Abstract
Phosphopantetheine adenylyltransferase (PPAT) regulates the key penultimate step in the essential coenzyme A (CoA) biosynthetic pathway. PPAT catalyzes the reversible transfer of an adenylyl group from Mg2+:ATP to 4′-phosphopantetheine to form 3′-dephospho-CoA (dPCoA) and pyrophosphate. The high-resolution crystal structure of PPAT complexed with CoA has been determined. Remarkably, CoA and the product dPCoA bind to the active site in distinct ways. Although the phosphate moiety within the phosphopantetheine arm overlaps, the pantetheine arm binds to the same pocket in two distinct conformations, and the adenylyl moieties of these two ligands have distinct binding sites. Moreover, the PPAT:CoA crystal structure confirms the asymmetry of binding to the two trimers within the hexameric enzyme. Specifically, the pantetheine arm of CoA bound to one protomer within the asymmetric unit displays the dPCoA-like conformation with the adenylyl moiety disordered, whereas CoA binds the twofold-related protomer in an ordered and unique fashion.
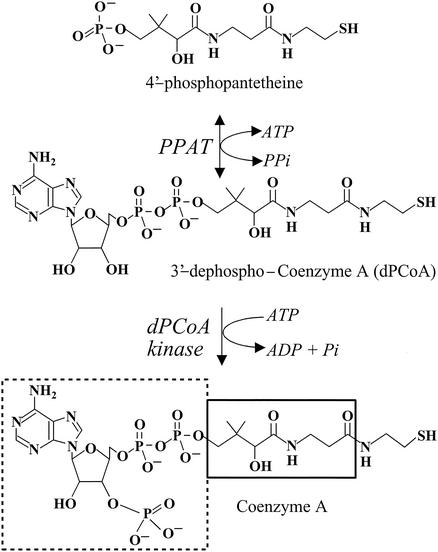
Coenzyme A (CoA) is synthesized in Escherichia coli in a series of five steps utilizing pantothenate (vitamin B5), cysteine, and ATP (29). Phosphopantetheine adenylyltransferase (PPAT) catalyzes the penultimate step, the reversible transfer of an adenylyl group from ATP to 4′-phosphopantetheine (Ppant), yielding 3′-dephospho-CoA (dPCoA) and pyrophosphate (21) (Fig. 1). PPAT is the second rate-limiting step in the CoA pathway and together with the first enzyme of the pathway, pantothenate kinase, regulates the cellular CoA content (18). Ppant is derived from the metabolism of pantothenate (17), degradation of CoA (18), or the turnover of the acyl carrier protein prosthethic group (31) and exits from cells when not utilized by PPAT (18). Biochemical regulation of PPAT has not been investigated, although CoA is tightly bound to the purified enzyme (12), suggesting that, like pantothenate kinase, PPAT is feedback regulated by CoA (32).
FIG. 1.
The final two steps in the biosynthetic pathway of CoA. CoA biosynthesis occurs in a series of five steps that utilize pantothenate (vitamin B5), cysteine, and ATP. PPAT catalyzes the penultimate step, the reversible transfer of an adenylyl group from ATP to 4′-phosphopantetheine to form dPCoA and pyrophosphate. Subsequent phosphorylation at the 3′-hydroxyl of the ribose ring by dPCoA kinase produces the acyl group carrier, CoA. For CoA, the 3′-phosphate ADP is shown by a dashed box, the pantothenic acid group of pantetheine is outlined by the solid box, and the β-mercaptoethylamine of pantetheine is not boxed.
