Abstract
To confirm that Mycobacterium tuberculosis chaperonin 10 (Cpn10) is secreted outside the live bacillus, infected macrophages were examined by electron microscopy. This revealed that the mycobacterial protein accumulates both in the wall of the bacterium and in the matrix of the phagosomes in which ingested mycobacteria survive within infected macrophages. To understand the structural implications underlying this secretion, a structural study of M. tuberculosis Cpn10 was performed under conditions that are generally believed to mimic the membrane environment. It was found that in buffer-organic solvent mixtures, the mycobacterial protein forms two main species, namely, a partially helical monomer that prevails in dilute solutions at room temperature and a dimer that folds into a β-sheet-dominated structure and prevails in either concentrated protein solutions at room temperature or in dilute solutions at low temperature. A partially helical monomer was also found and was completely associated with negatively charged detergents in a micelle-bound state. Remarkably, zwitterionic lipids had no effect on the protein structure. By using N- and C-truncated forms of the protein, the C- and N-terminal sequences were identified as possessing an amphiphilic helical character and as selectively associating with acidic detergent micelles. When the study was extended to other chaperonins, it was found that human Cpn10 is also monomeric and partially helical in dilute organic solvent-buffer mixtures. In contrast, Escherichia coli Cpn10 is mostly dimeric and predominately β-sheet in both dilute and concentrated solutions. Interestingly, human Cpn10 also crosses biological membranes, whereas the E. coli homologue is strictly cytosolic. These results suggest that dissociation to partially helical monomers and interaction with acidic lipids may be two important steps in the mechanism of secretion of M. tuberculosis Cpn10 to the external environment.
The Mycobacterium tuberculosis chaperonin10 antigen (M. tuberculosis Cpn10), also known as hsp10, or MPB57, or the 10-14 kDa antigen, is one of the most abundant proteins found in short-term culture filtrates of logarithmically growing M. tuberculosis (1). M. tuberculosis Cpn10 is also known to associate with cell wall components in mycobacterium preparations (4) and is therefore believed to be a secreted protein. Secretion of M. tuberculosis Cpn10 outside the cell may be important, since live mycobacteria are more effective than killed organisms in eliciting protective immunity, and this difference is thought to be due to protective antigens which are secreted only by the growing microorganism (28, 34). Consistent with this hypothesis is the powerful immune-stimulating activity observed with the mycobacterium and studies showing that M. tuberculosis Cpn10 can inhibit autoimmune models of disease (35).
Secretion is unusual for Cpn10s because they exert their physiological function of assisting the folding and refolding of other proteins in the cytosol (18). However, there are emerging examples of secreted Cpn10s, including the Helicobacter pylori and Campylobacter jejuni homologues, which have been detected in culture media (45, 47), and Cpn10-X, which accumulated in large amounts in the periplasm of E. coli transformed with a groE operon cloned from X bacteria (25). In contrast, E. coli Cpn10 is strictly intracellular. Mammalian and yeast Cpn10s also cross biological membranes as they are imported into the mitochondria in a fashion characteristic of archetypal mitochondrial precursors (40). Furthermore, recent evidences indicate that mammalian Cpn10 is also present in other cellular compartments, such as secretory granules of different cell types (41). The mechanism of transport of these proteins remains unclear because they do not contain a cleavable signal peptide.
In their path to secretion, proteins experience a variety of different environments, and this may have an effect on their structure. In the case of Cpn10s, structural changes may involve their quaternary structure, since at higher concentrations, Cpn10s form heptamers that fully dissociate to monomers at micro- to submicromolar concentrations (5, 16).
The aim of the work described here is threefold: (i) to confirm that M. tuberculosis Cpn10 is secreted from M. tuberculosis; (ii) to study the structural preferences of M. tuberculosis Cpn10 under conditions similar to those experienced by the protein when it approaches the lipid-water interface or crosses the hydrophobic lipid bilayer of the cytosolic membrane (7, 12, 19), and (iii) to understand whether any relationship at all exists between these structures and M. tuberculosis Cpn10 secretion. To achieve these aims, macrophages have been infected with virulent M. tuberculosis and the intracellular localization of the M. tuberculosis Cpn10 has been determined by immunogold labeling. Furthermore, by using three different Cpn10s, namely, those from M. tuberculosis, Homo sapiens, and E. coli, and synthetic peptides corresponding to N- and C-truncated forms of these proteins, a structural study was performed by circular dichroism (CD), nuclear magnetic resonance (NMR), and fluorescence spectroscopies and by mass spectrometry in order to study the structural preferences of these proteins in a hydrophobic environment and in the presence of detergents.
MATERIALS AND METHODS
Materials.
The recombinant and chemical syntheses of M. tuberculosis Cpn10 and its N-truncated fragments have been described elsewhere (11).
Peptides 1-25 and 1-15 were made using conventional peptide chemistry and purified by reversed-phase high-performance liquid chromatography (M+H+, expected for peptide 1-25, 2681; found, 2684; expected for peptide 1-15, 1705, found 1706).
Recombinant h-Cpn10 was obtained as described earlier (11), while E. coli Cpn10 was purchased from Stressgen.
The organic solvents used in this study were methanol (MeOH), trifluoroethanol (TFE), acetonitrile (AcCN), and hexafluoroacetone.
Anti-M. tuberculosis Cpn10 polyclonal antibodies.
New Zealand White rabbits (Charles River, Calco, Italy) were immunized subcutaneously twice with 250 μg of Cpn10 emulsified with complete (first immunization) or incomplete (second immunization) Freund's adjuvant. Ten days after the second injection, blood samples were collected from the ear vein. Sera were positive for the presence of anti-Cpn10 antibodies, as defined both by enzyme-linked immunosorbent assay and Western blot analysis, and the 50% titer was 1:50,000, as determined by enzyme-linked immunosorbent assay. It was determined that this polyclonal antiserum did not cross-react with human Cpn10 protein.
Morphological analysis.
The J774.2 murine monocyte cell line (obtained from the European Collection of Cell Cultures, Salisbury, United Kingdom) was infected in vitro with a clinical isolate of M. tuberculosis obtained from a patient with pulmonary tuberculosis. The isolate was acid fast, and the identification as M. tuberculosis was performed with standard procedures and confirmed by the Accuprobe technique (Gentile Probe, San Diego, Calif.). For each experiment, a subculture of frozen stock was made in 7H9 medium. After 9 days of cultivation at 37°C, a bacterial suspension was prepared in Dulbecco's modified Eagle medium to contain approximately 106 cells/ml. Six milliliters of this suspension was added to the J774 monolayer in a 100-mm-diameter culture dish. After incubation periods of 30 min and 1, 3, and 24 h at 37°C in the presence of 5% CO2, the monolayers were washed three times to remove extracellular bacteria and fixed in 4% paraformaldehyde for 20 min.
Immunogold labeling of M. tuberculosis Cpn10.
Pellets of J774-infected cells were embedded in acrylic resin (White resin; Polyscience Inc, Warrington, Pa.), 90 nm-thick sections were cut, and ultrastructural immunolocalization was performed using the Cpn10 rabbit polyclonal antisera (dilution, 1:5,000). The detection system consisted of goat anti-rabbit immunoglobulin G-coated colloidal gold particles, 10 nm in diameter (Ylem, Avezzano, Italy). Sections mounted on nickel grids were counterstained with uranyl acetate and observed in a Zeiss CEM 902 electron microscope. All appropriate controls for nonspecific binding were included.
Analytical ultracentrifugation (AUC).
Sedimentation velocity and equilibrium experiments were performed in a Beckman Optima XL-A analytical ultracentrifuge equipped with scanning absorbance optics. Samples were loaded into a centrifuge cell with a 12-mm optical path length; 400 μl of sample (concentration of protein, 0.8 μM in 155 mM PO43 −MeOH [3:7, vol/vol]) and 410 μl of buffer (166 mM PO43 −MeOH [3:7, vol/vol]) were used for a sedimentation velocity experiment, and between 70 and 120 μl of each were used for sedimentation equilibrium experiments.
Size exclusion chromatography (SEC).
Experiments were carried out using a Hewlett Packard 1090 high-performance liquid chromatography apparatus and a TSK-G3000 SWXL column (300 by 7.8 mm) (Toyo Pearl; Tosoh Corporation, Tokyo, Japan). The elution buffer was either 50 mM PO43 − or 166 mM PO43 − methanol (3:7, vol/vol). The flow rate was 0.38 ml/min, and monitoring was at 214 nm.
CD spectroscopy.
CD measurements were performed at 22°C using a Jasco J-600 spectropolarimeter calibrated with d-10 camphorsulphonic acid. Cells with 1- or 0.1-cm path lengths were used for dilute (i.e., 0.01 mg/ml) and concentrated protein solutions, respectively. Spectra were recorded at 0.1-nm intervals in the range 185 to 250 nm. They were the average of eight scans collected at 50 nm/min each, with a bandwidth set at 2 nm, and were baseline corrected by subtracting the corresponding blank.
