Abstract
Bioinformatic analyses of whole genome sequences highlight the problem of identifying the biochemical and cellular functions of many gene products that are at present uncharacterized. The open reading frame Rv3853 from Mycobacterium tuberculosis has been annotated as menG and assumed to encode an S-adenosylmethionine (SAM)-dependent methyltransferase that catalyzes the final step in menaquinone biosynthesis. The Rv3853 gene product has been expressed, refolded, purified, and crystallized in the context of a structural genomics program. Its crystal structure has been determined by isomorphous replacement and refined at 1.9 Å resolution to an R factor of 19.0% and Rfree of 22.0%. The structure strongly suggests that this protein is not a SAM-dependent methyltransferase and that the gene has been misannotated in this and other genomes that contain homologs. The protein forms a tightly associated, disk-like trimer. The monomer fold is unlike that of any known SAM-dependent methyltransferase, most closely resembling the phosphohistidine domains of several phosphotransfer systems. Attempts to bind cofactor and substrate molecules have been unsuccessful, but two adventitiously bound small-molecule ligands, modeled as tartrate and glyoxalate, are present on each monomer. These may point to biologically relevant binding sites but do not suggest a function. In silico screening indicates a range of ligands that could occupy these and other sites. The nature of these ligands, coupled with the location of binding sites on the trimer, suggests that proteins of the Rv3853 family, which are distributed throughout microbial and plant species, may be part of a larger assembly binding to nucleic acids or proteins.
The explosive growth in the number of fully sequenced genomes offers an unparalleled opportunity for the understanding of organisms at the molecular level. At the same time, it reveals the extent of our current ignorance. Bioinformatic analyses indicate that in most fully sequenced genomes, at least 40% of gene products are of unknown function. A significant proportion of the remaining ≈60% for which functional annotations have been made are, however, of imperfectly described or uncertain function (for example, described simply as putative dehydrogenases), and some are likely to be wrong because functional annotations are in most cases derived by inference rather than by experiment, through the observation of some level of sequence identity in a gene product with a characterized gene product from another organism. We present here the structural analysis of a gene product from Mycobacterium tuberculosis which indicates that the annotated function in this and other bacterial genomes is likely to be wrong.
The complete genome sequence for M. tuberculosis strain H37Rv was reported in 1998 (6). The global significance of this pathogen is immense. As the cause of tuberculosis, it kills two to three million people around the world each year, more than any other single infectious agent. It is further estimated that around one-third of the world's population is infected as a result of the ability of the organism to persist for many years inside activated macrophages in a semidormant or latent form (2, 3, 31). Although effective drugs are available, treatment regimens are long and difficult and multidrug resistance is rising (2, 3, 31). This has resulted in a resurgence of interest in the biology of the organism.
One initiative in worldwide efforts to understand the biology of tuberculosis and to characterize potential new drug targets has been the formation of the Tuberculosis Structural Genomics Consortium (http://www.doe-mbi.ucla.edu/TB), a group of collaborating laboratories in a number of countries whose aim is to coordinate and facilitate the determination of the three-dimensional structures of large numbers of proteins from M. tuberculosis.
Menaquinone (vitamin K) is an essential vitamin that is an obligatory component of the anaerobic electron transfer pathways that operate not only in strict anaerobes but also in aerobic gram-positive bacteria, including M. tuberculosis (25, 26). This function may be particularly important in M. tuberculosis under conditions of low oxygen and may thus play a role in the persistence of the bacteria within activated macrophages. Coupled with the observation that menaquinone is an essential nutrient that is not synthesized in animals, this makes enzymes of the menaquinone biosynthetic pathway attractive drug targets.
In Escherichia coli, the menaquinone biosynthetic pathway involves either seven or eight enzymes (26). Sequence comparisons have found homologues for seven of these enzymes in the M. tuberculosis H37Rv strain. One of these proteins, MenG, was identified with the open reading frame annotated Rv3853, based on sequence similarity with a gene product from the E. coli genome. The E. coli enzyme in turn had been annotated as MenG on the basis of its position adjacent to the menA gene in the genome and apparent sequence similarities with S-adenosylmethionine (SAM)-dependent methyltransferases (25); MenG was proposed to be the SAM-dependent methyltransferase that transfers a methyl group to demethylate menaquinone in this final step. Intriguingly, the MenG sequence, which comprises 157 amino acid residues and represents a polypeptide of 16.2 kDa, has none of the common methyltransferase motifs, and the M. tuberculosis genome encodes another protein, identified as UbiE (Rv0558), that could also catalyze this final step (22).
In order to clarify its function by revealing possible homologies that cannot be seen at the sequence level and to provide a template for possible drug design, determination of the structure of the Rv3853 gene product was undertaken in the context of the tuberculosis structural genomics initiative. While this work was in progress, the structure of the E. coli homolog of Rv3853 was independently determined (J. D. Robertus, personal communication); the two proteins were found to have essentially identical structures, in both cases suggesting an incorrect functional annotation.
MATERIALS AND METHODS
Cloning and expression.
Open reading frame Rv3853 was amplified by PCR from genomic DNA of the H37Rv strain of M. tuberculosis. Primers were chosen that introduced a 5′ NcoI restriction site and a 3′ HindIII site. The gene was subcloned into two expression vectors: pET42a-rTEV, which gives rise to a fusion protein in which glutathione S-transferase is attached to the N terminus of the protein of interest, joined by a cleavable linker; and pProEX Hta, which gives a protein with a cleavable N-terminal poly-His tag. Expression tests in E. coli BL21(DE3)/pRI592 showed that for both constructs, the expressed protein was insoluble in all conditions tested.
