Abstract
Chemically induced dimerization provides a general way to gain control over intracellular processes. Typically, FK506-binding protein (FKBP) domains are fused to a signaling domain of interest, allowing crosslinking to be initiated by addition of a bivalent FKBP ligand. In the course of protein engineering studies on human FKBP, we discovered that a single point mutation in the ligand-binding site (Phe-36 → Met) converts the normally monomeric protein into a ligand-reversible dimer. Two-hybrid, gel filtration, analytical ultracentrifugation, and x-ray crystallographic studies show that the mutant (FM) forms discrete homodimers with micromolar affinity that can be completely dissociated within minutes by addition of monomeric synthetic ligands. These unexpected properties form the basis for a “reverse dimerization” regulatory system involving FM fusion proteins, in which association is the ground state and addition of ligand abolishes interactions. We have used this strategy to rapidly and reversibly aggregate fusion proteins in different cellular compartments, and to provide an off switch for transcription. Reiterated FM domains should be generally useful as conditional aggregation domains (CADs) to control intracellular events where rapid, reversible dissolution of interactions is required. Our results also suggest that dimerization is a latent property of the FKBP fold: the crystal structure reveals a remarkably complementary interaction between the monomer binding sites, with only subtle changes in side-chain disposition accounting for the dramatic change in quaternary structure.
Many intracellular processes are mediated through inducible protein–protein interactions (1). Methods that allow such processes to be manipulated at will are powerful tools for understanding and controlling cellular activities (2). The use of chemical inducers of dimerization (“dimerizers”) has proved to be a particularly versatile approach (3). Cells are engineered to express chimeric proteins comprising a signaling domain fused to a drug-binding domain; treatment with bivalent ligands crosslinks the proteins and initiates signaling. This strategy has been used to create inducible alleles of numerous signaling proteins that are activated by oligomerization (see ref. 1 and references therein). Dimerizers can also be used to regulate transcription, through the controlled association of chimeric DNA-binding and activation domains (4, 5). These approaches allow protein function to be dissected inside cells or whole organisms, and also have therapeutic promise as a means for bringing cell and gene therapies under small-molecule control.
Most dimerizer applications to date have used the 12-kDa FK506-binding protein (FKBP12; herein called FKBP) and its ligands. FKBP is a monomeric and highly abundant cytosolic protein that serves as the primary receptor for the immunosuppressive ligands FK506 and rapamycin (6, 7). The physiological function of the protein remains unclear, although it also binds directly to several cell surface receptors, and exhibits peptidyl-prolyl cis-trans isomerase activity. Structural analysis reveals that a single binding site accounts for these diverse activities (8, 9). Dimerization of FKBP fusion proteins can be achieved by using FK1012, a semisynthetic dimer of FK506 (3), or simpler completely synthetic derivatives (10). Recently, we reported further modification of these ligands to improve specificity. A steric “bump” was added to prevent binding to wild-type FKBP, resulting in ligands that bind uniquely to FKBP fusion proteins equipped with a complementary “hole” mutation, Phe-36 → Val (11).
Use of dimerizers offers a general way to initiate processes that are activated by oligomerization. However, no general strategy is available for achieving the opposite mode of control, wherein proteins are constitutively associated until addition of a drug. Such control would be highly valuable for probing the consequences of rapidly abolishing oligomerization events inside cells—extinguishing activities that are activated by proximity, or conversely activating activities that are suppressed by particular protein–protein interactions. We describe here the discovery and characterization of F36M-FKBP (FM), a point mutant of FKBP that has the unusual property of forming discrete dimers that can be dissociated by ligand. We use this as the basis for a “reverse dimerization” system that should be broadly applicable as a disaggregation switch for intracellular processes.
Materials and Methods
Chemical Synthesis.
FM ligands AP21998 and AP22542 were prepared by using strategies analogous to those previously reported (10–12). Details of the syntheses will be described elsewhere and are available on request.
Two-Hybrid Analysis of FKBP Interactions.
The system for FKBP ligand-dependent transcriptional activation in mammalian cells has been described (10, 13, 14). Expression vectors were built by stepwise insertion of XbaI–BamHI fragments into pCGNN (15) and contained one or three copies of wild-type FKBP (10), F36V-FKBP (FV) (13) or FM (constructed analogously), fused to the ZFHD1 DNA-binding domain (16) or human p65 activation domain (amino acids 361–551). Vectors were transiently cotransfected (13) into HT1080L cells (5), and ligand-dependent secreted alkaline phosphatase (SEAP) production was assayed (13).