The first structure of enzymes involved in the CoA biosynthetic pathway reported was the crystal structure of E. coli PPAT in complex with dPCoA (14). To investigate the enzyme's reaction mechanism, the high-resolution crystal structures of PPAT in complex with either one of its substrates, ATP or Ppant, were subsequently determined (16). Recently, the crystal structures of E. coli pantothenate kinase in complex with 5′-adenylimido-diphosphate or CoA (36) and of the last enzyme in the pathway, dPCoA kinase, (from Haemophilus influenzae [23] and E. coli [24]) have also been determined. The PPAT: dPCoA crystal structure established that the enzyme displays a dinucleotide (or canonical Rossmann) binding fold (30) (Fig. 2). PPAT functions as a hexamer in solution (12), and the crystal structure is consistent with that of a hexamer having point group 32 (14). The asymmetric unit contains a dimer, and the crystallographic triad coincides with the oligomer's threefold axis. Furthermore, the PPAT:dPCoA crystal structure revealed that the PPAT hexamer consists of two distinct trimers with only one trimer binding dPCoA. Finally, the cooperative binding of the enzyme suggests that PPAT is an allosteric enzyme (14).
FIG. 2.
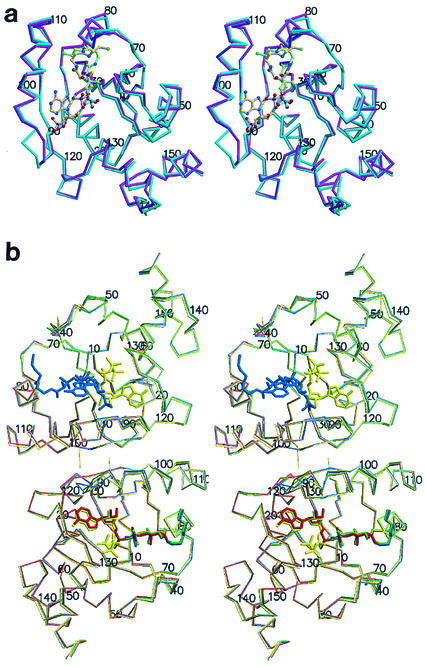
(a) Stereodiagram of the PPAT:CoA Cα trace of the two superimposed subunits within the asymmetric unit. Protomer A showing the entire inhibitor (magenta) and the twofold-related superimposed protomer B (teal) are shown. Ligands are shown in ball and stick representation. Oxygen (red) nitrogen (blue), sulfur (green), carbon (yellow), and phosphorus (black) atoms are shown. The overall rms deviation for the 314 equivalent Cα positions of the superposition of the two subunits in the asymmetric unit is 0.81 Å. This color coding of atoms is also used in panel b and in Fig. 3. (b) Stereodiagram of the PPAT Cα trace of the dimer within the asymmetric unit bound to CoA (blue), dPCoA (red) (Protein Data Bank identification code [PDB ID] 1B6T) (14), ATP (yellow) (PDB ID 1GN8) (16), and Ppant (green) (PDB ID 1QJC) (16). Protomer A (top subunit) binds CoA in the PPAT:CoA structure and ATP in the PPAT:ATP structure, while protomer B (bottom subunit) binds dPCoA (PPAT:dPCoA structure), ATP (PPAT:ATP structure), and Ppant (PPAT:Ppant structure) but shows only partially ordered binding to CoA (PPAT:CoA structure). This figure was produced with MolScript (19) or BobScript (11) and Raster3D (22) programs.
PPAT is a member of the nucleotidyltransferase α/β phosphodiesterase superfamily (2), which includes class I aminoacyl-tRNA synthetases (7, 9, 27), adenylylsulfate-phosphate adenylyltransferase (6, 34), glycerol-3-phosphate cytidylyltransferase (26, 35), nicotinamide mononucleotide adenylyltransferase (8), and pantothenate synthetase (33). This family is characterized by the presence of a mononucleotide binding fold and a conserved T/HXGH sequence motif, with a specific catalytic role for the second histidine within this motif. Evaluation of steady-state kinetics showed that the PPAT reaction proceeds by a ternary complex mechanism (12). The catalytic mechanism suggested by the PPAT crystal structures in complex with its substrates implicates transition state stabilization by PPAT, without the direct involvement of functional groups of the enzyme in acid-base or covalent catalysis (16). Similar mechanisms have been suggested for other members of the superfamily.