For the titration experiments, stock solutions of peptides and proteins were prepared in KH2PO4 buffer (pH 7.4) and then diluted with the organic solvent to the desired concentration. The concentration of phosphate ions in the buffer was kept at either 2.5 or 50 mM during titration of the organic solvent in buffer.
For the sodium dodecyl sulfate (SDS) or di-myristoylphosphatidylcholine (DMPC) titration experiments, a solution of either 0.1 or 0.01 mg/ml protein-peptide in 10 mM phosphate buffer, pH 7.4, was used.
Prior to use, the solutions were equilibrated for approximately 30 min at the desired temperature. The observed ellipticity was converted to mean residue weight ellipticity [Θ]\(deg · cm2 · d · mol−1). On some occasions, smoothing of the curves, using a mild function which increased the signal-to-noise ratio without altering the shape of spectra, was applied with the Jasco J-700 program. Superimposition of smoothed spectra with the raw curve was performed each time to check for artifacts.
Data were analyzed using the variable selection method developed by Manavalan and Johnson (30). Twenty-six reference proteins of known structure were used as the comparison set, and the results were judged as satisfactory when the following criteria were obeyed: (i) each secondary structure component was represented by positive values (or values larger than −0.05), (ii) the sum of the computed total secondary composition was in the range of 0.96 to 1.05, and (iii) the root mean square difference between reconstructed and experimental spectra was lower than 0.25.
The percentages of secondary structure composition discussed in the text are the averages of several satisfactory combinations obeying the above criteria. The standard deviation value between satisfactory solutions was 0.01 for α-helix and slightly larger for the other secondary structure components.
Mass spectrometry.
An Applied Biosystems-Sciex API 3000 equipped with the TurboIonSpray source was used for this study. The source operated under positive ion conditions at a needle voltage of 5,600 V with no turbo gas applied in view of the low flow rate entering the source. Mass calibration and resolution adjustments on the resolving quadrupoles were performed automatically by using a solution of polypropylene glycol standards (PE Applied Biosystems, Foster City, Calif.) introduced via a Model 22 Harvard infusion pump. The peak width was set at 0.7 atomic mass unit (measured at 1/2 height).
Orifice voltage was set at 40 V for full-scan experiments (from m/z 800 to 2,800). Some preliminary experiments were done in single ion monitoring over the most intense multiply charged ion of the dimeric form of M. tuberculosis Cpn10 (m/z 1,493) by ramping the orifice voltage from 5 to 120 V with increments of 2.5 V.
For the purpose of the present application, the mass spectrometer was used in single-quad mode. Samples were dissolved in aqueous methanol (1:1, vol/vol) containing trifluoroacetic acid at 0.05% and infused via the Harvard pump at a flow rate of 10 μl/min. Acquired data were processed using the BioToolBox proprietary software (Applied Biosystems-Sciex), and molecular weight deconvolution was made by using the “Bio-reconstruct” option.
NMR spectroscopy.
Samples contained peptide 1-25 (2 mM) in a mixture of either hexafluoroacetone, H2O, and D2O (5:4.5:0.5, vol/vol/vol) or CD3OH/CD3OD (9:1, vol/vol). Deuterated solvents (Cambridge Isotope Lab. Inc.) had 99.8% isotope enrichment. Spectra were acquired at 300 K using a Bruker DRX Avance 600 NMR spectrometer equipped with an SGI workstation (Silicon Graphics, Inc., Mountain View, Calif.) and analyzed with SwaN-MR 3.4.8 (3). The monodimensional proton spectrum was recorded with a spectral width of 10 ppm, 128 scans, and 64K data points. Two-dimensional TOCSY experiments with mixing time of 70 ms were carried out to assign each spin system. NOESY spectra with mixing times of 250 ms were used for sequence-specific assignments and the detection of dipolar couplings. A total of 512 blocks were collected in t1, with 1,024 data points and 64 scans in t2. Spectral width was 7 KHz in both dimensions. A 90-degree shifted sine-bell function was applied to t2 using 1,024 points. The same function was imposed on the 512 points in dimension t1, with a shift of 90 degrees. Zero filling was applied before two-dimensional Fourier transformation, yielding a final matrix of 2,048 by 2,048 real points. Interproton distance constraints were derived from the NOESY experiments by using the geminal Hβ-Hβ dipolar couplings of residue N17 as the calibration interproton distance. The experimental constraints were used in DYANA 1.5 (17) to generate 100 structures each for the two solvent mixtures. The structures were further relaxed through 10,000 cycles of energy minimization. For the two sets of calculations, a converged group of 30 refined structures was identified based on their lowest NOE violations and used to define the mean solution structure. The graphic interface was MOLMOL 2.5.1 (24).
RESULTS
Intracellular localization of M. tuberculosis Cpn10.
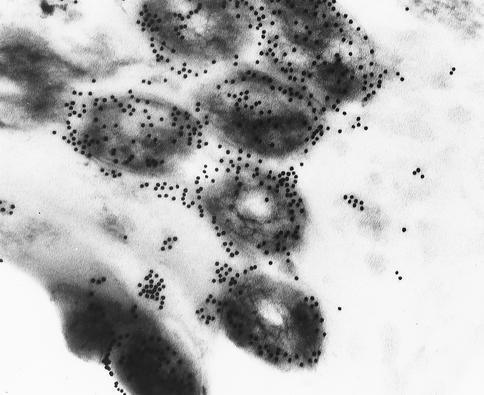
The invasion of the murine macrophage cell line J774 by virulent M. tuberculosis was examined by electron microscopy, and the localization of M. tuberculosis Cpn10 in invading bacteria was assessed by immunogold labeling using an antibody specific for the M. tuberculosis Cpn10 protein and which did not cross-react with the human, E. coli, or M. avium Cpn10 proteins (P. Mascagni, unpublished observations). A transverse section of M. tuberculosis in a phagosome shows the presence of gold particles in the interior of the cell. It also shows a preponderance of gold particles associated with the external surface of the bacterium and free within the host cell's vacuole (Fig. 1). No mitochondrial staining was noted, confirming the lack of cross-reactivity of this antibody with the human Cpn10.
FIG. 1.
Immunoelectronmicroscopy of M. tuberculosis-infected macrophages. The Cpn10 protein is revealed by using Cpn10-specific rabbit polyclonal antibodies. Shown are transverse sections of mycobacteria showing accumulation of the Cpn10 protein on the outer surface of the bacillus or immediately outside it in the surrounding vacuole space of the infected cell. Magnification, ×50,000.
These results confirm previous observations that M. tuberculosis Cpn10 is actively secreted by the live organism and pose the question as to whether structural changes occur in the heptameric form typical of these proteins during the path to secretion, where different environments, such as those of the lipid-cytosol interface of the membrane and the hydrophobic interior of the lipid bilayer, are encountered. Structural studies were therefore carried out in organic solvent-buffer mixtures and in the presence of detergents and phospholipids.
Structural studies in organic solvent-buffer mixtures. (i) M. tuberculosis Cpn10.
In Cpn10 heptamers, the association between subunits is quite labile, and dissociation to monomers takes place in dilute solutions (5, 11, 16). In another study (G. Fossati, G. Izzo, E. Rizzi, P. Cremonesi, G. Sandrone, S. Harding, N. Errington, C. Walters, B. Henderson, M. M. Roberts, A. R. M. Coates, and P. Mascagni, submitted for publication), it was shown that M. tuberculosis Cpn10 is predominantly monomeric in 50 mM PO43 − (pH 7.4) below the concentration of approximately 4.7 μM. Under similar conditions, the synthetic peptides corresponding to the N-terminally truncated forms of the protein, namely peptides 26-99, 46-99, 51-99, 54-99, 59-99, 65-99, and 75-99, were also monomeric (11). To verify whether protein and peptides maintain their monomeric state in organic solvent-buffer mixtures under similar concentration conditions, analytical ultracentrifugation was carried out in 166 mM PO43−-MeOH (3:7, vol/vol) mixtures, with methanol chosen as representative of the organic solvents used in this study.
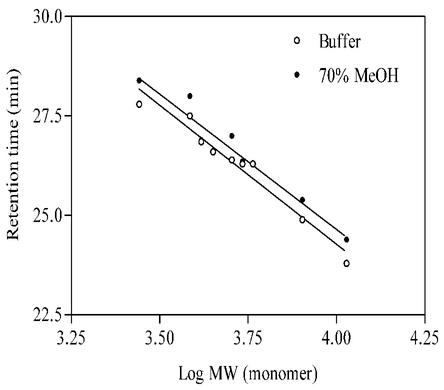
Sedimentation velocity and equilibrium experiments gave no meaningful signal corresponding to either monomeric or multimeric M. tuberculosis Cpn10. The absence of signal was not due to methanol-induced chemical modification of the protein, since under similar conditions of solvent composition, M. tuberculosis Cpn10 had a molecular weight of 10,673 by mass spectrometry, consistent with the value of 10,673.23 calculated for the monomer (see below and Fig. 5). Furthermore, there was no indication of protein fragmentation by SDS-polyacrylamide gel electrophoresis (not shown). Since our objective was to determine the sizes of the protein and peptides in mixed solvents, we did not further investigate the reasons for the lack of an AUC signal, and SEC was carried out in the same 166 mM PO43 −-MeOH (3:7, vol/vol) mixture. Under these conditions, the retention times of protein (2 μM) and peptides (approximately 10 μg of each/ml) were generally slightly longer than their corresponding values in 50 mM PO43 − (pH 7.4) alone (Fig. 2). Therefore, since peptides and protein are monomeric in buffer only, we concluded that they are also monomeric in dilute MeOH-buffer mixtures.
