Abstract
The crystal structure of a small, basic DNA binding protein, Sso10b2, from the thermoacidophilic archaeon Sulfolobus solfataricus was determined by the Zn multiwavelength anomalous diffraction method and refined to 1.85 Å resolution. The 89-amino-acid protein adopts a βαβαββ topology. The structure is similar to that of Sso10b1 (also called Alba) from the same organism. However, Sso10b2 contains an arginine-rich loop RDRRR motif, which may play an important role in nucleic acid binding. There are two independent Sso10b2 proteins in the asymmetric unit, and a plausible stable dimer could be deduced from the crystal structure. Topology comparison revealed that Sso10b2 is similar to several RNA-binding proteins, including IF3-C, YhhP, and DNase I. Models of the Sso10b2 dimer bound to either B-DNA or A-DNA have been constructed.
The families of small, abundant, basic DNA binding proteins in thermoacidophilic archaea of the genus Sulfolobus were first characterized by Reinhardt and colleagues in the 1980s (10, 13). These proteins can be grouped into three classes according to their molecular sizes (7, 8, and 10-kDa). Initially, those archaeal proteins, especially the 7-kDa and 10-kDa families, were thought to be histone-like in terms of their biochemical and structural properties.
Two members of the 7-kDa proteins, Sso7d from Sulfolobus solfataricus and Sac7d from Sulfolobus acidocaldarius, have been studied extensive (7, 9, 20), but little is known about the 8-kDa and 10-kDa proteins. Sulfolobus solfataricus Sso10b is one of the 10-kDa members. Unlike the 7-kDa protein, Sso10b is conserved in most archaeal genomes sequenced, including all of the thermophiles and hyperthermophiles whose genomes have been completed, and is distributed among both the euryarchaeota (which encode histone-like proteins) and the crenarchaeota (which do not) (8).
Sso10b is abundant (4 to 5% of total soluble protein) and binds double-stranded DNA tightly but without apparent sequence specificity (1, 10, 29). Sso10b may form dimers or oligomers in cells. Electron microscopic studies of SacI0b from Sulfolobus acidocaldarius suggest that it binds DNA duplexes without significant compaction, affording protection against degradation by the nuclease DNase I (15).
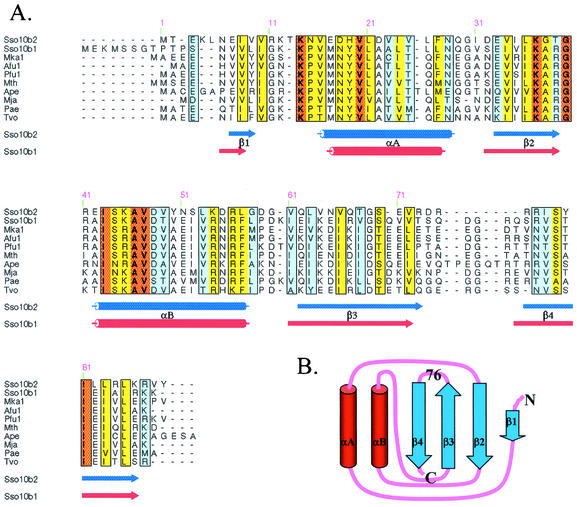
Several archaeal species, including Sulfolobus solfataricus, Archaeoglobus fulgidus, Aeropyrum pernix, and Methanopyrus kandleri, have two copies of the Sso10b genes (Sso10b1 and Sso10b2). The Sso10b1 gene is found ubiquitously. The Sso10b2-like gene exists in many Sulfolobus strains, for instance, S. solfataricus, S. shibatae, and S. tokodaii. Ssh10b1 was isolated from the fraction containing the DNA binding activity (16), but Ssh10b2 protein could not be found in S. shibatae cells. These two isoforms are different in size and sequence (Fig. 1A). Sso10b1 is larger, with 100 amino acids, and Sso10b2 is 89 amino acids long. Sso10b1 has a longer N-terminal end and a longer sequence between residues 71 and 80 (Sso10b2 numbering). Sso10b1 (pI = 11.11) is significantly more positively charged, with a net +7 charges, than Sso10b2 (pI = 9.39), with a net +3 charges.
FIG. 1.
(A) Sequence alignment of Sso10b proteins from different archaeal organisms, including Sso10b2 (GenBank accession no. P74762) and Sso10b1 (Q971T6) from Sulfolobus solfataricus, Mka1 from Methanopyrus kandleri (Q8TXF9), Afu1 from Archaeoglobus fulgidus (O28323), Pfu1 from Pyrococcus furiosus (Q8TZV1), Mth from Methanopyrus kandleri (Q8TWE6), Ape from Aeropyrum pernix (Q9YAX2), Mja from Methanococcus jannaschii (Q57665), Pae from Pyrobaculum aerophilum (Q8ZVL3), and Tvo from Thermoplasma volcanium (Q979S5), are aligned. Orange, yellow, and light blue indicate the level of homology (100, 70, and 50%, respectively) of these genes. The secondary structures of Sso10b2 and Sso10b1 are shown below the alignment. (B) Carton presenting the βαβαββ topology of Sso10b2.