In this report, the PPAT crystal structure in complex with CoA has been determined to a 1.78-Å resolution. The structure confirms the asymmetry of binding to the two trimers within the PPAT hexamer. Moreover, since PPAT catalyzes a rate-limiting step in this essential pathway, the enzyme is a plausible target for inhibiting bacterial growth by reducing the intracellular levels of CoA. Significantly, in contrast to bacteria, in mammalian systems PPAT and dPCoA kinase activities are present in a bifunctional enzyme called CoA synthase (1, 37). Thus, the PPAT:CoA structure presented herein could pave the way for the rational development of selective antibiotics.
MATERIALS AND METHODS
Crystallization and data collection.
PPAT crystals with bound CoA were grown as described previously (15) and were shown to belong to space group I23(cell edge = 135.5 Å) with a dimer in the asymmetric unit, a solvent content of 0.57, and a volume-to-protein mass ratio of 2.88 Å3 Da−1. In comparison, the cell edges for the previously reported structures are 135.1 Å (PPAT:ATP), 135.7 Å (PPAT:Ppant), and 135.6 Å (PPAT:dPCoA). X-ray data were collected at 100 K with a Rigaku RU-200 rotating anode generator with a copper anode, focusing mirrors, and an R-AXIS IV imaging plate. The data were processed using DENZO and SCALEPACK (25). Data statistics are given in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
| Parameter | Value |
|---|---|
| PPAT:CoA data collection | |
| Resolution [Å] | 1.78 |
| Total data | 815,367 |
| Unique data | 39,181 |
| Overall completeness | 0.99 |
| Completeness (1.84-1.78 Å) | 0.907 |
| F2 > 3σ(F2) [%] | 0.76 |
| Rmergea (overall) | 0.036 |
| Rmerge (1.84-1.78 Å) | 0.257 |
| Avg F2/σ(F2) | 41.5 |
| Crystallographic refinement | |
| No. of reflections | 39,134 |
| Final model parameters | |
| No. of amino acid residues | 315 |
| No. of protein atoms | 2,481 |
| No. of solvent molecules | 286 |
| No. of substrate (sulfate) molecules | 2 (4) |
| Resolution range [Å] | 20-1.78 |
| R factorb overall | 0.218 |
| R factor (1.89-1.78 Å) | 0.302 |
| Rfreec overall | 0.241 |
| Rfree (1.89-1.78 Å) | 0.323 |
| Avg main chain B factor [Å2] | 21.6 |
| Avg side chain B factor [Å2] | 22.9 |
| Avg water molecule B factor [Å2] | 35.7 |
| Avg ligand B factor (subunits A and B) [Å2] | 42.2, 54.2 |
| rms deviations from ideal geometry | |
| Covalent bond lengths [Å] | 0.008 |
| Bond angles [°] | 1.2 |
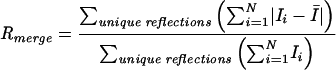 .
.
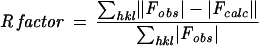 .
.
The free R factor is a cross-validation residual calculated by using 5% of the native data, which were randomly chosen and excluded from the refinement.
Structure determination and crystallographic refinement.
The refined model of the PPAT:dPCoA complex (14) was used as a starting model for refinement. The initial R factor was 0.31. The product dPCoA was omitted from the model. The model was refined with the program CNS (3, 4) using standard protocols. The free R value (5) was monitored throughout the refinement. The electron density was poorest for residues 41 and 42 in the A subunit within the asymmetric unit and for residues 92 to 94 in the B subunit (subunits A and B defined in Results). The electron density is not visible for the adenylyl moiety of CoA in the B subunit. Additional water molecules were initially identified in Fo − Fc maps and were screened for reasonable geometry and for a refined thermal factor of less than 50 Å2. Table 1 shows the overall crystallographic R factor and the free R factor for all observed reflections within the indicated resolution range. A Ramachandran plot analysis by the program PROCHECK (20) indicates that 90.9% of all the residues lie in most favorable regions and the remaining 9.1% lie in additional allowed regions. This structure analysis also showed that all stereochemical parameters are better than expected at the given resolution.
Coordinates.
The atomic coordinates have been deposited in the Protein Data Bank (identification code 1h1t).
RESULTS
Overall structure of PPAT:CoA.