Enhanced Green Fluorescent Protein (EGFP) Fusions.
Cytoplasmic and nuclear expression vectors in which multiple copies of FM are fused to EGFP (17) were constructed in a similar stepwise manner in pCGNN-derived expression vectors (15). Constructs were transiently transfected into HT1080 cells and analyzed by using fluorescence microscopy. Where indicated, AP22542 was added directly from a 100% ethanol stock at a final solvent concentration of <0.2%.
Protein Production.
Wild-type FKBP and FM were expressed in Escherichia coli BL21(DE3) as untagged proteins from vector pET20b, essentially as described for FV (11). Wild-type FKBP was purified as described (12). FM was purified similarly, except that cell paste was lysed by using a French press (Aminco); the S100 column was equilibrated with 20 mM potassium phosphate (KPi) buffer, pH 6.5, containing 200 mM NaCl, 2 mM EDTA, 5 mM DTT, and 0.02% NaN3 (S100 buffer); and the S100 pool was dialyzed against 20 mM Tris⋅HCl, pH 7.4/5 mM DTT/1 mM EDTA and then passed over MacroPrep High Q resin (Bio-Rad) equilibrated in the same buffer. The final yield of pure protein was approximately 6.7 mg/g of cell paste.
HPLC Gel Filtration.
Gel filtration HPLC was performed on TSK G2000SW columns (7.5 × 300 mm with 5-μm particle size, or 7.5 × 600 mm with 10-μm particle size; TosoHaas, Montgomeryville, PA). The columns were equilibrated in 50 mM KPi buffer, pH 6.8, containing 300 mM NaCl, 1 mM EDTA, and 0.05% NaN3, and run at flow rates of 0.8 and 1.0 ml/min, respectively. A set of 10 proteins was used for calibration of the column. FKBP samples contained 0.75 mg/ml protein in 25 mM KPi, pH 7/150 mM NaCl/1 mM EDTA and included a 3-fold molar excess of FK506 added from a 100% ethanol stock to a final solvent concentration of 0.8%, or ethanol alone. Samples were incubated on ice for 2 h, then spin-filtered through a 0.1-μm-pore membrane (Ultrafree MC; Millipore) before injection.
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were conducted at 20°C in a Beckman-Optima XL-I analytical ultracentrifuge using an An-60 Ti rotor and 1.2-cm path-length cells. FM was dialyzed against 50 mM KPi, pH 7.0/100 mM NaCl/2 mM EDTA/5 mM DTT/0.02% NaN3 and concentrated to ≈18 μM by using an Amicon 8MC ultrafiltration concentrator fitted with a 5-kDa-cutoff membrane (Millipore). Experiments were performed on ≈100-μl samples containing either a 5-fold molar excess of FK506 added from a 100% ethanol stock, or ethanol alone to give final solvent concentrations of 0.3%. Samples were equilibrated for ≈48 h at speeds ranging from 20,000 to 60,000 rpm, and sedimentation curves measured by absorbance at 280 nm were compared with a buffer reference cell. Multiple data sets were fitted simultaneously to yield the Kd and molecular mass values quoted, using Beckman-Optima XL-A/XL-I data analysis software (v4.0). Data fitting and simulations used the following calculated parameters: buffer density at 20°C = 1.01 mg/ml; monomeric molecular mass = 11,804 Da; and partial specific volume = 0.736.
X-Ray Crystallography.
The protein formed thick needle-shaped crystals upon concentration to 75 mg/ml in S100 buffer and incubation at 4°C (the crystals dissolved upon warming to room temperature). Crystals were tetragonal (P43212, a = b = 81.68 Å, c = 93.94 Å) with one dimer per asymmetric unit. Diffraction data were collected at −160°C with a Rigaku R-AXIS II area detector with graphite monochromated Cu Kα x-rays. Diffraction data were collected and processed, and the structure was solved by molecular replacement (using PDB structure 1bl4) and refined, using previously described methods (11). The current R value for reflections with F > 2σ from 10.0 to 2.2 Å is 0.23 and Rfree is 0.29. The rms deviations are 0.005 Å for bonds and 1.44° for angles. The model contains two molecules of FM with all residues from Gly-1 to Glu-107, and 191 water molecules. Coordinates have been deposited in the Protein Data Bank (PDB ID code 1EYM).
