Abstract
Background
BP1 is a novel homeobox gene cloned in our laboratory. Our previous studies in leukemia demonstrated that BP1 has oncogenic properties, including as a modulator of cell survival. Here BP1 expression was examined in breast cancer, and the relationship between BP1 expression and clinicopathological data was determined.
Methods
Total RNA was isolated from cell lines, tumors, and matched normal adjacent tissue or tissue from autopsy. Reverse transcription polymerase chain reaction was performed to evaluate BP1 expression. Statistical analysis was accomplished with SAS.
Results
Analysis of 46 invasive ductal breast tumors demonstrated BP1 expression in 80% of them, compared with a lack of expression in six normal breast tissues and low-level expression in one normal breast tissue. Remarkably, 100% of tumors that were negative for the estrogen receptor (ER) were BP1-positive, whereas 73% of ER-positive tumors expressed BP1 (P = 0.03). BP1 expression was also associated with race: 89% of the tumors of African American women were BP1-positive, whereas 57% of those from Caucasian women expressed BP1 (P = 0.04). However, there was no significant difference in BP1 expression between grades I, II, and III tumors. Interestingly, BP1 mRNA expression was correlated with the ability of malignant cell lines to cause breast cancer in mice.
Conclusion
Because BP1 is expressed abnormally in breast tumors, it could provide a useful target for therapy, particularly in patients with ER-negative tumors. The frequent expression of BP1 in all tumor grades suggests that activation of BP1 is an early event.
Keywords: BP1, breast cancer, distal-less, estrogen receptor, homeobox
Introduction
Breast cancer is predicted to be newly diagnosed in about 211,300 women in 2003 in the United States [1,2]. Twelve percent of all women will be diagnosed with breast cancer in their lifetime, and 3.5% will die of the disease. The cell transformation associated with breast cancer requires the coordinated activation or inhibition of specific intact or altered genes. Transcription factors frequently regulate these processes. An important type of transcription factor contains a conserved region of 60 amino acids called the homeodomain (HD), encoded by a DNA region, the homeobox (HB). HD proteins are involved in the control of development and differentiation in many organisms [3,4]. Initially discovered in Drosophila, mutations of the HB in homeobox genes result in vast morphologic abnormalities [3]. Subsequently, homologous HB genes have been cloned in many species including humans [5-7].
Vertebrate HB genes are divided into two classes: Class I genes (HOX genes) are organized into four clusters (HOX A, B, C, D) with a total of 39 members [8]. Members of Class II are also called divergent genes, including genes such as PAX, MSX, IRX, and DLX, named after their homologs in Drosophila (paired, muscle segment, Iroquois, and distal-less, respectively) [9,10]. Accumulating evidence suggests that homeobox genes are important in malignant transformation. Not only have altered expression levels of HB genes been detected in a variety of human malignancies [8,11-14], but the ectopic expression of certain HOX genes can induce tumors [15-17]. Abnormal HOX gene expression has been associated with breast cancer. For example, the murine Hoxa-1 gene is upregulated in neoplastic murine mammary glands [18]. In humans, the lack of HOXA5 expression results in the loss of p53 expression in breast cancer [19]. Enforced HOXB7 expression in SkBr3 breast cancer cells induces the expression of basic fibroblast growth factor and increases growth rate, serum-independent growth, and the ability of cells to form colonies in semisolid medium [20]. Nude mice, when injected with SkBr3/HOXB7 cells, developed tumors [21].
We have cloned a human homeobox cDNA called BP1, belonging to the DLX family. BP1 is apparently involved in the regulation of diverse pathways. In normal erythroid cells it acts as a repressor of the β-globin gene [22,23]. However, BP1 was also expressed in 47% of the adult and 81% of the pediatric acute myeloid leukemia patients we examined, and its overexpression increased the leukemogenic potential of K562 cells in vitro [11]. Molecular analysis revealed that BP1 expression is required for the survival of K562 leukemia cells, suggesting that BP1 might be part of an anti-apoptotic pathway (Davenport GJ, Fu S, Haga SB, Do K, Stevenson H, Pinzone J, Chase MB, Berg PE, unpublished data). Furthermore, these results imply that aberrant BP1 expression might also be involved in other malignancies. In the present study we analyzed BP1 expression in human breast cancer cell lines and assessed BP1 expression in invasive ductal carcinoma tumors, correlating its expression with age, tumor size, lymph node metastasis, histologic grade, estrogen receptor (ER) status, progesterone receptor (PR) status, and race.
Methods
Cell lines
The breast cancer cell lines Hs578T, MCF7, MDA-MB-231, MDA-MB-468, and T-47D were kindly given to us by Dr Angela Brodie; MCF7ADR and MDA-MB-435S were a gift from Dr Joseph Fontana; and MCF10A was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, except for MCF10A cells, which were grown as recommended by the ATCC.
Human breast tissues
Human breast tumor samples were obtained from The George Washington University Department of Pathology and the NCI Cooperative Human Tissue Network, Southern Division, all with Institutional Review Board approval, and as previously described [24]. Breast tissue samples, 0.5–1.0 cm in diameter, were obtained from fresh surgical resection specimens from patients with breast tumors, and were characterized pathologically in terms of their stage in malignancy. The samples were snap-frozen in liquid nitrogen and were kept frozen until use. Normal breast tissues were obtained from the periphery of specimens surgically resected for treatment of cancer or from an autopsy specimen obtained from a patient who had died of injuries sustained in an accident (not breast cancer); the latter was obtained from the Brain and Tissue Bank for Developmental Disorders at the University of Maryland.
RNA isolation and reverse transcription polymerase chain reaction (RT–PCR)
Total RNA was extracted from human breast cell lines, frozen breast tumor tissues and normal breast tissues with Trizol reagent (Invitrogen, Gaithersburg, MD, USA) in accordance with the manufacturer's protocol. Total RNA (1 μg) was used to synthesize cDNA with a SuperScript II reverse transcriptase system (Invitrogen). After synthesis of the first-strand cDNA, PCR was performed by initial denaturing at 94°C for 2 min followed by 28 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1.5 min. The PCR primers for BP1, which span an intron, were 5'-GTATG-GCCACCTCCTGTCTT-3' (forward) and 5'-GAGTA-GATGGTCCTCGGCTT-3' (reverse), giving a product of 225 base pairs. The internal control for all samples was β-actin.
Statistical analyses
Statistical analyses were used to determine the relationship between BP1 positivity, measured by RT–PCR, and clinicopathological parameters. All P values were calculated with Fisher's Exact Test. Tests were considered significant if their associated P value was 0.05 or less (two-tailed). All statistical analyses were performed with SAS 8.2 (SAS Institute, Cary, NC, USA).
Results
BP1 expression in breast cell lines
BP1 mRNA levels were analyzed in a series of breast cell lines by using RT–PCR. The cell lines are derived from breast cancer patients, with the exception of MCF10A, which is an immortalized, non-transformed epithelial cell line [25]. As shown in Fig. 1, the levels of BP1 expression varied widely between the cell lines, from barely detectable (Hs578T, MDA-MB-435S, and MCF10A; lanes 2, 6, and 9, respectively) to highly expressing (MCF7ADR, MDA-MB-468, and T-47D; lanes 4, 7, and 8, respectively); MCF7 and MDA-MB-231 (lanes 3 and 5, respectively) showed intermediate expression. For comparison, one normal breast tissue from an autopsy was included in the first lane; it was BP1-negative.
Figure 1.
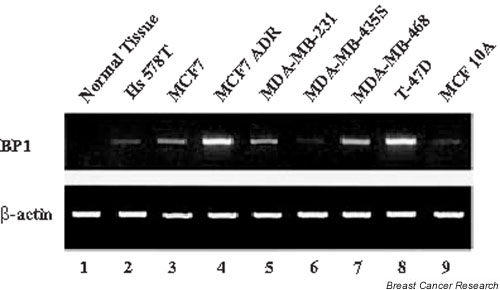
Expression of BP1 in breast cancer cell lines. Reverse transcription polymerase chain reaction was performed to analyze relative BP1 mRNA expression in cell lines and in one normal breast tissue. The loading control was β-actin.
There was a correlation between BP1 expression and tumorigenesis in mice (Table 1). The cell lines that express little or no BP1 (Hs578T, MDA-MB-435S, and MCF10A) are not tumorigenic, whereas MCF7ADR, MDA-MB-468, and MDA-MB-231 express BP1 and are tumorigenic. MCF7 and T-47D cells express variable BP1 levels and are tumorigenic only in the presence of estradiol [26,27]. BP1 expression might therefore be involved in tumorigenesis in mice. Transgenic studies would provide direct evidence for this hypothesis.
Table 1.
BP1 expression in breast cancer cell lines
| Cell line | ER | PR | Malignancy | Tumorigenic* | BP1 |
| Hs578T | - | - | Ductal carcinoma | No | +/- |
| MCF7 | + | + | Adenocarcinoma | † | ++ |
| MCF7ADR‡ | ADR-resistant | Yes | +++ | ||
| MDA-MB-231 | - | - | Adenocarcinoma | Yes | ++ |
| MDA-MB-435S | - | - | Ductal carcinoma | No | +/- |
| MDA-MB-468 | - | - | Adenocarcinoma | Yes | +++ |
| T-47D | + | + | Ductal carcinoma | † | +++ |
| MCF10A | + | + | Normal | No | +/- |
BP1 expression in breast tumor tissues
BP1 mRNA expression was examined in 46 invasive ductal breast tumors including grades I, II, and III tumors and, in some cases, compared with adjacent normal breast tissue. Of these tumors, 80% expressed BP1. In contrast, of seven normal breast tumors analyzed, six were BP1-negative and one showed low BP1 expression. A representative RT–PCR analysis of five tumors, three with matched normal tissue, is shown in Fig. 2. All of the tumors shown are ER-negative and PR-negative. Matched normal and tumor tissues from three patients are shown in lanes 1 and 2 (patient 1), 3 and 4 (patient 2), and 5 and 6 (patient 3). No BP1 expression was seen in the normal samples (lanes 1, 3, and 5), whereas BP1 was expressed in all of the tumors (lanes 2, 4, and 6). Two additional tumor tissues, shown in lanes 7 and 8, were positive for BP1; matched samples were not available.
Figure 2.
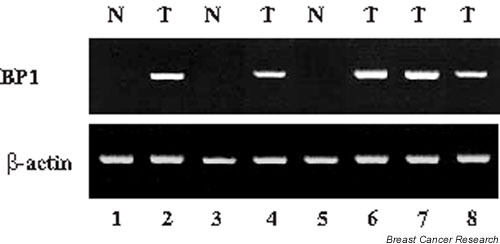
Expression of BP1 in breast tumors. Relative levels of BP1 mRNA were determined by analysis of breast ductal tumors (T) or normal controls (N) by reverse transcription polymerase chain reaction. The loading control was β-actin.
To determine the significance of BP1 expression in the tumor tissues, clinicopathological data were related to BP1 expression (Table 2). No significant relationship was detected between BP1 status and age, tumor size, or lymph node metastasis. Because 5-year survival data were not available for some patients, we were not able to evaluate survival outcome. BP1 expression was significantly more prevalent in ER-negative tumors, of which 100% were BP1-positive, compared with ER-positive tumors, of which 73% were BP1-positive (P = 0.03). There was also a significant difference in BP1 expression between PR-negative tumors, of which 96% were BP1-positive, and PR-positive tumors, of which 67% were BP1-positive (P = 0.02). BP1 expression was associated with race: it was expressed in 89% of the tumors of African American women but in only 57% of the tumors of Caucasian women (P = 0.04). There was no significant difference in BP1 expression between the three tumor grades: BP1 was expressed in 80% of grade I tumors, 73% of grade II tumors and 90% of grade III tumors.
Table 2.
Association between BP1 mRNA expression and clinicopathological data
| BP1 mRNA | |||
| Factors | Negative | Positive | P |
| Age | 1.00 | ||
| Premenopausal | 2 (17%) | 10 (83%) | |
| (<50 years) | |||
| Postmenopausal | 7 (21%) | 26 (79%) | |
| (≥ 50 years) | |||
| Tumor size | 1.00 | ||
| <2.0 cm | 1 (17%) | 5 (83%) | |
| ≥ 2.0 cm | 7 (19%) | 30 (81%) | |
| Lymph node metastasis | 1.00 | ||
| Negative | 4 (20%) | 16 (80%) | |
| Positive | 4 (18%) | 18 (82%) | |
| Histologic grade | 0.29 | ||
| I | 1 (20%) | 4 (80%) | |
| II | 4 (27%) | 11 (73%) | |
| III | 2 (9%) | 20 (91%) | |
| ER status | 0.03 | ||
| Negative | 0 (0%) | 18 (100%) | |
| Positive | 7 (27%) | 19 (73%) | |
| PR status | 0.02 | ||
| Negative | 1 (4.0%) | 24 (96%) | |
| Positive | 6 (33%) | 12 (67%) | |
| Race | 0.04 | ||
| Caucasian | 6 (43%) | 8 (57%) | |
| African American | 3 (11%) | 25 (89%) | |
ER, estrogen receptor; PR, progesterone receptor. Data were unavailable or indeterminate for parameters as follows: age (one tumor), tumor size (three tumors), lymph node metastasis (four tumors), histologic grade (four tumors), ER status (two tumors), PR status (three tumors), and race (four tumors).
Discussion
In the present study we found that 80% of invasive ductal breast tumors expressed BP1, a member of the DLX homeobox gene family. In contrast, only 14% of normal breast tissues expressed BP1. Our previous studies showed that BP1 is also expressed in leukemia and has characteristics of an oncogene: in acute myeloid leukemia, BP1 is highly expressed in the bone marrow of 63% of patients overall, with a preponderance of expression in pediatric patients [11]. When overexpressed in a leukemia cell line (K562), BP1 increased the ability of cells to grow in soft agar, an indicator of leukemogenic potential, up to 45-fold [11]. Consistent with a possible oncogenic role for BP1 in breast cancer was the observation that its expression was correlated with the ability of breast cancer cell lines to cause mammary tumors in mice.
Recent studies have shown that abrogation of BP1 expression in a leukemia cell line leads to apoptosis (Davenport GJ, Fu S, Haga SB, Do K, Stevenson H, Pinzone J, Chase MB, Berg PE, unpublished data). Coupled with the increased clonogenicity resulting from overexpression of BP1, these observations suggest that the expression of BP1 in breast tissue might also lead to increased survival and, conversely, that the abrogation of BP1 expression in malignant breast tissue could cause apoptosis of those cells expressing BP1.
Estrogen and progesterone, which stimulate the growth of breast epithelium, are two crucial hormones involved not only in normal breast development but also in carcinogenesis of breast epithelium and progression of breast cancer [28]. About 40% of breast cancers are ER-negative [28]. ER-negative tumors are unresponsive to anti-estrogen therapy, probably because of constitutive growth factor expression [28,29]. In general, these tumors have a higher histologic grade and a higher proliferative rate and are associated with poorer prognosis [28]. The finding that 100% of ER-negative tumors expressed BP1 might therefore be highly relevant to both the prognosis and therapy of those difficult-to-treat tumors.
It is well known that African American women have a higher mortality rate from breast cancer than Caucasian women (reviewed by Mancino and colleagues [30]). Although socioeconomic factors account for some of this difference, several recent studies have shown that the median survival of African American women presenting with Stage III or IV disease was significantly lower than that of Caucasian women diagnosed at the same stage of disease: in Stage III disease the median survival was 91.3 months for Caucasian patients and only 36.0 months for African American patients, whereas in Stage IV disease the median survival was reduced to 24.0 months for Caucasian patients and 8.9 months for African American patients [30,31]. African American women also have a higher risk of ER-negative tumors after adjustment is made for stage and age at diagnosis. These results have led to speculation that there could be biological differences between African American and Caucasian women that contribute to the differences in outcome. Interestingly, more African American women than Caucasian women were BP1-positive (P = 0.04). It will be important to expand further the number of women studied to determine whether BP1 expression might be one biological factor that underlies survival differences between African American and Caucasian women.
Because BP1 was expressed in all tumor grades, we propose that it is activated in an early stage of malignant development or even in a premalignant state. We have therefore begun to examine ductal carcinoma in situ (DCIS), which are preinvasive malignant lesions derived from epithelial cells, and fibroadenomas, which are non-malignant tumors composed of epithelial and stromal cells [32,33]. So far we have analyzed one DCIS, which was BP1-positive, and two fibroadenomas, both of which were BP1-positive, compared with no expression in the matched normal tissue for each fibroadenoma (data not shown). The sample number is very small, but the fact that any of the DCISs or fibroadenoma tumors are positive for BP1 demonstrates that activation of BP1 might be an early event, perhaps promoting cell survival as seems to occur in leukemia cells. This idea is also supported by the fact that BP1 is as likely to be expressed in small tumors (less than 2.0 cm) as it is in larger tumors (2.0 cm or more).
The mechanism for overexpression of BP1 in breast cancer is unknown. However, because of its map location at 17q21–22 [23], the BP1 gene is a potential candidate for DNA amplification, which could lead to its activation or overexpression. This hypothesis is based on the fact that two regions on chromosome 17q are sometimes amplified in breast cancer, one at 17q11–21 and another at 17q22–24 [34,35]. The locus of BP1 is also near BRCA1; the implications of this proximity are not yet known.
To begin to identify genes that might be targets of BP1, we have overexpressed BP1 in K562 cells and screened a microarray (Davenport GJ, Fu S, Haga SB, Do K, Stevenson H, Pinzone J, Chase MB, Berg PE, unpublished data). Although K562 cells are derived from leukemia, among the genes showing altered regulation were several that are also dysregulated in breast cancer, including c-myc and cyclin D2; their altered expression was verified by real-time PCR. c-myc was upregulated in K562 cells overexpressing BP1, and c-Myc protein is highly expressed in more than half of breast cancer patients [36]. Cyclin D2, downregulated on the array, is repressed in breast cancer [37]. It will be important to determine whether either of those genes is a target for BP1 in breast cancer.
These data strongly implicate BP1 in breast cancer. More case studies are underway to further strengthen our findings and to determine whether there is a statistical association between BP1 expression and prognosis or outcome of breast cancer patients.
Conclusion
These data demonstrate a statistically significant association between the expression of the transcription factor BP1, not previously implicated in breast cancer, and ER-negative breast tumors. This, together with other studies, strongly suggests that BP1 has oncogenic properties, making it a new potential target in breast tumors, especially the difficult-to-treat ER-negative tumors. Interestingly, BP1 is expressed in 73–90% of tumors, including all grades. This finding and the observation that fibroadenomas (non-malignant tumors) can express BP1 strongly suggest that its activation might be associated with loss of growth regulation, which is consistent with our molecular data.
Competing interests
None declared.
Abbreviations
DCIS = ductal carcinoma in situ; ER = estrogen receptor; HB = homeobox; HD = homeodomain; PR = progesterone receptor; RT–PCR = reverse transcription polymerase chain reaction.
Acknowledgments
Acknowledgements
We thank Dr Allan L. Goldstein for insightful discussions, and Dr Susanne Haga, Dr Kathryn Kennedy and Khanh Do for helpful comments on the manuscript. This research was supported by grants NIH CA89753 (to PEB) and CA91149 (to PEB) and by the Elaine Snyder Cancer Research Award (to PEB).
References
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA: Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- Cancer Facts and Figures 2003: American Cancer Society. http://www.cancer.org
- Gehring WJ. Homeoboxes in the study of development. Science. 1987;236:1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Magli MD, Largman C, Lawrence HJ. Effects of HOX homeobox genes in blood cell differentiation. J Cell Physiol. 1997;173:168–177. doi: 10.1002/(SICI)1097-4652(199711)173:2<168::AID-JCP16>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Levine M, Rubin GM, Tjian R. Human DNA sequences homologous to a protein coding region conserved between homeotic genes of Drosophila. Cell. 1984;38:667–673. doi: 10.1016/0092-8674(84)90261-7. [DOI] [PubMed] [Google Scholar]
- Akam M. Hox and HOM: homologous gene clusters in insects and vertebrates. Cell. 1989;57:347–349. doi: 10.1016/0092-8674(89)90909-4. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. Exploring the homeobox. Gene. 1993;135:215–221. doi: 10.1016/0378-1119(93)90068-E. [DOI] [PubMed] [Google Scholar]
- van Oostveen JW, Biji JJ, Raaphorst FM, Walboomers JJM, Meijer CJM. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia. 1999;13:1675–1690. doi: 10.1038/sj/leu/2401562. [DOI] [PubMed] [Google Scholar]
- Stein S, Fritsch R, Lemaire L, Kessel M. Checklist: vertebrate homeobox genes. Mech Dev. 1996;55:91–108. doi: 10.1016/0925-4773(95)00494-7. [DOI] [PubMed] [Google Scholar]
- Popovici C, Leveugle M, Birnbaum D, Coulier F. Homeobox gene clusters and the human paralogy map. FEBS Lett. 2001;491:237–242. doi: 10.1016/S0014-5793(01)02187-1. [DOI] [PubMed] [Google Scholar]
- Haga SB, Fu S, Karp JE, Ross DD, Williams DM, Hankins WD, Behm F, Ruscetti FW, Chang M, Smith BD, Becton D, Raimondi SC, Berg PE. BP1, a new homeobox gene, is frequently expressed in acute leukemias. Leukemia. 2000;14:1867–1875. doi: 10.1038/sj.leu.2401912. [DOI] [PubMed] [Google Scholar]
- Li H, Huang CJ, Choo KB. Expression of homeobox genes in cervical cancer. Gynecol Oncol. 2002;84:216–221. doi: 10.1006/gyno.2001.6498. [DOI] [PubMed] [Google Scholar]
- Park J, Park K, Kim S, Lee JH. Msx1 gene overexpression induces G1 phase cell arrest in human ovarian cancer cell line OVCAR3. Biochem Biophys Res Commun. 2001;281:1234–1240. doi: 10.1006/bbrc.2001.4474. [DOI] [PubMed] [Google Scholar]
- Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, Rosell R, Gemmill RM, Drabkin HA. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci USA. 2000;97:12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulbecker CC, Gruss P. The oncogenic potential of deregulated homeobox genes. Cell Growth Differ. 1993;4:431–441. [PubMed] [Google Scholar]
- Hawley RG, Fong AZC, Reis MD, Zhang N, Lu M, Hawley TS. Transforming function of the HOX11/TCL3 homeobox gene. Cancer Res. 1997;57:337–345. [PubMed] [Google Scholar]
- Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Over-expression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Y, Daniel CA, Strickland P, Daniel CW. Hox genes in normal and neoplastic mouse mammary gland. Cancer Res. 1994;54:5981–5985. [PubMed] [Google Scholar]
- Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- Care A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- Care A, Felicitti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, Peschle C, Colombo MP. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61:6532–6539. [PubMed] [Google Scholar]
- Chase MB, Fu S, Haga S, Davenport G, Stevenson H, Do K, Morgan D, Mah AL, Berg PE. BP1, a homeodomain-containing isoform of DLX4, represses the β-globin gene. Mol Cell Biol. 2002;22:2505–2514. doi: 10.1128/MCB.22.8.2505-2514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Stevenson H, Strovel JW, Haga SB, Stamberg J, Do K, Berg PE. Distinct functions of two isoforms of a homeobox gene, BP1 and DLX7, in the regulation of the beta-globin gene. Gene. 2001;278:131–139. doi: 10.1016/S0378-1119(01)00716-8. [DOI] [PubMed] [Google Scholar]
- Poola I, Clarke R, DeWitty R, Leffal L. Functionally active estrogen receptor isoform profiles are different in the breast tumors of African American and Caucasian women. Cancer. 2002;94:615–623. doi: 10.1002/cncr.10274. [DOI] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- Leung CK, Shiu RP. Required presence of both estrogen and pituitary factors for the growth of human breast cancer cells in athymic nude mice. Cancer Res. 1981;41:546–551. [PubMed] [Google Scholar]
- Tang CK, Concepcion XZ, Milan M, Gong X, Montgomery E, Lippman ME. Ribozyme-mediated down-regulation of ErbB-4 in estrogen receptor-positive breast cancer cells inhibits proliferation both in vitro and in vivo. Cancer Res. 1999;59:5315–5322. [PubMed] [Google Scholar]
- Dickson RB, Lippman ME. Growth factors in breast cancer. Endocr Rev. 1995;16:559–589. doi: 10.1210/edrv-16-5-559. [DOI] [PubMed] [Google Scholar]
- Ingvarsson S. Molecular genetics of breast cancer progression. Semin Cancer Biol. 1999;9:277–288. doi: 10.1006/scbi.1999.0124. [DOI] [PubMed] [Google Scholar]
- Mancino AT, Rubio IT, Henry-Tillman R, Smith LF, Landes R, Spencer HJ, Erkman L, Klimberg VS. Racial differences in breast cancer survival: the effect of residual disease. J Surg Res. 2001;100:161–165. doi: 10.1006/jsre.2001.6232. [DOI] [PubMed] [Google Scholar]
- Edwards MJ, Gamel JW, Vaughan WP, Wrightson WR. Infiltrating ductal carcinoma of the breast: the survival impact of race. J Clin Oncol. 1998;16:2693–2699. doi: 10.1200/JCO.1998.16.8.2693. [DOI] [PubMed] [Google Scholar]
- Iglehart JD, Kaelin CM. In: Sabiston Textbook of Surgery: The Biological Basis of Modern Surgical Practice. Townsend CM Jr, editor. Philadelphia: W.B. Saunders Company; 2001. p. 567. [Google Scholar]
- Abeloff MD, Lichter AS, Niederhuber JE, Pierce LJ, Love RR. In: Clinical Oncology. Abeloff MD, editor. Philadelphia, PA: Churchill Livingstone; 2000. pp. 2088–2093. [Google Scholar]
- Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R. Two distinct amplified regions at 17q11-q21 involved in human primary breast cancer. Cancer Res. 1996;56:3886–3890. [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi O-P, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, Buluwela L, Weitzman JM, Sukumar S. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001;61:2782–2787. [PubMed] [Google Scholar]


