Abstract
The expression of virulence determinants in Staphylococcus aureus is controlled by global regulatory loci (e.g., sarA and agr). One of these determinants, protein A (spa), is activated by sarS, which encodes a 250-residue DNA-binding protein. Genetic analysis indicated that the agr locus likely mediates spa repression by suppressing the transcription of sarS. Contrary to SarA and SarR, which require homodimer formation for proper function, SarS is unusual within the SarA protein family in that it contains two homologous halves, with each half sharing sequence similarity to SarA and SarR. Here we report the 2.2 Å resolution X-ray crystal structure of the SarS protein. SarS has folds similar to those of SarR and, quite plausibly, the native SarA structure. Two typical winged-helix DNA-binding domains are connected by a well-ordered loop. The interactions between the two domains are extensive and conserved. The putative DNA-binding surface is highly positively charged. In contrast, negatively charged patches are located opposite to the DNA-binding surface. Furthermore, sequence alignment and structural comparison revealed that MarR has folds similar to those of SarR and SarS. Members of the MarR protein family have previously been implicated in the negative regulation of an efflux pump involved in multiple antibiotic resistance in many gram-negative species. We propose that MarR also belongs to the winged-helix protein family and has a similar mode of DNA binding as SarR and SarS and possibly the entire SarA protein family member. Based on the structural differences of SarR, SarS, and MarR, we further classified these winged-helix proteins to three subfamilies, SarA, SarS, and MarR. Finally, a possible transcription regulation mechanism is proposed.
Staphylococcus aureus is a versatile bacterium capable of causing a wide spectrum of pathology in humans, ranging from superficial abscesses to pneumonia, endocarditis, and sepsis (3, 4). This versatility may be attributable to the impressive array of extracellular and cell wall-associated virulence determinants coordinately expressed during the infectious process (32). Many of these virulence factors can be generally classified into two groups. Secreted proteins such as hemolysins, enterotoxins, lipase, and proteolytic enzymes are responsible for invasion and tissue damage. Cell surface-associated proteins such as protein A, fibronectin-binding proteins, and collagen-binding protein mediate adhesion to host tissues (25). The coordinate expression of many virulence determinants in S. aureus has been shown to be regulated by global regulatory elements such as sarA and agr (9, 22). These regulatory elements, in turn, control the transcription of a wide variety of unlinked genes, many of which have been implicated in pathogenesis.
The global regulatory locus agr encodes a two-component, quorum-sensing system that is involved in the generation of two divergent transcripts, RNA II and RNA III, from two distinct promoters, P2 and P3, respectively. RNA III, initiated from the P3 promoter, is the regulatory molecule of the agr response and is hence responsible for the up-regulation in extracellular protein production and the down-regulation of cell wall-associated protein synthesis during the postexponential growth phase (19, 30). The RNA II molecule, driven by the P2 promoter, encodes a four-gene operon, agrBDCA. Additionally, AgrD, in conjunction with AgrB, participates in the generation of an octapeptide with quorum-sensing function (20, 27). This autoinducing peptide can stimulate the transcription of the agr regulatory molecule RNA III, which ultimately interacts with target genes to modulate transcription and possibly translation (28, 30).
Contrasting to agr, the sarA locus up-regulates the synthesis of selected extracellular and cell wall proteins. The sarA locus also represses the transcription of the protein A gene (8). The sarA locus is composed of three overlapping transcripts, sarA P1, sarA P3, and sarA P2. Each of these transcripts encodes the major 372-bp sarA gene, yielding the 14.5-kDa SarA protein (3). DNA footprinting studies revealed that the SarA protein binds to promoters of several target genes (13), including agr, hla, spa, and fnbA, implicating SarA as a regulatory molecule that can modulate target gene transcription via both agr-dependent and agr-independent pathways (7, 13, 14).