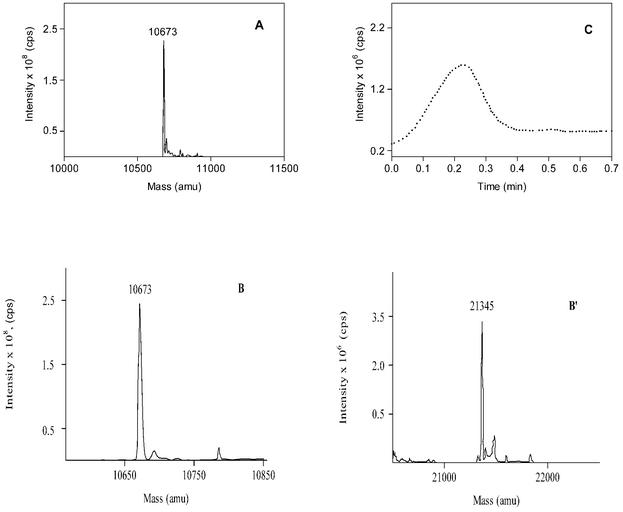
FIG. 5.
Mass spectra of M. tuberculosis Cpn10 in water-MeOH (1:1, vol/vol) at 4 μM (A) and 40 μM (B and B′), showing the existence of monomeric (B) and dimeric (B′) forms of the Cpn10 under these conditions. Panels B′ and B′′ are derived from the same spectrum and are separated for clarity due to the different intensities generated by the two molecular species. The solutions were infused into the spectrometer chamber as described in the experimental section. (C) Collision-induced dissociation of dimers to monomers obtained by increasing the mass spectrometer orifice voltage ramping from 5 to 120 V.
FIG. 2.
Retention times of M. tuberculosis Cpn10 monomer (1.5 μM) and its N-terminally truncated peptides (10 μM), as obtained by SEC in 50 mM KH2PO4, pH 7.4 (○), and in 166 mM KH2PO4-MeOH (30:70, vol/vol) (•). The N-terminally truncated synthetic peptides correspond to sequences 26-99, 54-99, 51-99, 59-99, 62-99, 65-99, and 75-99.
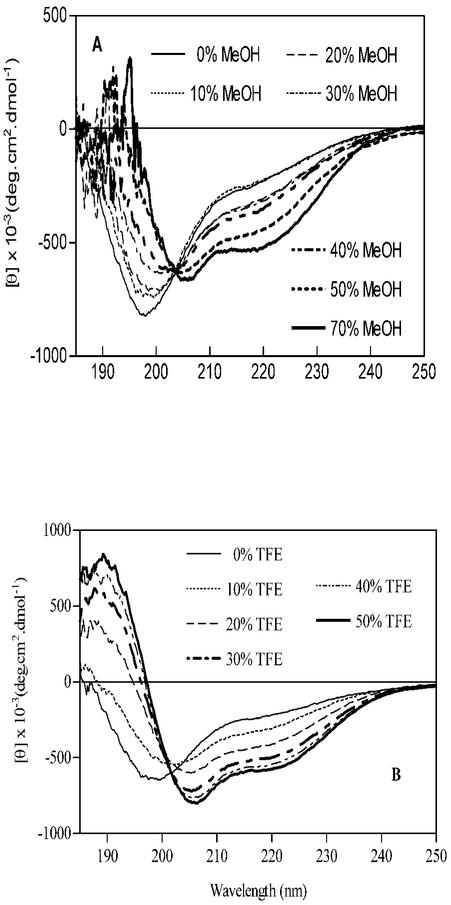
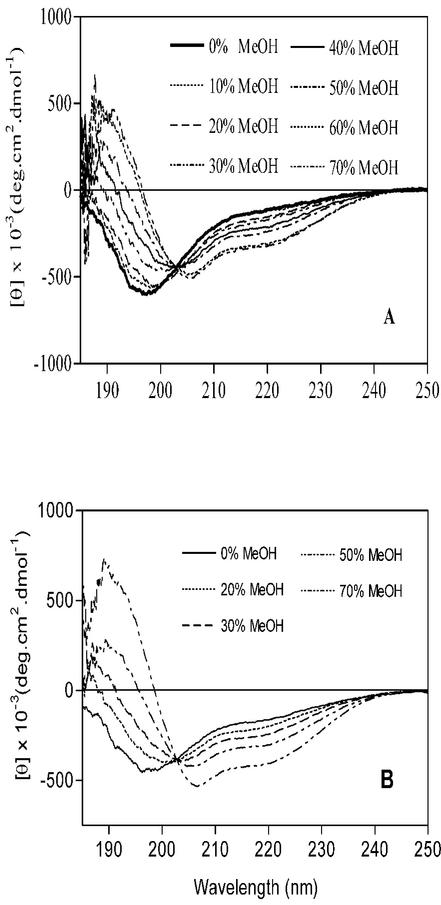
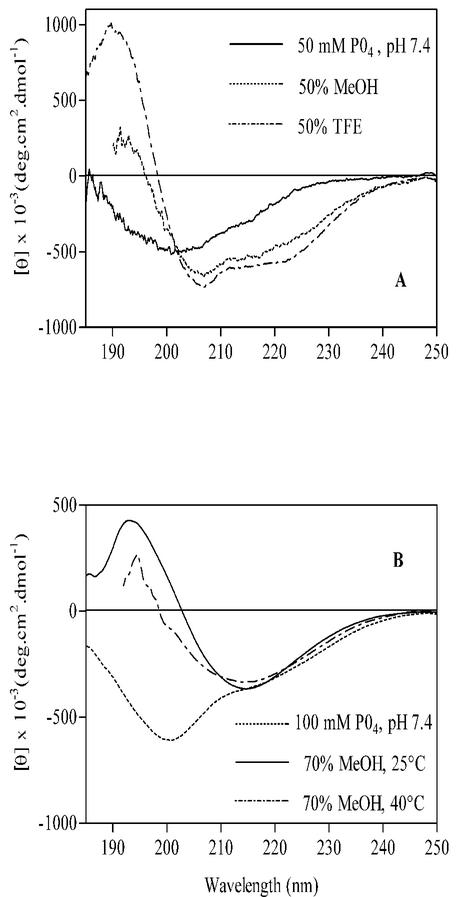
In the next study, titrations with MeOH, TFE, and AcCN were carried out in dilute aqueous solutions containing 2 μM protein where [KH2PO4] = 50 mM; changes in the secondary structure of the Cpn10 were measured by CD spectroscopy. Figure 3A and B show the CD spectra of protein at the various concentrations of MeOH and TFE. In the absence of organic solvent, the far-UV CD spectrum of monomeric Cpn10 was characterized by an intense negative ellipticity at 199 to 200 nm, corresponding to the random coil structure, and by a broad shoulder in the secondary structure region, between approximately 215 and 225 nm (Fig. 3A). MeOH titration induced a progressive decrease in the intensity of the negative ellipticity at 199 to 200 nm, its shift to longer wavelengths, and the increase in signal intensity in the secondary structure region (Fig. 3A). At 70% MeOH, the spectrum had a typical helical profile, with ellipticities at 206 and 220 nm and did not change upon further addition of MeOH (Fig. 3A). Deconvolution of this spectrum using the variable selection method (30) showed that approximately 25% of the residues (about 25 residues) are in an α-helix structure and 22% in a β-sheet.
FIG. 3.
CD spectra of monomeric M. tuberculosis Cpn10 in the presence of increasing concentrations of MeOH (A) and TFE (B). The organic solvents were added to a solution of protein in KH2PO4, pH 7.4. The concentration of KH2PO4 in these solvent mixtures was kept fixed at 50 mM, and that of the protein was kept at 4 μM.
Addition of TFE also induced the formation of helices, although less solvent (approximately 40%) was required for a full transition to a partially helical conformation than in the MeOH titration experiment (Fig. 3B). In 50% AcCN, the monomer had negative ellipticities at approximately 203 nm and 222 nm (not shown), indicating that the protein adopts a mixture of secondary structures, including α-helix, β-sheet, and random coil. The lack of solubility of protein where [AcCN] > 50% prevented us from verifying whether the helical content increases with increasing concentration of organic solvent.