Protein refolding and purification.
The N-terminally His-tagged fusion protein was denatured by cell lysis in a phosphate-Tris buffer containing 9 M urea at pH 8.0 and purified by Ni2+ affinity chromatography. The protein was then refolded by dialysis at room temperature through sequential transfers, first into refolding buffer (100 mM l-arginine, 100 mM sucrose, 50 mM morpholineethanesulfonic acid [MES], 10 mM NaCl, 0.4 mM KCl, 1 mM EDTA, 1 mM dithiothreitol) and then into storage buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA). The refolded protein was purified by size exclusion chromatography (Superdex 200; Pharmacia) and then further purified by anion exchange chromatography (Mono Q; Pharmacia). Light-scattering data for the final protein solution (2 mg of Rv3853 per ml) showed the protein to be a monodisperse solution of trimeric protein. The molecular mass calculated from the hydrodynamic radius was 64.5 kDa, compared with a monomer molecular mass for the His-tagged protein of 19.4 kDa.
Crystallization and soaking experiments.
Rv3853 crystals were grown by using hanging drops at 18° by mixing 4 to 5 μl of protein solution (50 mM Tris-HCl, 140 mM NaCl [pH 8.0], 2 mg of Rv3853 per ml) with 1 μl of precipitant solution (0.45 M potassium-sodium tartrate). Hexagonal blocks typically emerged after 2 to 3 days and grew larger over several weeks. The crystals were hexagonal, space group P63, with cell dimensions a = b = 102.5 Å and c = 117.5 Å. Three molecules occupy the asymmetric unit, corresponding to a solvent content of 63.9% and a Matthews coefficient of 3.43 Å3/Da.
Soaking experiments with heavy atom compounds and other ligands were conducted by soaking crystals at room temperature in an artificial mother liquor comprising 0.4 M potassium-sodium tartrate to which the appropriate compound was added. For heavy atom derivative preparation, crystals were soaked in 1 mM mercuric acetate for 9 days. Other soaking experiments were carried out with 1 mM and 10 mM SAM, 1 mM l-methionine, 1 mM ATP (chosen because of its adenosyl moiety), 1 mM menadione (equivalent to the product menaquinone but lacking the isoprenyl tail), and 1 mM Zwitergent 3-12, a detergent with a dodecyl group that might approximate the isoprenyl tail.
Data collection and processing.
Data collection was done at 110 K with crystals that had been soaked in cryoprotectant (mother liquor plus 35% glycerol) immediately prior to freezing in a stream of cold N2 gas. Native Rv3853 data and Hg derivative data were collected with CuKα radiation (λ = 1.5418 Å) from a Rigaku RU-H3R X-ray generator equipped with focusing mirrors and a Mar 345 imaging plate detector (Table 1). Subsequently, a high-resolution native data set was collected with synchrotron radiation (λ = 0.8452 Å) at DESY Hamburg, beamline BW7V. The raw data were processed with DENZO (30) and subsequently scaled with Scalepack (30).
TABLE 1.
Data collection and processing
| Parameter | Native | Mercuric acetate derivative |
|---|---|---|
| Space group | P63 | P63 |
| Cell dimensions (Å) | a = b = 102.5 | a = b = 102.3 |
| c = 117.5 | c = 117.5 | |
| Resolution range (Å) | 20.0-1.9 | 20.0-2.9 |
| Mosaicity (°) | 0.6 | 0.3 |
| Rmerge (%)a | 6.9 (31.2) | 18.1 (45.4) |
| No. of unique reflections | 54,630 | 15,453 |
| Mean I/σa | 14.4 (3.1) | 10.0 (2.6) |
| Multiplicitya | 3.35 | 6.18 |
| Completeness (%)a | 99.3 (96.1) | 99.4 (98.1) |
| Riso (%) | 23.7 | |
| Rano (%) | 8.6 | |
| RCullis (centric, acentric) | 0.77, 0.74 | |
| Phasing power (centric, acentric) | 1.06, 1.40 |
Values in parentheses are for the outermost shell of data.
Structure determination and refinement.
