Abstract
The crystal structure of the ketolide telithromycin bound to the Deinococcus radiodurans large ribosomal subunit shows that telithromycin blocks the ribosomal exit tunnel and interacts with domains II and V of the 23S RNA. Comparisons to other clinically relevant macrolides provided structural insights into its enhanced activity against macrolide-resistant strains.
Macrolide drugs, which have been used for decades, provide effective coverage against most pathogenic bacteria. Nevertheless, the rapid development of antibiotic resistance severely hampers modern medicine and has prompted a search for improved antimicrobial agents. Ketolides, a newer class of macrolides that exhibits a safety profile comparable to that of other antibiotics (15), were shown to be potent in vitro and in vivo against many bacterial strains, including those resistant to drugs such as macrolides-lincosamides-streptogramin B (10, 11).
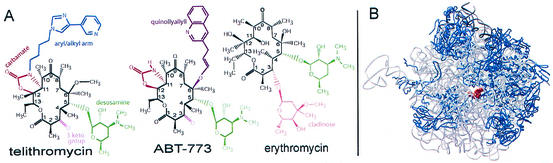
The ketolides are semisynthetic derivatives of erythromycin, the first macrolide in clinical use. The ketolides have a 14-member macrolactone ring, posses an extended arm, and lack the cladinose sugar. Their macrolactone ring has a keto group at position 3, and their hydroxyls in positions 11 and12 are replaced by a cyclic carbamate. Telithromycin has an alkyl-aryl extension that is bound to its cyclic carbamate, whereas ABT-773 has a quinolylallyl arm at the O-6 position (Fig. 1A).
FIG. 1.
(A) Chemical structures of the macrolide erythromycin and the ketolides telithromycin and ABT-773; (B) view into the D50S tunnel with a bound telithromycin molecule (red).
We present here the crystal structure of the ketolide telithromycin in complex with the large ribosomal subunit from Deinococcus radiodurans (D50S). Analysis of this structure shows that like other macrolides, telithromycin inhibits protein biosynthesis by blocking the passage of nascent proteins through the ribosome exit tunnel (Fig. 1B), but unlike them, it binds to two regions of the 23S RNA. This unique binding mode, observed also in the only available crystal structure of a ketolide (ABT-773) bound to the large ribosomal subunit (14), enabled correlation of the enhanced antimicrobial activity of ketolides against macrolide-resistant pathogens.
Methods. (i) Base numbering.
Nucleotides are numbered according to the D. radiodurans numbering. In the text, Escherichia coli numbering is given in parentheses (EC numbers).
(ii) Crystallization and data collection and processing.
Cocrystals were grown, as described in reference 8, from the complex of D50S and a clinical relevant concentration (0.01 mM) of telithromycin.
Data were collected at 90 to 100 K at ID29/ESRF/EMBL and ID19/APS/ANL and recorded on Quantum-210 and APS detectors, respectively. Data were processed and scaled (Table 1) with DENZO/SCALEPACK/HKL2000 (12).
TABLE 1.
Crystallographic data
| Parameter (unit) | Valuea |
|---|---|
| Space group | I222 |
| Wavelength (Å) | 1.038 |
| Unit cell parameters (Å) | 170.0, 414.5, 693.0 |
| Resolution range (Å) | 20-3.4 |
| Mosaicity (°) | 0.3 |
| Number of unique reflections | 273,922 |
| Completeness (%) | 82.7 (70.1) |
| Rmerge (%) | 18.1 (49.3) |
| 〈I〉/〈σ(I)〉 | 3.4 (1.5) |
| Redundancy | 3.4 (2.2) |
| Rfactor/Rfree (%) | 28.0/34.0 |
| Root mean square deviations from ideality | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.7 |
Values in parentheses refer to the highest-resolution shell (3.46 to 3.4 Å).
(iii) Antibiotic placement and structure refinement.
Overall and group rigid body refinements were performed with CNS (Crystallography & NMR System) (4) by using the 3.0-Å resolution structure of D50S as a starting model (8). A substantial improvement of the Fourier electron density (Fo-Fc and 2Fo-Fc) maps was achieved by solvent flattening by using SOLOMON (1). Telithromycin conformation was modeled interactively with O (9). Further refinement was carried out with CNS. Surface accessibilities were calculated with CCP4 (2). Figures were produced with RIBBONS (5) and LIGPLOT (16).