Collectively, these results indicated that the monomer consistently forms helix(ces)-containing conformations under conditions of reduced solvent polarity.
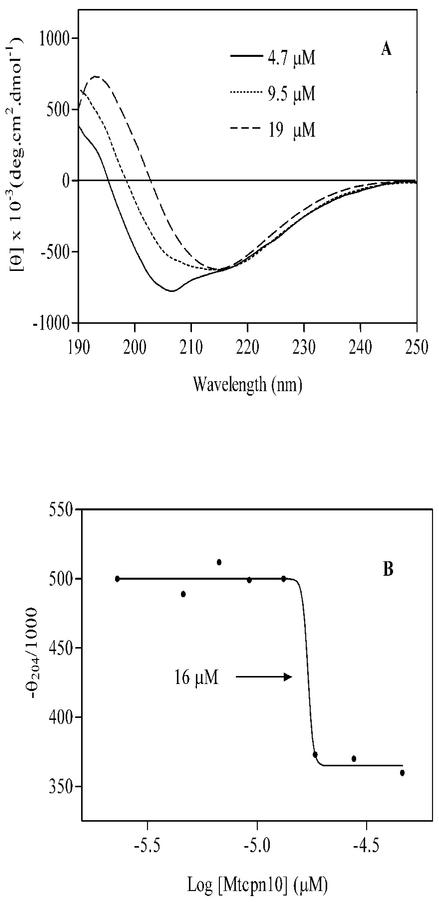
In a previous study (11), it was found that M. tuberculosis Cpn10 at a concentration of 0.1 mg/ml (9.3 μM) in 65% MeOH, it exhibits a typical β-sheet profile, with minimum ellipticity at 218 nm only. This is in contrast to the partially helical conformation seen here under similar conditions of solvent composition. To understand the reasons for this difference, CD spectra were measured in aqueous methanol at various concentrations of protein. In either 100 mM PO43 −-MeOH (1:1, vol/vol) or water-MeOH (1:1, vol/vol) mixtures, the structure of M. tuberculosis Cpn10 changed from partially helical to predominantly β-sheet (all-β) upon increasing the concentration of protein (Fig. 4A). The transition was sharp and occurred between approximately 9 and 18 μM (Fig. 4B). Mass spectrometry of a 4 μM protein sample in water-MeOH (1:1, vol/vol) gave only the signal of the monomer at 10,673 atomic mass units (Fig. 5A). In contrast, monomers and dimers at 21,345 atomic mass units were seen at higher concentrations of M. tuberculosis Cpn10 (Fig. 5B and B′). Furthermore, the concentration of dimers increased with increasing protein concentration (not shown), whereas collision-induced dissociation of dimers to monomers occurred upon increasing the instrument orifice voltage ramping from 5 to 120 V (Fig. 5C). Collectively, these data indicated that the signal at 21,345 atomic mass units was not due to instrumental artifacts (e.g., electrospray-induced dimerization) but likely originated from genuine oligomeric forms of protein existing in solution.
FIG. 4.
Protein concentration dependence of the CD structure of M. tuberculosis Cpn10 in water/MeOH (1:1, vol/vol) solutions. (A) CD spectra of M. tuberculosis Cpn10 at three different concentrations. (B) Concentration dependence of the molar ellipticity at 204 nm observed in 100 mM KH2PO4-MeOH (1:1, vol/vol) in the concentration range from about 2.4 to 47 μM. The midpoint of the structural transition, at 16 μm, is shown.
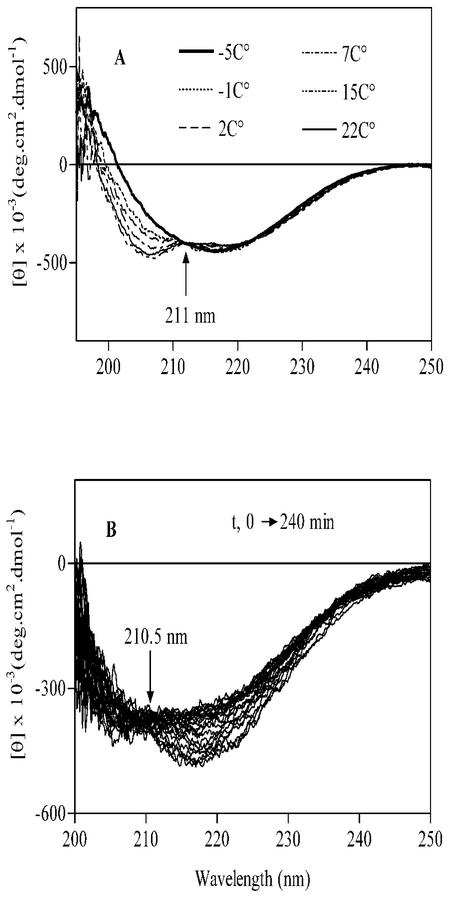
Therefore, the structural transition from a partially helical conformation to an all-β conformation is coupled to oligomerization. The transition is under thermodynamic control, as decreasing the temperature of a solution containing the partially helical monomer from 25 to −4°C induced a conformational change to a predominantly β-structure (Fig. 6A). The transition is reversible, as reheating the solution to 37°C restored the partially helical profile of the monomer (Fig. 6B). Upon both heating and cooling, the CD spectra intersect at an isodichroic point at approximately 211 nm, indicating the occurrence of an equilibrium between two components only, namely, the partially helical monomer and the β-sheet dimer.
FIG. 6.
Temperature dependence of the solution structure of M. tuberculosis Cpn10 (4 μM) in 70% MeOH. (A) Structural transition from α-helix to all-β that occurs at low temperature. (B) The all-β structure slowly converts back to the α-helix structure when the solution is kept at 37°C for 240 min.
(ii) N-terminally truncated peptides.
To identify the regions in the monomer that may be α-helical, the N-terminally truncated synthetic peptides were studied in mixed solvents. As with the full-length protein, peptides 26-99 and 51-99 were partially helical in MeOH- (Fig. 7A and B) or TFE-buffer (not shown) mixtures, and their helical content increased with increasing concentrations of the organic solvents. Spectral deconvolution in 70% MeOH assigned the helical structure to approximately 11 (15%) and 10 residues (21%) of peptides 26-99 and 51-99, respectively. Since in the whole protein there are approximately 25 residues in the α-helical structure, we concluded that one or more helices are contained at the N terminus of M. tuberculosis Cpn10. An additional helix is likely to be contained at the carboxyl end of the monomer, as suggested by a previous study of the peptide 75-99 in aqueous solutions (Fossati et al., submitted). This additional helix was assigned to sequence 83-99, based on secondary structure predictions and homology with the amphiphilic α-helices A (44%) and C (56%) of the cytokine leptin (Fossati et al., submitted).
FIG. 7.
CD spectra of the synthetic peptides (10 μM) corresponding to sequences 26-99 (A) and 51-99 (B) of M. tuberculosis Cpn10, showing the formation of a partially helical structure upon addition of methanol to solutions of protein in KH2PO4, pH 7.4. The concentration of the phosphate buffer in these mixtures was kept fixed at 50 mM.
(iii) Peptide 1-25 of M. tuberculosis Cpn10.
To verify whether the N terminus of M. tuberculosis Cpn10 contains the α-helix(ces) deduced from the above study, the synthetic peptide corresponding to sequence 1-25 of M. tuberculosis Cpn10 was studied in mixed solvents. In buffer only, the molecule was mostly random coil but acquired a partially helical structure both in MeOH-water (95:5, vol/vol) (Fig. 8) and hexafluoroacetone-water (1:1, vol/vol) (not shown). Details of the helical structure were obtained by NMR spectroscopy. In both solvent mixtures, the CαH chemical shifts of the peptide (Tables 1 and 2) were generally larger than those found in small peptides having the random coil structure, suggesting the existence of ordered structures in these solvent mixtures. 3J coupling constants (Tables 1 and 2) and NOE patterns (not shown) confirmed this conclusion and indicated that the ordered structures are mostly helical.
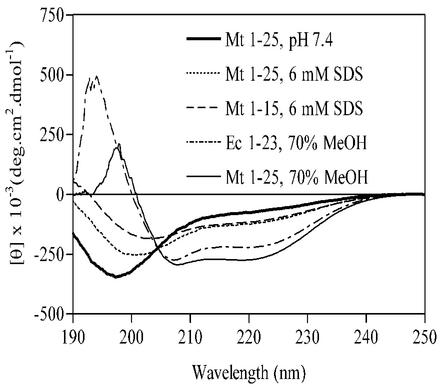
FIG. 8.
CD spectra of the synthetic peptides 1-15 and 1-25 of M. tuberculosis Cpn10 and 1-23 of E. coli Cpn10 in the presence of SDS micelles or in 166 mM KH2PO4-MeOH (0.3:0.7, vol/vol) mixtures.
TABLE 1.
