Abstract
Complete cellulose degradation is the first step in the use of biomass as a source of renewable energy. To this end, the engineering of novel cellulase activity, the activity responsible for the hydrolysis of the β-1,4-glycosidic bonds in cellulose, is a topic of great interest. The high-resolution X-ray crystal structure of a multidomain endoglucanase from Clostridium cellulolyticum has been determined at a 1.6-Å resolution. The endoglucanase, Cel9G, is comprised of a family 9 catalytic domain attached to a family IIIc cellulose-binding domain. The two domains together form a flat platform onto which crystalline cellulose is suggested to bind and be fed into the active-site cleft for endolytic hydrolysis. To further dissect the structural basis of cellulose binding and hydrolysis, the structures of Cel9G in the presence of cellobiose, cellotriose, and a DP-10 thio-oligosaccharide inhibitor were resolved at resolutions of 1.7, 1.8, and 1.9 Å, respectively.
Cellulases catalyze the hydrolysis of the β-1,4-glycosidic linkages in cellulose, the most abundant biopolymer. Varieties of microorganisms secrete these enzymes either individually or associated in a macromolecular complex referred to as the cellulosome (23). These different cellulases, acting synergistically, catalyze the complete hydrolysis of cellulose to glucose, which, under anaerobic conditions, is a highly fermentable fuel product. The ecological and economic advantages of plant biomass conversion have spawned considerable interest in the study and eventual use of recombinant cellulolytic complexes.
Clostridium cellulolyticum is an anaerobic, mesophilic, soil bacterium that is able to grow on cellulose as its sole carbon source. C. cellulolyticum secretes its cellulases in the form of a cellulosomal complex that has been extensively studied biochemically and genetically (2, 14). One of these cellulases, Cel9G, is a multidomain endoglucanase capable of hydrolyzing crystalline cellulose at an appreciable rate (13). The multiple domains of this cellulase include a catalytic domain (CD) responsible for the binding and hydrolysis of cellulose, a cellulose-binding domain (CBD) responsible for the attachment of the enzyme to a cellulose chain, and the dockerin domain, which is responsible for the attachment of the cellulase to the cellulosome (35). Interestingly, in the absence of cellulosomal interactions, the dockerin domain is autolyzed in a matter of a couple of weeks at 4°C. No true cellulolytic activity is attributed to the CBD and yet, in its absence, the CD of Cel9G is inactive toward carboxymethyl cellulose, Avicel, or phosphoric acid swollen cellulose (PASC), implying a possible role in catalysis (13). In addition, when expressed as a fusion protein fused to glutathione S-transferase, the CBD alone cannot bind to cellulosic substrates.
Based on sequence alignment studies, the CD of Cel9G belongs to family 9 of the glycoside hydrolase enzymes and uses a single displacement hydrolytic mechanism with net inversion of the substrate's anomeric C1 carbon. This mechanism involves general acid-base catalysis (21) mediated by the concerted action of two conserved acidic residues: glutamate and aspartate. The CBD belongs to family IIIc and is attached to the CD via a polypeptide linker.
Cellulase E4 from Thermomonospora fusca was the first multidomain cellulase structure containing both the CD and the CBD (42) and provided details into the tertiary organization of these domains. Functional studies on CelE4 combined with crystal structures of the enzyme complexed with cellodextrins ranging from cellobiose to cellopentaose provided insight into a possible mechanism of cellulose degradation. The CD and CBD provide an elongated, flat face that, hypothetically, linear strands of crystalline cellulose can adhere to. The binding energy is provided by stacking interactions between the aromatic glucose rings and aromatic residues lining the active-site cleft and flat face of the CBD. The work on CelE4 and prior work by Tormo et al. (46) on a family IIIa CBD of the cellulosomal scaffoldin subunit from Clostridium thermocellum proposed that the CBD not only provides a platform for binding but also disrupts the hydrogen bonding network of crystalline cellulose. This leads to the liberation of individual strands of cellulose that can enter the active-site cleft.
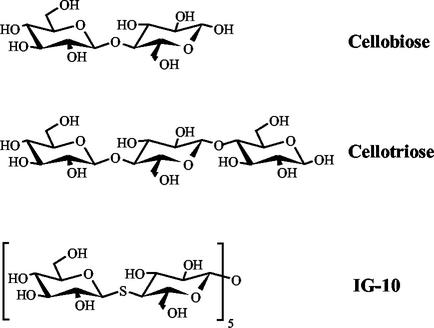
Sequence comparisons of CelE4 with Cel9G indicates 54% identity of the CDs and 42% identity of the CBDs, with an overall sequence identity of 51%. The X-ray crystal structure of Cel9G was solved by molecular replacement by using CelE4 as a phasing model. The native Cel9G structure was subsequently used to phase X-ray data collected on complexes of crystals soaked with natural and synthetic oligosaccharides (Fig. 1). The structures of these complexes offer additional insight into the catalytic machinery used by Cel9G to hydrolyze cellulose.
FIG. 1.
Chemical structures of the natural and synthetic inhibitors and substrates used for the crystallographic soaking experiments.
MATERIALS AND METHODS
Cel9G expression and crystallization and data collection.
Recombinant Cel9G was expressed in Escherichia coli BL21(DE3) cells and purified as previously described (13). The purified enzyme was dialyzed into 10 mM Tris (pH 7.5) and left at 4°C for 2 weeks during which, through autolysis, the C-terminal dockerin domain and His tag are cleaved off. The final form of Cel9G is composed of a CD and a CBD and has a molecular mass of 66 kDa (residues 36 to 649 of GUNG_CLOCE; SwissProt, P37700).