The superposition of the two PPAT:CoA subunits within the asymmetric unit is illustrated in Fig. 2a. Notably, within the hexamer, only one trimer showed electron density corresponding to all atoms of CoA and the electron density in the other trimer was visible only for the Ppant moiety of CoA. Thus, the two protomers in the asymmetric unit are distinct. The subunit showing density for the entire ligand is hereafter referred to as the A subunit (magenta subunit in Fig. 2a), while the twofold-related protomer is the B subunit (teal subunit in Fig. 2a).
CoA binding to PPAT.
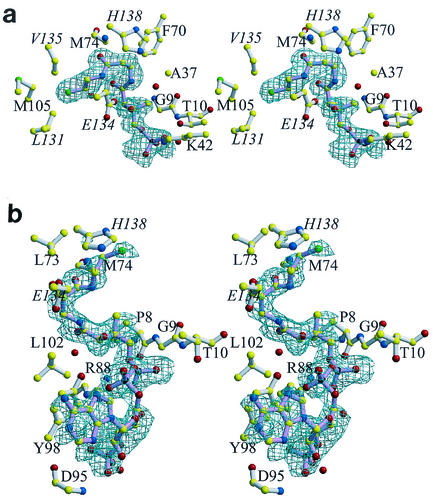
The active-site cleft in the B subunit binds to the pantetheine arm of CoA in the same fashion as seen for dPCoA (14). However, no electron density is observed for the adenylate group (Fig. 3a), and the nucleotide binding site is consistent with the binding of several ordered water molecules and with a sulfate anion. The sulfate anion's only interaction with the protein consists of bifurcated hydrogen bonds to the side chain of Arg-91 and is located near the β-phosphate position of ATP seen in the PPAT:ATP crystal structure (16). The lack of electron density found for the adenylate group could be due to its disorder or, alternatively to pyrophosphorylation of CoA by PPAT. However, the latter possible reaction has been examined biochemically, and it has been established that CoA is not a substrate for PPAT (12). Therefore, the lack of electron density for the adenylate group is indeed due to its disorder.
FIG. 3.
Stereo views of the binding of CoA to PPAT. For clarity, water molecules (red spheres) are not labeled, and only the groups that participate in binding (i.e., main chain or the side chain atoms) are shown. The protein structures shown in panels a and b are in similar orientations. Labels for residues belonging to a threefold-related subunit are italicized. Final σA (28) weighted difference omit electron density map for CoA bound to subunit B (a), where electron density is visible only for the Ppant moiety of CoA, and the twofold-related protomer A (b). The contour level of the electron density maps is 2.5σ, and the resolution is 1.78 Å. The bonds of CoA (pink) and the bonds of the enzyme (white) are shown. Color coding of atoms is explained in the legend to Fig. 2. This figure was produced with MolScript (19) or BobScript (11) and Raster3D (22) programs.
Strikingly, CoA binds to the A subunit in a unique fashion (Fig. 2 and 3b). Although it binds within the same pocket, the pantetheine arm bends in the opposite direction. Specifically, the β-mercaptoethylamine moiety of CoA engages in hydrophobic interactions with Leu-102, as well as His-138 and Glu-134 of a threefold-related subunit. The following carbonyl group is also hydrogen bonded to a water molecule and to the main chain of Met-74. The remainder of the pantothenic acid moiety makes hydrophobic contacts with the side chain of Leu-73 and the invariant Gly-9 residue and hydrogen bonds to an additional water molecule. The conserved Arg-88 residue has been identified as the crucial residue that orients the 4′-phosphate group of phosphopantetheine (16). In the PPAT:dPCoA complex, the side chain of Arg-88 participates in bifurcated hydrogen bonds to the nonesterified oxygen of the 4′-phosphate group. This phosphate makes two additional hydrogen bonds with a water molecule and with the amide of Thr-10. In contrast, the adenylyl moiety of CoA is only in loose contact with the enzyme. Specifically, the α-phosphate group forms a hydrogen bond with the hydroxyl group of conserved Tyr-98, whereas the contacts of the ribose are limited to the hydrogen bonds of the 3′-phosphate to the amide of invariant Asp-95 and to a water molecule. The adenine ring of CoA stacks on top of the phenyl ring of Tyr-98 and resides 6.5 Å away from the adenylate ring of a threefold-related CoA molecule within the PPAT hexamer. Therefore, similar to dPCoA, CoA is exposed to the solvent channel within the cavity of the oligomer.