Besides the SarA protein, a series of SarA homologs have been discovered within the S. aureus genome (11). Functional characterization of some of these proteins has led to the discovery of a complicated transcription regulation network (4, 11, 32). Structural elucidations of some family members at atomic resolution have been carried out. Novel structural features that are involved in DNA binding and a possible activation or repression mechanism of this transcription factor family have been revealed. We recently reported the dimeric SarR structure in the absence of DNA (24). Our data indicated that SarR belongs to the typical winged-helix DNA-binding protein family. In particular, SarR contains two atypical winged-helix motifs that recognize its target DNA not only through “helix-turn-helix major groove interaction” but also via the “wing-minor groove interaction” (24). We proposed that the two conserved acidic patches on the convex side of the SarR structure are possible activation motifs, while the concave surface, containing a tract of basic residues, is likely to be the DNA-binding domain (24). Moreover, we recently proposed that SarR, SarA, and SarS may have similar folds, contrary to the reported SarA structure that has a completely different topology from SarR and might represent a non-native-state structure of the SarA protein, as proposed by Schumacher et al. (35). Notably, the recently reported monomeric structure of MarR, despite its low sequence similarity to SarR, still has similar topological folds as the SarR protein. Nevertheless, the proposed mode of DNA binding of MarR is different from that of SarR (1).
SarS, originally described by Tegmark and colleagues and Cheung et al. (12, 36), is an activator for the expression of protein A (spa), a major surface protein presenting in over 90% of S. aureus strains. SarS is repressed by agr and sarA (12, 36). In particular, the repression of protein A (spa) by agr is mediated by the repression of sarS (12). In contrast to the smaller SarA or SarR, SarS encodes a 250-residue protein that contains two homologous halves, each of which is similar to SarA and other related homologs (12, 36). To address the similarities and differences in transcription regulation carried out by these divergent SarA family members, we determined the structure of the SarS protein at 2.2 Å resolution.
MATERIALS AND METHODS
Protein expression, purification, and crystallization.
The intact 750-bp sarS gene was amplified by PCR using chromosomal DNA from S. aureus strain RN6390 as the template and primers containing flanking restriction sites (XhoI and BamHI) to facilitate cloning into an expression vector (pET14b) (Novagen). The recombinant plasmid containing the sarS gene was transformed to Escherichia coli BL21(DE3)(pLysS). Enhanced expression of a SarS-His6 fusion was induced by adding isopropyl-1-thio-β-d-galactopyranoside to an 8-liter growing culture (37°C) at an optical density at 650 nm of 0.7. After 4 h of additional growth, cells were harvested, resuspended in buffer (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 5% glycerol), and subjected to cell lysis through a continuous-flow French press. After a 20,000 × g centrifugation step, the soluble fraction was loaded onto a 10-ml nickel resin affinity column and the SarS-His6 fusion protein was eluted with 250 mM imidazole. Thrombin was added to the SarS-His6 solution for overnight His6-tag cleavage. The solution containing thrombin-cleaved SarS was desalted and loaded onto a MonoS (Pharmacia) ion-exchange column. After elution with an NaCl gradient (0.1 to 0.5 M), the fraction containing the protein was found to be homogeneous as determined by a Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel. The concentration of the purified protein was determined with a Bio-Rad protein assay solution (Bio-Rad Laboratories, Richmond, Calif.), using bovine serum albumin as the standard. The SarS protein (15 mg/ml) was crystallized by vapor diffusion against a solution of 5 mM β-mercaptoethanol, 100 mM HEPES (pH 7.5), 3.5 to 4.0 M NaCl. For cryo-crystallography, crystals were soaked in steps of increasing glycerol concentration (5% each step every 30 min) and finally into 20% glycerol before flash-freezing.
Structure determination and refinement.