Crystals of Cel9G were grown by using a combination of the hanging-drop vapor diffusion technique (28) combined with microseeding (44). 2 μl of a reservoir solution containing 25% PEG 4000, 100 mM Tris-HCl (pH 8.4), 100 mM Mg(CH3COO)2, 5% isopropanol, and 15% glycerol were mixed with 2 μl of Cel9G at 6.25 mg/ml and allowed to equilibrate at 17°C. After 24 h, crushed Cel9G crystals were used as seeds to nucleate the equilibrated drops. Within 2 to 3 weeks of incubation at 17°C, the crystals were suitable for high-resolution X-ray diffraction experiments. The cellotriose cocrystallization experiment included 20 mM cellotriose (Sigma) in the protein drop and 100 mM HEPES (pH 7.0) instead of Tris-HCl. For the soaking experiments, 0.4 μl of 100 mM cellobiose or 20 mM IG-10 (i.e., hemithiocellodextrin) (Fig. 1) was added to drops containing crystals and allowed to incubate at 17°C for 20 h before the diffraction experiments were carried out. The synthesis and properties of the thio-oligosaccharide IG-10 have already been described (31).
Diffraction data for the native Cel9G structure were collected at the FIP/BM30 beamline at the European Synchrotron Research Facility (ESRF) (Grenoble, France) with a flash-frozen crystal at 100 K and at a wavelength of 0.980 Å.
Diffraction data for the cellobiose, cellotriose, and IG-10 (Fig. 1) crystal soaks were collected at 100 K on a Mar Research image-plate system 345 (Hamburg, Germany), coupled to a Nonius FU 581 rotating-anode X-ray generator, producing Cu Kα radiation focused with Osmic mirrors.
Data processing and structure solution.
All of the diffraction data sets were integrated and scaled with MarHKL (Claudio Klein, Mar Research), a GUI interface for DENZO and SCALEPACK (34). The crystals of the native enzyme, as well as those of its complexes with the substrate analogues, belong to the triclinic space group P1 with unit cell parameters of a = 56.9 Å, b = 57.7 Å, c = 86.3 Å, α = 94.2°, β = 100.9°, and γ = 99.6°. A Matthews coefficient (Vm) (26) of 2.1 A3/Da was calculated for two molecules in the asymmetric unit leading to an approximate solvent content of 39%.
The phase problem was solved by molecular replacement with AMoRe (33) using the crystal structure of cellulase E4 from T. fusca as the search molecule. The E4 structure was divided into a CD (residues 1 to 444) and a CBD (residues 462 to 605), and the rotation and translation searches with the different domains were performed independently. The molecular-replacement solutions, along with the 1.6 Å data, were used as input for the program ARP/wARP (24) to improve the overall map quality and to automatically trace the peptide chain, and subsequent restrained refinement was performed with REFMAC (32). This procedure allowed the tracing of >90% of the molecule. Multiple rounds of model rebuilding with the program TURBO-FRODO (41) were combined with the maximum-likelihood refinement routines in the CNS package (5). Initial refinements included a 5000 K simulated annealing with torsion angle dynamics (1, 40) and grouped B-factor refinement, treating the main chain and side chain atoms as separate groups. Then, 5% of the reflections were set aside as a test set for calculation of the free R factor (4). When the model was essentially complete, it was subjected to 60 cycles of energy minimization, followed by 30 cycles of restrained, individual, B-factor refinement, and finally the “waterpick” routine in CNS was used to automatically place water molecules in the (Fo-Fc) electron density map according to a 3σ cutoff level. For the cellodextrin complexes, the CNS refinements were carried out, followed by manual inspection of the electron density maps.
Stereochemical restraints for the individual glucosyl units and the β-1,4 glycosidic bonds were taken from the CNS library (19). In addition, the molecule 2,3,4,6-tetra-O-acetyl-1-S-benzhydroximoyl-d-glucopyranose (11) was used to take into account the stereochemistry of the thioglycosidic bonds in the IG-10 inhibitor. The complexes were refined by using a 1000 K slow-cool protocol with a harmonic restraint constant of 20 kcal/mol/A2 on the inhibitor. In order to characterize more precisely the bound ions (Ca2+ and Mg2+), anomalous-difference Fourier maps were calculated with the CCP4 suite of crystallography programs (6). Each final model was analyzed by the programs PROCHECK (25) and WHATCHECK (47). The data collections, refinement statistics, and model quality are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | Valuesa for:
|
|||
|---|---|---|---|---|
| Native enzyme | Cellobiose | Cellotriose | IG-10 | |
| Data | ||||
| Resolution (Å) | 30.0-1.6 | 45.0-1.7 | 40.0-1.8 | 50.0-1.9 |
| No. of unique reflections | 135,088 | 104,781 | 92,853 | 270,109 |
| Multiplicity | 2.1 | 3.0 | 4.4 | 2.6 |
| Completeness (%) | 97.0 (95.1) | 90.0 (70.0) | 94.1 (89.1) | 88.7 (87.4) |
| Rsym (%)b | 5.8 (45.1) | 4.2 (33.7) | 5.4 (37.0) | 4.8 (19.9) |
| 〈I〉/σ(I) | 14 (1.5) | 21.2 (1.9) | 25.6 (3.3) | 18.2 (3.8) |
| Refinement | ||||
| Rfree (%)c | 20.0 | 19.7 | 19.8 | 20.4 |
| Rcryst (%) | 17.4 | 17.0 | 16.6 | 16.7 |
| No. of nonhydrogen atoms | ||||
| Protein | 9,619 | 9,524 | 9,538 | 9,543 |
| Water molecules | 667 | 757 | 765 | 666 |
| rmsdd | ||||
| Bond length (Å) | 0.10 | 0.012 | 0.01 | 0.01 |
| Bond angle (°) | 1.5 | 1.5 | 1.5 | 1.4 |
| Dihedral angle (°) | 23.5 | 23.5 | 23.6 | 23.1 |
| Improper angle (°) | 0.96 | 0.95 | 1.8 | 0.9 |
| Avg B-factor (Å2) | 16.6 | 17.5 | 17.6 | 17.9 |
| Ramachandran plot statistics (chains A/B) | ||||
| Most favored regions | 87.8/87.5 | 87.2/87.0 | 88.1/87.0 | 87.7/87.2 |
| Additional allowed regions | 11.8/11.9 | 12.6/12.4 | 11.5/12.7 | 12.1/12.4 |
The corresponding values of the highest-resolution shell are given in parentheses.