Chemical shifts and scalar coupling constants of peptide 1-25 measured in 95% methanol solution
| Residue | NH | Hα | Hβ | Hγ | Hδ | Other | 3JΦa |
|---|---|---|---|---|---|---|---|
| 1A | 4.3 | 1.56 | b | ||||
| 2K | 8.56 | 4.43 | 1.74-1.89 | 1.47-1.53 | 1.72 | 2.97 | b |
| 3V | 8.05 | 3.73 | 2.08 | 0.89-0.97 | 7.7 | ||
| 4N | 8.57 | 4.83 | 2.64-2.70 | 7.07-7.73 | b | ||
| 5I | 8.25 | 4.4 | 2.13 | 1.35-1.49 | 0.99 | 1 | 7.9 |
| 6K | 8.53 | 4.3 | 1.95-2.09 | 1.56 | 1.68-1.81 | 3 | 2.8 |
| 7P | 4.43 | 2.10-2.35 | 1.81-1.94 | 3.65 | |||
| 8L | 7.18 | 4.15 | 1.74-1.86 | 0.90-0.97 | 4.5 | ||
| 9E | 8.09 | 4.02 | 2.15-2.28 | 2.43-2.52 | 3.2 | ||
| 10D | 8.76 | 4.41 | 2.61-2.91 | <3 | |||
| 11K | 7.81 | 3.95 | 2.06-2.10 | 1.32 | 1.52-1.67 | 2.96 | b |
| 12I | 8.04 | 4.19 | 2.04 | 1.16-1.29 | 0.9 | <3 | |
| 13L | 8.25 | 4.09 | 1.88-1.96 | 1.68 | 0.92-0.98 | <3 | |
| 14V | 8.3 | 3.73 | 2.25 | 0.97-1.15 | 3.4 | ||
| 15Q | 8.38 | 4.06 | 2.16-2.36 | 2.59-2.61 | 6.82-7.54 | 2.8 | |
| 16A | 8.81 | 4.11 | 1.55 | <3 | |||
| 17N | 8.4 | 4.51 | 2.81-3.00 | 6.88-7.60 | 4.2 | ||
| 18E | 8.69 | 4.12 | 2.10-2.37 | 2.42-2.72 | <3 | ||
| 19A | 8.56 | 4.15 | 1.58 | <3 | |||
| 20E | 8.38 | 4.18 | 2.24-2.33 | 2.51-2.66 | 3.0 | ||
| 21T | 8.12 | 4.12 | 4.33 | 1.3 | 4.5 | ||
| 22T | 7.94 | 4.43 | 4.43 | 1.48 | 3.1 | ||
| 23T | 7.93 | 4.21 | 4.34 | 3.2 | |||
| 24A | 7.77 | 4.31 | 1.28 | b | |||
| 25S | 7.8 | 4.5 | 3.93-3.96 | b |
b, broad.
TABLE 2.
Chemical shifts and scalar coupling constants of peptide 1-25 measured in hexafluoroacetone 50% solution
| Residue | NH | Hα | Hβ | Hγ | Hδ | Other | 3JΦ |
|---|---|---|---|---|---|---|---|
| 1A | 4.12 | 1.59 | ba | ||||
| 2K | 8.28 | 4.39 | 1.80-1.87 | 1.37-1.56 | 1.74 | ɛ3.06, NHɛ 6.53-7.3 | 6.0 |
| 3V | 7.63 | 4.16 | 2.05 | 0.89-0.94 | 7.7 | ||
| 4N | 8.18 | 4.86 | 2.75-2.91 | 6.53-7.34 | 6.8 | ||
| 5I | 7.57 | 4.34 | 2.01 | 1.21-1.46 | CH3 0.90-0.97 | 7.1 | |
| 6K | 7.81 | 4.38 | 2.08-2.01 | 1.30-1.34 | 1.66-1.78 | ɛ 2.33-2.3, NHɛ 6.3 7.11 | 6.2 |
| 7P | 0.3 | 4.37 | 1.84-2.37 | 2.0-2.08 | 3.74-3.76 | ||
| 8L | 7.1 | 4.25 | 1.73-1.76 | 1.65 | 0.94-0.97 | <3.5 | |
| 9E | 7.98 | 4.01 | 1.99-2.18 | 2.46-2.50 | <3.5 | ||
| 10D | 8.36 | 4.47 | 2.83-2.88 | <3.5 | |||
| 11K | 7.6 | 4.06 | 2.01-2.07 | 1.48-1.58 | 1.71-1.88 | ɛ 3.02, NHɛ 3 7.1 | <3.5 |
| 12I | 7.98 | 3.75 | 1.72 | 0.94-1.18 | 0.86 | <3.5 | |
| 13L | 8.03 | 4.13 | 1.58-1.93 | 1.81 | 0.89-0.93 | <3.5 | |
| 14V | 8.07 | 3.72 | 2.18 | 0.96-1.10 | <3.5 | ||
| 15Q | 8 | 4.09 | 2.23-2.29 | 2.39-2.58 | 6.53-6.98 | <3.5 | |
| 16A | 8.74 | 4.18 | 1.54 | <3.5 | |||
| 17N | 8.15 | 4.53 | 2.81-2.97 | 6.7-7.53 | <3.5 | ||
| 18E | 8.48 | 4.16 | 2.21-2.29 | 2.49-2.57 | <3.5 | ||
| 19A | 8.4 | 4.17 | 1.55 | <3.5 | |||
| 20E | 8.23 | 4.21 | 2.22-2.29 | 2.56-2.75 | <3.5 | ||
| 21T | 7.98 | 4.27 | 4.37 | 1.33 | <3.5 | ||
| 22T | 8.48 | 3.84 | 4.17 | 1.32 | <3.5 | ||
| 23T | 7.91 | 4.27 | 4.37 | 4.0 | |||
| 24A | 7.87 | 4.44 | 1.49 | 5.5 | |||
| 25S | 7.84 | 4.53 | 3.97-4.03 | 7.3 |
b, broad.
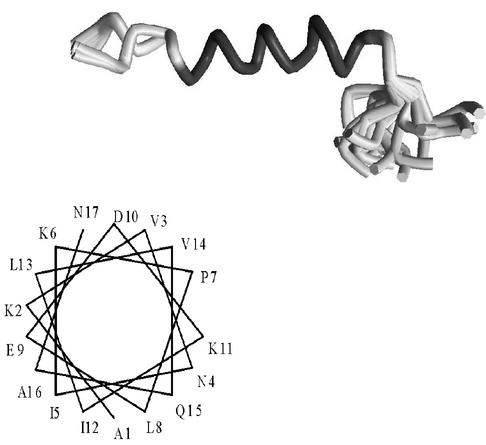
Using the DYANA distance geometry algorithm (17) and the experimental NMR constraints, 100 structures were generated for each of the two different solvent conditions and refined. A converging set of 30 refined structures was then extracted based on the lowest NOE violations and used to define the mean solution structure of the peptide. This contains a long α-helix between residues 1 and 16 which is compact between residues 6 and 16 and more frail at the N terminus (Fig. 9). It is amphiphilic, with the side chains of K2, K6, E9, D10, and N17 accumulating on the same side (Fig. 9). The residues at the carboxyl end of the peptide are disordered in this structure (Fig. 9).
FIG. 9.
Superimposition of the twenty lowest-energy NMR conformers of peptide 1-25 in 95% MeOH and helical wheel plot of the first 17 residues. Two main families are seen which differ in the torsional angles at the asparagine residue at position 4. With the exception of the C-terminal residues (sequence 17-25) which are mostly disordered, the rest of the peptide adopts an amphiphilic α-helix.
(iv) E. coli and human Cpn10s.
Since human Cpn10, but not E. coli Cpn10, is able to cross biological membranes (to enter the mitochondrion), we extended our investigation in organic solvent-buffer mixtures to include these additional chaperonins and verified whether or not a structural relationship exists with their different secretion patterns.
In dilute buffer solutions, human and E. coli Cpn10s had the same general spectral characteristics as monomeric M. tuberculosis Cpn10, i.e., a large random coil contribution at approximately 200 nm and a weak signal in the secondary structure region (Fig. 10A and B). Upon addition of either MeOH or TFE, the CD structure of human Cpn10 changed to partially helical (Fig. 10A), while that of E. coli Cpn10 changed to all-β (Fig. 10B). The all-β spectrum of E. coli Cpn10 was independent of protein concentration in the concentration range from 0.1 to 2 μM and of temperature in the temperature range from −4 to 40°C (Fig. 10B). As expected, E. coli Cpn10 was a mixture of monomers and dimers by mass spectrometry (not shown).
FIG. 10.
CD spectra of human Cpn10 (2 μM) in MeOH- or TFE-100 mM KH2PO4 mixtures (1:1, vol/vol) (A) and of E. coli Cpn10 (0.1 μM) in 166 mM KH2PO4-MeOH (0.3:0.7, vol/vol) at 25° and 40°C (B).
Thus, in the presence of organic solvents and at room temperature, M. tuberculosis and human Cpn10s are monomeric and partially helical, while E. coli Cpn10 is dimeric and has the structure of an all-β protein.
The absence of helical structures in buffer-organic solvent solutions of E. coli Cpn10 is not due to a reduced ability of its N-terminal tail to form α-helices, since the corresponding peptide 1-23 was partly helical in aqueous methanol (Fig. 8). This, together with a previous report of the N-terminal region of rat Cpn10 as forming an amphiphilic helix in aqueous TFE (22), suggests that the structural differences seen for the three proteins in dilute organic solvent-buffer mixtures cannot be accounted for by differences in α-helix propensity of their N-terminal sequences.
Structural studies in the presence of detergents.
In addition to protein components, several classes of lipids assist in the transport process, and acidic lipids are believed to interact with the positive charges of the signal peptide (46). The three Cpn10s considered in this work were therefore studied in the presence of SDS, which has a negatively charged head group, and DMPC, whose head group is instead zwitterionic.
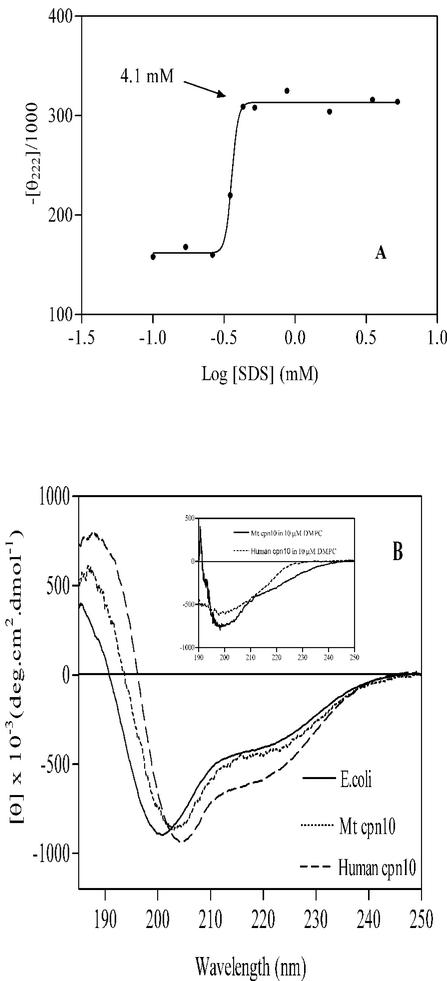
At low concentrations of SDS, the CD spectrum of the M. tuberculosis Cpn10 monomer was similar to that in phosphate buffer only, having a large minimum at about 200 nm (not shown). However, as the concentration of the detergent was increased above its critical micellar concentration (CMC) (6), the protein became partially helical (Fig. 11A and B). Further additions of SDS above 4.1 mM had no effect on the protein structure, suggesting that the partially helical monomer is completely associated with the detergent in a micelle-bound state (2). In a similar experiment, the other two chaperonins, human Cpn10 and E. coli Cpn10, were also partially helical above the CMC of SDS (Fig. 11B).
FIG. 11.
(A) Changes in the intensity of the molar ellipticity at 222 nm of M. tuberculosis Cpn10 (10 μM in 100 mM NaH2PO4) observed upon the addition of increasing amounts of SDS. Changes were maximal above [SDS] > 4.1 mM. (B) Comparison of the CD spectra of M. tuberculosis, human, and E. coli Cpn10s above the CMC of SDS. In the inset, CD spectra of M. tuberculosis and h-Cpn10s above the CMC (10−4 mM [23]) of the zwitterionic phospholipid DMPC.
The chemistry of the detergent head group affects the folding of Cpn10s, as in the presence of the zwitterionic DMPC, the three proteins were mostly random coil (negative ellipticity at approximately 200 nm) both below and above the CMC of the detergent (Fig. 11B, inset). So the transition to a partially helical structure appears to necessitate the presence in solution of a negatively charged surface such as that provided by SDS micelles. The interactions between monomer and the negative charges of the micelle head groups might involve the N-terminal region of the protein, where there are three positively charged residues, K-2, -6, and -11. Furthermore, peptide 1-16 forms an amphiphilic helix in aqueous organic solvents, and helices of this type are known to associate with negatively charged artificial phospholipid bilayers (20, 38, 39). The synthetic peptide 1-25 and its shorter form 1-15 were therefore studied in the presence of detergents. This confirmed that the two peptides have the ability to associate with the negatively charged SDS micelles and to form a mixture of random coil and α-helix when bound to them (Fig. 8). Furthermore, since the two peptides contain a similar amount of α-helix, as measured by the ellipticity at 222 nm (Fig. 8), we concluded that their α-helix is contained in the 1-15 region and may therefore be similar to that seen in aqueous organic solvents.
Peptides 26-99 and 75-99 were also partially helical in the presence of SDS micelles but not in the presence of DMPC, suggesting the existence of additional interactions between M. tuberculosis Cpn10 and the detergent. These additional interactions are likely to involve the C-terminal amphiphilic α-helix, previously identified as encompassing sequence 84-99, and are likely to be mediated by the side chains of R91 and K99, which, together with D92 and S98, are on the same side of the amphiphilic α-helix (Fossati et al., submitted).
DISCUSSION
The macrophage has evolved to phagocytose pathogenic bacteria and to kill them within the phagolysosome using a variety of toxic agents. However, a number of bacteria can survive within this environment. The most dangerous of such organisms is M. tuberculosis, the causative agent of tuberculosis, which is estimated to infect one-third of the world's population and to cause 3 to 4 million deaths each year (10). It is established that M. tuberculosis can prevent the maturation of the phagolysosome, thus preventing it from becoming a lethal environment (13). Exactly how M. tuberculosis evades the machinations of the lysososomal apparatus is not clear. One class of mycobacterial protein that has recently been shown to modulate myeloid cell behavior are the chaperonins, a specific class of molecular chaperone consisting of the oligomeric proteins, Cpn60 and Cpn10 (8, 27, 29, 32). It may be this ability to interact with myeloid cells that accounts for the immunogenicity of the mycobacterial chaperonins. A key requirement for these proteins to play some part in the pathology of tuberculosis is that they are selectively released by the mycobacterial cell.
Using immunogold labeling of M. tuberculosis Cpn10 with an antibody specific for the M. tuberculosis protein and with no cross-reactivity with the murine chaperonin, we show here that not only is M. tuberculosis Cpn10 present in the cytoplasm of the bacterium, but it is concentrated on the outer surface. In addition to this, significant amounts of label were found free in the vacuolar matrix. These results support previous suggestions that M. tuberculosis Cpn10 is actively secreted by the live bacillus in culture (1, 4). It is not clear if the Cpn10 contributes to the survival of the bacterium within the phagosome, but this hypothesis is currently being tested.
Having shown that M. tuberculosis Cpn10 is able to cross the mycobacterial cell wall, its solution structure was studied under conditions of reduced solvent polarity or in the presence of detergents to understand whether any structural changes might take place when the protein interacts with the cytosolic membrane phospholipids or goes through the highly hydrophobic interior of the mycobacterium cytosolic membrane. The study revealed that below the concentration of 2 μM, M. tuberculosis Cpn10 is monomeric in organic solvent-buffer mixtures, where it consistently forms stable, partially helical structures. In one of these solvent mixtures (i.e., 70% MeOH) taken as representative of the solvent-induced conformational changes, the number of α-helical residues was estimated to be 25. By using synthetic peptides corresponding to progressively shorter N-terminally truncated forms of the protein and some of their complementary C-terminally truncated peptides, one amphiphilic α-helix was identified and assigned to sequence 1-16 by NMR spectroscopy (Fig. 9A). The existence of stable helical monomers was not totally unexpected because in a previous study, evidence was produced to suggest that in aqueous solution, monomeric M. tuberculosis Cpn10 forms small amounts of a conformation which has residues in the α-helical structure (Fossati et al., submitted). Under the same conditions, peptide 75-99 was also partially helical. Furthermore, the existence of amphiphilic α-helices encompassing residues 2 to 13 and 83 to 99 of M. tuberculosis Cpn10 has been previously predicted based on sequence considerations (11; Fossati et al., submitted). Thus, the partially helical monomer is likely to contain two amphiphilic helices, one each at the N and C termini. The existence of N-terminal and C-terminal helices in the monomer is in contrast to the X-ray structure of the M. tuberculosis Cpn10 heptamer (37, 37a, 44), where each subunit folds into an irregular β-barrel which has no α-helices. Furthermore, the C- and N-terminal regions of the subunits contain two β-strands each, β1 (N4-L8) and β2 (K11-A16) and β6 (83-90) and β7 (93-98), respectively, with β1 from one subunit and β7′ from the adjacent subunit arranged in an anti-parallel β-sheet and providing the strongest intersubunit interactions (21, 31, 37, 44). Therefore, it appears from these data and those from a related study (Fossati et al., submitted) that in the absence of intersubunit interactions, a structural transition from β-strand to α-helix occurs in the N- and C-terminal regions of the monomer and that the stability of these helices increases as the hydrophobic or acidic environments increase (Fossati et al., submitted).