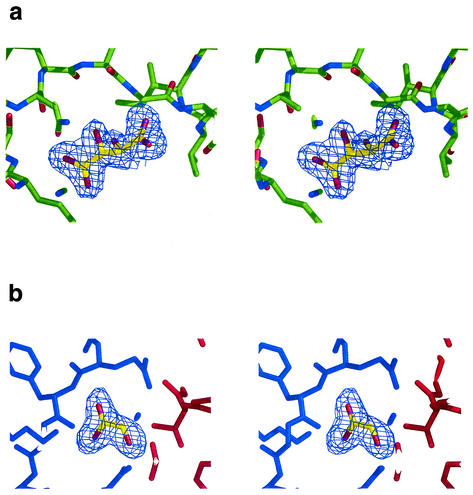
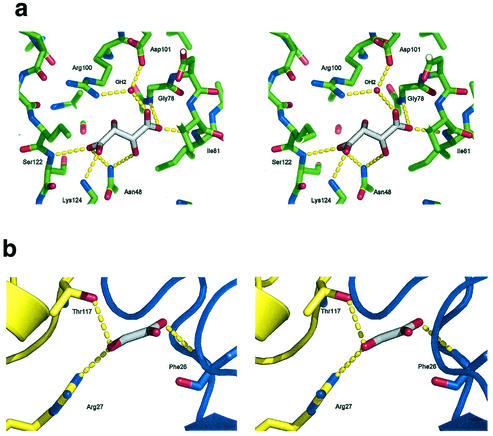
Phases were determined by single isomorphous replacement with anomalous scattering from the single mercury derivative with the program Solve (35). Two sites were found. Initial phases from Solve were calculated to 3.0 Å and gave a figure of merit of 30% and a z score of 8.18. These phases were extended to 1.9 Å resolution and modified by a maximum-likelihood density function with Resolve (35). The program wARP (18) was then used to autotrace 450 residues (150, 151, and 149 residues per respective chain). A further eight residues were built with O (16). Three iterations of manual building and refinement with O and CNS (5) were undertaken to complete the structure. Water molecules were added with CNS, taking a conservative approach so that only those with well-defined spherical density and hydrogen-bonded contacts of appropriate geometry were included in the model. In each monomer, two pieces of nonprotein density were found (Fig. 1). One was modeled as a tartrate ion, from the crystallization medium. The other was modeled first as urea, but refinement showed that it was larger, and it was modeled ultimately as glyoxalate (see below).
FIG. 1.
Stereo views showing the electron density for the two small molecules bound to each of the Rv3853 monomers, the putative tartrate molecule (a) and the putative glyoxalate molecule (b). Electron density is from a 2Fo-Fc electron density map, contoured at 1.0 σ. In b, the red and blue colors indicate adjacent monomers. Figure drawn with Pymol (8).
Model completeness and quality.
The final model consisted of residues 2 to 157 for chain A, 3 to 157 for chain B, and 4 to 157 for chain C together with three putative tartrate ions, three putative glyoxalate molecules, and 276 water molecules. For each chain, interpretable density was lacking for a small number of residues at the N terminus and for the attached the poly-His tag. The protein molecules conform well with standard geometry. Bond lengths and angles are restrained close to the standard values of Engh and Huber (12) (Table 2), and the polypeptide chain torsion angles conform with allowed regions of the Ramachandran plot; 89.4% of residues are in the most favored regions, as defined by Procheck (7, 19), and no residues are in disallowed regions.
TABLE 2.
Refinement and model detailsa
| Parameter | Value |
|---|---|
| Resolution limits (Å) | 25.0-1.9 |
| R factor (Rfree) (%) | 19.0 (22.0) |
| No. of reflections | 52,727 (5,344) |
| Model details | |
| No. of protein atoms | 3,384 |
| No. of other molecules, ions | 3 tartrate ions, 3 glyoxalate |
| No. of water molecules | 276 |
| Root mean square deviation from standard geometry | |
| Bond length (Å) | 0.005 |
| Bond angle (°) | 1.3 |
| Avg B factor (Å2) | |
| Protein (A, B, C chains) | 14.5, 17.3, 13.4 |
| Ligands (tartrate, glyoxalate) | 22.0, 29.1 |
| Water | 21.7 |
| % of residues in most-favored regions of Ramachandran plot | 89.4 |
See Table 1, footnote a.
In silico screening.
The tartrate and glyoxalate ligands were deleted from the model, which was then subjected to analysis with the SiteID method, as contained in Sybyl 6.8 (Tripos Inc.). Regions around the sites identified in this way were then subjected to in silico screening with Fred (http://www.eyesopen.com) with an in-house database of more than 520,000 small molecules. This library of compounds was assembled from databases available on the web, including those of the National Cancer Institute, Sigma-Aldrich, Maybridge, Interbioscreen, and Asinex, together with the complete set of small-molecule ligands present in the Protein Data Bank (http://www.rcsb.org/pdb/). Top-ranked hits were inspected with VIDA (http://www.eyesopen.com).
Atomic coordinates.
Atomic coordinates have been deposited with the Protein Data Bank, with accession code 1nxj.
RESULTS
Overview of structure.
The asymmetric unit of the crystal contains three monomers, but analysis of the crystal packing and the buried surface between monomers shows that these do not form a trimer. Instead, a symmetric homotrimeric unit is formed via the crystallographic symmetry, with each monomer packing with two other monomers related by the exact crystallographic threefold axis. The unit cell also contains three putative tartrate ions, one per protein monomer, and three other nonprotein molecules, tentatively modeled as glyoxalate. It is worth noting that the structure determined here was produced from protein that was refolded from insoluble material, but its validity is emphasized by the fact that it is essentially identical to that of its E. coli homolog, whose structure was determined in parallel (J. D. Robertus, personal communication).
Monomer fold.