Rsym is the factor for comparing the intensity of symmetry-related reflections given by Σ|In − 〈In〉|/ΣIn.
Rfree is the cross-validated R factor (4).
rmsd values were derived from ideal geometry as values as proposed by Engh and Huber (12).
Coordinates.
The coordinates for the native enzyme (PDB ID 1G87), the cellobiose complex (PDB ID 1GA2), the cellotriose complex (PDB ID 1K72), and the IG-10 complex (PDB ID 1KFG) have been deposited at the Research Collaboratory for Structural Bioinformatics Protein Databank.
RESULTS
Overall structure.
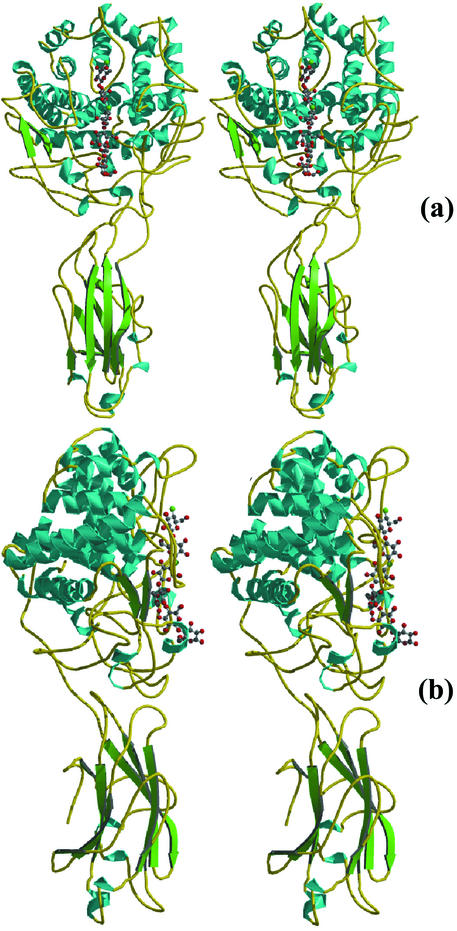
Like CelE4, Cel9G consists of two domains (Fig. 2): a catalytic N-terminal domain (i.e., the CD [residues 1 to 440]) displaying an (α/α)6-barrel topology and a CBD (residues 458 to 614) having an antiparallel β-sandwich fold. The two domains are separated by an 18-residue linker. The fold of the CD has already been observed in other family 9 glycoside hydrolases, such as Cel9D of C. thermocellum (20), and the CBD topology is typical of the family III CBD structures already described (42, 46).
FIG. 2.
Structure of Cel9G. Stereo pairs of the ribbon diagram, with the two domains colored according to secondary structure, are shown. Panels a and b are rotated ca. 90° with respect to one another. The six identified subsites in the active site are shown with glucose rings in ball-and-stick form. Figures 2, 4, and 6 were made with Molscript (22) and Raster-3D (29).
Apart from certain highly flexible loops, the electron density for the two copies of Cel9G (A and B) in the asymmetric unit was clear from residue Thr-3 to residue Pro-614. Except where noted, reference will be made to molecule A. One of the flexible loops (residues 245 to 248) is located at the far end of a cleft in the CD, the end opposite the CBD, which will hereafter be referred to as the nonreducing end of the cleft, to take into account the direction of cellulose cleavage. An other flexible (or disordered) segment (residues 590 to 594) is found in the CBD, near the C terminus. The electron density for two residues in Cel9G was found to be markedly different from the SwissProt data bank sequence (ID P37700). Arg-609 and Arg-610 (corresponding to residues 574 and 575 in the structure) were in fact modeled and refined as Thrs, and this discrepancy was later confirmed from the genetic sequence (A. Belaich, unpublished data).
Structural similarity searches were performed applying the DALI algorithm (17) to the refined structures (CD plus CBD) of Cel9G. The two highest-scoring structures for the CD were the family 9 cellulases CelE4 (a root mean square deviation [rmsd] of 0.9 Å and 55% sequence identity) and Cel9D (rmsd of 2.5 Å and 25% identity). For the CBD, the two highest-scoring structures were the family III CBDs of the CelE4 structure (rmsd of 1.5 Å and 44% sequence identity) and the cellulosomal scaffoldin subunit of C. thermocellum (rmsd of 2.1 Å and 18% sequence identity) (46). The aligned CD sequences of Cel9G, CelE4, and Cel9D are shown in Fig. 3. Due to these high similarities, the two domains of Cel9G will only be briefly described hereafter.