Results
FM Self-Interacts in a Mammalian Two-Hybrid Assay.
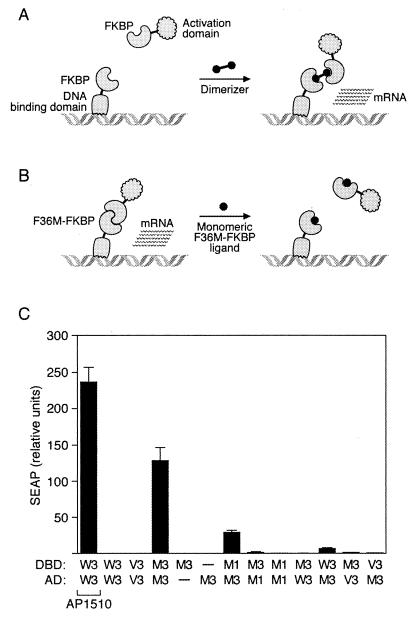
The unexpected properties of FM were discovered serendipitously by using a mammalian two-hybrid approach. Having previously engineered the F36V mutation into FKBP to allow specific binding of “bumped” ligands (see Introduction) (11), we tested the ability of this mutant to support dimerizer-mediated transcription, and we also included FKBPs with other substitutions at the 36 position, including methionine (Fig. 1A). Wild-type FKBP, FV, or FM was fused in one or three copies to DNA-binding and activation domains (see Materials and Methods). Constructs were transiently transfected into HT1080L cells (5), a human fibrosarcoma cell line containing a secreted alkaline phosphatase (SEAP) reporter gene, and SEAP production was assayed.
Figure 1.
Discovery of the self-dimerizing activity of FM by using a mammalian two-hybrid assay. (A) Assay configuration used to obtain dimerizer-dependent transcription. FKBP domains are separately fused to a DNA-binding domain and an activation domain. No transcription results until addition of a homodimeric small molecule that can crosslink the two chimeric proteins. (B) Performance of the “reverse dimerization” system in the same assay. If the FKBP domains can interact, transcription will be constitutively activated. Addition of a monomeric ligand should abolish transcription if FKBP self-association requires an intact ligand-binding site. (C) Results of a mammalian two-hybrid analysis of FKBP fusion proteins. Secreted alkaline phosphatase (SEAP) reporter cells were transiently cotransfected with vectors expressing DNA-binding domain (DBD) and activation domain (AD) fusion proteins fused to the indicated FKBP domains: W = wild-type, V = F36V mutant, M = F36M mutant; 1 and 3 indicate one or three copies of the domain. —, Empty vector substituted; AP1510 indicates that dimerizer AP1510 (10) was added to 100 nM. Mean values ± SD (n = 4) are plotted.
As previously observed (10), fusion proteins containing three copies of wild-type FKBP produced no transcription alone, and high level transcription in the presence of the homodimeric ligand AP1510, indicating a conventional ligand-dependent activation (Fig. 1C, lanes 1 and 2). Fusions to the F36V mutant (FV) also showed no interaction in the absence of ligand (lane 3) and potent dimerizer-dependent activation (13). However, transfection of constructs containing three copies of FM reproducibly led to high level transcription in the absence of any added ligand (lane 4). Activation was absolutely dependent on the presence of both fusion proteins: omitting either abolished transcription. Multimerization was required for maximal activity, with transcription reduced 5-fold when only one FM was included on the DNA-binding domain, and further reduced almost to background if the number of domains on the activation domain fusion protein was also reduced to 1. No significant transcription was seen when fusion proteins containing three FM domains were cotransfected with the proteins containing wild-type or FV domains. Taken together, these data indicate that the FM domain is able to specifically interact with itself, either directly (Fig. 1B) or indirectly, and that the interaction can be amplified by multimerizing the domain.
FM Self-Interaction Is Antagonized by Ligand.
Fluorescence polarization binding assays (11) showed that FM retains the ability to bind in vitro to FK506 and rapamycin, albeit with a 10-fold lower affinity (data not shown). This observation prompted us to investigate the effects of ligands on the apparent FM self-interaction in the two-hybrid assay. Strikingly, we observed that FM-dependent transcription was completely reduced to background levels in a concentration-dependent manner by addition of FK506 (Fig. 2B) or rapamycin (not shown). This antagonism was specific, as no effects were seen upon addition of cyclosporin A, a small molecule with no affinity for FKBPs (Fig. 2B). These data indicate that FM self-association involves the ligand-binding site.