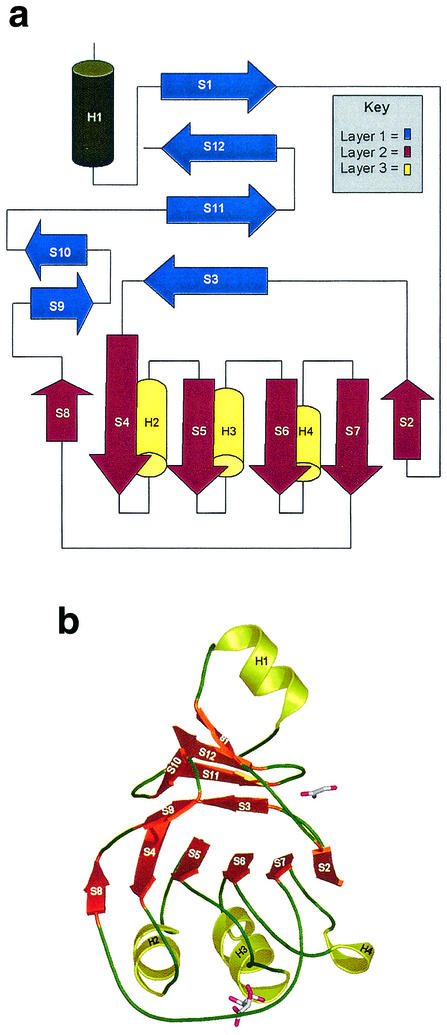
The monomer is folded into a single domain that can be described as a three-layer β/β/α structure (Fig. 2). The first layer consists of a four-stranded antiparallel β-sheet (strands S1, S12, S11, and S3) that sits adjacent to a two-stranded β-ribbon (S9 and S10). This layer packs against a central six-stranded, mostly parallel β-sheet (S8, S4, S5, S6, S7, and S2) that forms the second layer. The third layer of the “sandwich” comprises three parallel α-helices (H2, H3, and H4) that provide the S4-S5, S5-S6, and S6-S7 connections. A large extended loop region, comprising 20 residues, finishing with the short strand S8, wraps around layers 2 and 3 and leads back to layer 1. Located between the two β-sheet regions (layers 1 and 2) is a hydrophobic groove, which is “capped” by the loop region connecting strands S2 and S3 of the two sheets. Outside the main β/β/α domain, the N-terminal α-helix H1 packs against the first β-sheet and also forms an important part of the monomer-monomer interface in the trimer.
FIG. 2.
(a) Topology diagram for the Rv3853 monomer. The three layers in this β/β/α structure are shown in blue, red, and yellow. (b) Fold of the monomer, with β-strands shown as orange arrows and α-helices as yellow coils. The two bound ligands, tartrate (lower) and a putative glyoxalate (upper), are shown in stick mode.
Comparison of the MenG monomer with all other structures in the Protein Data Bank with DALI (15) revealed five structures with z scores of greater than 4. The highest match was to the phosphohistidine domain of pyruvate phosphate dikinase (14), with a z score of 7.4, and a root mean square difference in atomic positions of 3.0 Å for 106 matching Cα pairs. The fold shared by Rv3853 and this phosphohistidine domain is described in SCOP (27) as a “swiveling” β/β/α fold and in CATH (29) as a three-layer β/β/α sandwich. Other structures classified under this fold and also recognized as being related to Rv3853 by DALI (15) include the phosphohistidine domain of enzyme I of the E. coli phosphoenolpyruvate:sugar phosphotransferase system (23), the small subunit of carbamoyl phosphate synthase (36), and a domain from aconitase (20).
Quaternary structure.
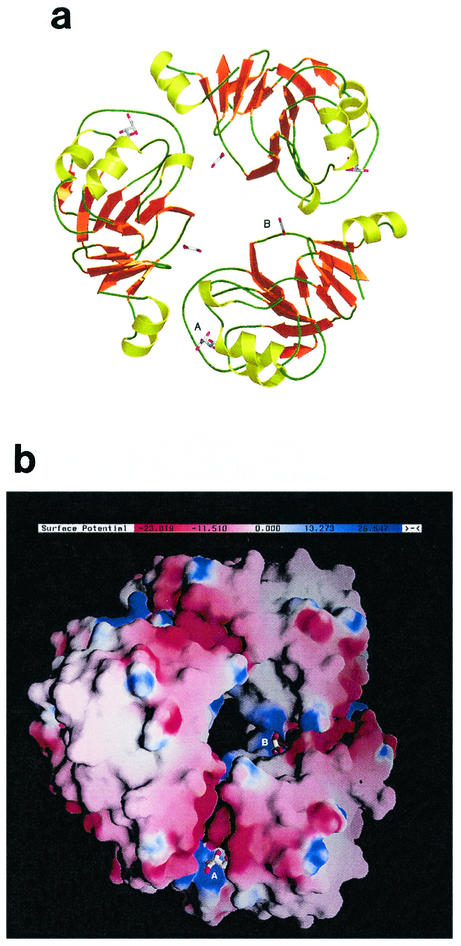
The Rv3853 trimer (Fig. 3) is donut shaped with a large hole (diameter approximately 8 to 10 Å) through the middle. At each monomer-monomer interface, the N-terminal helix (residues 7 to 16), the following H1-S2 connection (residues 21 to 27), and the C-terminal S11-S12 loop (residues 151 to 152) of one monomer pack into a cleft in the neighboring monomer that is formed between residues 115 to 121 of the extended loop joining S7 to S8 (Fig. 2) and the loops that connect the β-strands of layer 2 with their respective helices (the S4-H2, S5-H3, and S6-H4 loops). The total surface area buried at each of the three monomer-monomer interfaces is 530 Å2, meaning that 9.6% of the surface area of each monomer is buried. Trimer formation is stabilized by a number of hydrogen bonds and four salt bridges (Asp13-Arg100, Asp13-Lys121, Asp23-His73, and Asp23-Arg120). A striking feature of the trimer, highlighted by a GRASP (28) plot (Fig. 3b), is a groove that runs the whole length of each monomer-monomer interface, incorporating both the tartrate and glyoxalate binding sites (see below), and a canyon of negative charge in which are found Asp10, Asp13, Asp150, and Asp152.