FIG. 3.
Aligned sequences of the CDs of Cel9G of C. cellulolyticum, CelE4 of T. fusca, and Cel9D of C. thermocellum. The representative secondary structures are color coordinated with the sequence names. The putative catalytic base (Asp-55) and the proton donor (Glu-420) are represented by black stars under the sequences, and black triangles denote conserved aromatic residues involved in substrate stacking (see the text for an explanation of residue 208). The sequences were aligned with the CLUSTALW program (45), and the figure was prepared by using ESPript (16).
CD.
The CD comprises 14 helices, 12 of which form the central (α/α)6-barrel and range in length from 10 to 25 residues. Helix 14 is followed by the “linker” connecting the CD to the CBD. Hydrophobic residues predominate at the interior of the barrel, as well as at the space between the two helical layers, and the “burial” of these residues most likely dictates the (α/α)6-barrel motif. The most interesting feature of the CD is the shallow groove corresponding to the active site formed by the long loops connecting the N termini of the barrel helices. These loops range in size from 20 to 40 residues, except the loop between helices 13 and 14 comprising 68 residues. They form the flat “face” of the CD with a depression or cleft running down its center. The active site cleft contains the conserved catalytic residues: Asp-55, Asp-58, and Glu-420 (Fig. 3). This cleft shows the typical active site topology seen in structures spanning numerous glycosidase families (7). Notably, the carboxylate oxygens of Glu-420 and Asp-58 are situated 9.5 Å apart, an interatomic separation often observed for the acid-base catalysts in other inverting glycoside hydrolases (27). The cleft is lined by highly conserved aromatic and polar residues that interact with, and help bind, the cellulose substrate (Fig. 3).
A Ca2+ ion located near the nonreducing end of the cleft is coordinated to two water molecules (2.4 and 2.4 Å), Ser-209O (2.5 Å), Asp-259O (2.5 Å), Ser-209Oγ (2.6 Å), Asp-212Oδ2 (2.4 Å), Asp-212Oδ1 (2.6 Å), and Asp-213Oδ2 (2.4 Å) in a square antiprism geometry (15) and is found in an equivalent position in CelE4. Except for a Glu-to-Asp substitution at residue 213 of Cel9G, these side chain ligands appear to be perfectly conserved throughout the family 9 glycoside hydrolases, as has been already observed (42). Three Mg2+ ions were found associated to residues on the surface of Cel9G, their binding being most probably induced by the crystallization medium containing 100 mM magnesium acetate (see Materials and Methods). Toward the reducing end of the active site, a cis peptide, Pro-388, is situated in a loop forming a wall which is suggested to contribute to orient the substrate in the cleft.
CBD.
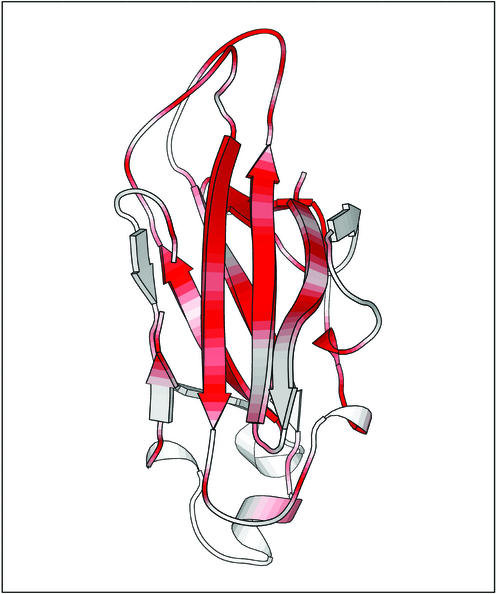
The 155 C-terminal residues of Cel9G make up the CBD. The linker connecting the CBD to the CD forms many hydrogen bonds with these domains and helps to rigidify the position of one domain relative to the other. The overall topology of the CBD is a nine-stranded antiparallel β-structure forming a jelly roll motif. The strands are arranged into two sets of parallel β-sheets adopting a β-sandwich topology, with the interior of the sandwich made up primarily of hydrophobic residues. One of the sheets, comprised of strands 1, 2, 4, and 8, forms an extended platform to the flat face of the CD. This platform is rich in charged and polar surface residues that are highly conserved throughout the family III CBDs (Fig. 6). Another Ca2+ ion was identified in the CBD and is octahedrally coordinated by ligands situated in the coils between β strands 3 and 4 and strands 6 and 7: Glu-503Oɛ1 (2.3 Å), Asp-582Oδ1 (2.4 Å), Asn-581Oδ1 (2.3 Å), Asp-500O (2.4 Å), Asn-578O (2.3 Å), and H2O (2.4 Å). Again, this calcium ion is in a similar location in CelE4, probably plays a structural role, and was previously suggested to be a conserved feature of the family III CBD members (46).
FIG. 6.
Sequence conservation in the family III CBDs. The figure shows Cel9G's CBD and is colored from white (low similarity score) to red (identity).
Cellodextrin-Cel9G complexes.