Drug binding site.
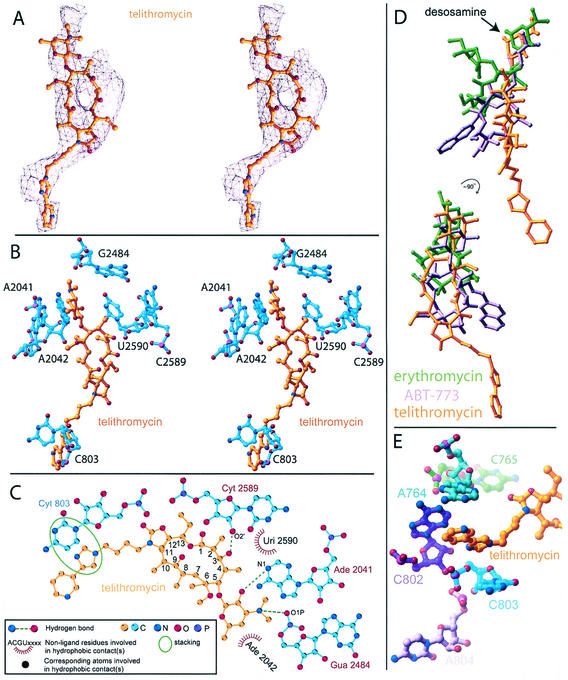
The crystal structure of the complex of D50S with telithromycin allowed a clear definition of the drug binding site (Fig. 2A and B). It showed that telithromycin interacts solely with the 23S RNA and that about 55% of the surface area of the bound telithromycin is buried by these interactions. We found that the mode of telithromycin binding to D50S (Fig. 2B and C) is consistent with biochemical results (7, 18).
FIG. 2.
(A and B) Stereo views of a telithromycin omit electron-density-map (A) and its binding site (B); (C) two-dimensional sketch of telithromycin interactions with D50S; (D) superposition of the positions of telithromycin (gold), ABT-773 (purple) (14), and erythromycin (green) (13) in D50S; (E) view of telithromycin along its extension arm showing its stacking interactions with the groove in domain II.
Telithromycin binds to 23S rRNA domain V.
The telithromycin macrolactone ring binds to domain V of the 23S rRNA through a hydrogen bond involving its 3-keto group and via hydrophobic interactions (Fig. 2C). Additional interactions with this domain are formed by telithromycin desosamine sugar, similar to the modes of binding of most macrolides to eubacterial large ribosomal subunits (13, 14). This sugar is involved in electrostatic interactions with the backbone of G2484 (G2502EC) and in hydrophobic interactions with A2042 (A2059EC). Its 2′ OH group is located within hydrogen bonding distance of A2041 (A2058EC). This nucleotide was shown to play a major role in binding of macrolides to eubacterial ribosomes (3, 13). It also plays a main role in resistance to macrolides from the erythromycin family, often acquired by steric hindrance caused by its methylation or A-to-G mutation (6, 17).
Ketolides anchor 23S rRNA domain II.
Telithromycin also interacts with domain II of the 23S rRNA (Fig. 2C), consistent with footprinting and mutation data (19) and with the binding mode of the ketolide ABT-773 (14) (Fig. 2D). Protection and mutation studies with E. coli indicated interactions between ketolides and nucleotide A752 (19). In their complexes with D50S, neither ABT-773 (14) nor telithromycin interacts with the corresponding D50S base, C765, but both are located in close proximity to this base (Fig. 2E). As with ABT-773 (14), telithromycin contacts to domain II are confined to the nucleotide C803 (C790EC), although the extension arms of these two ketolides have different structures and positions on the macrolactone rings.
These two ketolides interact with the same regions of the 23S RNA, but a comparison of the binding modes shows some subtle, albeit significant, differences. For example, G2484 (G2502EC) forms a hydrogen bond in the telithromycin complex whereas it is involved is less intensive coulombic interactions in the ABT-773 complex (14). Similar considerations apply to the O-6 atom, which can act as both a hydrogen bonding acceptor and donor. We observed that the alkyl-aryl extension of telithromycin reaches further than and is rather ordered compared to the quinolylallyl arm of ABT-773 (14) (Fig. 2D). We attributed the lower flexibility of telithromycin extension to the groove formed by nucleotides A764 (A751EC), A802 (A789EC), and C803 (U790EC), which surrounds its extended arm (Fig. 2E), compared to the larger space that is available for conformational mobility of the ABT-773 quinolylallyl group (14).