Interestingly, Sso10b1 is located at the 5′ end of the reverse gyrase gene, which is also ubiquitous in the Sulfolobus genomes, whereas Sso10b2 overlaps the last part of the reverse gyrase gene in the reverse orientation. Taken together, those data suggest that Sso10b may be involved in the regulation of DNA packing, but the two isoforms may have different biological functions in cells.
A subfraction of the proteins from Sulfolobus solfataricus cell extracts were found in a stable complex with the silencing protein Sir2 (1). A protein tightly bound to Sir2 was identified as Alba, which turned out to be identical to Sso10b1. Lys16 and the adjacent Lys17 of Alba are known to be important for DNA binding (1). It was shown that Lys16 of Alba underwent deacetylation by Sir2, which affected the DNA-binding affinity of Alba.
The crystal structures at 2.8 Å (tetragonal) and 2.6 Å (hexagonal) resolution of Sso10b1 have been solved recently (25). To further explore the function and diversity of the Sso10b1 and Sso10b2 proteins, we present the crystal structure of Sso10b2 from Sulfolobus solfataricus at a substantially higher resolution, 1.85 Å, than those of Sso10b1.
MATERIALS AND METHODS
Purification, crystallization, and X-ray analysis.
The Sulfolobus solfataricus P2 (EMBL accession number CAC23286) (21) Sso10b2 (89 amino acids) gene was cloned into the PET30 (Novagen) vector and expressed in Escherichia coli BL21(DE3). The purification procedure followed that described previously (26). Finally, the pure fraction was dialyzed against deionized water and lyophilized for longer storage. Mass spectrometry identified that the first methionine residue was removed.
For crystallization experiments, the Sso10b2 protein was dissolved in deionized water to make a 10-mg/ml solution. The hanging-drop vapor diffusion method was used, with the protein-to-reservoir ratio being 1 μl:1 μl. With Hampton Research Crystallization kits for the initial screening, we obtained plated crystals in 2 to 3 days, with the reservoir containing 500 μl of 25% polyethylene glycol MME 550-10 mM ZnSO4-100 mM morpholineethanesulfonic acid (MES), pH 6.5.
We performed electrophoretic mobility shift assays on the binding of Ssob2 to more than 10 different self-complementary DNA oligonucleotides. Our results showed that Sso10b2 effectively retarded the mobility of all DNAs tested. Thus, Sso10b2 has the ability to bind to double-helical DNA, just like Sso10b1. Sso10b2 mixed with different sequences and lengths of DNA fragments were also tested for crystallization at the same time, but no crystal could be obtained.
A high-quality crystal was crystallized with the drop containing an additional 7% sucrose. The crystal was soaked in the reservoir solution for cryodata collection. The preliminary X-ray analysis was performed with an in-house MicroMax002 X-ray generator with a Rigaku R-Axis IV++ image plate system. The high-resolution data were collected at beamline BL17B2 in the National Synchrotron Radiation Research Center, Hsinchu, Taiwan. The Sso10b2 crystal belongs to the P21212 space group, with unit cell dimensions of a = 36.02 Å, b = 134.98 Å, and c = 35.86 Å. There are two Sso10b2 molecules in an asymmetric unit.
Since the crystallization setup contained ZnSO4, we surmised that the crystals might have Zn2+ ions bound to protein molecules at specific positions, which may be used as anomalous scattering atoms. Although X-ray fluorescence scanning near the Zn absorption edge confirmed the presence of a Zn2+ ion, we were not certain whether the signal was merely due to the Zn2+ ion from solution. Nevertheless, we proceeded to collect the Zn-multiwavelength anomalous diffraction (MAD) data at Taiwan beamline BL12B2 in SPring-8, Japan, with the wavelengths of peak (1.2824 Å), edge (1.2830 Å), and high remote (1.2802 Å). The ADSC Quantum 4R charge-coupled device and Oxford Cryostream cooler were used for data collection. In total, 120 degrees of reflection data were collected by the oscillation method, and the oscillation range was 1 degree. The crystal-to-detector distance was 150 mm, and the exposure time was 30 s for each image. The data were processed and integrated with HKL2000 (19). The detailed statistics are listed in Tables 1 and 2.
TABLE 1.
