR dimer. However, this binding does not provide sufficient affinity for the biological effects of PurR. One PurBox is strong due to an outer A track that kinks the DNA, thereby allowing a longer DNA stretch to bind nonspecifically to the positively charged surface of the PRT domain. This nonspecific binding provides essential affinity. Additional affinity is provided by the second PurBox and by a weak dimer-dimer interaction similar to the common lattice contact in PurR crystals. PRPP binding does not cause a large allosteric structural change to PurR. Instead, PRPP, which binds without an Mg2+ counterion, alters the electrostatic surface of key regions of the PRT domain, reducing their DNA affinity. In this model, many factors contribute to PurR binding to the pur control region. Control of gene expression is achieved by slight alterations in the balance of these factors.
Comparison to other transcription regulators.
B. subtilis responds to changes in nucleotide availability via the intracellular pool of PRPP. Thus, many purine inputs affect the response of a single output effector molecule because the effector is PRPP, a central nucleotide metabolite. The result is fine control of target gene expression. This differs substantially from the familiar prokaryotic repressor-operator system in which a dimeric protein binds with a high affinity to a short palindromic DNA operator, and an effector molecule alters DNA binding via protein allosteric changes. E. coli PurR, a member of the LacI family with an HTH DNA-binding domain, is a typical example of the classic prokaryotic repressor in which only the HTH domain contacts DNA and purine bases are the effector molecules (14).
A more complex system has evolved to fit the intracellular conditions pertinent to the range of environmental conditions encountered by B. subtilis. PurR regulates transcription initiation through a network of weak interactions: PurR-DNA, PurR-PRPP and PurR-PurR. The PurR structural change wrought by PRPP is small relative to other winged-helix and HTH proteins. The PurR-DNA interaction is strikingly different from other winged-helix and HTH protein-DNA complexes with respect to the large separation within the DNA inverted repeat, length of DNA protected by the protein, interaction of more than one protein dimer with a DNA inverted repeat, formation of protein multimers on DNA, low affinity of the effector molecule for the protein, and fixed association of subunits in the protein dimer. Each of these features has precedent in the HTH superfamily, but not simultaneously in any other family member. Thus, PurR represents a mechanism not seen previously for other transcription regulators.
Finally, some aspects of DNA binding may differ in organisms having PurR homologs. For example, L. lactis PurR activates transcription from genes having only one PurBox in the upstream control region (5). This molecule is expected to have high affinity for a single PurBox and not to form multimers on DNA. Consistent with this idea, the small hydrophobic contact seen in crystals of B. subtilis PurR would be prevented by replacement of Leu60 by Glu60 in L. lactis PurR. No data are available for the function of other PurR homologs.
Acknowledgments
This work was supported by grants DK42303 and GM24658 from the U.S. Public Health Service.
REFERENCES
- 1.Ebbole, D. J., and H. Zalkin. 1989. Bacillus subtilis pur operon expression and regulation. J. Bacteriol. 171:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebbole, D. J., and H. Zalkin. 1989. Interaction of a putative repressor protein with an extended control region of the Bacillus subtilis pur operon. J. Biol. Chem. 264:3553-3561. [PubMed] [Google Scholar]
- 3.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110-116. [DOI] [PubMed] [Google Scholar]
- 4.Kilstrup, M., S. G. Jessing, S. B. Wichmand-Jørgensen, M. Madsen, and D. Nilsson. 1998. Activation control of pur gene expression in Lactococcus lactis: proposal for a consensus activator binding sequence based on deletion analysis and site-directed mutagenesis of purC and purD promoter regions. J. Bacteriol. 180:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 180:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krahn, J. M., J. H. Kim, M. R. Burns, R. J. Parry, H. Zalkin, and J. L. Smith. 1997. Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry 36:11061-11068. [DOI] [PubMed] [Google Scholar]
- 7.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 8.Navaza, J. 1994. AMoRe—an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 9.Nelson, H. C., J. T. Finch, B. F. Luisi, and A. Klug. 1987. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature 330:221-226. [DOI] [PubMed] [Google Scholar]
- 10.Nygaard, P. 1993. Purine and pyrimidine salvage pathways, p. 359-378. In A. L. Sonenstein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington D.C.
- 11.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode., Methods Enzymol. 276:307-325. [DOI] [PubMed] [Google Scholar]
- 12.Rappu, P., B. S. Shin, H. Zalkin, and P. Mäntsälä. 1999. A role for a highly conserved protein of unknown function in regulation of Bacillus subtilis purA by the purine repressor. J. Bacteriol. 181:3810-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxild, H. H., K. Brunstedt, K. I. Nielsen, H. Jarmer, and P. Nygaard. 2001. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J. Bacteriol. 183:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher, M. A., K. Y. Choi, H. Zalkin, and R. G. Brennan. 1994. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by α helices. Science 266:763-770. [DOI] [PubMed] [Google Scholar]
- 15.Shin, B. S., A. Stein, and H. Zalkin. 1997. Interaction of Bacillus subtilis purine repressor with DNA. J. Bacteriol. 179:7394-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha, S., J. M. Krahn, B. S. Shin, D. R. Tomchick, H. Zalkin, and J. L. Smith. 2003. The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation. J. Bacteriol. 185:4087-4098. [DOI] [PMC free article] [PubMed]
- 17.Sinha, S. C., and J. L. Smith. 2001. The PRT protein family. Curr. Opin. Struct. Biol. 11:733-739. [DOI] [PubMed] [Google Scholar]
- 18.Tan, S., Y. Hunziker, L. Pellegrini, and T. J. Richmond. 2000. Crystallization of the yeast MATα2/MCM1/DNA ternary complex: general methods and principles for protein/DNA cocrystallization. J. Mol. Biol. 297:947-959. [DOI] [PubMed] [Google Scholar]
- 19.Weng, M., P. L. Nagy, and H. Zalkin. 1995. Identification of the Bacillus subtilis pur operon repressor. Proc. Natl. Acad. Sci. USA 92:7455-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng, M., and H. Zalkin. 2000. Mutations in the Bacillus subtilis purine repressor that perturb PRPP effector function in vitro and in vivo. Curr. Microbiol. 41:56-59. [DOI] [PubMed] [Google Scholar]