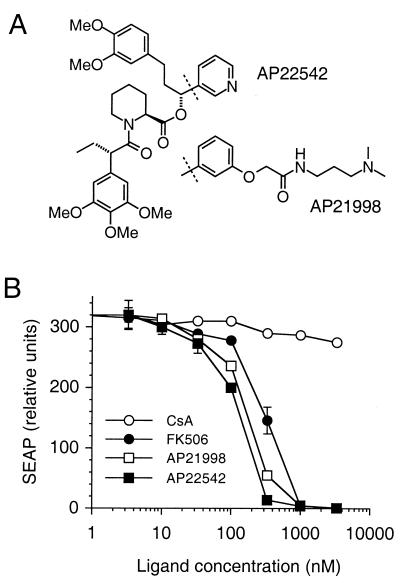
Figure 2.
FM self-association is specifically inhibited by FKBP ligands. (A) Chemical structures of synthetic FM ligands. (B) Effects of ligands on FM-dependent transcription in the two-hybrid assay (Fig. 1B). HT1080L reporter cells expressing DNA-binding domain and activation domain fusions to three copies of FM were treated with increasing concentrations of ligand and assayed as in Fig. 1. CsA, cyclosporin A. Mean values ± SD (n = 3) are plotted.
FM also retains the ability to bind with high affinity (Kd ≈ 1 nM; data not shown) to the “bumped” fully synthetic ligands we previously designed for the F36V mutant (11, 12). Because these ligands bind minimally to the endogenous wild-type FKBP (11, 12) they are preferable for intracellular applications. To test the ability of these ligands to antagonize transcription, we first modified them to improve their cellular permeance by 1–2 orders of magnitude (not shown). The resulting compounds, AP22542 and AP21998 (Fig. 2A), were 3- to 5-fold more potent than FK506 at inhibiting transcription (Fig. 2B). Of interest, even for the most potent ligand (AP22542) the concentration required for 50% inhibition was 100 nM, whereas the in vitro binding affinity is ≈1 nM, suggesting that a higher ligand concentration is required to overcome the avidity-driven interaction of the two multivalent FM fusion proteins. These experiments demonstrate that FM fusion proteins and monomeric synthetic ligands can be used to create a dimerization-based off switch for transcription, in which transcription is abolished by addition of ligand.
Purified FM Behaves as a Dimer in Gel Filtration Analysis.
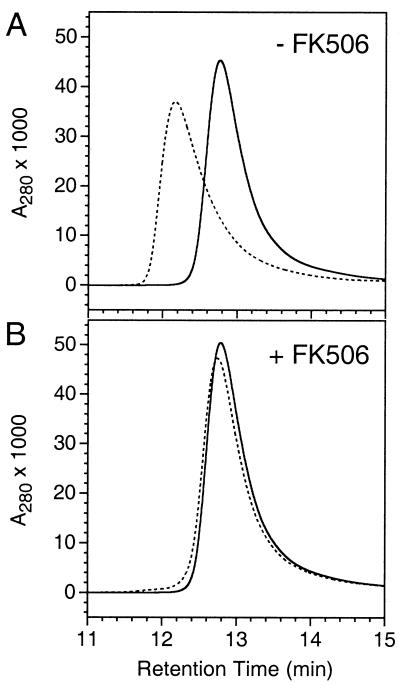
The simplest explanation for the unexpected behavior of FM is direct formation of homodimers. Therefore we expressed a single copy of the mutant in E. coli, purified it to homogeneity, and compared its behavior in gel filtration analysis to that of the wild-type protein. Wild-type FKBP (Fig. 3) and the F36V mutant (not shown) migrated as a single peak of approximate molecular mass 10 kDa (the true molecular mass is 11.8 kDa). However, FM migrated substantially faster, with an apparent size that increased in a saturable manner from 1.2 to 1.8-fold that of wild type on increasing the concentration of the injected sample from 7 to 600 μM. By contrast the wild-type protein showed almost no shift in retention time over the same range of concentrations (data not shown). Also, at injection concentrations below 1 mg/ml, FM showed significant asymmetric peak broadening relative to wild-type FKBP (Fig. 3A). The concentration dependence and peak broadening are consistent with a monomer–dimer equilibrium (18, 19). Strikingly, addition of FK506 caused FM to migrate identically to the wild-type protein (Fig. 3B). These data demonstrate that the intracellular behavior can indeed be explained by a direct dimerization model.