Ketolide binding to macrolide-resistant pathogens.
Comparisons of the binding modes of telithromycin and the macrolides from the erythromycin family (13) to domain V of the 23S RNA indicated that telithromycin binding should be less hindered by A2058EC modifications. Not only should the additional domain II interactions of the ketolides (14) lead to tighter binding, as observed in pharmacological studies (15), but they are also likely to compensate for the alterations of the binding site A2058EC due to bacterial resistance. Further stabilization of telithromycin binding seems to be achieved by the unique stacking arrangement of its long arm (Fig. 2E). It is therefore likely that drug interactions with domain II allow binding to ribosomes of resistant pathogens, even when the macrolide binding site in domain V is perturbed (6). This finding is consistent with previous observations showing that troleandomycin, a macrolide with a desosamine sugar lacking hydroxyls, binds nevertheless to the large ribosomal subunits, and its binding seems to benefit from its interactions with domain II (3). Hence, studies on the binding of telithromycin and of ABT-773 (14) provide a structural basis for the efficiency of ketolides against macrolide-resistant pathogens.
Conclusions.
Analysis of the crystal structure of the D50S-telithromycin complex showed binding to both domains V and II of the 23S RNA. The latter seem to compensate for perturbations in domain V binding interactions induced by methylation or mutation, thus providing structural clues to the elevated antimicrobial action of telithromycin against several macrolide resistant strains. The structural model for the unique binding mode of telithromycin paves the way for further improved drugs, capable of bypassing alterations in the drug binding sites occurring arising from bacterial resistance.
Protein Data Bank identification code.
The coordinates reported here have been deposited in the Protein Data Bank with the code 1P9X.
Acknowledgments
We thank R. Albrecht, T. Auerbach, D. Baram, A. Bashan, H. Bartels, W. S. Bennett, M. Kessler, C. Liebe, and A. Wolff for participating in this work and the staff at ID19/SBC/APS and ID29/ESRF/EMBL.
The Max Planck Society, the National Institutes of Health (GM34360), the German Science & Technology Ministry (BMBF/05-641EA), and the Kimmelman Center for Macromolecular Assembly provided support. A.Y. holds the Hellen and Martin Kimmel Professorial Chair.
REFERENCES
- 1.Abrahams, J. P., and A. G. W. Leslie. 1996. Methods used in the structure determination of bovine mitochondrial F-1 ATPase. Acta Crystallogr. D 52:30-42. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, S. 1994. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 3.Berisio, R., F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram, and A. Yonath. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10:366-370. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Carson, M. 1997. Ribbons. Macromol. Crystallogr. B 277:493-505. [PubMed] [Google Scholar]
- 6.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 7.Goldman, R. C., S. W. Fesik, and C. C. Doran. 1990. Role of protonated and neutral forms of macrolides in binding to ribosomes from gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 34:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 9.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2):110-119. [DOI] [PubMed] [Google Scholar]
- 10.Liu, M., and S. Douthwaite. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto, H., S. Miyazaki, K. Tateda, Y. Ishii, and K. Yamaguchi. 2001. In vivo efficacy of telithromycin (HMR3647) against Streptococcus pneumoniae and Haemophilus influenzae. Antimicrob. Agents Chemother. 45:3250-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307-326. [DOI] [PubMed] [Google Scholar]
- 13.Schluenzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 14.Schluenzen, F., J. M. Harms, F. Franceschi, H. A. Hansen, H. Bartels, R. Zarivach, and A. Yonath. 2003. Structural basis for the antibiotic activity of ketolides and azalides. Structure 11:329-338. [DOI] [PubMed] [Google Scholar]
- 15.Shain, C. S., and G. W. Amsden. 2002. Telithromycin: the first of the ketolides. Ann. Pharmacother. 36:452-464. [DOI] [PubMed] [Google Scholar]
- 16.Wallace, A. C., R. A. Laskowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127-134. [DOI] [PubMed] [Google Scholar]
- 17.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong, L., P. Kloss, S. Douthwaite, N. M. Andersen, S. Swaney, D. L. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]