Although it could be argued that the structural transition to α-helix seen here may be due to the α-helical stabilizing effects of organic solvents such as TFE (33), there are a number of observations that support the conclusion of a genuine propensity for the M. tuberculosis Cpn10 monomer to form partially helical structures. Firstly, regions without helical propensity do not form helices even in 100% TFE (26, 43), and other structures (e.g., β-turn and β-sheet) are supported by this solvent (14, 15, 42). Secondly, a partially helical monomer exists also in the absence of organic solvents, albeit at a very low concentration (Fossati et al., submitted). Thirdly, the partially helical monomer accumulates in the presence of all the organic solvents used in this study, including AcCN, whose α-helix-stabilizing effects are less known. Finally, under the same conditions of solvent composition (i.e., 70% MeOH), M. tuberculosis Cpn10 folds into two different structures, namely, the dimer, where the β-sheet structure predominates, and the monomer, which contains both the β-sheet and the α-helix structures (Fig. 4A), arguing against a solvent effect as the only explanation for the structural changes seen in this study. Therefore, it seems reasonable to conclude that reducing the solvent polarity stabilizes the partially helical monomer already present in aqueous solutions, thus increasing its concentration.
Stabilization of the partially helical monomer was not restricted to organic solvent-buffer mixtures only, as similar structures were seen when M. tuberculosis Cpn10 associates with SDS micelles. Remarkably, the presence of zwitterionic lipids had no effect on the protein secondary structure, indicating that an interaction between the negatively charged head group of SDS and positively charged or hydrophilic regions of the protein occurs at the detergent-water interface. The selective association with negatively charged lipids has been shown for yeast Cpn10 (9). Here we extended this observation to the other Cpn10s. Furthermore, using truncated forms of M. tuberculosis Cpn10, we provide evidence that the positive charges of the N- and C-terminal amphiphilic helices may be involved in the association of M. tuberculosis Cpn10 with SDS micelles.
The ability to form positively charged amphiphilic helices and to bind to negatively charged detergents may be important in the mechanism of secretion of M. tuberculosis Cpn10 because it is well established that transportation needs acidic lipids (36, 45), and signal peptides of transported proteins have in common the ability to form positively charged amphiphilic helices. In mammalian Cpn10, the N-terminal sequence is both necessary and sufficient for mitochondrial targeting in vitro (22) and forms a stable positively charged amphiphilic helix in membrane mimetic solvent mixtures (22). Strangely, human Cpn10 is not processed after secretion, while signal peptides are generally removed by membrane-located proteases after transport. Therefore, it seems that this protein contains a noncleavable signal peptide. A similar situation may exist for M. tuberculosis Cpn10, which also contains an N-terminal amphiphilic helix and is not processed after transport. However, the M. tuberculosis Cpn10 monomer has an additional α-helix at the carboxyl end that is also amphiphilic and selectively associates with SDS micelles, suggesting that the mechanism of secretion of M. tuberculosis Cpn10 may be more complex and that if α-helicity is the route to secretion, then the N- and C-terminal sequences may act jointly to traverse the lipid membrane.
The partially helical monomer is in equilibrium with a dimeric form of the protein, where the predominant secondary structure is the β-sheet. The association to dimers is reversible and is favored by either the increase in protein concentration above approximately 9 μM or by reduction of the temperature below 0°C (Fig. 4 and 6). E. coli Cpn10 also self-associates to a predominantly β-sheet dimer in 70% MeOH. However, unlike the mycobacterial or human homologues, which are monomeric and partially helical in dilute mixed solvents at room temperature, the all-β structure of the E. coli Cpn10 dimer is independent of protein concentration in the concentration range 0.1 to 2 μM or of solution temperature between approximately 0 and 40°C. These different structural patterns do not appear to depend on the N-terminal sequence alone, since synthetic peptides corresponding to the N-terminal region of the three proteins were helical in aqueous organic solvents or in the presence of SDS (Fig. 8) (22). On the other hand, in aqueous solution, M. tuberculosis Cpn10 and human Cpn10 are predominantly monomeric (>95%) below approximately 4.7 μM (Fossati et al., submitted) and 1 μM (16), respectively, whereas complete dissociation of E. coli Cpn10 occurs well below the concentration of 0.1 μM (48). Furthermore, unlike E. coli Cpn10, which is either heptameric or monomeric only, other oligomers, such as tetramers, trimers, and dimers, have been detected in solutions of M. tuberculosis and human Cpn10s (11). Therefore, the increased stability to dissociation exhibited by the E. coli Cpn10 heptamer may be a plausible explanation for the lack of partially helical monomers in dilute mixed solvents. The ability to dissociate to monomers may be important in the mechanism of transport of these proteins because the N-terminal and C-terminal regions are engaged in intersubunit interactions in the heptamer and therefore are not available for interaction with the transport apparatus or membrane acidic lipids. So, the increased propensity to dissociate to monomers may represent an additional requirement that these proteins need to interact with the biological membrane.
In conclusion, we have shown that invading M. tuberculosis organisms present in the phagosomes of murine macrophages are able to transport the Cpn10 protein on to the cell surface and into the environment of the phagosome. Since this protein is able to modulate macrophage behavior, such a release may facilitate bacterial survival. It is not clear how such secretion is mediated. However, physiochemical analysis has established that M. tuberculosis Cpn10 has the ability to dissociate to monomers in a hydrophobic environment and then to form stable partially helical monomers that interact with acidic lipids. We propose that these two properties are key for the secretion of M. tuberculosis Cpn10.
REFERENCES
- 1.Abou-Zeid, C., I. Smith, J. M. Grange, T. L. Ratliff, J. Steele, and G. A. Rook. 1988. The secreted antigens of Mycobacterium tuberculosis and their relationship to those recognized by the available antibodies. J. Gen. Microbiol. 134:531-538. [DOI] [PubMed] [Google Scholar]
- 2.Bairaktari, E., D. F. Mierke, S. Mammi, and E. Peggion. 1990. Conformations of bombolitins I and III in aqueous solutions: circular dichroism, 1H NMR, and computer simulation studies. Biochemistry 29:10090-10096. [DOI] [PubMed] [Google Scholar]
- 3.Balacco, G. 1994. SwaN-MR: a complete and expansible NMR software for the Macintosh. J. Chem. Inf. Comput. Sci. 34:1235-1241. [Google Scholar]
- 4.Barnes, P. F., V. Mehra, G. R. Hirschfield, S. J. Fong, C. Abou-Zeid, G. A. Rook, S. W. Hunter, P. J. Brennan, and R. L. Modlin. 1989. Characterization of T cell antigens associated with the cell wall protein-peptidoglycan complex of Mycobacterium tuberculosis. J. Immunol. 143:2656-2662. [PubMed] [Google Scholar]
- 5.Boudker, O., M. J. Todd, and E. Freire. 1997. The structural stability of the co-chaperonin GroES. J. Mol. Biol. 272:770-779. [DOI] [PubMed] [Google Scholar]
- 6.Chattopadhyay, A., and E. London. 1984. Fluorimetric determination of critical micelle concentration avoiding interference from detergent charge. Anal. Biochem. 139:408-412. [DOI] [PubMed] [Google Scholar]
- 7.Chupin, V., J. A. Killian, J. Breg, H. H. de Jongh, R. Boelens, R. Kaptein, and B. de Kruijff. 1995. PhoE signal peptide inserts into micelles as a dynamic helix-break-helix structure, which is modulated by the environment. A two-dimensional 1H NMR study. Biochemistry 34:11617-11624. [DOI] [PubMed] [Google Scholar]
- 8.Coates, A. R., B. Henderson, and P. Mascagni. 1999. The unfolding story of the chaperonins. Biotechnol. Genet. Eng. Rev. 16:393-405. [DOI] [PubMed] [Google Scholar]
- 9.de Jongh, H. H., S. Rospert, and C. M. Dobson. 1998. Comparison of the conformational state and in vitro refolding of yeast chaperonin protein cpn10 with bacterial GroES. Biochem. Biophys. Res. Commun. 244:884-888. [DOI] [PubMed] [Google Scholar]
- 10.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO global surveillance and monitoring project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 11.Fossati, G., P. Lucietto, P. Giuliani, A. R. Coates, S. Harding, H. Colfen, G. Legname, E. Chan, A. Zaliani, and P. Mascagni. 1995. Mycobacterium tuberculosis chaperonin 10 forms stable tetrameric and heptameric structures. J. Biol. Chem. 270:26159-26167. [DOI] [PubMed] [Google Scholar]
- 12.Garavito, R. M., and S. Ferguson-Miller. 2001. Detergents as tools in membrane biochemistry. J. Biol. Chem. 276:32403-32406. [DOI] [PubMed] [Google Scholar]
- 13.Goebel, W., and R. Gross. 2001. Intracellular survival strategies of mutualistic and parasitic prokaryotes. Trends Microbiol. 9:267-273. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, M., F. Naider, and C. Toniolo. 1971. Circular dichroism studies of isoleucine oligopeptides in solution. Biopolymers 10:1719-1730. [DOI] [PubMed] [Google Scholar]
- 15.Greff, D., S. Fermandjian, P. Fromageot, M. C. Khosla, R. R. Smeby, and F. M. Bumpus. 1976. Circular-dichroism spectra of truncated and other analogs of angiotensin II. Eur. J. Biochem. 61:297-305. [DOI] [PubMed] [Google Scholar]
- 16.Guidry, J. J., C. K. Moczygemba, N. K. Steede, S. J. Landry, and P. Wittung-Stafshede. 2000. Reversible denaturation of oligomeric human chaperonin 10: denatured state depends on chemical denaturant. Protein Sci. 11:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guntert, P., C. Mumenthaler, and K. Wuthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 18.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 19.Houbiers, M. C., R. B. Spruijt, C. J. Wolfs, and M. A. Hemminga. 1999. Conformational and aggregational properties of the gene 9 minor coat protein of bacteriophage M13 in membrane-mimicking systems. Biochemistry 38:1128-1135. [DOI] [PubMed] [Google Scholar]
- 20.Hoyt, D. W., and L. M. Gierasch. 1991. Hydrophobic content and lipid interactions of wild-type and mutant OmpA signal peptides correlate with their in vivo function. Biochemistry 30:10155-10163. [DOI] [PubMed] [Google Scholar]
- 21.Hunt, J. F., A. J. Weaver, S. J. Landry, L. Gierasch, and J. Deisenhofer. 1996. The crystal structure of the GroES co-chaperonin at 2.8 A resolution. Nature 379:37-45. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis, J. A., M. T. Ryan, N. J. Hoogenraad, D. J. Craik, and P. B. Hoj. 1995. Solution structure of the acetylated and noncleavable mitochondrial targeting signal of rat chaperonin 10. J. Biol. Chem. 270:1323-1331. [DOI] [PubMed] [Google Scholar]
- 23.Kleinschmidt, J. H., M. C. Weiner, and W. L. Tamm. 1999. Outer membrane protein A of E. coli folds into detergent micelles, but not in the presence of monomeric detergent. Protein Sci. 8:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koradi, R., M. Billeter, and K., and Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structure. J. Mol. Graphics 14:51-55. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. E., and T. I. Ahn. 2000. Periplasmic localization of a GroES homologue in Escherichia coli transformed with groESx cloned from Legionella-like endosymbionts in Amoeba proteus. Res. Microbiol. 151:605-618. [DOI] [PubMed] [Google Scholar]
- 26.Lehrman, S. R., J. L. Tuls, and M. Lund. 1990. Peptide alpha-helicity in aqueous trifluoroethanol: correlations with predicted alpha-helicity and the secondary structure of the corresponding regions of bovine growth hormone. Biochemistry 29:5590-5596. [DOI] [PubMed] [Google Scholar]
- 27.Lewthwaite, J. C., A. R. Coates, P. Tormay, M. Singh, P. Mascagni, S. Poole, M. Roberts, L. Sharp, and B. Henderson. 2001. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a cd14-binding domain. Infect. Immun. 69:7349-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackaness, G. B. 1967. The relationship of delayed hypersensitivity to acquired cellular resistance. Br. Med. Bull. 23:52-54. [DOI] [PubMed] [Google Scholar]
- 29.Maguire, M., A. R. M. Coates, and B. Henderson. 2002. Chaperonin 60 unfolds its secrets of cellular communication. Cell Stress Chaperones 7:317-329. [DOI] [PMC free article] [PubMed]
- 30.Manavalan, P., and W. C. Johnson, Jr. 1987. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 167:76-85. [DOI] [PubMed] [Google Scholar]
- 31.Mande, S. C., V. Mehra, B. R. Bloom, and W. G. Hol. 1996. Structure of the heat shock protein chaperonin-10 of Mycobacterium leprae. Science 271:203-207. [DOI] [PubMed] [Google Scholar]
- 32.Meghji, S., P. A. White, S. P. Nair, K. Reddi, K. Heron, B. Henderson, A. Zaliani, G. Fossati, P. Mascagni, J. F. Hunt, M. M. Roberts, and A. R. M. Coates. 1997. Mycobacterium tuberculosis chaperonin 10 stimulates bone resorption: a potential contributory factor in Pott's disease. J. Exp. Med. 186:1241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutter, M., F. Maser, K. H. Altmann, C. Toniolo, and G. M. Bonora. 1985. Sequence-dependence of secondary structure formation: conformational studies of host-guest peptides in alpha-helix and beta-structure supporting media. Biopolymers 24:1057-1074. [DOI] [PubMed] [Google Scholar]
- 34.Orme, I. M. 1988. Characteristics and specificity of acquired immunologic memory to Mycobacterium tuberculosis infection. J. Immunol. 140:3589-3593. [PubMed] [Google Scholar]
- 35.Ragno, S., V. R. Winrow, P. Mascagni, P. Lucietto, F. Di Pierro, C. J. Morris, and D. R. Blake. 1996. A synthetic 10-kD heat shock protein (hsp10) from Mycobacterium tuberculosis modulates adjuvant arthritis. Clin. Exp. Immunol. 103:384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rietveld, A. G., M. C. Koorengevel, and B. de Kruijff. 1995. Non-bilayer lipids are required for efficient protein transport across the plasma membrane of Escherichia coli. EMBO J. 14:5506-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, M. M., A. R. Coker, G. Fossati, P. Mascagni, A. R. Coates, and S. P. Wood. 1999. Crystallization, X-ray diffraction and preliminary structure analysis of Mycobacterium tuberculosis chaperonin 10. Acta Crystallogr. D Biol. Crystallogr. 55:910-914. [DOI] [PubMed] [Google Scholar]
- 37a.Roberts, M. M., A. R. Coker, G. Fossati, P. Mascagni, A. R. M. Coates, and S. P. Wood. 2003. Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops. J. Bacteriol. 185:4172-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roise, D., S. J. Horvath, J. M. Tomich, J. H. Richards, and G. Schatz. 1986. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 5:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozek, A., G. W. Buchko, and R. J. Cushley. 1995. Conformation of two peptides corresponding to human apolipoprotein C-I residues 7-24 and 35-53 in the presence of sodium dodecyl sulfate by CD and NMR spectroscopy. Biochemistry 34:7401-7408. [PubMed] [Google Scholar]
- 40.Ryan, M. T., N. J. Hoogenraad, and P. B. Hoj. 1994. Isolation of a cDNA clone specifying rat chaperonin 10, a stress-inducible mitochondrial matrix protein synthesised without a cleavable presequence. FEBS Lett. 337:152-156. [DOI] [PubMed] [Google Scholar]
- 41.Sadacharan, S. K., A. C. Cavanagh, and R. S. Gupta. 2001. Immunoelectron microscopy provides evidence for the presence of mitochondrial heat shock 10-kDa protein (chaperonin 10) in red blood cells and a variety of secretory granules. Histochem. Cell. Biol. 116:507-517. [DOI] [PubMed] [Google Scholar]
- 42.Siligardi, G., A. F. Drake, P. Mascagni, P. Neri, L. Lozzi, N. Niccolai, and W. A. Gibbons. 1987. Resolution of conformation equilibria in linear peptides by circular dichroism in cryogenic solvents. Biochem. Biophys. Res. Commun. 143:1005-1011. [DOI] [PubMed] [Google Scholar]
- 43.Sonnichsen, F. D., J. E. Van Eyk, R. S. Hodges, and B. D. Sykes. 1992. Effect of trifluoroethanol on protein secondary structure: an NMR and CD study using a synthetic actin peptide. Biochemistry 31:8790-8798. [DOI] [PubMed] [Google Scholar]
- 44.Taneja, B. and S. C. Mande. 2001. Three-dimensional structure of Mycobacterium tuberculosis chaperonin-10 reveals a partially stable conformation of its mobile loop. Curr. Sci. 81:87-91. [Google Scholar]
- 45.Vanet, A., and A. Labigne. 1998. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect. Immun. 66:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Voorst, F., and B. De Kruijff. 2000. Role of lipids in the translocation of proteins across membranes. Biochem. J. 347:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, Y. L., L. H. Lee, D. M. Rollins, and W. M. Ching. 1994. Heat shock- and alkaline pH-induced proteins of Campylobacter jejuni: characterization and immunological properties. Infect. Immun. 62:4256-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zondlo, J., K. E. Fisher, Z. Lin, K. R. Ducote, and E. Eisenstein. 1995. Monomer-heptamer equilibrium of the Escherichia coli chaperonin GroES. Biochemistry 34:10334-10339. [DOI] [PubMed] [Google Scholar]