Abstract
Gene Dr1184 from Deinococcus radiodurans codes for a Nudix enzyme (DR-CoAse) that hydrolyzes the pyrophosphate moiety of coenzyme A (CoA). Nudix enzymes with the same specificity have been found in yeast, humans, and mice. The three-dimensional structure of DR-CoAse, the first of a Nudix hydrolase with this specificity, reveals that this enzyme contains, in addition to the fold observed in other Nudix enzymes, insertions that are characteristic of a CoA-hydrolyzing Nudix subfamily. The structure of the complex of the enzyme with Mg2+, its activating cation, reveals the position of the catalytic site. A helix, part of the N-terminal insertion, partially occludes the binding site and has to change its position to permit substrate binding. Comparison of the structure of DR-CoAse to those of other Nudix enzymes, together with the location in the structure of the sequence characteristic of CoAses, suggests a mode of binding of the substrate to the enzyme that is compatible with all available data.
Nudix hydrolases, pyrophosphatases that hydrolyze nucleoside diphosphates linked to another moiety (x), are widely distributed enzymes characterized by the Nudix box, a highly conserved consensus sequence, GX5EX7REUXEEXGU (where U is usually Ile, Leu, or Val) (4). A large number of sequences containing the Nudix motif have been found in the genomes of species from all three kingdoms, in organisms ranging in complexity from viruses to humans. Many of these proteins have been enzymatically characterized; their substrates include nucleoside triphosphates (5, 8, 27, 28), coenzymes (2, 14, 17, 29, 39), sugar nucleotides (25, 39, 40), and dinucleoside polyphosphates (1, 9, 10, 18, 29, 33). These substrates are either potentially toxic compounds, cell signaling molecules, regulators of cellular metabolism, or metabolic intermediates whose concentrations require modulation in different stages of the cell cycle. Recently, coenzyme A (CoA) and its derivatives were identified as Nudix substrates in Deinococcus radiodurans (39), yeast, mice (Nudt7-M), and humans (Nudt7-H) (17). Interestingly, all of them have higher levels of activity for oxidized disulfide CoA than for CoA. An additional conserved motif, identified by the code name UPF0035 (13), was found just N-terminal to the Nudix motif in all of these enzymes (Fig. 1). The UPF0035 motif (LLTXR(SA)X3RX3GX3FPGG; hereafter the NuCoA motif) is present in CoA-hydrolyzing Nudix enzymes and is postulated to be involved in CoA recognition (Fig. 1). CoA plays a central role in lipid metabolism. In the cytosol it is used mainly in the initial steps of fatty acid synthesis; in mitochondria it is required for oxidation of fatty acids and for the citric acid cycle. In peroxisomes it functions in the initial oxidation of very long-chain fatty acids prior to their complete oxidation in mitochondria. Peroxisomes are also involved in bile acid, cholesterol, and plasmalogen synthesis and in amino acid and purine metabolism. In most of these processes, CoA has the very important role of activating fatty acids for further modification into key biological signaling molecules.
FIG. 1.
Alignment of the sequences of characterized Nudix CoAses. Conserved residues are shown as white letters in black boxes. Conservative substitutions are shown with grey letters in open boxes. Stars indicate residues of the UPF0035 (NuCoA) motif; triangles indicate residues of the Nudix motif. GenBank GI numbers for sequences are as follows: DR1184, D. radiodurans, gi 15806203; Nudt7_H, human, gi 1848966; Nudt7_M, mouse, gi 12746409; Pcdlp, yeast, gi 6323180.
Pcd1p, the yeast CoA pyrophosphatase, has the C-terminal peroxisome targeting signal sequence PTS1, SKL; targeting was confirmed by subcellular fractionation (2). (In yeast, peroxisomes are the sole site of fatty acid β-oxidation.) The mouse homolog of Pcd1p (Nud7_M) (Fig. 1) was also reported to be targeted to the peroxisome (17). The highly homologous human enzyme (Nud7_H) (Fig. 1) is also expected to be targeted to this organelle. Peroxisomes provide an oxidizing environment which favors conversion of CoA to its disulfide form. The postulated function of CoA pyrophosphatases (CoAses) is the elimination of oxidized, inactive CoA, which can inhibit CoA-utilizing enzymes. In bacteria, this enzyme could have a similar function. Since the bacterial cytoplasm is usually a reductive environment, the need of a CoA pyrophosphatase would arise mainly under conditions of oxidative stress. This scenario is fully compatible not only with the general function of Nudix enzymes but also with the biological idiosyncrasy of D. radiodurans—high resistance to radiation and other stresses.