DISCUSSION
Comparison of the PPAT:CoA complex to the substrate- and product-bound structures.
Within the dimer in the asymmetric unit, the PPAT:CoA structure adopts a backbone conformation that is virtually identical to those of the enzyme bound to its product or substrates (Fig. 2b). The overall root mean square (rms) deviations for the 314 equivalent Cα positions of either the complexed PPAT:dPCoA, PPAT:Ppant, or PPAT:ATP structure to the PPAT:CoA structure are 0.44, 0.41, and 0.49 Å, respectively. The most dramatic discrepancies between the PPAT:CoA structure compared to the substrate- or product-bound crystal structures are caused by the disordered adenyl moiety of CoA in the B subunit (Fig. 2b). Specifically, in the PPAT:CoA structure, residues 91 to 95 lining the binding pocket have moved almost 2.5 Å towards the binding cleft. These residues reside on the N terminus of the α-helix, which is sandwiched between the twofold-related helix and the binding site of the adenylate of dPCoA. This region of PPAT is involved in subunit communication (14). On the opposite site of the binding pocket, residues 38 to 43 as well as the sulfate anion (not shown) interacting with Ser-41 have shifted almost 1 Å away from the binding pocket. These movements are not seen in the substrate-bound PPAT:ATP and PPAT:Ppant structures or in the product-bound PPAT:dPCoA structure. However, it should be noted that residues 91 to 95 make up an ill-defined region with weaker electron density in all structures except in the PPAT:ATP structure, whereas residues 39 to 42 show weak electron density in all but the PPAT:CoA structure. The ordering of these residues is consistent with the presence of the adenyl moiety in both subunits in the asymmetric unit as seen in the PPAT:CoA and PPAT:ATP structures, whereas both regions are more mobile in the absence of adenyl binding as seen in the B subunit of the PPAT:dPCoA structure and both subunits of the PPAT:Ppant structure. Thus, CoA binding to the adenyl binding pocket of PPAT, but not to the pantetheine binding pocket, orchestrates these movements.
The B subunit in PPAT:CoA binds to the phosphopantetheine moiety of CoA in a manner similar to the binding of dPCoA or Ppant in the PPAT:dPCoA (magenta subunit in Fig. 2a) or in the PPAT:Ppant structure, respectively. By contrast, the A subunit in PPAT:CoA, which binds CoA in a distinct fashion, resembles the ligand-free subunit in PPAT:dPCoA (teal subunit in Fig. 2a) or the ligand-free subunit in PPAT:Ppant (Fig. 2b). Movements of up to 2 Å are also found for residues 71 to 74 among the four PPAT-complexed crystal structures in the A subunit. However, the PPAT:CoA structure is the only structure where the ligand binds in subunit A within the pantetheine binding pocket. In particular, Asp-72 is reoriented and ordered upon CoA binding in the PPAT:CoA structure compared to all of the other liganded PPAT structures.
In the twofold-related protomer (the A protomer), residues 40 to 42 show very weak electron density in the PPAT:dPCoA structure (where there is no ligand binding) but almost continuous electron density in the PPAT:CoA structure, where CoA binding is observed. Although these residues are not in direct contact with CoA, this loop, together with the loop comprising residues 71 to 74 on the other side, sandwich the pantetheine moiety of CoA. In the presence of CoA, three water molecules bind between these two loop regions, creating a water-mediated hydrogen bonding network between the amides of Ser-39, Asp-72, and the carbonyl of Asp-72. These water molecules are not found in the PPAT:dPCoA structure. Thus, CoA binding causes reorientation of Asp-72, which allows binding of water molecules, which then order residues 40 to 42.
CoA conformation and binding site.