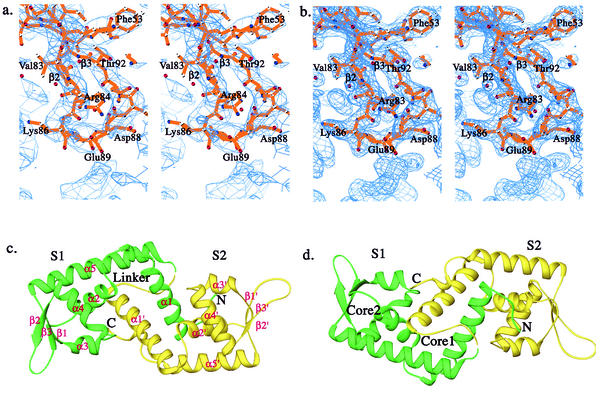
Native crystals diffracted to 2.8 Å with an in-house Rigaku R-axis IV system. Two heavy atom derivatives were prepared by adding 1 mM HgCl or 10 mM trimethyl lead acetate in 100 mM HEPES (pH 7.5), 4.0 M NaCl crystal soaking solutions for 24 h. Both derivative crystals diffracted to 3.0 Å at an in-house X-ray facility. A 2.8 Å native data set and derivative data sets were collected in the laboratory on a Rigaku R-axis IV system. Another 2.2 Å resolution native data set was collected at BM-19 of SBC at Advanced Photon Resource of Argonne National Laboratory. Data processing was performed with DENZO and SCALEPACK (31). The space group of the crystal was P6 (1)22 or P6 (5)22 with cell dimensions of a = 84.887 Å, b = 84.887 Å, c = 195.743 Å, α = 90°, β = 90°, and γ = 120° and with one SarS molecule in the asymmetry unit. The initial phases were obtained by multiple isomorphous replacement (MIR) using the program SOLVE (37) and the electron density modification program SOLOMON (14a), with the three data sets collected at home (one native set, two derivative sets) at 3.0 Å (Table 1 and Table 2). Both space groups were tested during the initial phase calculation. The final space group resolved by the output map from final solvent flattening was P6 (5)22. The quality of the output map was good with all main chains connected, secondary structure resolved, and some of the ordered side chains revealed (Fig. 1a and b). Map interpretation and model building were performed using the program O (21) aided by the SarR model (24) and residues with big side chains. Model refinement was performed with the Crystallography & NMR system (5) using the 2.2 Å resolution native data set collected at 19-BM of SBC with root mean square distance (RMSD) bonds = 0.008 and RMSD angle = 1.58 (Table 1). The final model contains the complete SarS (residues 1 to 250) and 121 water molecules (Fig. 1c and d). Stereochemical values are all within or better than the expected ranges for a 2.2 Å structure, as determined using PROCHECK (23).
TABLE 1.
Experimental data on crystal structure determinationa
| Data set | Resolution (Å) | Rmerge (%) | No. of unique reflections | Total observations | Completeness (%) | Phasing power | No. of sites | <I>/<Isigma> |
|---|---|---|---|---|---|---|---|---|
| Native | 2.2 | 8.1 (50.0) | 22,016 | 576,575 | 99.4 (98.2) | 44 (2.0) | ||
| HgCl2 | 3.0 | 10.0 (39.8) | 8,706 | 59,020 | 97.2 (93.0) | 0.50 | 3 | 25 (1.9) |
| Trimethyl-Pb | 3.0 | 9.6 (40.3) | 8,619 | 55,739 | 96.0 (64.5) | 0.67 | 3 | 23 (2.3) |
Rmerge = Σ|Ij −<I>|/ΣIj, with Bijvoct pairs treated as equivalent for native and as different for derivatives. Total observations, the number of full and partial observations measured with non-negative intensity to the indicated resolution. Completeness, the percentage of possible unique reflections measured with I/σ(I) ≥ 0 to the indicated resolution. Phasing power = <FH>/Erms. No. of reflections, the number of reflections used in refinement for each resolution bin. R factor = Σ|FO − FC|/ΣFO for all amplitudes with F/σ(F) ≥ 0 measured in the indicated resolution bin; the free R factor is calculated with 5% of the data in each bin. Numbers in parentheses are values in the highest resolution shell. There are 121 water molecules in the current model.
TABLE 2.
Crystal structure refinement data
| Parameter | Value |
|---|---|
| Mean figure of merit | 0.25a |
| Refinement resolution | 20-2.2 Å |
| No. of reflections | 21,142 |
| R factor | 24.50% |
| Free R factor | 28.20% |
| Ramachandran plot | |
| Residues in most-favored regions | 89.7% |
| Residues in additional allowed regions | 9.5% |
| Residues in generously allowed regions | 0.8% |
For 8,681 phased reflections.
FIG. 1.