FIG. 3.
Rv3853 trimer, shown (a) as a ribbon diagram, drawn with Pymol (8), and (b) in a surface representation, drawn with GRASP (27), showing the distribution of surface charge. In both diagrams, the putative tartrate (A) and glyoxalate (B) molecules are shown in stick representation, bound to each of the three monomers. Adjacent to the tartrate binding site is a prominent, negatively charged canyon at the subunit interface that contains several residues conserved in all Rv3853 homologs.
Sequence conservation in Rv3853 homologues.
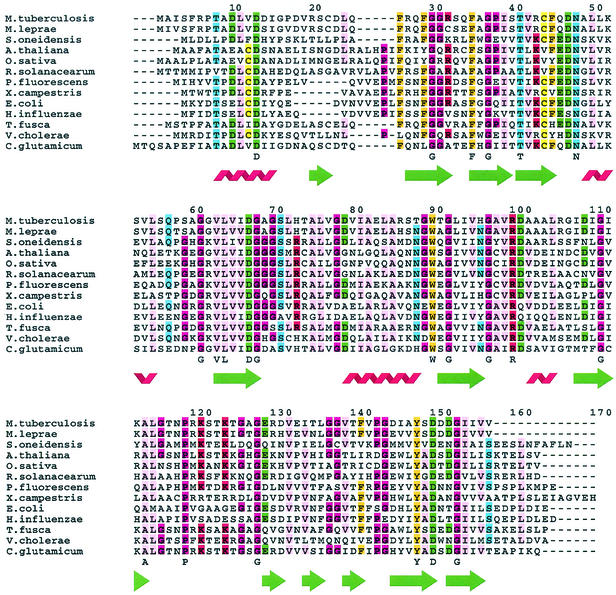
Searches of the current nonredundant NCBI sequence database with Blast (1) reveal a large number of sequences that are homologous with Rv3853. Most are from bacterial genomes, but some are from plants, with the Arabidopsis thaliana genome having no fewer than three Rv3853 homologues. Sequence alignments of the top hits given by Blast (Fig. 4) show that there is only one position at which insertions or deletions are found within the polypeptide, although the N and C termini have extensions of various lengths. This is consistent with a tightly folded, conserved domain.
FIG. 4.
Sequence alignment of 13 representative Rv3853 homologs chosen from both bacteria and plants, including Mycobacterium leprae, Shewanella oneidensis, Arabidopsis thaliana, Oryza sativa, Ralstonia solanacearum, Pseudomonas fluorescens, Xanthomonas campestris, Escherichia coli, Haemophilus influenzae, Thermobifida fusca, Vibrio cholerae, and Corynebacterium glutamicum. Fully conserved residues in these 13 sequences are indicated below each alignment, as are the locations of secondary-structure elements. The alignment was generated with FarOut.
Comparison of the top 32 sequences homologous with Rv3853 shows 20 residues that are totally conserved. These include one Pro and nine Gly residues (probably conserved for structural reasons) and four hydrophobic residues from the structural core (see Fig. 4, where the top 13 sequences are aligned). The remaining six invariant residues, Asp13, Thr41, Asn48, Asp67, Arg100, and Asp150, are potential candidates for involvement in substrate binding and/or catalysis. Particularly notable are Asp13 and Asp150, both projecting into the negatively charged cleft at the subunit interface, and Asn48 and Arg100, both associated with the bound tartrate ion nearby. In contrast, the side chains of Thr41 and Asp67 are remote from the other invariant residues and are extensively involved in hydrogen bonds that link together different parts of the fold, suggesting that their role is in stabilization of the structure.
Small-molecule binding sites.
None of the soaking experiments with ligands related to the presumed substrate (menaquinone) or cofactor (SAM) showed any evidence of binding. However, every electron density map, for either native or soaked crystals, showed two well-defined pieces of nonprotein density that must represent bound small-molecule ligands. Both were present for all three independent monomers in the asymmetric unit of the crystal.
The first (Fig. 5a) occupies a shallow pocket between the N terminus of helix H3 and a portion of the long S7-S8 loop (Fig. 2) near the monomer-monomer interface. This density was interpreted as a bound tartrate ion on the basis of the excellent fit of tartrate to the density (Fig. 1a) and the presence of 0.45 M tartrate in the crystallization medium. Refinement supported this assignment, as all atoms assumed B factors that were similar to each other and similar to atoms in the surrounding protein structure (15 to 25 Å2). One carboxylate group is nicely positioned at the N terminus of helix H3, hydrogen bonded to the free peptide NH groups of residues 78 and 81 and to a conserved, well-defined water molecule that bridges to Arg100 and Asp101 (Fig. 5a), which are located in the loop region connecting S6 to H4. The other carboxylate group receives hydrogen bonds from Asn48 ND2, the peptide NH of Ser122, and the amino group of Lys124. On the other hand, the two tartrate hydroxyl groups make few or no hydrogen bonds. One is hydrogen bonded to Asn48 ND2 and (in two out of three monomers) to a water molecule that bridges to Lys52 NZ. The other makes no hydrogen-bonded interactions.
FIG. 5.
Stereo views of the two binding sites for small-molecule ligands. In a, the binding site for the tartrate ion is shown, with its protein ligands and a conserved water molecule found for all three monomers, and in b that for the tentatively assigned glyoxalate molecule is shown. In each case, hydrogen bonds are shown with broken yellow lines. In b, the two adjacent monomers that form the binding site are shown in blue and yellow, respectively.
The second bound ligand (Fig. 5b) was first modeled as a urea molecule on the basis of its planar, somewhat triangular shape and the use of urea in the refolding of the expressed protein. Refinement as urea left residual positive electron density, however, that suggested that this species was at least one atom longer. When it is modeled as glyoxalate, the density is nicely accounted for, B values for all atoms are normal, and good hydrogen bond contacts are made with Arg27 NH1, Thr117 OG1, the peptide NH of Phe26, and several well-defined water molecules. Whether or not this species is indeed glyoxalate, perhaps derived from degradation of tartrate, it clearly has very similar shape and size. The site it occupies is deep and highly positively charged at its entrance, suggesting a strong preference for anionic species, and is located at the inner end of the monomer-monomer interface, close to the hole through the center of the trimer (Fig. 3).
In silico analysis.
Nine binding sites were found by the SiteID analysis (Tripos Inc.), distributed symmetrically round the trimer, three per monomer. Each set of three sites was found to be located in the groove at the monomer-monomer interface (Fig. 3b). The largest (site 1, volume 28 Å3) is at the inner end of the interface, adjacent to the hole through the center of the trimer. In the crystal structure, this pocket is occluded by the glyoxalate molecule, which sits in the site entrance, leaving the majority of the volume unoccupied. This site is bounded by residues 22 to 28, 113, 117, and 151 to 154. Site 2 (volume 16 Å3) corresponds to the acidic canyon, with contributing residues including Phe6, Asp10, Gln34, Asp101, Ala102, Ala103, Asp150, Asp151, and Asp152. Site 3 (volume 14 Å3) is adjacent to site 2 and is completely filled by the tartrate ion described above.
When the regions around the above three sites were screened against our in-house database of potential ligands, there was a strong preference for planar, fused-ring systems, most containing at least one nitrogen atom, such as indole or nucleoside base derivatives. The best 50 hits for the region around site 1 gave ScreenScore values (34) from −39.7 down to −35.7, suggesting affinities in the submicromolar range. All of the binding orientations bury a substantial region of the ligand within the deep site 1 pocket, although a proportion extend beyond the pocket and interact with residues in site 2 as well. Hits in the region around sites 2 and 3 suggest a lower affinity (ScreenScore values of −37.6 to −27.9), but still high enough to suggest significant in vitro affinity of some ligands. Again there is a preference for planar, fused-ring compounds. Interestingly, the tartrate pocket (site 3) is poorly filled by many of the compounds, which prefer to bind beneath the pocket in a continuation of the groove between the monomers, where it wraps under the protein.
DISCUSSION
The annotation of Rv3853 and its homologs as methyltransferases and as the terminal enzyme in the menaquinone biosynthetic pathway, MenG, seems to have originated from the E. coli protein. In E. coli, the gene for this was found to be adjacent to the menA gene, and its sequence was reported to contain a “characteristic” (but not specified) SAM binding motif (25). The deposition of this sequence in GenBank (accession code U56082), annotated as MenG, seems to have led to the annotation of all subsequent homologs and now persists through more than 30 genomes in which such homologs can be found. Several lines of evidence, most strikingly the molecular structure, now leave little doubt that this annotation is incorrect.
Two families of SAM-dependent methyltransferases have been characterized structurally. The predominant family has a conserved α/β fold whose defining feature is a seven-stranded β-sheet that has six parallel strands and one antiparallel and carries the SAM binding site (24). Although sequence identity is very low across the whole superfamily, structure-based alignment of the sequences of 28 family members shows that conserved amino acid sequence patterns are associated with SAM binding (24). These methyltransferases act on a wide variety of substrates, including nucleic acids, proteins, lipids, and small molecules, with diverse binding sites being created by a variety of extrusions from the canonical seven-stranded β-sheet. The Rv3853 gene product has neither the fold nor the sequence patterns that are characteristic of this superfamily of methyltransferases.
The second family of SAM-dependent methyltransferases acts on histones, methylating lysine residues, and is defined by a conserved domain called the SET domain (37, 38). This is a small domain (≈130 residues) with several small antiparallel β-sheets, a relatively exposed SAM binding site, and several conserved sequence motifs (37, 38). Again, neither fold nor sequence motifs are shared by the Rv3853 gene product. Other SAM-binding proteins, such as the C-terminal domain of methionine synthase (11) and the cobalt precorrin-4 methyltransferase CbiF (33), also have folds very distinct from that of the Rv3853 gene product.
Bioinformatic analysis of the M. tuberculosis genome further suggests that Rv3853 is not MenG, the terminal enzyme in menaquinone biosynthesis. Homologs of five of the other enzymes from this biosynthetic pathway (MenA, MenB, MenC, MenD, and MenE) can be found clustered in close proximity in the genome, between Rv0534 and Rv0555, far removed from Rv3853. Also in this portion of the genome is a gene (Rv0558) that is annotated as ubiE, encoding a SAM-dependent methyltransferase that is the terminal enzyme in ubiquinone biosynthesis. Given that it has been shown experimentally in several bacterial species that UbiE can carry out the MenG reaction (17, 22) and that no other ubiquinone biosynthetic enzymes can be found in the M. tuberculosis genome, it is highly probable that it is the gene product of Rv0558 that performs this final methyl transfer step in menaquinone biosynthesis. Unlike Rv3853, Rv0558 does contain SAM-dependent methyltransferase sequence motifs and has homologs in several bacterial species that are actually annotated as MenG, with functional support (17, 32).
What, then, is the biochemical and cellular function of Rv3853 and its homologs in other organisms? Searches of the current structural database show that the closest structural relationships with Rv3853 involve the phosphohistidine domains of several proteins involved in phosphate transfer. In these proteins,a phosphate group is transferred from one substrate to another (the substrates being either small molecules or entire protein domains) via an active-site histidine residue that is transiently phosphorylated. Other, weaker matches are found with the transferrin receptor apical domain (z = 4.0) (21), part of the thermosome (z = 3.6) (10), subdomain 4 of the AICAR (5-aminoimidazole-4-carboxamide-ribonucleotide) transferase domain of AICAR transformylase, which is involved in purine biosynthesis (z = 3.2) (13), the substrate binding domain of d-2-hydroxyisocaproate dehydrogenase (z = 3.2) (9), and the apical domain of GroEL (z = 3.0) (4).
In the two closest matches, the phosphohistidine domains of pyruvate phosphate dikinase (14) and enzyme I of the E. coli phosphoenolpyruvate:sugar phosphotransferase system (23), the active-site histidine is at the N terminus of a helix corresponding to H4 in Rv3853 and is preceded by a loop that contains several conserved residues. Rv3853 does not have a histidine residue in this position, precluding a similar histidine-mediated phosphoryl transfer. It is intriguing to note, however, that in Rv3853 the equivalent loop carries Arg100, which is one of the few residues that is totally conserved in the all the homologous sequences that we have been able to examine. Moreover, this arginine lies between sites 2 and 3 at the monomer-monomer interface. Site 2 also contains two fully conserved residues, both aspartate, suggesting a functional importance for these two pockets and for Arg100 between them.
Considering the structure as a whole, the trimer exhibits a potential ligand-binding groove that encompasses most of each monomer-monomer interface. Within this groove, a series of obvious pockets exist which may represent the primary ligand-binding sites on the protein. Two of these sites are occupied in the crystal structure; a small pocket on the outside of the protein is filled by a molecule of tartrate, and the largest pocket on the protein on the inside of the ring is occluded by a single small molecule that is tentatively modeled as glyoxalate. These bound ligands must come from the purification and crystallization media, since the urea denaturation that preceded refolding of the protein should have dislodged any cell-derived metabolites or cofactors.
In silico screening of the three sites on the protein identified by SiteID analysis indicated that the larger pocket (site 1) has a predilection for heterocyclic fused 5,6 ring systems such as indoles and purines. This may indicate that the protein function is related to nucleotide manipulation, with this site being involved in base binding, and if this is the case, the positive charge at its opening is potentially relevant. The tartrate pocket is apparently a fairly poor binding site, with the 78% of hits in the region including the pocket actually binding elsewhere.
The location of the binding sites at the subunit interfaces of the trimer, coupled with its apparent stable association, suggests that this is the physiologically relevant entity. The extended grooves at each interface and probable binding pocket in the internal face of the ring suggest the possibility that the protein binds to its ligand in a manner analogous to the interaction of a sliding clamp with a nucleic acid. The hole through the center of the trimer is large enough (diameter ≈8 to 10 Å) to accept a single-stranded (but not double-stranded) polynucleotide or a polypeptide strand. These observations suggest that it could be part of a larger system, again analogous to the multimeric DNA polymerase complex that includes a sliding clamp. Alternatively, it may be part of a system for binding linear peptide sequences, with the peptide lying along the groove and some specificity for aromatic side chains, possibly tryptophan, arising from interactions in the internal pocket.
Acknowledgments
We thank Shaun Lott for help with bioinformatic analyses and with protein expression, Heather Baker for advice on crystallization, Peter Haebel and staff at DESY Hamburg, beamline BW7V, for help with synchrotron data collection, and members of the Tuberculosis Structural Genomics Consortium for their encouragement and interest.
This work was supported by the New Economy Research Fund of New Zealand, the Health Research Council of New Zealand, and the Foundation for Research, Science and Technology for the award of a Bright Futures Scholarship to J.M.J.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barry, C. E., III. 1997. New horizons in the treatment of tuberculosis. Biochem. Pharmacol. 54:1165-1172. [DOI] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, R. A. Slayden, A. E. Sampson, and R. E. Lee. 2000. Use of genomics and combinatorial chemistry in the development of new antimycobacterial drugs. Biochem. Pharmacol. 59:221-231. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, D. C., J. Wang, Z. Otwinowski, A. L. Horwich, and P. B. Sigler. 1996. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATPγS. Nat. Struct. Biol. 3:170-177. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLeah, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Soeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrett. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Collaborative Computational Project Number Four. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed]
- 8.DeLano, W. L. 2002. Pymol. DeLano Scientific, Houston, Tex.
- 9.Dengler, U., K. Niefind, M. Kiess, and D. Schomburg. 1997. Crystal structure of a ternary complex of d-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 Å resolution. J. Mol. Biol. 267:640-660. [DOI] [PubMed] [Google Scholar]
- 10.Ditzel, L., J. Löwe, D. Stock, K.-O. Stetter, H. Huber, R. Huber, and S. Steinbacher. 1998. Crystal structure of the thermosome, the archael chaperonin and homolog of CCT. Cell 93:125-138. [DOI] [PubMed] [Google Scholar]
- 11.Dixon, M. M., S. Huang, R. G. Matthews, and M. Ludwig. 1996. The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure 4:1263-1275. [DOI] [PubMed] [Google Scholar]
- 12.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. Sect. A 47:392-400. [Google Scholar]
- 13.Greasley, S. E., P. Horton, J. Ramcharan, G. P. Beardsley, S. J. Benkovic, and I. A. Wilson. 2001. Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis. Nat. Struct. Biol. 8:402-406. [DOI] [PubMed] [Google Scholar]
- 14.Herzberg, O., C. C. H. Chen, G. Kapadia, M. McGuire, L. J. Carroll, S. J. Noh, and D. Dunaway-Mariano. 1996. Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites. Proc. Natl. Acad. Sci. USA 93:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., J. Y. Zou, and S. W. Cowan. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 17.Koike-Taeshita, A., T. Koyama, and K. Ogura. 1997. Identification of a novel gene cluster participating in menaquinone (vitamin K-2) biosynthesis. Cloning and sequence determination of the 2-heptaprenyl-1,4-naphthoquinone methyltransferase gene of Bacillus stearothermophilus. J. Biol. Chem. 272:12380-12383. [DOI] [PubMed] [Google Scholar]
- 18.Lamzin, V. S., and K. S. Wilson. 1997. Automated refinement for protein crystallography. Methods Enzymol. 277:269-305. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 20.Lauble, H., and C. D. Stout. 1995. Steric and conformational features of the aconitase mechanism. Proteins Struct. Funct. Genet. 22:1-11. [DOI] [PubMed] [Google Scholar]
- 21.Lawerence, C. M., S. Ray, M. Babyonyshev, R. Galluser, D. W. Borhani, and S. C. Harrison. 1999. Crystal structure of the ectodomain of the human transferrin receptor. Science 286:779. [DOI] [PubMed] [Google Scholar]
- 22.Lee, P. T., A. Y. Hsu, H. T. Ha, and C. F. Clarke. 1997. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: Isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179:1748-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao, D.-I., E. Silverton, Y.-J. Seok, B. R. Lee, A. Peterkofsky, and D. R. Davies. 1996. The first step in sugar transport: Crystal structure of the amino terminal domain of enzyme I of the E. coli phosphoenolpyruvate:sugar phosphotransferase system and a model of the phosphotransfer complex with HPr. Structure 4:861-872. [DOI] [PubMed] [Google Scholar]
- 24.Martin, J. L., and F. M. McMillian. 2002. SAM (dependent) I am: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12:783-793. [DOI] [PubMed] [Google Scholar]
- 25.Meganathan, R. 1996. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q), p. 642-656. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 26.Meganathan, R. 2001. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic mechanisms. Vitamins Hormones 61:173-218. [DOI] [PubMed] [Google Scholar]
- 27.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:537-540. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11:281-296. [DOI] [PubMed] [Google Scholar]
- 29.Orengo, C. A., A. D. Michie, S. Jones, D. T. Jones, M. B. Swindells, and J. M. Thornton. 1997. CATH—a hierarchic classification of protein domain structures. Structure 5:1093-1108. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 31.Pym, A. S., and S. T. Cole. 1999. Post DOTS, post genomics: the next century of tuberculosis control. Lancet 353:1004-1005. [DOI] [PubMed] [Google Scholar]
- 32.Sakuragi, Y., B. Zybailov, G. Shen, A. D. Jones, P. R. Chitnis, d. E. A. van, R. Bittl, S. Zech, D. Stehlik, J. H. Golbeck, and D. A. Bryant. 2002. Insertional inactivation of the menG gene, encoding 2-phythyl-1,4-naphthoquinone methyltransferase of Synechocystis sp. PCC 6803, results in the incorporation of 2-phythyl-1,4-naphthoquinone into the A1 site and alteration of the equilibrium constant between A1 and FX in photosystem I. Biochemistry 41:394-405. [DOI] [PubMed] [Google Scholar]
- 33.Schubert, H. L., K. S. Wilson, E. Raux, S. C. Woodcock, and M. J. Warren. 1998. The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase. Nat. Struct. Biol. 5:585-592. [DOI] [PubMed] [Google Scholar]
- 34.Stahl, M., and M. Rarey. 2001. Detailed analysis of scoring functions for virtual screening. J. Med. Chem. 44:1035-1042. [DOI] [PubMed] [Google Scholar]
- 35.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoden, J. B., F. M. Raushel, M. M. Benning, I. Rayment, and H. M. Holden. 1999. The structure of carbamoyl phosphate synthetase determined to 2.1 angstrom resolution. Acta Crystallogr. Sect. D 55:8-24. [DOI] [PubMed] [Google Scholar]
- 37.Trievel, R. C., B. M. Beach, L. M. A. Dirk, R. L. Houtz, and J. A. Hurley. 2002. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111:91-103. [DOI] [PubMed] [Google Scholar]
- 38.Yeates, T. O. 2002. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell 111:5-7. [DOI] [PubMed] [Google Scholar]