Data collectiona
| Data type | Wavelength (Å) | Resolution (Å) | No. of unique reflections | Redundancy | Completeness (%) | Rmerge (%) | <I>/σ<I> |
|---|---|---|---|---|---|---|---|
| λ1 (min.f′) | 1.2830 | 2.1 | 10,380 | 4.2 | 97.0 (80.5) | 8.9 (17.1) | 20.4 (5.6) |
| λ2 (max.f") | 1.2824 | 2.1 | 10,392 | 4.2 | 96.9 (80.7) | 8.9 (16.9) | 20.5 (5.8) |
| λ3 (remote) | 1.2802 | 2.1 | 10,399 | 4.2 | 97.2 (81.9) | 9.1 (17.5) | 20.2 (5.5) |
| Native | 1.1163 | 1.85 | 15,639 | 4.5 | 99.5 (92.6) | 7.6 (22.9) | 15.9 (5.8) |
All MAD data sets were collected from a single crystal. Values in parentheses are for the outermost shell.
TABLE 2.
Refinement of X-ray data
| Parameter | Value |
|---|---|
| Resolution range (Å) | 31.78-1.85 |
| Data cutoff (σF) | 2.0 |
| Completeness of reflections used (%) | 93.0 |
| No. of reflections used | 14610 |
| Rfree test set size (%) | 4.9 |
| R/Rfree (%) | 23.9/27.4 |
| No. of nonhydrogen atoms, protein/solvent/Zn2+ | 1,408/150/1 |
| B values (Å2), overall/protein/solvent/Zn2+ | 34.8/34.2/41.2/17.9 |
| Root mean square deviations, bond length (Å)/bond angle (°) | 0.005/1.1 |
MAD phasing.
The location of the Zn2+ ion was easily determined by the anomalous Patterson map. MAD phasing based on the Zn2+ anomalous data was performed by Solve (24) with the data from the 20 to 2.2 Å range, and further density modification and model building were done with Resolve (23) and XtalView (17). The initial model contained amino acids 5 to 89 of chain A and 6 to 88 of chain B. The Crystallography and NMR System (CNS) program (3) was used for structural refinement, including simulated annealing procedure, positional, and B-factor refinements. Bulk solvent correlation was estimated in order to omit the solvent signal. Improvement of the model was guided by the sigma A-weighted 2Fo-Fc electron density maps. The composite omit map was also calculated to prevent model bias, and a maximum 5% of the model was omitted for each cycle.
The final model contains the residues from 2 to 89 in chain A, 4 to 89 in chain B, 150 water molecules, and one Zn2+ ion. Figure 2 shows the electron density map surrounding the Zn2+ ion, which reveals the tetrahedrally coordinated Zn2+ ion. The detailed coordination structure is discussed later. The first two residues in chain B could not be determined because the unclear electron density. Ramachandran plot (nonglycine and nonproline) shows 144 (91%) residues in the most-favored regions, 12 (7.6%) in the additional allowed regions, and 2 (1.3%) in generously allowed regions. No residue is located in the disallowed regions. The refinement statistics are listed in Tables 1 and 2.
FIG. 2.
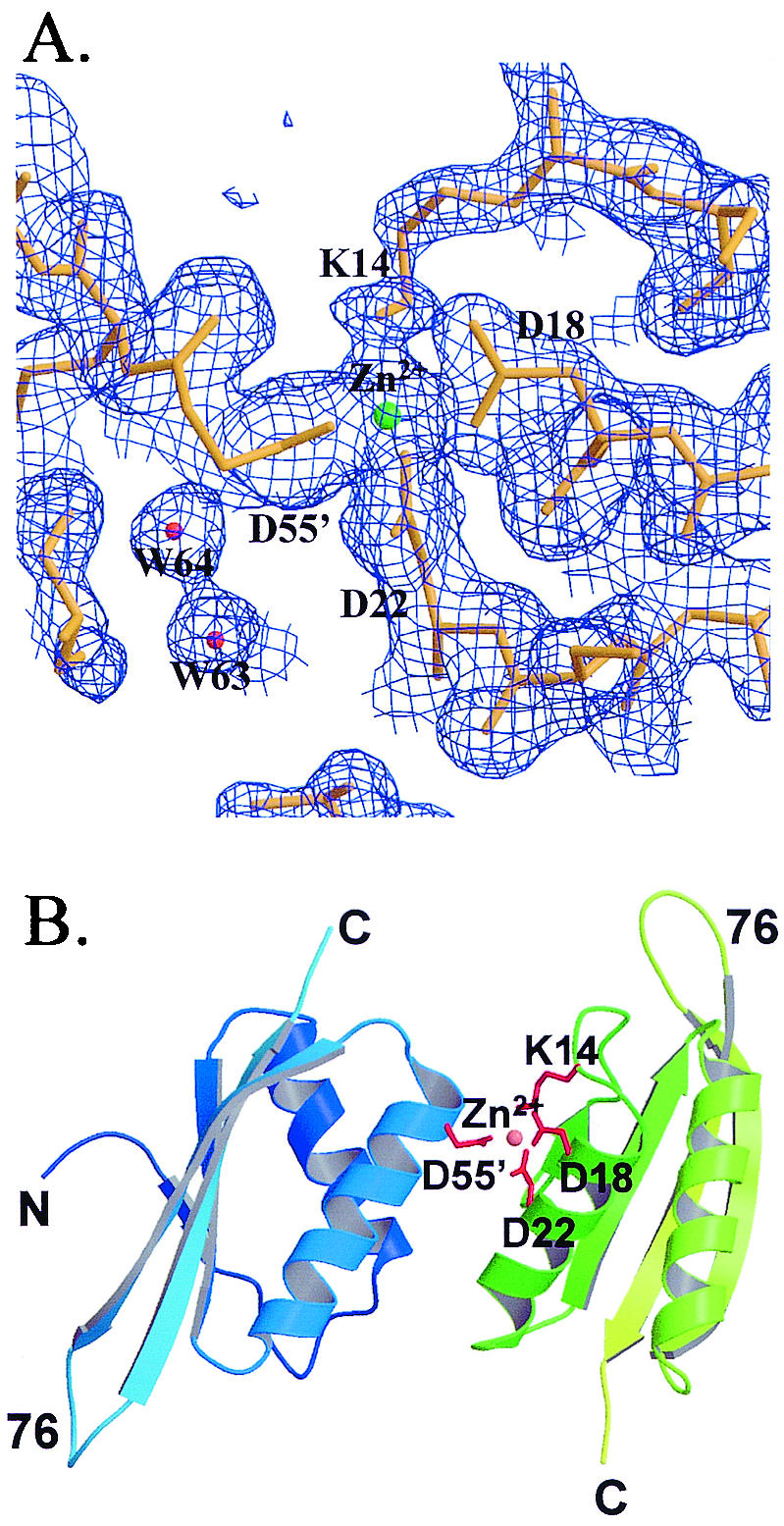
(A) 2Fo-Fc electron density maps surrounding the Zn2+ ion at the 1σ level. (B) Dimer structure mediated by the Zn2+ ion.
The atomic coordinates of Sso10b2 have been deposited in the Protein Data Bank (code 1 udv).
RESULTS AND DISCUSSION
Monomer structure.
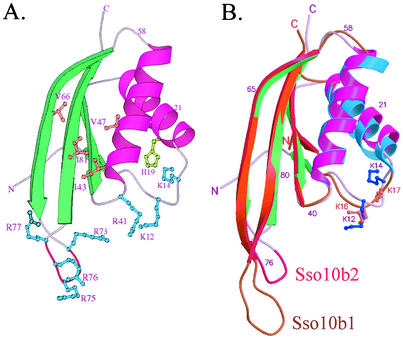
The Sso10b2 monomer has a βαβαββ topology (Fig. 1B) and comprises two parallel α-helices (αA and αB) packed against a four-β-stranded (β1 to β4) sheet (Fig. 3A). The first two β strands are parallel, and the third strand is antiparallel to the other strands. The fold does not resemble any of the histone-like families. The two monomers in an asymmetric unit are almost identical, with an root mean square distance of 0.60 Å and 81 Cα atoms, but has a slight variation in two loop regions from 29 to 33 which is formed by helix αA and strand β2, and 71 to 77, connected with strands β3 and β4, respectively.
FIG. 3.
Crystal structure of Sso10b2 monomer. (A) Ribbon drawing of Sso10b2 monomer. The α-helices are colored in magenta, and the β-strands are green. The conserved basic residues are colored light blue, and the hydrophobic residues are coral. The RDRRR motif is shown in red. (B) Superposition of Sso10b2 and Sso10b1. The conserved Lys12 and Lys14 residues in Sso10b2 (Lys16 and Lys17 in Sso10b1) that may be acetylated are highlighted, with their side chains drawn as ball and stick bonds.
Recently, the crystal structure of Alba (Sso10b1) was determined at 2.6 Å resolution by Wardleworth et al. (25). The folding of the two structures is similar (root mean square distance between the two structures being 0.92 Å, with 75 Cα atoms) (Fig. 3B), but Sso10b2 has a shorter hairpin loop between strands β3 and β4 and contains an arginine-rich RDRRR motif. The higher B-factor of the hairpin loop in chain A but not in chain B, which is stabilized with the sym metry-related molecules in the crystal, implies that this loop is flexible in solution.
The study of Alba and Sir2 suggested that Alba bound to DNA targets, and the binding between Alba and DNA was regulated by Sir2 via the deacetylation of the conserved Lys16 residue of Alba (1). Sequence alignment of various archaeal Sso10b gene products indicated that the conserved acetylated Lys residue also existed in Sso10b2 (Fig. 1A and Fig. 3B). The conserved acetylated Lys12 (Lys16 in Alba) and Lys14 (Lys17 in Alba, which is not acetylated) are located at the center of the loop connected by the first β-strand, β1, and helix αA, which is stabilized by the interaction network composed by His19 and the neighboring residues. This loop, which contains two negatively charged residues, Glu17 and Asp18, is less hydrophobic than Alba and all archaeal Sso10b1-like proteins. The conserved positively charged Arg41 and the neighboring residues all pointing toward the same space as Lys12 confirmed that the ability to bind to nucleic acid targets is likely located along this surface.
Dimer structure.
The two independent molecules (molecule A and molecule B) in the asymmetric unit of the P21212 space group make contacts with each other in three directions, reflecting three kinds of possible dimer forms. The surface areas of the two Sso10b2 molecules in the asymmetric unit are 5,510 Å2 and 5,597 Å2. We found 1,220 Å2, 942 Å2, and 401 Å2 buried in the interface of three possible dimer forms (form 1, form 2, and form 3, respectively). Form 1, having the largest contact surface area, resembles a body with outstretched arms (the β-hairpin loops), which is the same as the proposed Alba dimer (25). The 13 hydrogen bonds (Table 3) involved in the dimerization are located at the second helix (αB) and last two β strands (β3 and β4). Extensive interactions between conserved hydrophobic residues Ile43, Val47, Val66, and Ile81 also stabilize the dimer. Those hydrophobic interactions would be the major driving force for dimer formation in solution.
TABLE 3.
Distances between hydrogen bonds and salt bridges in the Sso10b2 dimer
| Molecule A | Molecule B | Distance (Å) |
|---|---|---|
| G40 O | S44 Oγ | 3.13 |
| R41 O | S44 Oγ | 3.09 |
| S44 Oγ | G40 O | 3.32 |
| S44 Oγ | R41 O | 3.18 |
| S44 Oγ | S44 Oγ | 2.62 |
| D48 Oδ1 | S79 Oγ | 2.52 |
| N51 Nδ2 | G69 O | 3.02 |
| N51 Nδ2 | S79 Oγ | 2.92 |
| G69 O | N51 Nδ2 | 3.25 |
| R77 Nɛ | D55 Oδ1 | 2.58 |
| R77 Nη2 | D55 Oδ1 | 2.65 |
| S79 Oγ | D48 Oδ2 | 2.82 |
| S79 Oγ | N51 Nδ2 | 2.79 |
| Molecule A | Molecule A | |
| K14 Nζ | D18 Oδ1 | 2.85 |
| R56 Nη1 | E17 Oɛ1 | 2.69 |
| R56 Nη2 | E17 Oɛ2 | 2.78 |
| R87 N | D32 O | 2.73 |
| Molecule B | Molecule B | |
| K14 Nζ | D22 Oδ2 | 2.70 |
| K37 Nζ | E71 Oɛ2 | 2.81 |
| R84 Nη2 | E33 Oɛ1 | 3.07 |
| R84 Nɛ | E33 Oɛ2 | 3.10 |
| R87 N | D32 O | 2.43 |
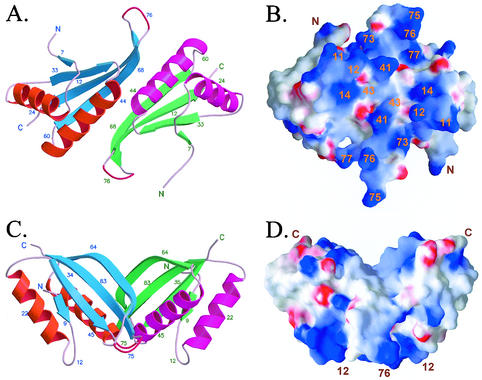
Figure 4A and 4B show a ribbon view and electrostatic potential surface view, respectively, looking into the pseudo-dyad axis of the putative dimer. Although Sso10b2 has only +3 net charges (seven lysines and nine arginines, and seven aspartates and six glutamates), the charge distribution is highly uneven. It can be seen that the frontal surface (Fig. 4B) is highly positively charged with several conserved amino acids, including Arg11, Lys12, Lys14, Arg41, Lys45, Arg73, Arg75, Arg76, and Arg77, broadly distributed on this surface. The negatively charged Asp74, sandwiched between Arg73 and Arg75 in the sequence, is pointing away from the surface. It is likely that this surface interacts with the phosphates of nucleic acids. Figures 4C and 4D correspond to Fig. 4A and 4B, respectively, but looking from a 90-degree angle. The dimer now has a trapezoid shape, with the putative DNA-binding surface pointing toward the bottom. Note that the DNA-binding surface is flat.
FIG. 4.
Structure and surface potential of Sso10b2 dimer. On the predicted surface for DNA binding, there are 18 (R11, K12, K14, R41, K45, R73, R75, R76, and R77 on each molecule) basic residues.
Form 2 is also found in the hexagonal and tetragonal crystals of Alba, which has the two molecules related by crystallographic twofold axis. Two symmetry-related αA helices provide 10 hydrogen bonds, and the conserved hydrophobic Leu21 and Ile24 amino acids participate in the formation of the dimer. This interface also appears in the structure of Sso10b1 (25).
In the third form, the interface of the dimer is also contributed by the two helices (αA from molecule A and αB from molecule B), but it is unique in the Sso10b2 crystal formed along the 21 axis. There are only six hydrogen bonds between two monomers. However, unlike the first two dimers, a strong ionic force is provided by Asp18, Asp22, and, surprisingly, Lys14 in one monomer and Asp55 in the other via a Zn2+ ion, as described before (Fig. 2A and B). The coordination distances surrounding the Zn2+ ion are listed in Table 4. The coordination involving the NH2 group of the Lys14 side chain implies that this amino group is not protonated. Since Lys14 is likely to be involved in the acetylation-deacetylation process, based on its homology to Alba, its ability to coordinate the Zn2+ ion raises an interesting question, whether the Zn2+ ion participates in the acetylation-deacetylation process of Lys14 in Sso10b2.
TABLE 4.
Distances around the Zn2+ ion
| Atom | Residue | Atom | Distance (Å) |
|---|---|---|---|
| Zn2+ | D18 | Oδ1 | 3.11 |
| Oδ2 | 2.10 | ||
| D22 | Oδ1 | 1.87 | |
| Oδ2 | 2.51 | ||
| D55′ | Oδ1 | 1.97 | |
| Oδ2 | 2.53 | ||
| K14 | Nζ | 2.13 | |
| K14 Nζ | D18 | Oδ1 | 2.98 |
Biological implication.
So far, it is not clear how Sso10b proteins bind to DNA. In the study of Alba, a model was proposed in which Sso10b1 binds at the minor groove based on the 4′,6′-diamidino-2-phenylindole (DAPI) (a minor-groove-binding ligand) displacement experiment (25). Alba has a longer hairpin loop than Sso10b2, suggesting different binding properties between Sso10b1 and Sso10b2. If the hairpin is indeed the major DNA binding motif of Sso10b, Sso10b1 and Sso10b2 should have different binding directions to the DNA target. The surface potentials of Sso10b2 and Alba are also different between these two isoforms. Previous data suggested that Ssh10b formed a dimer, or even an oligomer, under high-protein concentration conditions (estimated >10 mg/ml) in cells (29).
Interestingly, Sso10b2 has a unique arginine-rich motif, which is known to be important for RNA binding (27). RNA-binding proteins containing arginine-rich motifs function in transcription, translation, RNA trafficking, and packing. The arginine-rich motifs exhibit diverse secondary structures. The side chain of arginine can make specific contacts with the phosphate and enlarges the major groove of RNA. The charged guanidinium group has the potential to form pentadentate hydrogen bonds and is flexibly tethered to the protein main chain by a long aliphatic side chain. In a previous study, the Ssh10b2 protein could not be identified from the fraction containing DNA binding activity. The purified DNA binding fraction of S. solfataricus also identified the existence of Alba only (1). It is possible that Sso10b2 is not expressed at a high enough level to be detected or is not expressed at all. If this is the case, why and how these two isoforms are differentially expressed are interesting questions to pursue. These data seem to suggest that Sso10b2 may have a distinct role from Sso10b1, as discussed below.
A three-dimensional alignment search of the DALI server (11) showed that the topology of Sso10b is similar to that of two RNA-binding proteins, the C-terminal domain of E. coli translation initiation factor IF3 (2) and cell division protein YhhP (12). But the hairpin loop has large variations. All of them adopt a βαβαββ topology (Fig. 5). In IF3, the conserved Arg89, Lys102, and Phe99 are important for the binding of rRNA (6, 18). Arg89 and Lys102 may interact with rRNA directly, and Phe99 stabilizes the Arg139-Glu96 salt bridge by the stacking interaction with Arg139. These three residues are close to the position of Lys12, Lys14, and His19 in Sso10b2. The Glu21 important for cell elongation in Yhhp (30) is also located near the acetylated Lys14 in Sso10b2. Sso10b2 also has a high level of structural homology to the N-terminal domain of the DNase I protein (22) (Fig. 5A). The structure of DNase I with a nicked DNA duplex defined the DNA binding site of DNase I, whereby a protruding β-hairpin loop sits in the minor groove of duplex in the distorted B-DNA form, and another study (28) indicated that DNase I can interact with but cannot cleave DNA in the A conformation. The highly homologous conformations, including the length of the hairpin, between Sso10b2 and DNase I suggest that the DNA-binding mode may be similar as well.
FIG. 5.
Structural alignment of Sso10b2 with DNase I (2DNJ), YhhP (1DCJ), and IF3-C (1TIG). The root mean square deviation values of the main chain atoms are 1.4 Å (180 atoms), 1.6 Å (212 atoms), and 1.5 Å (200 atoms), respectively. The root mean square deviation values of the Cα atoms are 2.8 Å (70 atoms), 2.5 Å (69 atoms) and 2.3 Å (69 atoms), respectively.
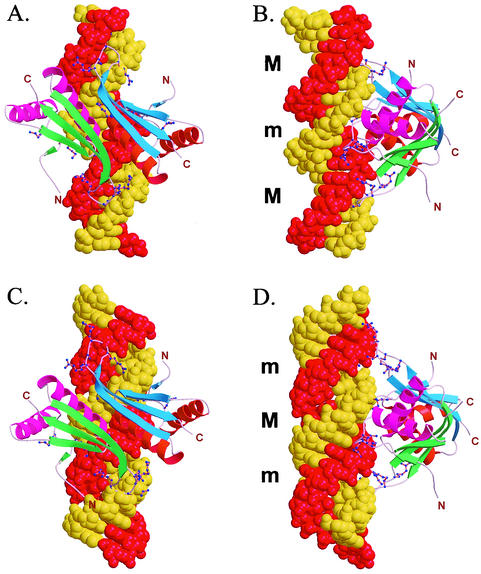
The comparisons of Sso10b2 with three RNA (or A-DNA) binding proteins, IF3 (4), YhhP (12), and DNase I (22), prompted us to consider the possibility that Sso10b2 may be an RNA-binding protein. We constructed models of Sso10b2 bound to both B-DNA and A-DNA (or RNA), shown in Fig. 6.
FIG. 6.
Models of Sso10b2 dimer bound to DNA. The conserved positively charged amino acids Lys12, Lys14, Arg41, Arg73, Arg75, Arg76, and Arg77 are close to the phosphate groups along the minor groove. (A and B) Model of an Sso10b2 dimer bound to B-DNA. (C and D) Model of an Sso10b2 dimer bound to A-DNA.
In the Sso10b2-B-DNA complex model, which is essentially similar to that of the Alba-B-DNA model (25), the Sso10b2 dimer covers about 10 to 12 bp. The two loops containing Lys12 and Lys14 bind to the phosphates across the major groove, whereas the loops containing the RDRRR motif (R73 to R77) extend away from each other and bind to the phosphates over the minor groove at a distance nearly one turn of the helix apart. The two αA helices of the dimer in this model are prominently located at opposite sides of the DNA double helix. It is conceivable that one Sso10b2 dimer could interact with an adjacent dimer through two αA helices involving the conserved hydrophobic Leu21 and Ile24 amino acids and other hydrogen bonds, as seen in form 2 (see above) dimer interactions. Such interactions extending in both directions up and down the DNA would allow the DNA double helix to be completely covered by Sso10b proteins, thermally stabilizing the DNA duplex and protecting DNA from DNase attacks. Whether these interactions are cooperative is not clear.
In another model, the Sso10b2 dimer binds on the relatively shallow surface of the minor groove of A-DNA (or RNA). The two loops containing Lys12 and Lys14 bind to the phosphates across the major groove, whereas the loops containing the RDRRR motif (R73 to R77) extend away from each other and bind to the phosphates over the narrow entrance of the deep major groove (and beyond), also at a distance nearly one turn of the A-DNA helix apart.
In both models, the loops and the side chains of Arg and Lys are presumably flexible enough to adjust their conformations to avoid any possible unfavorable steric contacts. Of course, the exact binding mode of Sso10b2 to DNA or RNA will need to be resolved by structural analysis of an Sso10b2-nucleic acid complex and biochemical and biophysical studies of Sso10b proteins. It is of interest that Sac7d and Sso7d produced a large DNA kink with the local A-DNA conformation at the protein binding sites (9, 20). Therefore, it would not be surprising that the actual DNA conformation induced by Sso10b1 and Sso10b2 binding will not occur as suggested in either of the hypothetical models proposed here or elsewhere (25).
There remain a number of interesting issues to be addressed in the future. Why are there two Sso10b proteins, Sso10b1 and Sso10b2? Do they have different biological functions? It is not clear whether the native Sso10b2 protein isolated from S. solfataricus contains acetylated lysines or not. What are the relationships between the three classes of 7-kDa, 8-kDa, and 10-kDa proteins in terms of their biological functions? Like Sso7d, Sso10b2 is a small hyperthermophilic protein. What is the structural basis for its extraordinary heat stability? Those issues will be pursued.
Acknowledgments
This work was supported by grants from the Academia Sinica and the National Science Council (NSC91-3112-P-001-019-Y) to A.H.-J.W. The synchrotron data collections were conducted with the Biological Crystallography Facilities (Beamline BL17B2 at NSSRC in HsinChu and Taiwan Beamline BL12B2 at SPring-8) supported by the National Science Council (NSC).
REFERENCES
- 1.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 2.Biou, V., F. Shu, and V. Ramakrishnan. 1995. X-ray crystallography shows that translational initiation factor IF3 consists of two compact α/β domains linked by an α-helix. EMBO J. 14:4056-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. GrosseKunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54:905-921. [DOI] [PubMed]
- 4.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50:760-763. [DOI] [PubMed]
- 5.Cousins, R. J. 1998. A role for zinc in the regulation of gene expression. Proc. Nutr. Soc. 57:307-311. [DOI] [PubMed] [Google Scholar]
- 6.De Bellis, D., D. Liveris, D. Goss, S. Ringquist, and I. Schwartz. 1992. Structure-function analysis of Escherichia coli translation initiation factor IF3: tyrosine 107 and lysine 110 are required for ribosome binding. Biochemistry 31:11984-11990. [DOI] [PubMed] [Google Scholar]
- 7.Edmondson, S. P., and J. W. Shriver. 2001. DNA-binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 334:129-145. [DOI] [PubMed] [Google Scholar]
- 8.Forterre, P., F. Confalonieri, and S. Knapp. 1999. Identification of the gene encoding archaeal-specific DNA-binding proteins of the Sac10b family. Mol. Microbiol. 32:669-670. [DOI] [PubMed] [Google Scholar]
- 9.Gao, Y.-G., S. Y. Su, H. Robinson, S. Padmanabhan, L. Lim, B. S. McCrary, S. P. Edmondson, J. W. Shriver, and A. H.-J. Wang. 1998. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Struct. Biol. 5:782-786. [DOI] [PubMed] [Google Scholar]
- 10.Grote, M., J. Dijk, and R. Reinhardt. 1986. Ribosomal and DNA binding proteins of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 873:405-413. [Google Scholar]
- 11.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh, E., T. Hatta, H Shindo, Y. Ishii, H. Yamada, T. Mizuno, and T. Yamazaki. 2000. High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division. J. Mol. Biol. 304:219-229. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, M., J. Kimura, P. Davie, R. Reinhardt, and J. Dijk. 1984. The amino acid sequence of a small DNA binding protein from the archaeabacterium Sulfolobus solfataricus. FEBS Lett. 176:176-178. [DOI] [PubMed] [Google Scholar]
- 14.Knapp, S., A. Karshikoff, K. D. Berndt, P. Christova, B. Atanasov, and R. Ladenstein. 1996. Thermal unfolding of the DNA-binding protein Sso7d from the hyperthermophile Sulfolobus solfataricus. J. Mol. Biol. 264:1132-1144. [DOI] [PubMed] [Google Scholar]
- 15.Lurz, R., M. Grote, J. Dijk, R. Reinhardt, and B. Dobrinski. 1986. Electron microscopic study of DNA complexes with proteins from the Archaebacterium Sulfolobus acidocaldarius. EMBO J. 5:3715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mai, V. Q., X. Chen, R. Hong, and L. Huang. 1998. Small abundant DNA binding proteins from the thermoacidophilic archaeon Sulfolobus shibatae constrain negative DNA supercoils. J. Bacteriol. 180:2560-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 18.Ohsawa, H., and C. Gualerzi. 1981. Structure-function relationship in Escherichia coli initiation factors. J. Biol. Chem. 256:4905-4912. [PubMed] [Google Scholar]
- 19.Otwinowski, Z., and W. Minor. 1997. Processing of the X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 20.Robinson, H., Y.-G. Gao, B. S. McCrary, S. P. Edmondson, J. W. Shriver, and A. H.-J. Wang. 1998. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392:202-205. [DOI] [PubMed] [Google Scholar]
- 21.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, Chan- C. C. Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suck, D., A. Lahm, and C. Oefner. 1988. Structure refined to 2 Å of a nicked DNA octanucleotide complex with DNase I. Nature 332:464-468. [DOI] [PubMed] [Google Scholar]
- 23.Terwilliger, T. C. 2001. Map-likelihood phasing. Acta Crystallogr. D57:1763-1775. [DOI] [PMC free article] [PubMed]
- 24.Terwilliger, T. C., and J. Berendzen,1999. Automated MAD and MIR structure solution. Acta Crystallogr. D55:849-861. [DOI] [PMC free article] [PubMed]
- 25.Wardleworth, B. N., R. J. Russell, S. D. Bell, G. L. Taylor, and M. F. White. 2002. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 21:4654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wardleworth, B. N., R. Russell, M. F. White, and G. L. Taylor. 2001. Preliminary crystallographic studies of the double-stranded DNA binding protein Sso10b from Sulfolobus solfataricus. Acta Crystallogr. D57:1893-1894. [DOI] [PubMed]
- 27.Weiss, M. A., and N. Narayana. 1999. RNA recognition by arginine-rich peptide motifs. Biopolymers 48:167-180. [DOI] [PubMed] [Google Scholar]
- 28.Weston, S. A., A. Lahm, and D. Suck. 1992. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 Å resolution. J. Mol. Biol 226:1237-1256. [DOI] [PubMed] [Google Scholar]
- 29.Xue, H., R. Guo, Y. Wen, D. Liu, and L. Huang. 2000. An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 182:3929-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashino, T., M. Isomura, C. Ueguchi, and T. Mizuno. 1998. The YhhP gene encoding a small ubiquitous protein is fundamental for normal cell growth of Escherichia coli. J. Bacteriol. 180:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]