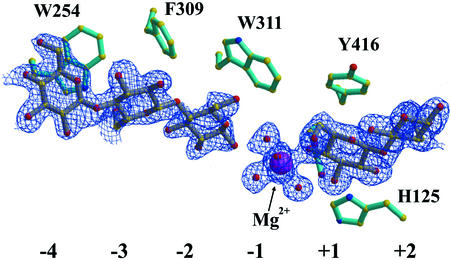
Crystals of Cel9G were soaked with cellodextrins ranging in size from cellobiose (DP-2) to a DP-10 thio-oligosaccharide inhibitor (Fig. 1). For the cellobiose substrate, the difference Fourier maps allowed the positioning of two cellobiose molecules in the active-site cleft (Fig. 4). From comparisons with the complexes of CelE4 (42), the occupied subsites corresponded to −4/−3 and +1/+2. A weaker electron density was found in the −2 subsite and was used to position another glucose molecule bonded to Glc(−3). After refinement of the structure, the electron density in the active site became much clearer, especially in subsite −2. Therefore, the electron density observed in subsites −4 to −2 is best interpreted in terms of two overlapping positions for a cellobiose unit, which were introduced and confirmed in the final refinement stages. Stabilization of Glc(+2) is provided by interactions with three residues from a symmetry-related molecule B (Fig. 5). For the cellotriose (DP-3) substrate, the electron density (data not shown) is consistent with a substrate cleaved into cellobiose and glucose with inversion of the glucose-O1 into an α-configuration. The cellobiose moiety occupied subsites +1/+2, and the electron density for the glucose molecule was found well defined ∼4.0 Å away from the +2 subsite, toward the CBD. The electron density for Glc(+2)-O1 was seen to be equally distributed between the alpha and beta configurations and was modeled in a double conformation, as was the O6 of the same glucose molecule. For the complex with DP-10, continuous, well-defined density was found for only four glucosyl moieties corresponding to subsites −4 to −1. The proper register of the IG-10 ligand was determined by increasing the signal-to-noise ratio of the electron density difference map. At the 6σ contour level, the electron density for the thio-glycosidic bond was still evident, whereas the oxygen's density had disappeared. The inhibitor's sulfur positions were also later confirmed by inspection of an anomalous difference Fourier map at the 3σ contour level to distinguish between the O- and S-glycosidic bonds. The fact that only four subsites are visible may be consistent either with a hydrolytic cleavage of the IG-10 into cellohexaose and cellotetraose or with the nonoccupancy of the subsites beyond subsite −1, toward the reducing end. In the latter case the geometrical distorsions introduced by the sulfur interglycosidic atoms are most probably preventing complete binding of the IG-10 molecule, and the remaining hexaose might be disordered in the external medium. It should also be mentioned that there was no visible electron density pertaining to glucosyl units found in the CBD of Cel9G.
FIG. 4.
Electron density (blue grid) for cellobiose bound in the active site cleft of Cel9G. Binding sites −4 to +2 are indicated, as are the corresponding aromatic stacking partners for the substrate. An Mg2+ (purple) hydrated by water (red) occupies the −1 site and interacts with cellobiose in the +1/+2 subsites.
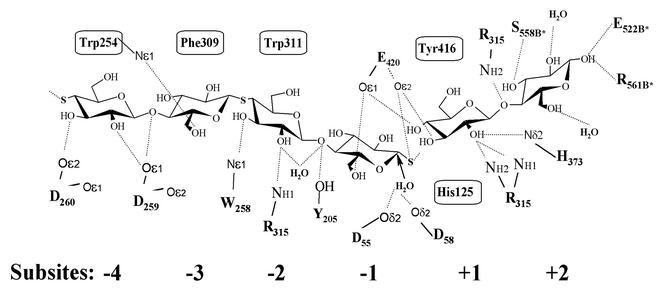
FIG. 5.
Schematic and composite diagram of the interactions between Cel9G and natural and synthetic cello-oligosaccharides in subsites −4 to +2 of the active site. The boxed residues correspond to the aromatic stacking partners, and interatomic hydrogen bonds (<4.0 Å) are indicated by dashed lines. The putative nucleophilic water's attack on the anomeric C1 carbon in subsite −1 is indicated with an arrow. Symmetry-related residues are indicated with a an asterisk.
A schematic and composite representation of the stacking and hydrogen bonding interactions between Cel9G and its substrates or inhibitors in subsites −4 to +2 is shown (Fig. 5). Both cellobiose and IG-10 have identical binding patterns in subsites −4 to −2, involving four aromatic residues—His-125, Trp-254, Trp-311, and Tyr-416—that are highly conserved throughout the family 9 glycoside hydrolases. Also, a large number of residues are forming hydrogen bonding interactions with the ligands, namely, Asp-259, Asp-260, Trp-254, Arg-315, Tyr-204, Tyr-205, Glu-420, His-373, and Arg-375.
Interestingly, when cellobiose occupies the +1/+2 subsites of Cel9G, an Mg2+ ion is observed in the −1 site and binds to the O4 atom of Glc(+1) (Fig. 4). Five water molecules solvate the remainder of the octahedrally coordinated Mg2+ ion. The cellobiose-bound Mg2+ ion is found in both the cellobiose- and cellotriose-soaked structures. Three other magnesium ion surface-binding sites, already localized in the uncomplexed structure, are also present in these enzyme complexes.
For substrate binding, residues in the active site cleft of Cel9G undergo significant conformational changes. The segment from residues 415 to 419 forms a type II hairpin loop that moves ∼1.5 Å into the active site. This movement brings the Cβ of Tyr-416 2.0 Å into the cleft and a rotation about the χ1 bond base stacks Tyr-416 with Glc(+1). The aromatic rings of Trp 311 rotate about their χ2 bond to stack with Glc(−2). In the native structure, the electron density for Glu-420 stops at the Cβ and multiple conformers can be modeled in the remaining electron density. In the cellobiose and cellotriose structures, the carboxylate oxygens of Glu-420 hydrogen bond with O3 (2.6 Å) and O4 (2.6 Å) of Glc(+1) (Fig. 5), and the totality of the electron density for this residue is well defined. These H-bonds force the Cβ 1.0 Å into the cleft and cause a rotation of 180° about the χ1 bond.