The observation that the adenylate group of CoA does not bind to the adenylate binding site of PPAT was at first unexpected. However, modeling of the phosphate group at the 3′ position of the ribose of dPCoA revealed that major conformational changes need to occur to prevent steric hindrance of the phosphate group with the backbone of Gly-89 (2.2 Å) and Arg-88 (2.6 Å) and with the side chains of Arg-88 (1.2 Å) and Glu-99 (2.2 Å; not shown). In fact, the invariant His-18 of PPAT is a key catalytic residue in the T/HXGH motif described for many enzymes belonging to the superfamily of nucleotidyltransferase α/β phosphodiesterases. However, given its distinct binding position, His-18 does not interact with CoA. Interestingly, the adenine moieties of ATP and CoA also bind in very different orientations to pantothenate kinase, the first enzyme of the CoA biosynthetic pathway, where this also involves sets of nonoverlapping residues (36).
The reorientation of the pantetheine arm of CoA in the A subunit of PPAT:CoA is remarkable given that is still binds to the same binding pocket as dPCoA in the twofold-related subunit (Fig. 2a). dPCoA cannot bind the A subunit due to the steric hindrance imposed by residues 72 to 74. However, the pantetheine arm of CoA escapes this constraint by binding in its novel conformation.
The exclusive nature of CoA binding, but not dPCoA binding, to the A subunit is due to steric constraints. The adenylate group of CoA only weakly interacts with the A subunit. Although electron density is clearly visible for all atoms of CoA (Fig. 3b), the electron density is weaker for the adenylate group than for the remainder of the ligand. This critical difference results from the hydrogen bonding interactions made by the 3′-phosphate group of CoA, which lacks dPCoA. The crystal structure presented here, together with those previously determined for the PPAT:dPCoA (14) and PPAT:ATP (16) structures, demonstrate that the 3′-phosphate on the ribose is required for the binding of CoA in its novel conformation. Inspection of the crystal contacts shows that these contacts are not responsible for the conformational change that would force CoA to bind differently to either PPAT subunit within the asymmetric unit.
PPAT:CoA crystals were obtained from a solution of 1.1 M ammonium sulfate, 0.2 M sodium chloride, 100 mM sodium acetate (pH 5), and 10 mM CoA (15). The fact that the PPAT:CoA crystals were grown by cocrystallization rather than by CoA soaking into apo-PPAT crystals indicates that the observed hexamer population is not a crystallization artifact. Sulfate efficiently competes with phosphate or phosphate-containing compounds, such as nucleotides, for binding to proteins. For example, crystals of chloramphenicol phosphotransferase (CPT) were obtained from 1.6 M ammonium sulfate (10). In the absence of sulfate, CPT is active as a homodimer, whereas in ammonium sulfate, CPT exists as an active tetramer. Indeed, the crystal structure of the nucleotide-free protein showed that the binding of a sulfate anion coincides with the β-phosphate location of ATP in the CPT:ATP crystal structure (13). To address possible effects of the high sulfate concentration used for crystallization of PPAT, the sulfate binding sites were therefore inspected. Of the PPAT structures determined in complex with either dPCoA (14), ATP (16), Ppant (16), or CoA, only the PPAT:Ppant and PPAT:CoA crystal structures showed a bound sulfate anion that coincides with the β-phosphate location of ATP in the PPAT:ATP crystal structure. However, PPi binds to PPAT weakly, with a Km only in the millimolar range (12), compared to the tighter binding of ATP, which has an apparent Km in the micromolar range. Therefore, it is very unlikely that the sulfate anion that is present 1.1 Å near the β-phosphate location of ATP can compete with the nucleotide for binding to PPAT. Indeed, the PPAT:CoA crystal structure revealed only a single interaction of the enzyme to the sulfate anion located in the active site, which is consistent with a weak anion binding.
The PPAT:CoA and PPAT:dPCoA structures establish how dPCoA, but not CoA, is a substrate for PPAT. Comparison of the PPAT:dPCoA and PPAT:CoA structures shows that CoA mimics the dPCoA and Ppant substrates in the B subunit. Specifically, the binding conformation of CoA's pantetheine moiety resembles that of dPCoA or Ppant and causes similar conformational changes in the twofold-related protomer. These changes prevent CoA binding in a dPCoA- or Ppant-like conformation to the A protomer. Importantly, the structural differences found in the binding of the enzyme to its substrates, product, and now CoA can be exploited in the development of new antibiotics.