Overall structure of SarS. (a) The initial MIR map at the beta-hairpin region of S1 with the final SarS model, which is one of the most flexible parts in the entire SarS structure. (b) The final 2Fo-Fc map with the final SarS model at the same region as in panel a. (c) Ribbon diagram of the three-dimensional structure of the SarS protein. The first domain, S1, is shown in green. The second domain, S2, is yellow. (d) Orientation of the panel, 180°. Two hydrophobic cores are labeled. All figures were prepared using RIBBONS (6), except for Fig. 1a and b, which were prepared using BobScript (15).
Data deposition
The atomic coordinates and reflections have been deposited in the Protein Data Bank as 1P4X.
RESULTS
Overall structure of the SarS protein.
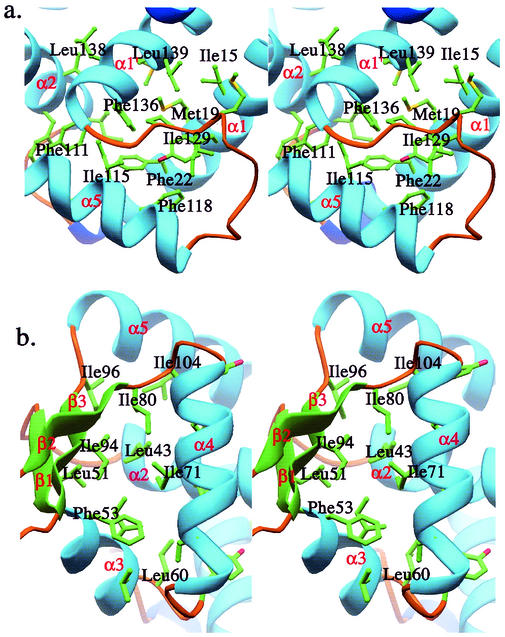
The structure of the SarS protein reported here has a similar topology to the dimeric SarR structure. Overall, there are five alpha-helices and three beta-strands in each SarS homologous domain, designated S1 and S2, respectively (Fig. 1). There are three coil regions, which are located at the N terminus, C terminus, and the gap region connecting S1 and S2. Briefly, α1, α2, α5, and α1′ are primary helices that form the main interaction interface to bring two domains together by extensive hydrophobic contacts. The interaction between the two domains is further strengthened by the linker that connects S1 and S2, which makes the relative movement between the two domains difficult. There are in total 19 residues involved in the interactions, including residues Ile15, Tyr18, Met19, Phe22, and Val22 from α1, residues Ile34, Phe37, and Ile38 from α2, residues Phe111, Ile114, Ile115, and Phe118 from α5, residues Met128, Ile129, and Pro130 from the linker region, and residues Phe136, Leu137, Leu139, and Met140 from α1′ (Fig. 2a).
FIG. 2.
The detailed hydrophobic cores of SarS. (a) The hydrophobic core 1, which brings S1 and S2 tightly together. (b) The hydrophobic core 2, which stabilizes the winged-helix motif.
Interestingly, the helix-turn-helix motifs and the beta-hairpins in both S1 and S2 are surrounded by a very rigid environment consisting of a large number of hydrophobic residues, including residues from α2, α3, α4, and α5, and residues from three beta-strands. In S1, residues Leu39, Leu40, and Leu43 from α2, residue Leu51 from β1, residues Phe53, Ile56, Val57, and Leu60 from α3, residues Tyr62, Leu67, Ile71, and Leu74 from α4, residues Tyr79 and Ile80 from β2, residues Ile94 and Ile96 from β3, and residue Ile104 from α5 build up the huge hydrophobic environment (Fig. 2b). This huge hydrophobic core leads to a compact structure for the entire winged-helix motif. For example, α3, α4, and the beta-hairpin (β2 and β3) are well ordered compared to their flexible counterparts in SarR. In SarR, the tip of the hairpin is completely disordered. This is also in contrast to other winged-helix proteins, such as CAP, which has quite a different conformation of the entire winged-helix motif with or without DNA. However, we did not have the DNA-protein complex structure yet, but it is reasonable to predict that SarS may have subtle conformational changes with DNA. It may be argued that the beta-hairpin in S1 has crystal contact with the symmetry-related molecule, and this could stabilize the beta-hairpin to some extent. However, the corresponding part in S2 does not have the packing environment and is well ordered, although we just see a slight conformation difference.