DISCUSSION
Of the different types of cellulases characterized up to now in C. cellulolyticum, Cel9G displays the highest activity toward bacterial microcrystalline cellulose (3). One is tempted to propose that, after cellulosomal attachment to crystalline cellulose, Cel9G's principal role would be the initial disruption of the tertiary structure of cellulose and subsequent endolytic attack on the liberated chains. Indeed, work by Tormo et al. (46) has already suggested that the three-dimensional structure of a family III CBD is ideal for disrupting the intrinsic H-bonding network of cellulose. This would then open the way for the synergistic attack by accompanying cellulases on the cellulosome. The X-ray crystal structure of Cel9G allows a detailed examination of a family 9 CD attached to an intact family IIIc CBD. It is the second multidomain cellulase structure to harbor both a CD and a CBD. In both CelE4 and Cel9G structures, the CBD appears as an extended flat surface that could eventually bind and guide cellulose strands toward the CD (Fig. 2).
Role of the CBD.
When looking at the distribution of homologous residues among the different CBDs, the highest concentration of conserved residues is along the planar strip of residues carpeting the β-strands and in-line with the catalytic cleft (Fig. 6). Biochemically, cellulose binding by the family IIIc CBDs is weak. For example, in Cel9G, when expressed alone, the CBD cannot bind to Avicel or PASC (13). In CelE4 the catalytic module cannot bind to bacterial microcrystalline cellulose, whereas an inactive mutant recovers significant binding to this substrate, indicating that the CD plays an important role in substrate binding (18). However, the CBD's role in processivity cannot be discounted. In both CelE4 and Cel9G, there is a significant, if not total, loss of activity when the CD is expressed alone.
To prove this crystallographically does not seem to be possible at present because a soluble natural cello-oligosaccharide cannot exceed a DP-6 (cellohexaose). To form an enzyme substrate complex spanning the CD and CBD would require an inactive mutant and a minimum DP-15 or longer. Synthetic, water-soluble, thio-oligosaccharides have been used elsewhere for crystallographic studies of substrate binding but have shown differential binding characteristics to the natural substrate (38). Modeling experiments on the structure of CelE4 (42) have demonstrated a feasible path along the flat CBD surface that is strewn with polar residues that could weakly interact with glycoside residues and is in-line with the catalytic cleft. The family IIIc CBDs of Cel9G and CelE4 act in conjunction with other CBD family members to bind and hydrolyze cellulose (18). It is clear now that the family IIIa CBDs play a different role from the family IIIc CBDs even though the overall topologies are extremely similar. Cellulosome integrating protein A (CipA) and CipC, which contain two family IIIa CBDs, bind very tightly to both Avicel or PASC (30, 36) whereas, under the same conditions, the CBD of Cel9G alone does not bind to these substrates. These Cip CBDs are found attached to the noncatalytic, scaffoldin protein of the cellulosome. Their function would be to bind and bring into close proximity the catalytic module with cellulose. Thus, different CBD members cooperate to bind and eventually disrupt the crystalline structure of cellulose and guide the chains into the catalytic cleft.
Currently, two family IIIa CBD structures exist: the structures of C. thermocellum (46) and C. cellulolyticum (43). A noticeable structural difference between the family IIIa and family IIIc CBDs lies in the loops connecting the beta strands. In CelE4 and Cel9G, the loops which lie at the interface of the CD and CBD are much longer than their equivalent structures in the family IIIa CBD structures. When looking at the molecular surfaces of the CD and CBD structures, one notes complementarities between the surfaces at the interface. These loops, in conjunction with H-bonding interactions stemming from the linker, serve to stabilize the conformation of the CBD vis à vis the CD to form an extended, flat platform capable of interacting with the flat crystalline surface of cellulose.
Active-site and magnesium ion binding.
Six glucose-binding subsites have been identified crystallographically in the active site cleft of Cel9G. These subsites range from +2 to −4, from the reducing to the nonreducing end of the substrate according to standard nomenclature for glucosyl unit binding sites (9). The cellobiose soak allowed identification of subsites +1/+2 and −2 to −4. The cellotriose soak confirmed both the identity of subsites +1/+2 and the inverting mechanism of Cel9G by cleaving the substrate into β-cellobiose and α-glucose. This also exemplified the preferential binding of cellotriose in subsites +2 to −1 rather than subsites +1 to −2, a logical extension to the cellobiose complex in which no binding was seen in subsites −2/−1.
For both the cellobiose and cellotriose Cel9G complexes, but not in the free enzyme, a magnesium ion binds in subsite −1, whereas three others bind in various surface sites far from the active-site region. However, if these surface binding sites are most probably due to the crystallization medium, the magnesium ion bound in the active site, as observed in the presence of a short substrate, may have a functional role. Being bound in subsite −1, this magnesium ion obviously participates also in stabilizing the negative charges of important catalytic residues, Glu-420 and Asp-55, with which it interacts via its coordinated water molecules. Moreover, one is tempted to propose that divalent metal cations, such as Mg2+ or Mn2+, could play a role in enhancing or modulating substrate binding in Cel9G and related cellulases. However, this has not yet been reported with CelE4, and it is essential that the functional significance of this metal site be verified experimentally in the presence or absence of substrates.