Acknowledgments
Thanks to Arie Geerlof (Heidelberg, Germany) for expert assistance with biochemistry and crystallization, Jurgen Sygusch (Montréal, Canada) for helpful discussions, and John Cleveland (St. Jude Children's Research Hospital) for helpful comments on the manuscript.
This work was supported in part by the Cancer Center Support (CORE) grant CA 21765 and by the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Aghajanian, S., and D. M. Worrall. 2002. Identification and characterization of the gene encoding the human phosphopantetheine adenylyltransferase and dephospho-CoA kinase bifunctional enzyme (CoA synthase). Biochem. J. 365:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bork, P., L. Holm, E. V. Koonin, and C. Sander. 1995. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Proteins 22:259-266. [DOI] [PubMed] [Google Scholar]
- 3.Brünger, A. T., J. Kuriyan, and M. Karplus. 1988. Crystallographic R-factor refinement by molecular dynamics. Science 235:458-460. [DOI] [PubMed] [Google Scholar]
- 4.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kunszeweski, N. Nilges, M. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. [DOI] [PubMed] [Google Scholar]
- 6.Bruser, T., T. Selmer, and C. Dahl. 2000. “ADP sulfurylase” from Thiobacillus denitrificans is an adenylylsulfate:phosphate adenylyltransferase and belongs to a new family of nucleotidyltransferases. J. Biol. Chem. 275:1691-1698. [DOI] [PubMed] [Google Scholar]
- 7.Cusack, S. 1997. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 7:881-889. [DOI] [PubMed] [Google Scholar]
- 8.D'Angelo, I., N. Raffaelli, V. Dabusti, T. Lorenzi, G. Magni, and M. Rizzi. 2000. Structure of nicotinamide mononucleotide adenylyltransferase: a key enzyme in NAD+ biosynthesis. Structure Fold. Des. 8:993-1004. [DOI] [PubMed] [Google Scholar]
- 9.Delarue, M. 1995. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 5:48-55. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, J., D. J. Campopiano, and T. Izard 1999. Cubic crystals of chloramphenicol phosphotransferase from Streptomyces venezuelae. Acta Crystallogr. Sect. D 55:1086-1088. [DOI] [PubMed] [Google Scholar]
- 11.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. Sect. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 12.Geerlof, A., A. Lewendon, and W. V. Shaw. 1999. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J. Biol. Chem. 274:27105-27111. [DOI] [PubMed] [Google Scholar]
- 13.Izard, T., and J. Ellis. 2000. The crystal structures of chloramphenicol phosphotransferase reveal a novel inactivation mechanism. EMBO J. 19:2690-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izard, T., and A. Geerlof. 1999. The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity. EMBO J. 18:2021-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izard, T., A. Geerlof, A. Lewendon, and J. J. Barker. 1999. Cubic crystals of phosphopantetheine adenylyltransferase from Escherichia coli. Acta Crystallogr. Sect. D 55:1226-1228. [DOI] [PubMed] [Google Scholar]
- 16.Izard, T. 2002. The crystal structures of phosphopantetheine adenylyltransferase with bound substrates reveal the enzyme's catalytic mechanism. J. Mol. Biol. 315:487-495. [DOI] [PubMed] [Google Scholar]
- 17.Jackowski, S., and C. O. Rock. 1981. Regulation of coenzyme A biosynthesis. J. Bacteriol. 148:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackowski, S., and C. O. Rock. 1984. Metabolism of 4′-phosphopantetheine in Escherichia coli. J. Bacteriol. 158:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 20.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 21.Martin, D. P., and D. G. Drueckhammer. 1993. Separate enzymes catalyze the final two steps of coenzyme A biosynthesis in Brevibacterium ammoniagenes: purification of pantetheine phosphate adenylyltransferase. Biochem. Biophys. Res. Commun. 192:1155-1161. [DOI] [PubMed] [Google Scholar]