This rigid feature remains true for the α5 helix (or w2) as well, which is loosely attached to the left part of the molecule in SarR but tightly associated in S1 and S2. Residues Phe22 and Val26 from α1, residues Val30, Met32, Ile34, and Phe37 from α2, residues Leu110, Phe111, Ile114, Ile115, and Phe118 from α5, and residue Phe136 from α1′ form the hydrophobic core (data not shown), which stabilizes helix α5.
Overall, the above special arrangement renders the entire SarS structure a tightly packed domain, different from all other winged-helix proteins, including SarR. This kind of special structure could underlie a specific transcription regulation mechanism for the SarS protein and its subfamily.
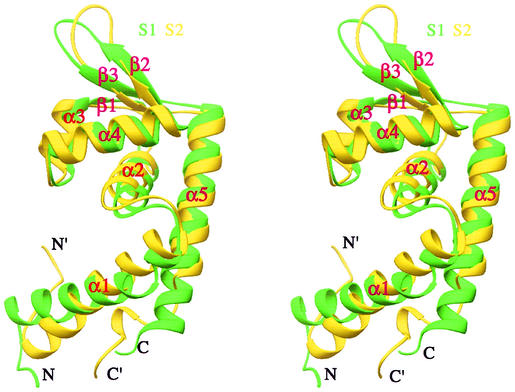
The superposition of the two domains.
As mentioned above, the two halves of SarS (S1 and S2) are each similar to the SarR structure. They are similar to each other, with an RMSD of 2.8 Å. Superimposition of S1 and S2 showed some differences (Fig. 3). First, the relative orientations of α1 and α1′ in their corresponding domains are quite different. Second, α5 is much longer than α5′. Otherwise, the two domains are extremely similar. If only the helix-turn-helix and w1 (the beta-hairpin) are considered (without α1 and α5), the RMSD is 1.6 Å.
FIG. 3.
Comparison of the two SarS domains. S1 is shown in green, and S2 is yellow.
Putative DNA-binding and activation surfaces.
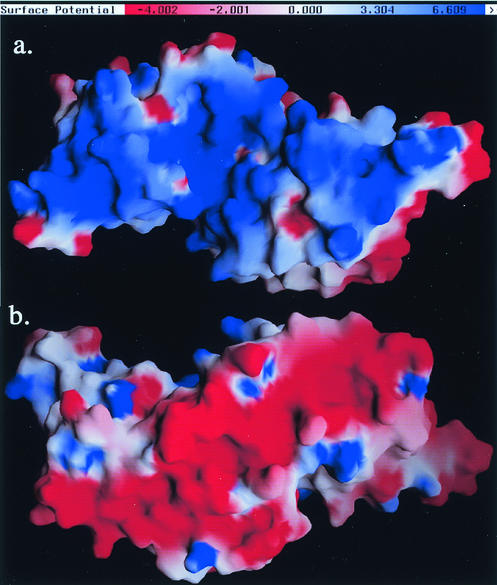
Like SarR, SarS is a highly positively charged protein with a calculated pI of 9.4 (12, 36). The positively charged residues are randomly distributed in the primary sequence but accumulated on the concave side of the structure consisting of the two winged-helix motifs. The electrostatic potential on the surface of SarS, calculated by the GRASP program (29), revealed a positively charged track on this side. Thus, the concave side, similar to SarR, is the most likely site for DNA binding (24) (Fig. 4a).
FIG. 4.
The electrostatic potential surface of SarS calculated by GRASP (29), with charges of +1 for Lys and Arg, −1 for Glu and Asp, and zero for all other residues. The color bar from red to blue represents the potential from negative to positive, defined as in GRASP. (a) The putative DNA-binding surface; (b) the possible activation motifs.
In contrast, a negatively charged surface track was noted on the opposite side of the putative DNA-binding side. Similar to SarR, which has acidic patches on its convex surface, we assume the continuous negatively charged track might be the activation motifs (24). Further characterization of these patches is necessary to confirm our speculation (Fig. 4b).
Comparison to SarR.
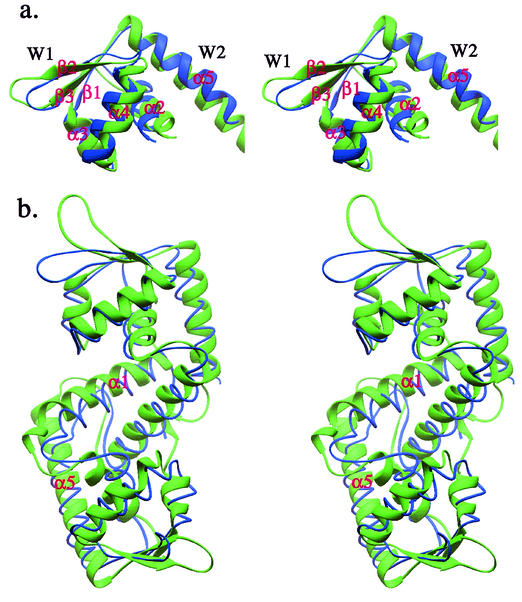
To reveal the structural similarities and differences between SarS and SarR, superimposition of SarS on the SarR dimer was carried out. The RMSD of the winged-helix motif was 2.7 Å (Fig. 5a). This is much bigger than that of S1 and S2 (1.6 Å). The overall main chain RMSD between SarS and SarR is 3.2 Å, which is close to that of S1 and S2 (2.8 Å), with a structural similarity Z-score of 9.1 from the program Dali (18) (Fig. 5b). The major differences are in the following regions. First, the SarS structure looks a little more stretched than that of SarR. Second, the helix-turn-helix motifs of SarS are not highly flexible, contrary to other winged-helix proteins (e.g., SarR) in the absence of DNA. As we mentioned above, SarS has a very compact structure, including the two hairpins, which are well ordered in the initial MIR map. Third, the lengths of the two helices, the so-called second wing (w2) in the protein family, are different in both length and orientation between the two structures.
FIG. 5.
Comparison of SarS with SarR. (a) The overlap of winged-helix motifs from SarS and SarR. (b) The overlap of entire SarS over SarR homodimer. SarS is shown in green, and SarR is blue.
Comparison to MarR.
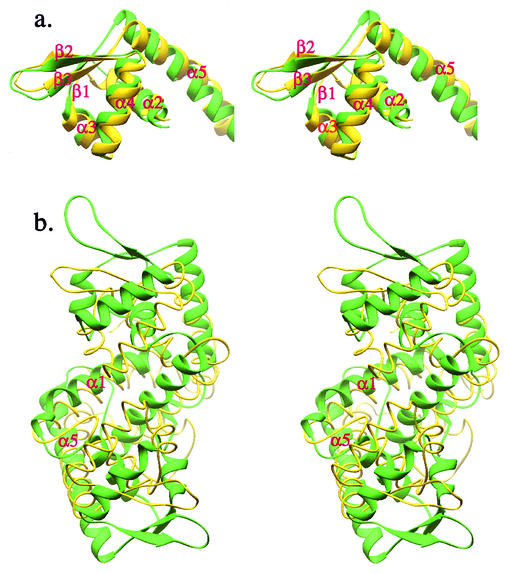
The structure of MarR, the regulatory element that negatively controls an efflux pump involved in multiple antibiotic resistance in E. coli, has been elucidated (1). Although Alekshun et al. did not mention that MarR is a winged-helix protein, it has a very similar topology to SarR and SarS. Except for the helix at its C terminus, all other secondary structures have corresponding counterparts in SarR or SarS. When comparing the MarR monomer to S1, the RMSD is 3.2 Å, which is similar to those of S1 with S2 and SarS with SarR. Interestingly, MarR also has a very long beta-hairpin, similar to those of SarS and SarR, which is possibly involved in its interaction with the minor groove of target DNA. If only the winged-helix motif is considered, the RMSD between S1 and MarR is 2.3 Å, which is smaller than that between SarS and SarR (2.7 Å) and bigger than that between S1 and S2 (1.6 Å), with a structural similarity Z-score of 9.3 (Fig. 6a). The similarity of these two subdomains between SarS and MarR is obvious. It is safe to say that MarR also belongs to the winged-helix protein family. Therefore, we speculate that MarR has a DNA-binding feature and mode of binding similar to those of SarS or SarR, albeit different from the known modes of DNA binding described for winged-helix proteins (16). Interestingly, when the putative MarR functional dimer was superimposed onto a single SarS molecule or a SarR homodimer, big discrepancies with RMSDs of 8.2 Å were observed (Fig. 6b). While MarR has two symmetric winged-helix motifs, the arrangement of the two motifs is quite different from those of SarS and SarR. The relative width of the putative DNA-binding surface in MarR (56 Å) is much narrower than that in SarS (86 Å) or SarR (80 Å). This narrowness could be caused by crystal packing, since the two motifs are relatively flexible in the absence of DNA for most winged-helix family members. Alternatively, it may also reflect the real DNA-binding state, requiring a shorter DNA fragment to fit the structural feature. Overall, the entire MarR structure looks more loosely packed than SarR, and especially than SarS.
FIG. 6.
Comparison of SarS and MarR. (a) The overlap of winged-helix motifs from SarS and MarR. (b) The overlap of the entire SarS over the MarR homodimer. SarS is shown in green, and SarR is yellow.
DISCUSSION
The subfamily classification.
As discussed above, the homodimers of SarR and MarR and the monomer of SarS have very similar overall structures. The differences among them are also obvious enough for additional classification into three subfamilies. In the first subfamily, five helices and three short beta-strands are needed to build up a global structural domain for SarR, SarA, and possibly SarT, a repressor of α-hemolysin expression (34). Two identical domains form the functional homodimer. In the second subfamily (SarS, SarU, and SarY) (26), two heterologous but similar domains, connected by a well-ordered linker, are harbored in each monomer. Each half of the molecule has the exact topology of individual members in the first subfamily. In the third subfamily, there is an additional helix at the C terminus, for example, the MarR monomer. Besides the normal five helices and three beta-strands, this additional helix also further strengthens the monomer-monomer interaction. Interestingly, in searching the S. aureus genome (www.ncbi.nlm.nih.gov/genomics), we came across two MarR-like homologs (A. L. Cheung et al., unpublished data). Structural elucidation of these two MarR-like proteins will provide us with insightful information on the structure and function relationship of this subfamily. It is possible that transcriptional regulation of the three aforementioned groups could be controlled by different association features, especially when we consider the SarR homodimer to have the lowest stability, MarR in the middle, and SarS as the most stable functional unit.
The putative MarR DNA-binding mode.
All three groups of proteins belong to the winged-helix family. Although the original prediction of the DNA-binding surface for MarR is at an unusual surface (1), based on our structural alignment we speculate that MarR should have a similar DNA-binding surface as that of SarR or SarS. The helix-turn-helix should bind to the major groove of DNA, and the beta-hairpin binds to the minor groove of the bended double helix DNA. The latter interaction is unique for these three groups of proteins and the BmrR protein (17). Furthermore, this interaction should be nearly symmetric.
Possible regulation mechanisms.
As discussed in the literature (2, 8-14), members of the SarA protein family characterized so far have dual functions, acting as activators in one transcription pathway and then as a repressor in another. How these transcription factors carry out their specific and individual functions is still unknown. Recently, Arvidson and his colleagues proposed a universal transcription regulation theory: all these subfamily members primarily act as a repressor, so the repressor's repressor actually functions as an activator, as derived from the accumulating data on the regulation of hla by the SarA protein (2). While this repressor hypothesis represents a very attractive transcription regulation mechanism, it remains to be determined if this model can be applied to other family members. Accordingly, additional in vitro transcription assays on a putative activator, such as SarS on the spa promoter, will be needed to verify this model. Regardless of the repressive or activating roles, we do not have a clear idea of how these transcription factors carry out their actual function. As mentioned in the previous section, there are acidic patches on the convex side of the SarR and SarS structures; these patches possibly act as activation motifs to recruit RNA polymerase to the promoter region via direct electrostatic interaction with a positively charged surface of the RNA polymerase subunits, such as the C-terminal domains of the alpha-subunit. Schumacher et al. proposed a bending and shrinking model based on their complex structure of SarA with or without DNA (35). We propose that the multiple bending points (at least four bending points) of target DNA that result from binding by the winged-helix protein might lead to the foreshortening of DNA, resulting in activation (24).
Besides the above plausible regulation mechanisms, there is an additional but very interesting DNA-binding feature of this family, possibly providing a hint of a novel regulation mechanism. Chien et al. and Rechtin et al. have previously reported the existence of multiple binding sites on many target promoter regions for corresponding family members, based on accumulating protein-promoter DNA gel-retarding assays and DNase I footprinting experiments (14, 33). For example, there may be multiple binding sites on the agr promoter region for the SarA protein (33). We also found that multiple SarS proteins could bind to the spa promoter region (12). This binding property has been reflected on the laddering protein-DNA bands on gel-shift assays. These findings indicate that one homodimer of SarA or SarR, or one SarS monomer, may not be enough to activate or repress the corresponding gene transcription. How many actual copies of SarA and SarR homodimer or SarS monomers are required for their proper function and how they are spatially arranged within the promoter region are not clear. In inspecting the SarS packing environment, we do find extensive interactions between two neighboring SarS molecules. The beta-hairpin of one SarS molecule interacts with that of the other (the corresponding side). Several hydrophobic residues, such as Ile174, Leu176, and Leu217 from both molecules, are involved (data not shown). In addition to the hydrophobic interactions, there are also multiple hydrogen bonds (data not shown). At the present time, we do not have data to suggest if this dimerization underlies some of the functional requirements for the SarS protein. Further characterization of the oligomeric state of SarS in solution is required to answer this question.
From the above discussion, we classified the SarA and MarR protein families into three subfamilies. The subtle but distinct structural differences could cause diverse regulation outputs. From the structural comparison standpoint, we speculated that MarR could have a DNA-binding surface similar to those of SarA, SarR, and SarS. Furthermore, solving the structure of the putative MarR members in S. aureus would shed light on our hypothesis.
Acknowledgments
We thank R. Zhao for reading our manuscript; Howard Hughes Medical Institute, Zuckerman/Canyon Ranch, and Allen Lapporte for support of our X-ray and computing facility; SBC (BM-19) at APS for high-resolution data; and Philippa C. Marrack, James D. Crapo, and other members at the National Jewish Medical and Research Center for their kind support.
G.Z. is supported by a Pew Scholar Award. This work was supported by a start-up fund from the National Jewish Medical and Research Center and NIH grant AI50678 (A.C. and G.Z.).
REFERENCES
- 1.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyce, J. M. 1997. Epidemiology and prevention of nosocomial infections, p. 309-329. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Carson, M. 1996. Wavelets and molecular structure. J. Comput. Aided Mol. Des. 10:273-283. [DOI] [PubMed] [Google Scholar]
- 7.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., K. Eberhardt, and J. H. Heinrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., and G. Zhang. 2001. Do SarA and SarR possess similar structures? Trends Microbiol. 9:570-573. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:D1825-D1842. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, L. C., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphlococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 14.Chien, Y. T., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 31:991-1001. [DOI] [PubMed] [Google Scholar]
- 14a.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 15.Esnouf, R. M. 1997. BobScript v2.4 changes (C). J. Mol. Graphics 15:132-134. [Google Scholar]
- 16.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110-116. [DOI] [PubMed] [Google Scholar]
- 17.Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]
- 18.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 19.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 22.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH, New York, N.Y.
- 23.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y., A. Manna, R. Li, W. E. Martin, R. C. Murphy, A. L. Cheung, and G. Zhang. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6877-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowy, F. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 26.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 30.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski, Z., and W. Minor. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 32.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 33.Rechtin, T. M., A. F. Gillaspy, M. A. Schumacher, R. G. Brennan, M. S. Smeltzer, and B. K. Hurlburt. 1999. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33:307-316. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a prepressor of α-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher, M. A., B. K. Hurlburt, and R. G. Brennan. 2001. Crystal structures of SarA, a pleiotropic regulator of virulence genes in S. aureus. Nature 409:215-219. (Erratum, 414:85.) [DOI] [PubMed]
- 36.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. J. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]