The aromatic stacking residues in the active-site cleft of the family 9 glycoside hydrolases are extremely well conserved. In Cel9G these residues correspond to Trp-254, Phe-309, and Trp-311 for subsites −4 to −2, respectively. Subsite +1 is doubly stacked by residues His-125 and Tyr-416. Phe-309 in Cel9G is an exception to this conservation rule. Interestingly, in this position, only Cel9G and Cel9D contain aromatic side chains; the remainders of family 9 enzymes contain an Asp at this position. Structurally, this substitution at Glc(−3) is compensated for in the remainder of the family 9 enzymes by a highly conserved Trp or His at residue 208 (Fig. 3). Additionally, in CelE4, a two-residue insertion between helices 5 and 6 pushes a Trp in this position closer to its stacking partner.
Active-site and conformational changes.
Protein residues involved in substrate binding adopt new, more-stable, conformations upon substrate binding. In the native structure, except for Trp-254, the electron density for the aromatic residues mentioned above appears disordered, and the B-factor values are higher than for the atoms of surrounding residues. Upon substrate binding, the electron density for these residues becomes more organized and complete and allows a precise repositioning of the residues into their density. Consequently, the corresponding B values drop significantly. Thus, the active-site cleft is flexible in nature and adapts and rigidifies when the substrate is bound, and this is observed with the three substrate analogues used in the present study. In contrast, Trp-254 at the nonreducing end of the cleft does not undergo any conformational rearrangement or stabilization upon substrate binding. Rather, it is poised for substrate binding, implying that this end of the active-site cleft is predisposed to substrate binding. Therefore, one may suggest that cellulose binding proceeds via a mechanism whereby the cellulose substrate is bound by the extremities of the Cel9G molecule, the CBD, and the nonreducing end of the cleft at Trp-254. Subsequently, the inserted part of the cellulose chain is bound by the aromatic residues that guide it into the active-site cleft for hydrolysis.
Additional hydrogen bonding occurs as the substrate descends into the cleft and is forced to eventually “kink” between Glc(−1) and Glc(+1). Such a kink of the cellulose substrate between subsites −1/+1 has already been noted in other cellulase structures such as Cel48F (37). For the Cel9G structures, an extrapolation has to be made between the cellobiose and IG-10 complexes since the IG-10 substrate was only present in subsites −4 to −1. The lack of electron density in subsites +1/+2 in this complex does not discount the possibility of very low occupancies and/or disorder at these subsites. The sulfur atom, Glc(−1)-S1, intended to prevent substrate hydrolysis, may have prevented the glucose molecule in this position from adopting a conformation necessary to accommodate substrate binding in subsites +1/+2. Indeed, substrate deformation prior to hydrolysis has already been described in Cel5A of Bacillus agaradhaerens with a fluorinated cellobioside derivative spanning the −1/+1 active site (8), with the −1 subsite sugar being distorted into a 1S3 skew-boat conformation. This type of conformation leads to an axial configuration of the glycosidic oxygen, an orientation that is known to be rate enhancing for the hydrolysis of the scissile bond (10). This distortion also serves to displace the otherwise sterically hindering C1 hydrogen from its axial orientation. Furthermore, it is likely that the S-glycosidic bond of the IG-10 substrate prevents the skewing of the pyranose ring in the −1 subsite, thus displacing the proper position of the substrate in subsites +1 and +2, leading to ineffective substrate stacking interactions. The electron density for the IG-10 substrate was better defined in molecule B and electron density for Glc(−1)-S1 could be seen in both the α and β orientations with the subsequent density for Glc(+1) too disjointed to position a glucose molecule.
The differential behavior of substrate binding by synthetic oligosaccharides has already been seen in the crystal structures of complexes between thio-oligosaccharide inhibitors and Cel48F (38). Contrary to the relatively few bound sites identified in the enzyme-inhibitor complex seen in Cel9G, the complexes of Cel48F display well-occupied subsites from −6 to +4. Cel48F resembles the family 9 enzymes in that its catalytic module is an (α/α)6-helix architecture with long N-terminal loops that form not only the active-site cleft but also a “closed” tunnel through which the substrate passes to be hydrolytically processed. The tunnel, which forms subsites −6 to −1, provides a large number of substrate-binding interactions that favor the endoprocessive nature of the enzyme, that is, to perform an endocellulolytic cleavage of the cellulose chain and then to continue sliding along the substrate, processively cleaving off cellodextrins ranging from G4 to G2 (39).
The conclusions from our studies confirm and complete the views of the cellulose recognition and degradation by cellulases from family 9, and the structural analysis presented here contributes to providing a rational framework for future functional and biochemical studies of this type of enzyme. Especially, the high level of activity of Cel9G on crystalline cellulose can in part be explained by the three-dimensional architecture of its catalytic module (CD plus CBD), suggesting a complementarity of surfaces between the flat surface of crystalline cellulose and Cel9G. This type of endolytic and eventually processive activity is critical for the initial disruption of the cellulose structure and the eventual synergistic action with other cellulases on the cellulosome for the complete degradation of cellulose.
Acknowledgments
This work was supported by an EU contract (Fourth Framework, Biotechnology Program, BIO4-97-2303) and by the Centre National de la Recherche Scientifique.
We thank Jean-Luc Ferrer for help at beamline BM30 at the ESRF (Grenoble, France). We also thank Michel Juy for discussions and Patrice Gouet for help, especially for the use of his program ESPript.