- 22.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 23.Obmolova, G., A. Teplyakov, N. Bonander, E. Eisenstein, A. J. Howard, and G. L. Gilliland. 2001. Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae. J. Struct. Biol. 136:119-125. [DOI] [PubMed] [Google Scholar]
- 24.O'Toole, N., J. A. R. Barbosa, Y. Li, L.-W. Hung, A. Matte, and M. Cygler. 2003. Crystal structure of a trimeric form of dephosphocoenzyme A kinase from Escherichia coli. Protein Sci. 12:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 26.Park, Y. S., P. Gee, S. Sanker, E. J. Schurter, E. R. Zuiderweg, and C. Kent. 1997. Identification of functional conserved residues of CTP:glycerol-3-phosphate cytidylyltransferase. Role of histidines in the conserved HXGH in catalysis. J. Biol. Chem. 272:15161-15166. [DOI] [PubMed] [Google Scholar]
- 27.Perona, J. J., M. A. Rould, and T. A. Steitz. 1993. Structural basis for RNA aminoacylation by E. coli glutaminyl-tRNA synthetase. Biochemistry 32:8758-8771. [DOI] [PubMed] [Google Scholar]
- 28.Read, R. J. 1986. Improved Fourier coefficients for maps using phases from partial structures with error. Acta Crystallogr. Sect. A 42:140-149. [Google Scholar]
- 29.Robinshaw, J. D., and J. R. Neely. 1985. Coenzyme A metabolism. Am. J. Physiol. 248:E1-E9. [DOI] [PubMed] [Google Scholar]
- 30.Rossmann, M. G., A. Liljas, C. I. Brändén, and L. J. Banaszak. 1975. Evolutionary and structural relationships among dehydrogenases, p. 62-102. In P. D. Boyer (ed.), The enzymes, vol. XI, 3rd ed. Academic Press, New York, N.Y.
- 31.Vallari, D. S., and S. Jackowski. 1988. Biosynthesis and degradation both contribute to the regulation of coenzyme A content in Escherichia coli. J. Bacteriol. 170:3961-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallari, D. S., S. Jackowski, and C. O. Rock. 1987. Regulation of pantothenate kinase by coenzyme A and its thioesters. J. Biol. Chem. 262:2468-2471. [PubMed] [Google Scholar]
- 33.van Delft, F., A. Lewendon, V. Dhanaraj, T. L. Blundell, C. Abell, and A. G. Smith. 2001. The crystal structure of the E. coli pantothenate synthetase confirms it as a member of the cytidylyltransferase superfamily. Structure 9:439-450. [DOI] [PubMed] [Google Scholar]
- 34.Venkatachalam, K. V., H. Fuda, E. V. Koonin, and C. A. Strott. 1999. Site-selected mutagenesis of a conserved nucleotide binding HXGH motif located in the ATP sulfurylase domain of human bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase. J. Biol. Chem. 274:2601-2604. [DOI] [PubMed] [Google Scholar]
- 35.Weber, C. H., Y. S. Park, S. Sanker, C. Kent, and M. L. Ludwig. 1999. A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from Bacillus subtilis. Structure Fold. Des. 7:1113-1124. [DOI] [PubMed] [Google Scholar]
- 36.Yun, M., C. G. Park, J. Y. Kim, C. O. Rock, S. Jackowski, and H. W. Park. 2000. Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A. J. Biol. Chem. 275:28093-28099. [DOI] [PubMed] [Google Scholar]
- 37.Zhyvoloup, A., I. Nemazanyy, A. Babich, G. Panasyuk, N. Pobigailo, M. Vudmaska, V. Naidenov, O. Kukharenko, S. Palchevskii, L. Savinska, G. Ovcharenko, F. Verdier, T. Valovka, T. Fenton, H. Rebholz, M. L. Wang, P. Shepherd, G. Matsuka, V. Filonenko, and I. T. Gout. 2002. Molecular cloning of CoA synthase. The missing link in CoA biosynthesis. J. Biol. Chem. 277:22107-22110. [DOI] [PubMed] [Google Scholar]