D. radiodurans is an organism that can withstand gamma-radiation fluxes 200-fold greater than those withstood by Escherichia coli and is also highly resistant to UV radiation and to desiccation (38). It has recently been proposed that an unusual DNA topology makes DNA repair more efficient in this organism, thus contributing to its resistance (23). Another source of this remarkable resistance to stress may be the presence in the genome of D. radiodurans of an uncommonly large number (21) of Nudix hydrolases, contributing to the elimination of deleterious compounds (4). The product of gene Dr1184 of D. radiodurans (hereafter referred to as DR-CoAse), a Nudix hydrolase, contains the NuCoA motif (Fig. 1) and was shown to hydrolyze CoA (39). In this paper we report the structure of DR-CoAse, alone and in complex with Mg2+, its activating cation. These structures, the first of a Nudix CoAse, are a major step toward understanding the mechanism and the specificity of CoA pyrophosphatases.
MATERIALS AND METHODS
Protein expression and purification.
pET24a, containing the Dr1184 gene under the control of the T7 promoter, was used to transform the E. coli expression host, TUNER (DE3). A single colony was inoculated into 3 ml of Luria-Bertani medium containing 30 μg of kanamycin/ml and grown overnight at 37°C. The cells were transferred to 100 ml of Luria-Bertani medium and grown to exponential phase. The 100 ml of culture was transferred to 2 liters of culture medium and grown to an optical density at 600 nm of 0.5. The culture was induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside and grown for an additional 4 h.
The induced cells were harvested, washed by suspension, resuspended in 25 ml of buffer A (50 mM Tris [pH 7.5], 1 mM EDTA), sonicated, and centrifuged, and the precipitate was discarded. The protein concentration of the supernatant was adjusted to 10 mg/ml with buffer A (fraction I). A typical preparation yielded approximately 300 mg of total protein from 3 g of cells.
Nucleic acids were precipitated by addition of 1/10 volume of 10% streptomycin sulfate, and after 15 min on ice, the precipitate was removed by centrifugation. The supernatant was collected, ammonium sulfate was added to 30% saturation, the sample was centrifuged, and the supernatant was brought to 45% saturation of ammonium sulfate. The precipitate was dissolved in 10 ml of buffer A and dialyzed overnight against 2 liters of buffer B (50 mM Tris [pH 7.5], 1 mM EDTA, 300 mM NaCl). The solution was then loaded at 0.5 ml/min onto a HiPrep 26/60 Sephacryl S-200 high-resolution gel filtration column preequilibrated with buffer B at 4°C. After this step the protein was at least 85% pure. The eluted fractions, dialyzed against 2 liters of buffer A, were loaded onto a mono-Q anion exchange column and eluted using a continuous NaCl gradient (0 to 0.5 M) in the same buffer at a rate of 1 ml/min, collecting 2-ml fractions. Protein concentrations were determined by absorbance at 280 nm. The purified DR-CoAse was stable for months at −80°C.
Crystallization of the DR-CoAse.
DR-CoAse, at a concentration of 7 mg/ml in buffer A, was crystallized at 20°C in hanging drops using a precipitant solution consisting of 27% PEG 1500-100 mM sodium citrate (pH 6.0). In 1 month, overlapped plate-shaped crystals appeared in the drop. After 3 months, these crystals grew to a size of 0.5 by 0.5 by 0.02 mm. For data collection, the multiple crystal conglomerate was broken into several single crystals. Crystals were cryoprotected in a solution containing 30% PEG 1500, 100 mM sodium citrate (pH 6.0), and 20% glycerol and flash frozen at 100 K.
Structure determination.
Native data were collected to a 2.09 Å resolution at beamline X25 of the National Synchrotron Light Source, Brookhaven National Laboratory. Diffraction data were processed with HKL_2000 (30). Consideration of systematically absent reflections revealed that the crystals belong to space group P21212 with the following unit-cell parameters: a = 40.6 Å, b = 94.0 Å, c = 43.7 Å. These cell dimensions and space group symmetry are compatible with one DR-CoAse molecule in the asymmetric unit. Molecular replacement trials with Amore (26) and Molrep (34, 35) using either MutT (37) (Protein Data Bank accession code 1TUM) or the dimeric ADP-ribose pyrophosphatase from E. coli (EC-ADPRase) (15) (Protein Data Bank accession code 1GQ9) were unsuccessful. Thus, structure determination was carried out using heavy atom derivatives.