REFERENCES
- 1.Adams, P. D., N. S. Pannu, R. J. Read, and A. T. Brunger. 1997. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. USA 94:5018-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J. P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 3.Belaich, A., G. Parsiegla, L. Gal, C. Villard, R. Haser, and J. P. Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brünger, A. T. 1992. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 5.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, R. W. P. Gros, Grosse-Kunstleve, J. J.-S. Jiang, Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 7.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 8.Davies, G. J., L. Mackenzie, A. Varrot, M. Dauter, A. M. Brzozowski, M. Schulein, and S. G. Withers. 1998. Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase. Biochemistry 37:11707-11713. [DOI] [PubMed] [Google Scholar]
- 9.Davies, G. J., K. S. Wilson, and B. Henrissat. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321:557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deslongchamps, P. 1983. Stereoelectronic effects in organic chemistry. Pergamon Press, Oxford, England.
- 11.Durier, V., H. Driguez, P. Rollin, E. Duee, and G. Buisson. 1992. Stereochemical investigation of 2,3,4,6-tetra-O-acetyl-1-S-benzhydroximoyl-d-glucopyranose. Acta Crystallogr. C 48:1791-1794. [Google Scholar]
- 12.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47:392-400. [Google Scholar]
- 13.Gal, L., C. Gaudin, A. Belaich, S. Pages, C. Tardif, and J. P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed]
- 14.Gal, L., S. Pages, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J. P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glusker, J. P. 1991. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 42:1-76. [DOI] [PubMed] [Google Scholar]
- 16.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 17.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 18.Irwin, D., D. H. Shin, S. Zhang, B. K. Barr, J. Sakon, P. A. Karplus, and D. B. Wilson. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffrey, G. A. 1990. Crystallographic studies of carbohydrates. Acta Crystallogr. B 46:89-103. [DOI] [PubMed] [Google Scholar]
- 20.Juy, M., A. G. Amit, P. M. Alzari, R. J. Poljak, M. Claeyssens, P. Beguin, and J. P. Aubert. 1992. Three-dimensional structure of a thermostable bacterial cellulase. Nature 357:89-91. [Google Scholar]
- 21.Koshland, D. E. 1953. Stereochemistry and the Mechanism of enzymatic reactions. Biol. Rev. 28:416-436. [Google Scholar]
- 22.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 23.Lamed, R., E. Setter, R. Kemig, and E. A. Bayer. 1983. The cellulosome: a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic cellulose binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13:163-181. [Google Scholar]
- 24.Lamzin, V. S., and K. S. Wilson. 1993. Automated refinement of protein models. Acta Crystallogr. D 49:129-147. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291.
- 26.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 27.McCarter, J. D., and S. G. Withers. 1994. Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4:885-892. [DOI] [PubMed] [Google Scholar]
- 28.McPherson, A. 1999. Crystallization of biological macromolecules. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Merritt, E. A., and D. J. Bacon. 1997. Raster-3D. Photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed]
- 30.Morag, E., A. Lapidot, D. Govorko, R. Lamed, M. Wilchek, E. A. Bayer, and Y. Shoham. 1995. Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreau, V., and H. J. Driguez. 1996. Enzymatic synthesis of hemithiocellodextrins. Chem. Soc. Perkin Trans. 1:525-527. [Google Scholar]
- 32.Murshudov, G. N., V. A. A., and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 33.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D. Biol. Crystallogr. 57:1367-1372. [DOI] [PubMed]
- 34.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 35.Pages, S., A. Belaich, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J. P. Belaich. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pages, S., L. Gal, A. Belaich, C. Gaudin, C. Tardif, and J. P. Belaich. 1997. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. J. Bacteriol. 179:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsiegla, G., M. Juy, C. Reverbel-Leroy, C. Tardif, J. P. Belaich, H. Driguez, and R. Haser. 1998. The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 Å resolution. EMBO J. 17:5551-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsiegla, G., C. Reverbel-Leroy, C. Tardif, J. P. Belaich, H. Driguez, and R. Haser. 2000. Crystal structures of the cellulase Cel48F in complex with inhibitors and substrates give insights into its processive action. Biochemistry 39:11238-11246. [DOI] [PubMed] [Google Scholar]
- 39.Reverbel-Leroy, C., S. Pages, A. Belaich, J. P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice, L. M., and A. T. Brunger. 1994. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins 19:277-290. [DOI] [PubMed] [Google Scholar]
- 41.Roussel, A., and C. Cambillau. 1991. Turbo Frodo. In Silicon Graphics geometry partners directory. Silicon Graphics, Mountain View, Calif.
- 42.Sakon, J., D. Irwin, D. B. Wilson, and P. A. Karplus. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4:810-818. [DOI] [PubMed] [Google Scholar]
- 43.Shimon, L. J. W., S. Pagès, A. Belaich, J.-P. Belaich, E. A. Bayer, R. Lamed, S. Y., and F. Frolow. 2000. Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 Å resolution. Acta Crystallogr. D 56:1560-1568. [DOI] [PubMed] [Google Scholar]
- 44.Stura, E. A. 1999. Seeding, p. 139-153. In T. M. Bergfors (ed.), Protein crystallization: techniques, strategies, and tips: a laboratory manual. International University Line, La Jolla, Calif.
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tormo, J., R. Lamed, A. J. Chirino, E. Morag, E. A. Bayer, Y. Shoham, and T. A. Steitz. 1996. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15:5739-5751. [PMC free article] [PubMed] [Google Scholar]
- 47.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed]