Abstract
The N-terminal regulatory domains of bacterial response regulator proteins catalyze phosphoryl transfer and function as phosphorylation-dependent regulatory switches to control the output activities of C-terminal effector domains. Structures of numerous isolated regulatory and effector domains have been determined. However, a detailed understanding of regulatory interactions among these domains has been limited by the relative paucity of structural data for intact multidomain response regulator proteins. The first multidomain structures determined, those of transcription factor NarL and methylesterase CheB, both revealed extensive interdomain interfaces. The regulatory domains obstruct access to the functional sites of the effector domains, indicating a regulatory mechanism based on inhibition. In contrast, the recently determined structure of the OmpR/PhoB homologue DrrD revealed no significant interdomain interface, suggesting that the domains are tethered by a flexible linker and lack a fixed orientation relative to each other. To address the generality of this feature, we have determined the 1.8-Å resolution crystal structure of Thermotoga maritima DrrB, providing a second structure of a multidomain response regulator of the OmpR/PhoB subfamily. The structure reveals an extensive domain interface of 751 Å2 and therefore differs greatly from that observed in DrrD. Residues that are crucial players in defining the activation state of the regulatory domain contribute to this interface, implying that conformational changes associated with phosphorylation will influence these intramolecular contacts. The DrrB and DrrD structures are suggestive of different signaling mechanisms, with intramolecular communication between N- and C-terminal domains making substantially different contributions to effector domain regulation in individual members of the OmpR/PhoB family.
The use of modular phosphorylation-regulated switch domains to control effector protein activity is a prevalent theme in both eukaryotic and prokaryotic signal transduction pathways. In eukaryotes, the modular switch is often a small GTPase of the ras superfamily, in which a single phosphate moiety of a bound guanine nucleotide (GTP versus GDP) dictates the activation state of the protein and its intermolecular associations with downstream targets (for a review, see reference 43). In prokaryotes, the switch is typically the regulatory domain of a multidomain response regulator protein that is paired with a histidine protein kinase within a two-component signaling system (for reviews, see references 34 and 45). Phosphorylation of a specific aspartyl residue in the regulatory domain of the response regulator protein results in activation of an attached effector domain and generation of the output response of the signaling pathway. In both families of switch proteins, phosphorylation-induced conformational changes alter molecular surfaces that are exploited for protein-protein interactions specific to the unphosphorylated or phosphorylated states. This provides a versatile strategy that is compatible with many different mechanisms of regulation that can be tailored to the specific requirements of individual signaling proteins.
Biochemical analyses have indicated that response regulator proteins employ many different mechanisms for regulation of effector domain activity including inhibition, allosteric activation, dimerization or higher-order oligomerization, and interactions with heterologous partners. Numerous structures of isolated regulatory domains in their unphosphorylated and phosphorylated (or otherwise activated) states have provided details of the conformational changes associated with activation. However, a complete description of how these structural perturbations control effector domain activity has been limited by the relatively small number of structures of intact multidomain response regulator proteins available to provide information about interactions between regulatory and effector domains.
Response regulator proteins can be classified by similarities in their effector domains (39). The majority of response regulators function as transcription factors and have DNA-binding effector domains. They are subdivided into three groups termed the OmpR/PhoB, NarL/FixJ, and NtrC/DctD subfamilies. The remaining response regulator proteins contain miscellaneous effector domains, such as enzymes, and are included as a fourth subfamily. Representative multidomain structures are available for three of the four subfamilies: transcription factor DrrD (an OmpR/PhoB family member) (8), transcription factor NarL (3), and methylesterase CheB (11).
Members of the OmpR/PhoB subfamily, comprising approximately 40% of all response regulators, have been relatively resistant to crystallization. As has been the case for many recent structural projects, OmpR/PhoB homologues from thermophilic bacteria have proved more amenable to crystallization than their mesophilic counterparts (8). The genome of Thermotoga maritima encodes four response regulators of the OmpR/PhoB subfamily, designated DrrA to -D for DNA-binding response regulators (8, 30). The lack of a genetic system in T. maritima has precluded identification of the genes regulated by Drr proteins, and nothing is known of the two-component systems in which they operate. With this caveat, the Drr proteins have served as useful models for structural analysis.
The structure of DrrD provided the first structural information for an intact OmpR/PhoB family member (8). Here we report the crystal structure of DrrB. The interdomain interfaces of DrrB and DrrD are remarkably different, providing direct structural evidence for different mechanisms of regulation in two members of the same response regulator subfamily.
MATERIALS AND METHODS
Protein expression and purification.
The gene sequence encoding DrrB (The Institute for Genomic Research accession no. TM0126) was amplified by PCR from genomic T. maritima DNA with flanking restriction sites and was inserted into the NdeI and BamHI sites of the T7 promoter-based expression vector pJES307 (40) to yield plasmid pDB1. The plasmid was transformed into Escherichia coli strain BL21(DE3). To create an expression vector for the regulatory domain of DrrB, the DNA encoding residues 1 to 117 of DrrB (DrrB-N) followed by a stop codon and BamHI restriction site was amplified by PCR from plasmid pDB1 and inserted into the NdeI and BamHI sites of the polylinker region of the pJES307 vector, producing plasmid pEF38, which was subsequently transformed into BL21(DE3). Cells were grown in Luria-Bertani medium supplemented with 100 μg of ampicillin/ml at 37°C to mid-log phase, and DrrB expression was induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After further growth for 5 h at 30°C, cells were harvested by centrifugation for 30 min at 5,000 × g and washed with 50 mM Tris-Cl (pH 8.0)-2 mM 2-mercaptoethanol (βME) (buffer A). Cell pellets were resuspended in buffer A, and cells were lysed by sonication. The lysate was clarified by centrifugation at 40,000 × g for 60 min.
For purification of full-length DrrB, the soluble portion of the lysate was incubated in a 70°C water bath for 30 min to denature endogenous E. coli proteins. The denatured proteins were removed by centrifugation at 4,900 × g for 30 min. All subsequent steps were carried out at 25°C. The remaining thermostable protein was precipitated by addition of 1.5 volumes of a saturated solution of ammonium sulfate (60% saturation, final) and collected by centrifugation. The ammonium sulfate pellet was resuspended in buffer A, filtered through a 0.45-μm-pore-size filter, loaded onto a 10-ml HiTrap Blue column (Amersham Biosciences), and eluted with a 0 to 1.0 M gradient of NaCl in buffer A. DrrB eluted at ∼0.4 M NaCl. Fractions containing DrrB were pooled and precipitated by addition of a saturated solution of ammonium sulfate to 60% saturation. The ammonium sulfate pellet was resuspended in 12.5 mM Tris-Cl (pH 8.0)-1.0 M NaCl-2 mM βME (buffer B). Ammonium sulfate (4 M) was added dropwise to a final concentration of 0.8 M, and the solution was stirred gently at room temperature for 30 min. The protein was filtered and applied to a HiLoad phenyl Sepharose 16/10 column Amersham Biosciences) equilibrated in 25 mM sodium phosphate (pH 7.0)-1.0 M ammonium sulfate-2 mM βME. DrrB was eluted with 25 mM sodium phosphate (pH 7.0)-2 mM βME. Fractions containing DrrB were pooled, concentrated, and subjected to gel filtration chromatography over a Superdex 75 26/60 column (Amersham Biosciences) in buffer B.
For purification of DrrB-N, the soluble portion of the sonicated lysate was incubated at 64°C for 30 min. The ammonium sulfate pellet was then resuspended in 20 mM Bis-tris [Bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane] (pH 6.0)-5 mM βME, filtered, and applied to a 70-ml MonoQ column (Amersham Biosciences). The protein was eluted with a 0.1 to 0.9 M NaCl gradient. Fractions containing DrrB-N were pooled, concentrated, and loaded onto a Superdex 75 26/60 column equilibrated in 20 mM Bis-tris (pH 6.0)-100 mM NaCl-5 mM βME.
A selenomethionine-derivatized DrrB protein was prepared in cells grown as described previously (17, 35). Protein expression was induced with 1.0 mM IPTG, and cell growth was continued at 20°C for 12 h. Cells were harvested by centrifugation, and the protein was isolated and purified as described above for the native protein except that 10 mM βME was added to all solutions.
Size exclusion chromatography.
Fast performance liquid chromatography (FPLC) experiments were conducted with an AKTA FPLC system (Amersham Biosciences). Chromatography was performed at 4°C at a flow rate of 1.0 ml/min with a Superdex 75 10/30 column equilibrated in 12.5 mM Tris-Cl (pH 7.5)-100 mM NaCl-2 mM βME. Absorbance was monitored at 280 nm. For each analysis, a 100-μl sample containing approximately 30 μM DrrB or DrrB-N in 12.5 mM Tris-Cl (pH 7.5)-200 mM NaCl-2 mM βME was injected onto the column. Phosphorylated DrrB and DrrB-N were prepared by incubating proteins with 100 mM ammonium hydrogen phosphoramidate (synthesized as described in reference 36)-12.5 mM Tris-Cl (pH 7.5)-100 mM NaCl-20 mM MgCl2-2 mM βME for 10 min at 60°C prior to injection onto the column.
Crystallization.
DrrB was concentrated at 37°C to 46.5 mg/ml in 12.5 mM Tris-Cl (pH 8.0)-1.0 M NaCl-10 mM dithiothreitol with a BioMax-10 concentrator. Crystals of DrrB were grown at 37°C by the hanging drop method of vapor diffusion. The protein solution was mixed with equal volumes of the reservoir solution composed of 1.2 to 1.4 M sodium phosphate-potassium phosphate, pH 6.3 (NaH2PO4/K2HPO4 ratio, 65:35 [mol/mol]). Selenomethionine-derivatized protein crystals were grown under similar conditions. Once crystals had formed, trays containing crystals were moved to 25°C. Cryopreservation was achieved by transferring the crystals into reservoir solution containing 25% ethylene glycol. Crystals were then cooled in a 100°K nitrogen cryostream. The crystals belonged to space group P212121, with cell constants a = 57.89 Å, b = 59.39 Å, and c = 77.34 Å, corresponding to one molecule per asymmetric unit. Multiwavelength anomalous dispersion data were collected from a single crystal of selenomethionine-substituted DrrB at beamline X4A at the National Synchrotron Light Source at Brookhaven National Laboratory, Upton, N.Y. Data were processed and scaled with DENZO and SCALEPACK (32).
Structure determination and refinement.
Three of five selenomethionines in DrrB were identified with the SAD phasing protocol in the Crystallography & NMR System software suite (CNS) (7), with a mean figure of merit of 0.326 and phasing power of 1.4. Initial phase estimates were improved with solvent flipping density modification in CNS. The resulting high-quality electron density maps permitted automatic model building of DrrB to 2.0-Å resolution with the program ARP/WARP (33). Refinement was carried out by iterative cycles of maximum likelihood, simulated annealing, and temperature factor refinement in CNS and manual rebuilding in O (20) until convergence. The resolution of the model was gradually extended to 1.8 Å and 174 water molecules were added in positive-difference Fourier peaks greater then 3σ. The final model, containing 217 residues, is missing the three C-terminal residues that have no discernible density. The density was also weak for residues 199 through 201, located in the loop region extending from the C-terminal end of helix α3. The final model has a crystallographic R factor of 0.198 and an Rfree value of 0.224. All residues are in the allowed region of the Ramachandran plot and exhibit favorable stereochemistry as defined by PROCHECK (23). Data collection and refinement statistics for DrrB are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Statistic | Valuea |
|---|---|
| Data collection | |
| Resolution limits (Å) | 25.0-1.8 (1.86-1.8) |
| No. of observations | 424,245 |
| Wavelength (Å) | 0.9786 |
| No. of unique reflections | 44,556 |
| Completeness (%) | 97.4 (87.6) |
| I/σ (I) | 25.0 (2.35) |
| Redundancy | 7.2 |
| Rsymmb | 0.066 (0.32) |
| Refinement | |
| Resolution limits (Å) | 20.0-1.8 |
| No. of molecules in the asymmetric unit | 1 |
| No. of protein atoms | 1,785 |
| No. of solvent molecules | 174 |
| No. of reflections (work/test) | 40,239/4,324 |
| Crystallographic Rwork/Rfreec | 0.198/0.224 |
| rmsd bond length (Å) | 0.006 |
| rmsd bond angle (°) | 1.22 |
| Mean temp factor (Å2) | 25.83 |
Values in parentheses correspond to the highest-resolution shell.
Rsymm = Σ|Iobs − Iavg|/Σ Iavg.
Crystallographic R factor = Σ||Fobs|(hkl) − |Fcalc|(hkl)|/Σ|Fobs|(hkl). The crystallographic R factor and Rfree values were computed for 90 and 10% of the data, respectively.
PDB accession code.
Coordinates and structure factors have been deposited in the Protein Data Bank (PDB), accession code 1P2F.
RESULTS AND DISCUSSION
Crystal structure of DrrB.
The structure of DrrB was solved by using SAD phasing with data collected from a single crystal of selenomethionine-substituted protein. Three of the five selenomethionines in DrrB were identified with CNS, and these sites were used in initial phasing. The other two methionines, including the N-terminal methionine that exhibits alternate conformations, were readily identified in the experimental maps. After density modification, the initial model was constructed by using the automatic chain-tracing program ARP/WARP (33). The final model of DrrB, consisting of residues 1 to 217, was refined to 1.8-Å resolution with a crystallographic R factor of 0.198 and an Rfree of 0.224 (Table 1).
In DrrB, the α/β fold of the regulatory domain is analogous to that observed in other response regulators (Fig. 1A). Likewise, the topology of the winged-helix effector domain resembles that of the previously characterized winged-helix proteins including DrrD (8), OmpR (22, 26), and PhoB (5, 31). The two domains are connected by a short, well-ordered linker and pack together, forming an extensive buried interface.
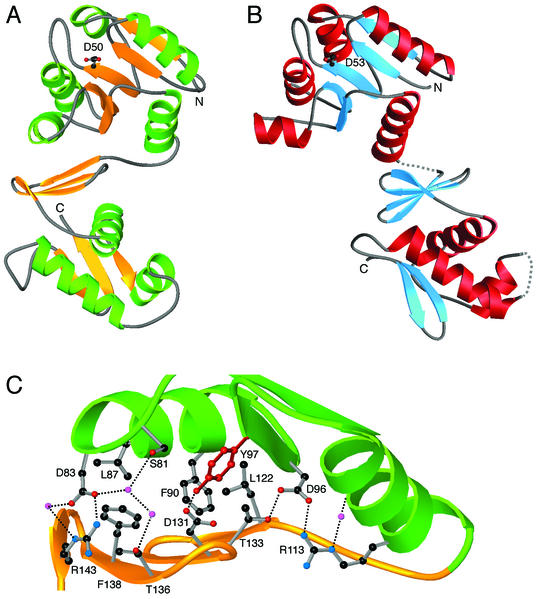
FIG. 1.
(A and B) Structures of T. maritima DrrB (A) and DrrD (B). Shown are ribbon diagrams, with the site of phosphorylation on the β3 strand delineated by a ball-and-stick representation of the conserved aspartate residue. (C) Interactions across the interdomain interface of DrrB. Green, α4-β5-α5 face of the regulatory subunit of DrrB; gold, β-sheet platform of the winged-helix domain. Residues at the interface that are involved in important interactions are shown in ball-and-stick representation. Small magenta spheres, water molecules; dotted lines, hydrogen bonds. The interface can be described as having a large hydrophobic core surrounded by a network of polar interactions, some of which are mediated by well-ordered water molecules.
Interdomain interface of DrrB.
The first detailed information about interdomain contacts in an OmpR/PhoB subfamily member was provided by the structure of T. maritima DrrD (8). The DrrD interface is small, burying only 245 Å2 of surface area and involving exclusively polar contacts between a small number of residues (Fig. 1B). Disorder in the linker region implies that a flexible tether connects the two domains. These structural features suggest that the regulatory and effector domains of DrrD have no fixed orientation with respect to one another.
The domain arrangement of DrrB is unlike that of DrrD. The individual regulatory and effector domains of DrrB and DrrD superimpose well, with root mean square deviations, rmsd, of 2.4 and 1.8 Å, respectively. However, when the two full-length proteins are aligned by superposition of their regulatory domains, the winged-helix domains occupy very different positions (Fig. 1A and B). In DrrD, only the distal end of helix α5 of the regulatory domain contacts the antiparallel β-sheet platform of the DNA-binding domain. In DrrB, the domains are positioned such that the α4-β5-α5 face of the regulatory domain packs squarely against the β-sheet platform of the effector domain.
The interdomain interface of DrrB is 751 Å2, three times the size of the interdomain surface of DrrD. The nature of the interdomain interface of DrrB is typical of that commonly observed in other protein-protein interactions (19, 42) and is quite distinct from that seen in DrrD. The hydrophobic core of the DrrB interface comprises L87, F90, Y97, L122, and F138 (Fig. 1C). Surrounding this core are a series of polar interactions, some mediated by highly ordered water molecules with B factors of 20 to 25 Å2. The central component of this interface, and perhaps its most interesting feature, is the aromatic residue Y97.
The presence of Y97 at the domain interface of inactive DrrB is intriguing, as this residue corresponds to one of the highly conserved residues that exhibit dramatic reorientations in structures of activated regulatory domains (for reviews, see references 34 and 45). Specifically, in phosphorylated or otherwise-activated regulatory domains, a highly conserved Ser/Thr residue on β4 reorients to allow formation of a hydrogen bond to the phospho-Asp. A conserved Tyr/Phe residue, located near the center of β5, is repositioned to an inward orientation, occupying the space vacated by the reoriented Ser/Thr. In DrrB, this conserved residue is Y97. Presumably, DrrB activation involves the repositioning of Y97 to an inward position, thus altering prominent components of the DrrB interface.
It is logical to question whether DrrD could adopt the domain arrangement seen in the DrrB crystal structure. To assess this, the isolated regulatory and winged-helix domains of DrrD were superimposed onto the DrrB crystal structure. The interdomain interface in the resulting model was then evaluated with regard to steric complementarity, hydrogen bonds, and hydrophobic interactions. The newly created DrrD interface is 460 Å2, one-half the size of the DrrB interface. An analysis of protein-protein contacts across this interface provided no indication of juxtapositioning of hydrophobic residues or occurrence of hydrogen bonds or salt bridges in spite of the very polar nature of the domain surfaces. Shape complementarity is yet another issue. Not only does this model lack surface specificity, but also there are steric clashes across the interface where helix α4 from the regulatory domain contacts the loop between β3 and β4 of the winged-helix domain. These collisions are the direct result of the protrusion of the α4 helix in the DrrD regulatory domain. Together, the results of this analysis imply that unactivated DrrD is unable to adopt the same domain arrangement as that observed in the DrrB crystal structure.
Comparison of interdomain interfaces in response regulators.
There are now four full-length response regulators for which high-resolution structural data are available: NarL (2, 3), CheB (11), DrrD (8), and DrrB. Despite the overall structural similarities in the regulatory domains, the interfaces differ (Fig. 2). In all four proteins, a distinct subset of the conformationally variable α4-β5-α5 face of the regulatory domain is used for interdomain contacts. Moreover, the biologically relevant residues of each protein are in a different environment with respect to this interdomain interface, providing a basis for diverse strategies of regulation by phosphorylation. For example, the interdomain interface in CheB is 800 Å2 and spans the α4-β5-α5 face of the regulatory domain. The regulatory domain occludes the methylesterase catalytic triad, suggesting that relief of inhibition plays a role in regulation. The interdomain interface of NarL is also large, greater than 900 Å2. Contacts between the regulatory and effector domains in NarL consist of the N terminus of β5 and the C-terminal regions of the α4 and α5 helices of the regulatory domain. In NarL, residues of the recognition helix participate in the interdomain interface, blocking this functional site in the inactivated structure. Phosphorylation disrupts interactions across this interface, promoting DNA binding (12, 47). As mentioned previously, the interdomain interfaces of DrrB and DrrD differ greatly in their sizes and natures. Whereas DrrB has a large, stable interface, interdomain contacts in DrrD are minimal. Additionally, as shown in Fig. 1, the recognition helices in both DrrB and DrrD are distal to the interdomain interfaces, implying that inhibition is not a result of direct obstruction of the functionally important region of the effector domain.
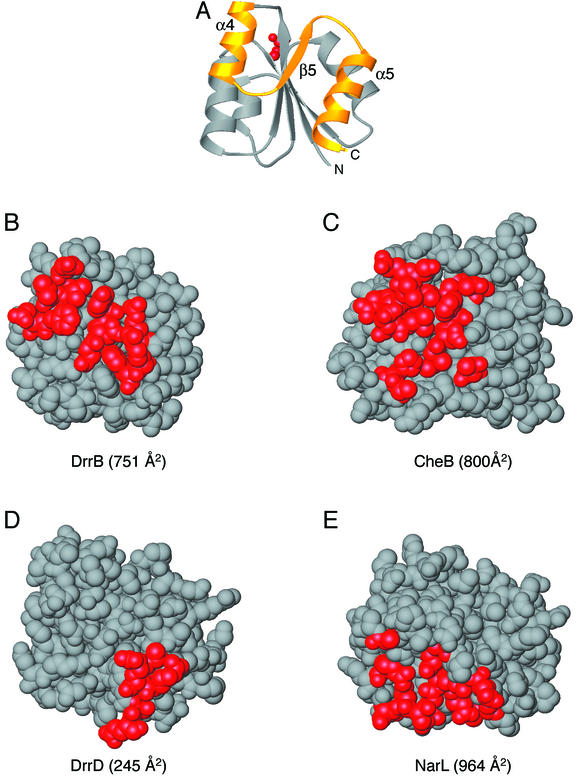
FIG. 2.
Regulatory domain surfaces involved in interdomain interfaces. (A) Ribbon diagram of a representative regulatory domain indicating the orientation of the regulatory domains depicted in panels B to E. Red, active-site Asp; gold, α4-β5-α5 face of the regulatory domain that undergoes conformational changes in response to phosphorylation. (B to E) Space-filling models of the regulatory domains of DrrB (B), CheB (PDB accession code 1A2O [11]) (C), DrrD (1KGS [8]) (D), and NarL (1A04 [2]) (E). Red, residues involved in interdomain associations in each protein. In DrrB, this interface extends across the entire α4-β5-α5 face of the regulatory domain and is 751 Å2. The CheB interdomain interface is ∼800 Å2 and also spans the majority of this surface. The DrrD interface is much smaller, 245 Å2, and is localized to the C-terminal region of helix α5. The NarL interdomain interface, 964 Å2, utilizes some residues in the α4-β5-α5 region of the domain but also includes residues from the α3 helix and surrounding loops.
Winged-helix domain.
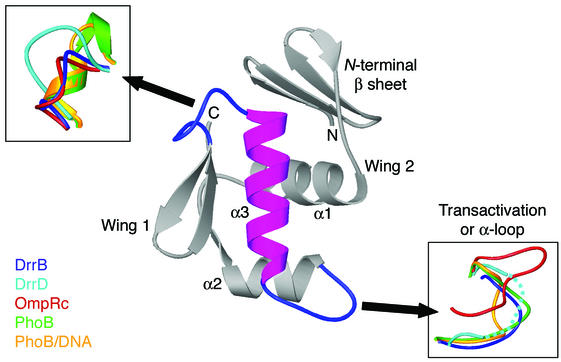
Members of the OmpR/PhoB subfamily are classified by their C-terminal winged-helix domains that contain the structural and sequence elements responsible for DNA recognition. The fold of the OmpR C-terminal domain is unique among the larger family of winged-helix proteins by virtue of its N-terminal four-stranded, antiparallel β-sheet platform and the presence of a large loop, known as the transactivation or α-loop, between the positioning helix, α2, and the recognition helix, α3 (6, 22, 27). The novelty of the β-sheet platform previously led to postulation of a role specific to response regulators, specifically, mediating interactions with the regulatory domain. It is apparent from the structure of DrrB that this is the case. The entire surface of the platform interfaces with the regulatory domain.
There are structures available for several winged-helix domains from the OmpR/PhoB subfamily including DrrD (8), OmpR (22, 26), and PhoB, both alone (31) and bound to DNA (5). The cores of the winged-helix domains in all these proteins superimpose, with an rmsd for Cα atoms of ∼2.0 Å. Furthermore, as previously noted, significant structural flexibility in the loop regions surrounding the recognition helix has been observed (Fig. 3). The PhoB/DNA crystal structure (5) indicates that these flexible loops are used for DNA binding and recognition and also for mediating protein-protein associations. The increased flexibility in these regions perhaps contributes to the optimal fit of the surfaces involved in protein-DNA or protein-protein contacts.
FIG. 3.
OmpR/PhoB subfamily winged-helix domains. Gray, ribbon diagram of a representative winged-helix domain that defines the OmpR/PhoB subfamily of response regulators; magenta, recognition helix that binds to the major groove of DNA. Effector domains of DrrB, DrrD (PDB accession code 1KGS [8]), OmpR (1OPC [26]), PhoB (1GXQ [26]), and PhoB bound to DNA (1GXP [5]) were superimposed based on the core region of the protein, defined as the three α helices and six β strands. In all proteins, the two loops flanking the recognition helix exhibit considerable conformational flexibility, as shown in the insets. The α loop of DrrD (dotted line) is disordered.
Mechanisms of regulation used by the OmpR/PhoB subfamily.
Response regulators of the OmpR/PhoB subfamily exert positive or negative effects on transcription by binding as dimers to tandemly arranged half sites upstream of σ70 promoters. While repression of transcription presumably can be achieved simply by sterically blocking proper positioning of RNA polymerase, activation is thought to involve positive contacts between response regulators and the σ subunit and/or α subunit (C-terminal domain) of polymerase. Thus, while DNA binding is necessary for response regulator function, it may or may not be sufficient. Nonetheless, most in vitro characterization of the OmpR/PhoB subfamily has been limited to analysis of DNA binding.
A variety of studies focused on different OmpR/PhoB subfamily members have addressed the role of the regulatory domain and its phosphorylation state on DNA-binding activity. No unified theme has emerged. It is clear that, for some OmpR/PhoB homologues, the isolated winged-helix domain is capable of binding to DNA as a dimer (5, 15, 21, 25). However, in all studied cases, the regulatory domain influences this interaction.
Inhibition.
For some OmpR/PhoB family members such as PhoB, the isolated DNA-binding domain has been found to have higher affinity for DNA than the unphosphorylated intact protein (13), leading to the conclusion that the N-terminal regulatory domain exerts an inhibitory effect on the DNA-binding domain. However, note that no such inhibitory interaction has been documented for OmpR (21, 41), suggesting that this mechanism may not be common to all subfamily members.
The structures of the intact multidomain response regulators DrrB and DrrD provide no structural explanation for an inhibitory role for the N-terminal domain. The crystal structure of DrrD has only a small region of interdomain contact, most likely the consequence of crystal packing, and the N- and C-terminal domains are not expected to have a fixed orientation in solution. Although it has been noted that steric collisions between regulatory domains of DrrD are observed when intact DrrD is superimposed onto the DNA-binding domain of PhoB bound to DNA (5), there is no reason to believe that such collisions would actually occur in solution or represent a hindrance to DNA binding. For DrrB, which has a large domain interface, which is presumably altered by phosphorylation, the multidomain response regulator can be superimposed onto the DNA-bound C-terminal domain of PhoB without extensive steric collisions (Fig. 4). Contacts between the proteins occur in only one small region involving the β3-α4 loop of the regulatory domain of the upstream monomer and the C-terminal end of helix α1 of the effector domain of the downstream monomer. Furthermore, the N-terminal domain does not interfere with any functional surface of the C-terminal domain such as the recognition helix, the transactivation loop, or the dimerization surfaces.
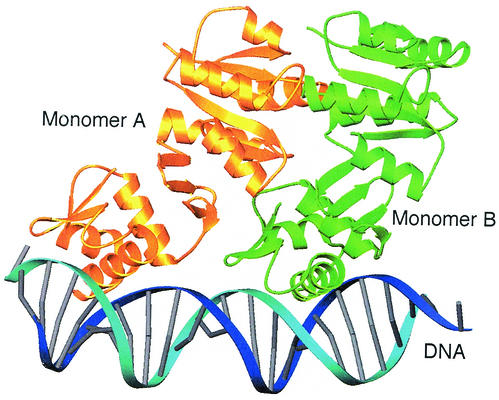
FIG. 4.
Model of DrrB bound to DNA. Shown is a ribbon schematic of the superposition of two full-length DrrB monomers onto the crystal structure of the C-terminal domain of PhoB bound to DNA (1GXP [5]). Monomers A and B are gold and green, respectively, and DNA is blue. There is only one small region of steric conflict between the β3-α4 loop of the regulatory domain of monomer A (residues 80 to 83) and the C-terminal end of helix α1 of the DNA-binding domain of monomer B (residues 160 to 163). Thus the model provides no structural basis for a mechanism of inhibition of DNA binding by the unphosphorylated regulatory domain. This model is not expected to approximate the structure of phosphorylated DrrB bound to DNA. Surfaces of the regulatory domain are presumably altered by phosphorylation, and these alterations would be expected to affect the domain interface, as well as promote specific interactions among regulatory domains within the dimer.
Dimerization.
All OmpR/PhoB subfamily members form dimers or higher-order oligomers. In many cases, regulatory domains have been shown to participate in these associations. Although dimers of OmpR are not detected in solution, binding to DNA occurs exclusively in dimeric units (16). PhoB dimerizes upon phosphorylation (14, 29), while other response regulators such as Bacillus subtilis PhoP and Salmonella enterica PmrA dimerize and bind to DNA in their unphosphorylated states, with phosphorylation merely enhancing DNA binding (24, 46). Unphosphorylated ArcA exists as a dimer and, upon phosphorylation, forms octamers with a 1:1 ratio of unphosphorylated to phosphorylated protein in the hetero-oligomer (18).
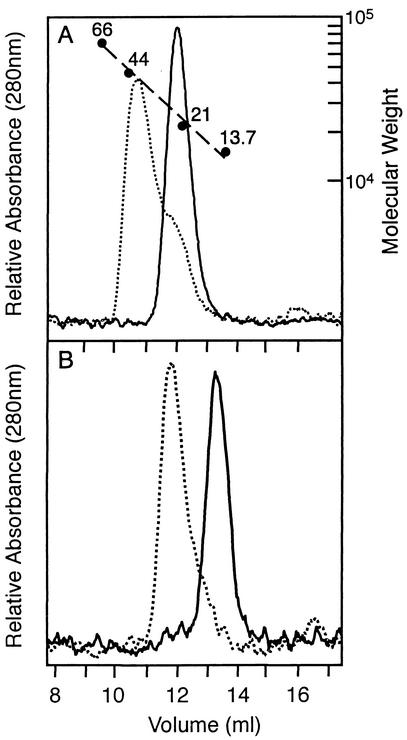
Oligomerization of DrrB was examined by using size exclusion chromatography (Fig. 5). Unphosphorylated DrrB, a 25.5-kDa protein, migrated with an apparent molecular mass of 22.2 kDa, consistent with the monomeric state observed in the crystal structure. Phosphorylated DrrB, generated by using phosphoramidate as the phosphodonor, migrated more rapidly, with an apparent molecular mass of 42.1 kDa, indicative of a dimer. An analogous monomer-to-dimer transition with the isolated regulatory domain of DrrB was observed. Gel filtration analysis of DrrB-N, consisting of residues 1 to 117 with a calculated molecular mass of 13.6 kDa, showed apparent molecular masses of 13.8 and 27.9 kDa in its unphosphorylated and phosphorylated states, respectively. Thus the regulatory domain of DrrB appears to be sufficient to mediate phosphorylation-induced dimerization.
FIG. 5.
Phosphorylation of DrrB and DrrB-N induces dimerization. Size exclusion chromatography of DrrB proteins in the presence and absence of phosphoramidate was carried out with a Superdex 75 column as described in Materials and Methods. The column was calibrated (A) with albumin (66 kDa), ovalbumin (44 kDa), trypsin inhibitor (21 kDa), and RNase A (13.7 kDa) as molecular mass standards. (A) Profile of full-length DrrB and phosphorylated DrrB, which eluted at positions corresponding to molecular masses of 22.2 and 42.1 kDa, respectively. (B) Profile of DrrB-N and phosphorylated DrrB-N, which migrated with apparent molecular masses of 13.8 and 27.9 kDa, respectively.
The direct rather than inverted repeat orientation of DNA recognition sites bound by OmpR/PhoB subfamily transcription factors raises a question about the symmetry of regulatory domain dimerization. Few structural data are available to address this question. The recent structure of a dimer of N-terminal domains of B. subtilis PhoP and the accompanying biochemical characterization have unequivocally established that unphosphorylated PhoP regulatory domains interact as an asymmetric dimer, consistent with the tandem arrangement of PhoP DNA-binding domains bound to direct repeat target sequences (4, 10). In contrast, the regulatory domains of unphosphorylated E. coli PhoB were observed to be associated as a rotationally symmetric dimer in the crystal structure (38), but the physiological relevance of this dimer has not been established.
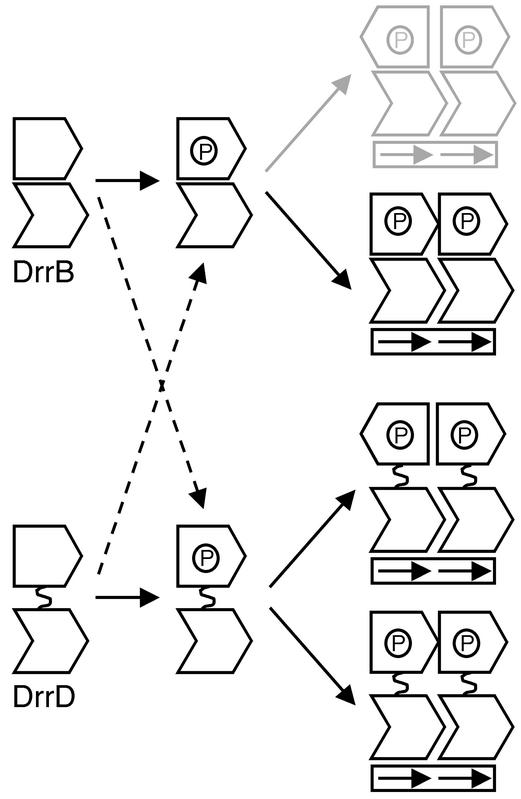
Interestingly, the different domain arrangements observed in DrrB and DrrD are potentially compatible with different symmetries of N-terminal domain dimerization (Fig. 6). If the extensive interface in DrrB is maintained, at least to some extent, in the phosphorylated dimer, then the N-terminal domains would be expected to associate in a head-to-tail fashion, as dictated by the tandemly arranged C-terminal domains. The domain arrangements in DrrD are possibly more varied. If the N- and C-terminal domains of the phosphorylated protein are free to rotate about a flexible linker, as they presumably are in the unphosphorylated protein, then no restrictions on the symmetry of regulatory domains are imposed by the tandem orientation of the DNA-binding domains. The regulatory domains could dimerize with either twofold rotational or translational symmetry. While the existence of different symmetries of individual domains within a dimer is an unusual occurrence, it is not without precedent, having been previously observed in the E. coli transcription factor AraC (9, 37). The symmetry of regulatory domain dimerization in DrrB and DrrD remains to be determined, but the potential exists for significant diversity in domain arrangements among these subfamily members.
FIG. 6.
Possible symmetries of dimerization of OmpR/PhoB subfamily transcription factors when bound to direct repeat DNA recognition sequences. Response regulators, such as DrrB, that contain an extensive domain interface, most likely dimerize with an asymmetric orientation of their regulatory domains. Twofold rotational symmetry of the regulatory domains is unlikely, as it requires nonequivalent monomers within the dimer (gray). Response regulators such as DrrD, in which the regulatory and DNA-binding domains lack a fixed orientation, can potentially dimerize with either rotationally symmetric or asymmetric orientations of their regulatory domains. If the nature of the interface changes upon phosphorylation, then additional combinations of dimerization mechanisms are possible (dashed arrows).
Mechanistic diversity.
Several studies have addressed the role of the interdomain linker sequence and length and the functionality of chimeric response regulators composed of heterologous OmpR/PhoB subfamily domains (1, 28, 44). The structures of DrrB and DrrD have implications for the interpretation and generalization of such experiments. It would be expected that response regulators such as DrrD, which lack specific interactions between regulatory and effector domains, would function well as chimeras with domains of other OmpR/PhoB homologues that utilize a similar mechanism. On the other hand, chimeric proteins constructed of domains from response regulators such as DrrB, which contain extensive specific interdomain contacts, would not generally be expected to be functional. Similarly, the role of the linker may vary. In proteins such as DrrD, the only constraints on the linker may be that it have sufficient flexibility and length to allow the proper orientation of the N- and C-terminal domains, while in proteins such as DrrB, the linker may contribute residues in a specific manner to the domain interface. Differences in mechanism among OmpR/PhoB subfamily members have previously been attributed to differences in lengths of interdomain linkers. It should be noted that DrrB and DrrD, which appear to function by significantly different mechanisms, have interdomain linkers of 3 and 5 residues, respectively, and cluster together at the shorter end of the 3- to 21-residue range of linker length. Thus linker length alone has little predictive value in ascribing mechanisms of regulation.
Regardless of the presence or absence of an inhibitory function, the existence of the unphosphorylated protein as a monomer or dimer, or the ability of the unphosphorylated protein to bind to DNA, phosphorylation of the regulatory domain has been found to enhance DNA binding in all OmpR/PhoB family members that have been studied. However, a detailed understanding of the mechanisms through which this regulation is achieved is lacking. Accumulated biochemical and molecular genetics data suggest that different subfamily members may employ different mechanisms of activation. The structures of DrrB and DrrD provide structural evidence for this hypothesis. The presence of an extensive domain interface in DrrB is suggestive of mechanisms involving intramolecular communication between regulatory and DNA-binding domains, while the absence of such an interface in DrrD points to an intermolecular regulatory mechanism. Thus, fundamentally different strategies appear to be used for regulation within the OmpR/PhoB subfamily. Indeed, for response regulators, as for their eukaryotic counterparts, the small GTPases of the ras superfamily, there may be a limit to the extent that similar sequences and structures of modular domains can be used to predict specific modes of function.
Acknowledgments
We thank E. Fox for technical assistance with DrrD expression vectors and proteins. We also thank Y. Volovik for help with purification and crystallization trials and R. Abramowitz and X. Yang for assistance with data collection at Howard Hughes Medical Institute beamline X4A at the National Synchrotron Light Source at Brookhaven National Laboratory.
This work was supported by a grant from the National Institutes of Health (GM47958). A.M.S. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ames, S. K., N. Frankema, and L. J. Kenney. 1999. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc. Natl. Acad. Sci. USA 96:11792-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baikalov, I., I. Schröder, M. Kaczor-Grzeskowiak, D. Cascio, R. P. Gunsalus, and R. E. Dickerson. 1998. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry 37:3665-3676. [DOI] [PubMed] [Google Scholar]
- 3.Baikalov, I., I. Schröder, M. Kaczor-Grzeskowiak, K. Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053-11061. [DOI] [PubMed] [Google Scholar]
- 4.Birck, C., Y. Chen, F. M. Hulett, and J. P. Samama. 2003. The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface. J. Bacteriol. 185:254-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco, A. G., M. Sola, F. X. Gomis-Ruth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701-713. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, R. G. 1993. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74:773-776. [DOI] [PubMed] [Google Scholar]
- 7.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Buckler, D. R., Y. Zhou, and A. M. Stock. 2002. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Structure 10:153-164. [DOI] [PubMed] [Google Scholar]
- 9.Carra, J. H., and R. F. Schleif. 1993. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 12:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y., C. Birck, J. P. Samama, and F. M. Hulett. 2003. Residue R113 is essential for PhoP dimerization and function: a residue buried in the asymmetric PhoP dimer interface determined in the PhoPN three-dimensional crystal structure. J. Bacteriol. 185:262-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djordjevic, S., P. N. Goudreau, Q. Xu, A. M. Stock, and A. H. West. 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 95:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldridge, A. M., H. S. Kang, E. Johnson, R. Gunsalus, and F. W. Dahlquist. 2002. Effect of phosphorylation on the interdomain interaction of the response regulator, NarL. Biochemistry 41:15173-15180. [DOI] [PubMed] [Google Scholar]
- 13.Ellison, D. W., and W. R. McCleary. 2000. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J. Bacteriol. 182:6592-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiedler, U., and V. Weiss. 1995. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 14:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlocker, S. 1996. Characterization of the binding of OmpR, a transcription factor in Escherichia coli, to the upstream regulatory sequences of ompF. Ph.D. dissertation. Rutgers, The State University of New Jersey, and Robert Wood Johnson Medical School, New Brunswick, N.J.
- 16.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon, Y., Y. S. Lee, J. S. Han, J. B. Kim, and D. S. Hwang. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 276:40873-40879. [DOI] [PubMed] [Google Scholar]
- 19.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 21.Kato, M., H. Aiba, S. Tate, Y. Nishimura, and T. Mizuno. 1989. Location of phosphorylation site and DNA-binding site of a positive regulator, OmpR, involved in activation of the osmoregulatory genes of Escherichia coli. FEBS Lett. 249:168-172. [DOI] [PubMed] [Google Scholar]
- 22.Kondo, H., A. Nakagawa, J. Nishihira, Y. Nishimura, T. Mizuno, and I. Tanaka. 1997. Escherichia coli positive regulator OmpR has a large loop structure at the putative RNA polymerase interaction site. Nat. Struct. Biol. 4:28-31. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., M. W. McArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:282-291. [Google Scholar]
- 24.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, K., M. Amemura, T. Kawamoto, S. Kimura, H. Shinagawa, A. Nakata, and M. Suzuki. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15-26. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structure of a winged-helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 28.Mattison, K., R. Oropeza, and L. J. Kenney. 2002. The linker region plays an important role in the inter-domain communication of the response regulator OmpR. J. Biol. Chem. 277:32714-32721. [DOI] [PubMed] [Google Scholar]
- 29.McCleary, W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155-1163. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleishmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 31.Okamura, H., S. Hanaoka, A. Nagadoi, K. Makino, and Y. Nishimura. 2000. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J. Mol. Biol. 295:1225-1236. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276(Pt. A):307-326. [DOI] [PubMed] [Google Scholar]
- 33.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 34.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:628-633. [DOI] [PubMed] [Google Scholar]
- 35.Robinson, V. L., J. Hwang, E. Fox, M. Inouye, and A. M. Stock. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10:1649-1658. [DOI] [PubMed] [Google Scholar]
- 36.Sheridan, R. C., J. F. McCullough, and Z. T. Wakefield. 1971. Phosphoramidic acid and its salts. Inorg. Synth. 13:23-26. [Google Scholar]
- 37.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 38.Solà, M., F. X. Gomis-Rüth, L. Serrano, A. González, and M. Coll. 1999. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 285:675-687. [DOI] [PubMed] [Google Scholar]
- 39.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tate, S., M. Kato, Y. Nishimura, Y. Arata, and T. Mizuno. 1988. Location of DNA-binding segment of a positive regulator, OmpR, involved in activation of the ompF and ompC genes of Escherichia coli. FEBS Lett. 242:27-30. [DOI] [PubMed] [Google Scholar]
- 42.Veselovsky, A. V., Y. D. Ivanov, A. S. Ivanov, A. I. Archakov, P. Lewi, and P. Janssen. 2002. Protein-protein interactions: mechanisms and modification by drugs. J. Mol. Recognit. 15:405-422. [DOI] [PubMed] [Google Scholar]
- 43.Vetter, I. R., and A. Wittinghofer. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299-1304. [DOI] [PubMed] [Google Scholar]
- 44.Walthers, D., V. K. Tran, and L. J. Kenney. 2003. Interdomain linkers of homologous response regulators determine their mechanism of action. J. Bacteriol. 185:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 46.Wösten, M. M., and E. A. Groisman. 1999. Molecular characterization of the PmrA regulon. J. Biol. Chem. 274:27185-27190. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, J. H., G. Xiao, R. P. Gunsalus, and W. L. Hubbell. 2003. Phosphorylation triggers domain separation in the DNA binding response regulator NarL. Biochemistry 42:2552-2559. [DOI] [PubMed] [Google Scholar]