Three derivatives were prepared by soaking preformed crystals under the conditions shown in Table 1. After the heavy atom sites were determined by visual inspection of difference Patterson maps, phases calculated to 3.5 Å resolution with the program SOLVE (32) had an overall figure of merit of 0.47 (20 to 3.5 Å; 0.35 in the last resolution shell) using all the data and including anomalous differences for the gadolinium derivative. Phases were improved by solvent flattening, histogram matching, and multiresolution modification using the program DM (11). An electron density map calculated with these phases was used to trace the chain using the program O (19, 20) (residues 1 to 22, 33 to 56, and 63 to 190). Refinement was performed using CNS v1.1 (7) with a residual target. Rebuilding and correction of the model was guided by σA-weighted 2Fo-Fc electron density maps. R and R-free (calculated with randomly selected 10% of the reflections) were used to monitor refinement of the model (6). The structure of the Mg2+ complex was refined in a similar manner using data to a 1.7 Å resolution. Residue 4, not visible in the native structure, had clear electron density in this complex and was built and refined.
TABLE 1.
Statistics for data collection and refinement
| Data set | Value for groupa
|
||||
|---|---|---|---|---|---|
| Native Apo enzyme | Heavy atom derivatives
|
Complexed with MgCl2 | |||
| Gd(Ac)3 | Pt(NH)2Cl2 | PtPIPb | |||
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 |
| Resolution | 30-2.09Å | 30-2.5Å | 30-3.0Å | 30-3.0Å | 30-1.7Å |
| Soaking conditions | |||||
| Metal (mM) | 0.5 | 5 | 1 | 50 | |
| Time (h) | 9 | 20 | 20 | 18 | |
| Data collection and phasing statistics | |||||
| Observed reflections | 161,533 | 43,453 | 11,377 | 11,091 | 100,105 |
| Unique reflections | 10,529 | 6,005 | 3,685 | 3,747 | 19,112 |
| Completenessc | 99.2% (94.3) | 93.0% (87.7) | 72.7% (71.7) | 66.6% (56.5) | 99.6% (97.5) |
| I/σd | 8.9 (8.0) | 23.0 (8.6) | 4.8 (2.3) | 6.6 (2.0) | 15.4 (5.5) |
| Rsyme | 0.061 | 0.071 | 0.092 | 0.097 | 0.062 |
| R cullisf | 0.67 | 0.88 | 0.62 | ||
| Figure of merit | 0.47 | ||||
| Refinement | |||||
| R crystal/R free | 0.200/0.279 (0.248/0.295) | 0.216/0.258 (0.313/0.325) | |||
| Model compositiong | |||||
| Amino acids (atoms) | 170 (1,318) | 171 (1,326) | |||
| Metal | 1 | ||||
| Water molecules | 96 | 124 | |||
| Total atoms | 1,414 | 1,451 | |||
| Stereochemistryh | |||||
| rmsi bond length (Å) | 0.006 | 0.012 | |||
| rms angles (°) | 1.37 | 1.38 | |||
| Temp factors | |||||
| 〈B-factor protein〉 | 20.6 | 20.81 | |||
| 〈B-factor metal〉 | 29.61 | ||||
| 〈B-factor water〉 | 24.3 | 29.54 | |||
Cell constants: a = 40.6Å, b = 94.0Å, c = 43.7Å.
PtPip, Di-(mu)-iodobis(ethylenediamine)-di-platinum(II) nitrate PIP.
Completeness in the highest resolution shell in parentheses.
I/σ in the highest resolution shell in parentheses.
Rsym = Σh (Σj |Ij,h−<Ih>|/ΣIj,h), where h is set of Miller indices and j is set of observations of reflection h.
Rcullis = Σ||Fh| - (|Fph|−|Fp|)|/Σ|(|Fph|−|Fp|)|, where Fph is observed derivative structure factor, Fp is observed native structure factor, and Fh is calculated heavy atom structural factor.
Model composition shows one monomer in asymmetric unit.
Over 90% of main chain dihedrals fall within the “most allowed regions” of the Ramachandran plot.
rms, root mean square.
Other computations.
The quality of the structures was assessed with the program PROCHECK (22). Figures were drawn with MOLSCRIPT (12, 21), BOBSCRIPT, and RASTER3D (24).
Coordinates.
Coordinates of the structures have been deposited in the Protein Data Bank (accession codes 1NQY and 1NQZ).
RESULTS AND DISCUSSION
Overall structure.
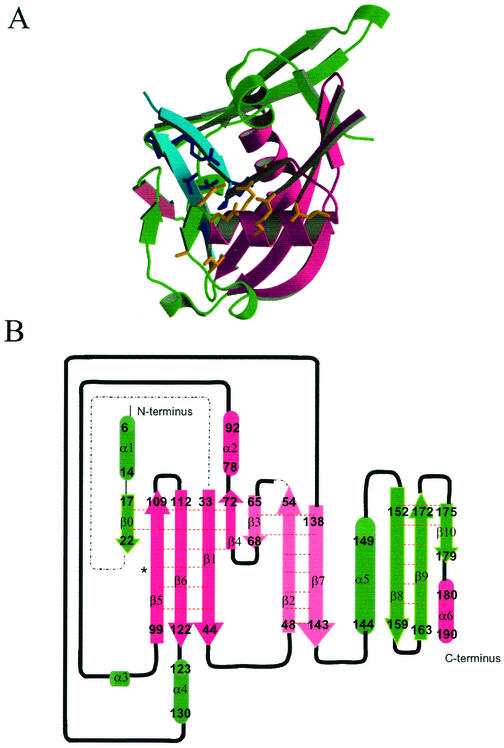
The structure of DR-CoAse was determined by multiple isomorphous replacement (Table 1). The final model, comprising residues 5 to 22, 33 to 56, and 63 to 190 (the complete DR-CoAse has 194 residues), was refined using data to 2.09 Å to an R value of 20.0% (R-free = 27.9%) with excellent geometry (Table 1). The molecule consists of a single globular monomer formed by 6 α-helices (α1 to α6) and 11 β-strands (β0 to β10). Its core is similar to the Nudix fold observed in other members of the Nudix family: a mixed β-sheet formed by four β-strands with two α-helices, one on each side (15) (Fig. 2A). In DR-CoAse this core is formed by strands β1, β4, β5, and β6 and helices α2 and α6. Strands β2, β3, and β7 form another antiparallel β-sheet that is also present in all Nudix enzymes of known structure (see below). As in other Nudix enzymes, the Nudix motif of DR-CoAse (residues 71 to 92) folds as a loop-helix-loop structure (helix α2 in DR-CoAse). Gly-71, the first residue of the motif, is the last residue of strand β4. Before the Nudix fold, DR-CoAse has an α-helix (α1) and a β-strand (β0). Helix α1 partially occupies the site that is used for substrate binding in other Nudix enzymes.
FIG. 2.
Structure of the DR-CoAse. (A) Schematic representation of the three-dimensional structure of DR-CoAse. The regions corresponding to the Nudix fold are magenta, those of NuCoA are turquoise (side chains in dark blue), and the rest of the structure is emerald green. The side chains of the Nudix motif are shown in orange. (B) Secondary structure diagram of the DR-CoAse. Strands are shown as arrows, helices are shown as cylinders, and connecting regions are shown as black lines. Dashed lines show regions that were not built in the structure. The beginning and the end of each element are shown.
The NuCoA motif occurs in residues 51 to 71 and comprises strands β2, β3, and β4 of the structure (Fig. 2A and B). Strand β-2 is at the center of an antiparallel β-sheet and makes hydrogen bonds with strands β3 and β7. Strand β4 starts immediately following β3 at a 90° bend that occurs at Pro-69. It makes hydrogen bonds with strand β1 and forms the edge of a β-sheet. Phe-68 and Leu-51 pack toward the inside of the molecule and form a core that pins the NuCoA motif to the rest of the structure. Gly-70 and Gly-71, the two last residues of the sequence Phe-Pro-Gly-Gly of the NuCoA motif, occur in an extended conformation. Gly-71 is also the first residue of the Nudix motif.
Two antiparallel β-strands, β8 and β9, are inserted into the Nudix C-terminal region. This C-terminal extension is not present in other Nudix hydrolases (see below).
Metal binding site.
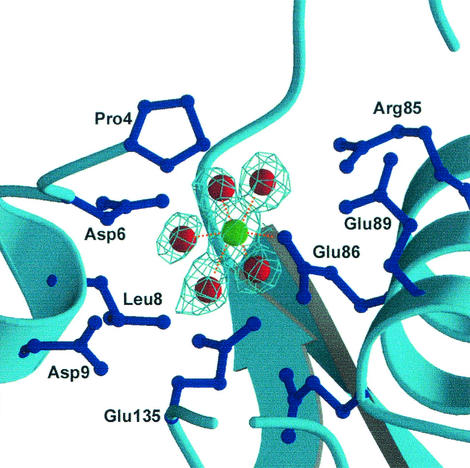
All Nudix enzymes require a divalent cation, such as Mg2+ or Mn2+, for activity (4). To identify the metal binding site(s) of DR-CoAse, we determined the structure of the enzyme in the presence of 50 mM Mg2+. After inclusion of the Mg2+ ion identified in a difference electron density map, the structure refined against the DR-CoAse/Mg2+ data (1.7 Å resolution) to an R value of 21.6% (R-free = 25.8%) with excellent geometry (Table 1). The cation is coordinated by five water molecules and the side chain carboxylate of Glu-86 (Fig. 3). The Mg2+-O distances vary between 2.1 and 2.4 Å. The position of this ion is similar to that of the central ion of the three-Mg2+ (or Mn2+) cluster found in the structure of the EC-ADPRase (16). The presence of this ion binding site establishes that the catalytic site of the DR-CoAse is formed by the same residues of the Nudix motif as those of other Nudix enzymes.
FIG. 3.
Electron density of the coordination of the Mg2+ ion. The Mg2+ ion has a near-perfect octahedral coordination formed by five water molecules and the side chain of Glu 86 of the Nudix motif. Other side chains surrounding the position of the ion are also shown.
Comparison with other Nudix enzymes
As mentioned above, all Nudix hydrolases share the conserved Nudix fold. Within the fold, residues of the Nudix motif bind the activating cation and are directly involved in catalysis (15, 16). Differences in specificity for diverse substrates are determined mainly by the N-terminal extension or by residues in variable loop regions. With the structure reported here, the first Nudix enzyme with CoAse activity, the structures of Nudix enzymes belonging to four different subfamilies have now been determined. Their preferred substrates are the following: CoA, ADP-ribose (ADPR) (15), nucleoside triphosphates (3), and Ap4A. (31) (A fifth structure has been determined [36], but the specificity of this enzyme remains unknown.) Alignment of the three-dimensional structures of these enzymes reveals both common and distinctive features (Fig. 4). All Nudix enzymes are built around the Nudix fold, a core formed by four β-strands and two α-helices. We call the secondary structure elements of this fold Nu-β1, Nu-β2, Nu-α1, Nu-β3, Nu-β4, and Nu-α2, independent of the numbers they are assigned in the individual structures. An additional antiparallel β-sheet formed by three short strands (Nu-β′1, Nu-β′2, and Nu-β′3) is also present in all the structures, but its length, geometry, and position are highly variable (Fig. 4). Helix Nu-α1 and the loops preceding and following it contain the Nudix motif in all the enzymes. MutT, the simplest Nudix enzyme, is about 130 residues long and is essentially just a Nudix fold with connecting loops (Fig. 4B) (37). EC-ADPRase has, in addition to the Nudix fold, a 54-residue N-terminal extension formed by three antiparallel β-strands (Fig. 4C) (15) that is involved in the formation of the dimer by domain swapping. The N-terminal extension of each monomer makes contact with the substrate bound to the other monomer and is a major contributor to the specificity of the enzyme. Ap4A hydrolase (Fig. 4D) (31) contains all the secondary structure elements of the Nudix fold, plus an additional α-helix that connects strands Nu-β3 and Nu-β4 (equivalent to strands β5 and β6 of DR-CoAse). This helix, which is away from the binding site in the free enzyme, moves to close over the substrate and provides additional contacts during the catalytic cycle (31).
FIG. 4.
Comparison of the Nudix hydrolases of known structure. (A) Sequence comparison based on the alignment of the three-dimensional structure of the four enzymes. No attempt was made to align the sequences of the regions not structurally conserved (black letters). Secondary structure elements are indicated: helices with thick lines, strands with arrows. Elements common to all four enzymes are shown in red. Elements characteristic of individual enzymes are shown in blue. Bold characters show the helices. (B, C, D, and E) Schematic representation of the four known structures of Nudix hydrolases. The Nudix fold (magenta), including the additional three-stranded sheet (pink), is highlighted in all the structures. (B) MutT. This enzyme consists of only these elements. (C) EC-ADPRase. Additional portions of the structure, including the N-terminal extension involved in dimer formation by domain swapping, are shown in turquoise. (D) Ap4Ase. Additional regions, including the α-helical insertion, are shown in orange. (E) DR-CoAse. Additional regions, including the N-terminal and the C-terminal extensions, are shown in green.
DR-CoAse has, in addition to the C-terminal extension mentioned above, a 32-residue N-terminal extension, consisting of one α-helix (α1) and one β-strand (β0) (Fig. 4E) (15). Helix α-1 overlaps the position occupied by the ADPR in EC-ADPRase and by AMPCPP in E. coli MutT.
CoA recognition.
Alignment of the structures of EC-ADPRase,MutT, and Ap4A in complex with their substrates shows that the position of the diphosphate with respect to the helix of the Nudix motif is highly conserved. Alignment of the structures of the EC-ADPRase and the DR-CoAse shows that the divalent cation site observed in DR-CoAse is in a position similar to that occupied by one of the Mg2+ ions of the EC-ADPRase (15, 16). These two observations make it possible to model the position of the diphosphate of CoA in the binding site of the DR-CoAse.
In CoA, the diphosphate has substituents at both phosphates: an adenosine-3′-phosphate moiety and an N-mercaptoethyl-pantothenylamide (Fig. 5A). With the diphosphate in the predicted position, it is clear that there is no room in the binding site to accommodate both of these groups. The culprit is helix α1, which overlaps the volume at one side of the diphosphate. This volume is used by the other Nudix enzymes to bind one side of the substrate. In the case of DR-CoAse, the substrate, CoA, has groups of significant size on both sides of the diphosphate (Fig. 5A). It is therefore reasonable to assume that helix α1 has to move away from its position in the apo enzyme for the substrate to bind. Movement of helix α1 would free the second side of the binding site for CoA binding.
FIG. 5.
Binding of CoA to DR-CoAse. (A) Chemical structure of CoA. (B) Schematic representation of the portions of the DR-CoAse structure that interact with CoA. (C) Surface representation of the binding site. Positive regions are in blue; negative regions are in red. (In panels B and C, helix α1 was omitted.).
The remaining problems are to find the conformation and the actual position of CoA in the binding site. One might think that predicting which side is occupied by the adenosyl is straightforward. Unfortunately, this is not the case. Ap4A has adenosine moieties at both sides of the diphosphate, and the nucleoside moieties of AMPCPP and ADPR bind to MutT and ADPRase at opposite sides of the binding site (15). It can be argued that the reversal in the position of the nucleoside in the EC-ADPRase with respect to MutT is a consequence of the dimeric nature of the protein: a loop from the other monomer partially occludes one side of the binding site, limiting the volume of this side to one that can accommodate only a ribose (15). Furthermore, the bend at the end of this loop that packs directly against the ribose has a proline residue that is characteristic of the ADPR-hydrolyzing subfamily of Nudix enzymes (15).
Based on these considerations, it seems likely that the adenosine-3′-P moiety of CoA binds to the DR-CoAse in the position occupied by AMPCPP in MutT. Placing the adenosyl moiety in this position orients the pantothenate and the mercaptoethanolamine in the direction of the strands that contain the NuCoA motif. With CoA in the resulting orientation, it is possible to find a conformation and a position for this half of the substrate that result in specific contacts with residues of the NuCoA motif (Fig. 5B and C). In this model, the diphosphate makes a hydrogen bond with the side chain of Arg-55 and with the carbonyl oxygen of Gly-70. It makes also a close contact with the Cα of Gly-71. The phosphate at the 3′ position of the ribose makes an H-bond with the side chain oxygen of Ser-72. The carbonyl of the pantothenyl moiety forms an H-bond with the main chain NH of Gln-65. Additional close contacts are made by other portions of the substrate. The interactions of adenine and the ribose in the model are similar to those found in MutT and Ap4A. One interesting feature of this model is that the substrate has an L-shaped conformation that conforms to the 90° bend of the second and third strands of the NuCoA motif fostering contacts between the pantothenyl-mercaptoethylamine moiety and the NuCoA motif. Additional contacts may be formed between the substrate bound in this position and the new position of helix α1. In this binding arrangement, a hydrophobic surface in the binding site of the free enzyme is covered by helix α1. This makes it possible for the enzyme to provide a hydrophobic surface for interaction with the extensive hydrophobic portions of the substrate without having this surface exposed to solvent in the free enzyme. This characteristic provides a mechanism for increasing the specificity of the DR-CoAse: only substrates that can make these hydrophobic interactions are able to displace the helix and become hydrolyzed by the enzyme.
The need to move helix α-1 to bind the substrate would also explain the failure of CoA or parts of this molecule to bind to the DR-CoAse crystals. Extensive attempts involving diffusion into preformed crystals and cocrystallizations failed to yield crystals of the complexes.
Summary and conclusions.
The structure of the DR-CoAse presented here is the first of a Nudix enzyme with CoAse activity. DR-CoAse shares the basic fold of the other Nudix enzymes of known structure. In addition to the Nudix motif, DR-CoAse has two inserted sequences: an N-terminal helix followed by a strand and a C-terminal, long hairpin formed by two strands. The NuCoA motif, characteristic of CoA-hydrolyzing Nudix enzymes, folds as a two-stranded hairpin followed by a 90° turn of the second strand at a conserved proline residue.
Alignment of the structure of DR-CoAse with that of other Nudix hydrolases shows that helix α-1 occupies part of the site required for substrate binding. Thus, substrate binding must involve movement of this helix. With helix α1 out of the way, a model for binding of CoA can be proposed in which the substrate interacts with the NuCoA motif characteristic of Nudix CoAses.
Acknowledgments
Support was provided by NIH grants GM51362 and GM66895 (to L.M.A.) and GM18649 (to M.J.B.). The personnel of beamline X25 of the National Synchrotron Light Source, Brookhaven National Laboratory, are gratefully acknowledged.
REFERENCES
- 1.Abdelghany, H. M., L. Gasmi, J. L. Cartwright, S. Bailey, J. B. Rafferty, and A. G. McLennan. 2001. Cloning, characterisation and crystallisation of a diadenosine 5′,5‴-P(1),P(4)-tetraphosphate pyrophosphohydrolase from Caenorhabditis elegans. Biochim. Biophys. Acta 1550:27-36. [DOI] [PubMed] [Google Scholar]
- 2.AbdelRaheim, S. R., and A. G. McLennan. 2002. The Caenorhabditis elegans Y87G2A.14 Nudix hydrolase is a peroxisomal coenzyme A diphosphatase. BMC Biochem. 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abeygunawardana, C., D. J. Weber, A. G. Gittis, D. N. Frick, J. Lin, A. F. Miller, M. J. Bessman, and A. S. Mildvan. 1995. Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry 34:14997-15005. [DOI] [PubMed] [Google Scholar]
- 4.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 5.Bhatnagar, S. K., L. C. Bullions, and M. J. Bessman. 1991. Characterization of the mutT nucleoside triphosphatase of Escherichia coli. J. Biol. Chem. 266:9050-9054. [PubMed] [Google Scholar]
- 6.Brunger, A. T. 1997. Free R value: cross-validation in crystallography. Methods Enzymol. 277:366-396. [DOI] [PubMed] [Google Scholar]
- 7.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. Read, L. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Bullions, L. C., V. Mejean, J. P. Claverys, and M. J. Bessman. 1994. Purification of the MutX protein of Streptococcus pneumoniae, a homologue of Escherichia coli MutT. Identification of a novel catalytic domain for nucleoside triphosphate pyrophosphohydrolase activity. J. Biol. Chem. 269:12339-12344. [PubMed] [Google Scholar]
- 9.Cartwright, J. L., P. Britton, M. F. Minnick, and A. G. McLennan. 1999. The IalA invasion gene of Bartonella bacilliformis encodes a (de)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem. Biophys. Res. Commun. 256:474-479. [DOI] [PubMed] [Google Scholar]
- 10.Cartwright, J. L., and A. G. McLennan. 1999. The Saccharomyces cerevisiae YOR163w gene encodes a diadenosine 5′,5‴-P1,P6-hexaphosphate (Ap6A) hydrolase member of the MutT motif (Nudix hydrolase) family. J. Biol. Chem. 274:8604-8610. [DOI] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project, no. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Esnouf, R. M. 1999. Further additions to Molscript version 1.4, including reading and countouring of electron density maps. Acta Crystallogr. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 13.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, H. K., and B. A. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frick, D. N., and M. J. Bessman. 1995. Cloning, purification, and properties of a novel NADH pyrophosphatase. Evidence for a nucleotide pyrophosphatase catalytic domain in MutT-like enzymes. J. Biol. Chem. 270:1529-1534. [DOI] [PubMed] [Google Scholar]
- 15.Gabelli, S. B., M. A. Bianchet, M. J. Bessman, and L. M. Amzel. 2001. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 8:467-472. [DOI] [PubMed] [Google Scholar]
- 16.Gabelli, S. B., M. A. Bianchet, Y. Ohnishi, Y. Ichikawa, M. J. Bessman, and L. M. Amzel. 2002. Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase. Biochemistry 41:9279-9285. [DOI] [PubMed] [Google Scholar]
- 17.Gasmi, L., and A. G. McLennan. 2001. The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives. Biochem. J. 357:33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaywee, J., W. Xu, S. Radulovic, M. J. Bessman, and A. F. Azad. 2002. The Rickettsia prowazekii invasion gene homolog (invA) encodes a Nudix hydrolase active on adenosine (5′)-pentaphospho-(5′)-adenosine. Mol. Cell Proteomics 1:179-185. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjelgaard. 1991. Improved methods for binding protein models to electron density maps and the location of errors in these models. Acta Crystallogr. A 42:110-119. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. A., and M. Kjeldgaard. 1997. Electron-density map interpretation. Methods Enzymol. 277:173-208. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis, J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 22.Laskowski, R., M. MacArthur, D. Moss, and J. Thornton. 1993. PROCHECK: a program to check the sterochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 23.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minsky. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 24.Merrit, E., and D. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 25.Moreno-Bruna, B., E. Baroja-Fernandez, F. J. Munoz, A. Bastarrica-Berasategui, A. Zandueta-Criado, M. Rodriguez-Lopez, I. Lasa, T. Akazawa, and J. Pozueta-Romero. 2001. Adenosine diphosphate sugar pyrophosphatase prevents glycogen biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:8128-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A50:157-163.
- 27.O'Handley, S. F., C. A. Dunn, and M. J. Bessman. 2001. Orf135 from Escherichia coli is a Nudix hydrolase specific for CTP, dCTP, and 5-methyl-dCTP. J. Biol. Chem. 276:5421-5426. [DOI] [PubMed] [Google Scholar]
- 28.O'Handley, S. F., D. N. Frick, L. C. Bullions, A. S. Mildvan, and M. J. Bessman. 1996. Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins. Cloning, purification, and characterization of the enzyme. J. Biol. Chem. 271:24649-24654. [DOI] [PubMed] [Google Scholar]
- 29.O'Handley, S. F., D. N. Frick, C. A. Dunn, and M. J. Bessman. 1998. Orf186 represents a new member of the Nudix hydrolases, active on adenosine(5′)triphospho(5′)adenosine, ADP-ribose, and NADH. J. Biol. Chem. 273:3192-3197. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 277:307-326. [DOI] [PubMed] [Google Scholar]
- 31.Swarbrick, J. D., T. Bashtannyk, D. Maksel, X. R. Zhang, G. M. Blackburn, K. R. Gayler, and P. R. Gooley. 2000. The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L. J. Mol. Biol. 302:1165-1177. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorne, N. M., S. Hankin, M. C. Wilkinson, C. Nunez, R. Barraclough, and A. G. McLennan. 1996. Human diadenosine 5′,5‴-P1,P4-tetraphosphate pyrophosphohydrolase (Ap4A hydrolase) possesses a MutT motif. Biochem. Soc. Trans. 24:209S. [DOI] [PubMed]
- 34.Vagin, A., and A. Teplyakov. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56:1622-1624. [DOI] [PubMed] [Google Scholar]
- 35.Vagin, A. A., and M. N. Isupov. 2001. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 57:1451-1456. [DOI] [PubMed] [Google Scholar]
- 36.Wang, S., C. Mura, M. R. Sawaya, D. Cascio, and D. Eisenberg. 2002. Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets. Acta Crystallogr. D Biol. Crystallogr. 58:571-578. [DOI] [PubMed] [Google Scholar]
- 37.Weber, D. J., C. Abeygunawardana, M. J. Bessman, and A. S. Mildvan. 1993. Secondary structure of the MutT enzyme as determined by NMR. Biochemistry 32:13081-13088. [DOI] [PubMed] [Google Scholar]
- 38.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, W., J. Shen, C. A. Dunn, S. Desai, and M. J. Bessman. 2001. The Nudix hydrolases of Deinococcus radiodurans. Mol. Microbiol. 39:286-290. [DOI] [PubMed] [Google Scholar]
- 40.Yang, H., M. M. Slupska, Y. F. Wei, J. H. Tai, W. M. Luther, Y. R. Xia, D. M. Shih, J. H. Chiang, C. Baikalov, S. Fitz-Gibbon, I. T. Phan, A. Conrad, and J. H. Miller. 2000. Cloning and characterization of a new member of the Nudix hydrolases from human and mouse. J. Biol. Chem. 275:8844-8853. [DOI] [PubMed] [Google Scholar]