Figure 3.
Ligand-reversible dimerization of purified FM monitored by HPLC gel filtration. Samples (50 μl) of wild-type FKBP (solid line) or FM (dotted line) at 0.75 mg/ml were injected onto a TSK G2000SW column, either alone (A) or after incubation with a 3-fold molar excess of FK506 (B). A280, absorbance measured at 280 nm.
Analytical Ultracentrifugation.
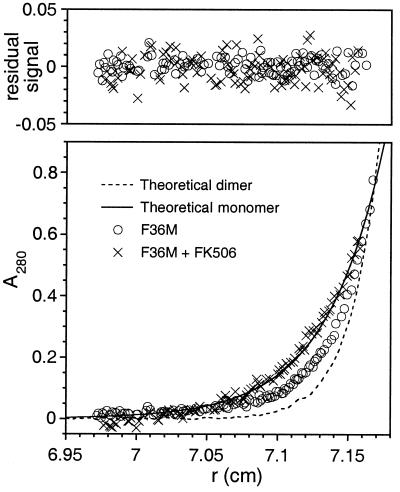
To confirm and quantitate the proposed dimerization event we subjected samples of the purified FM protein to analytical ultracentrifugation. Equilibrium data sets obtained with the protein alone were intermediate between theoretical traces that were calculated assuming ideal single species with molecular weights equal to one or two FM units (Fig. 4). However the data could be fitted with high confidence to an equation describing a monomer—dimer equilibrium. The calculated Kd for the monomer—dimer transition was 30 μM, with 95% confidence limits of 24 and 40 μM. A variety of other fitting models either returned results consistent with these fits or did not give stable solutions. No evidence of higher-order aggregation was seen in these experiments. Inclusion of a 5-fold molar excess of FK506 altered the sedimentation so that the data corresponded exactly with the predicted behavior of a unique monomeric species (molecular weight = 10,354, with 95% confidence limits of 9,292 and 11,404). These data demonstrate that FM indeed undergoes discrete and dynamic self-dimerization.
Figure 4.
Dimerization of purified FM quantitated by analytical ultracentrifugation. A representative data set, recorded at 50,000 rpm, is shown and provides curves typical of sedimented and equilibrated species. (Lower) Protein concentration (measured as A280) versus distance (r) from the center of the centrifuge rotor. The starting protein concentration was ≈18 μM (A280 ≈ 0.2). Experimental data points are shown for sedimentation of FM alone (circles) or in the presence of a 5-fold molar excess of FK506 (crosses). Also shown are simulated curves calculated assuming a completely monomeric (solid line) or dimeric (broken line) protein. (Upper) The residual signals, calculated as the difference between the experimental and fitted data, show no systematic errors, indicating that the fits are robust.
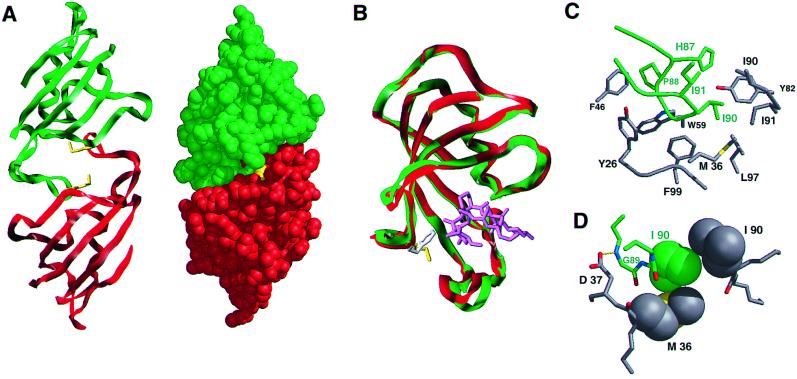
X-Ray Crystal Structure of the FM Dimer Complex.
To analyze in detail the basis for the self-interaction, we crystallized the homodimeric FM complex and determined the x-ray structure at 2-Å resolution (Fig. 5). The two proteins interact symmetrically in the manner of two clasping hands. The 80s loop of each protein engages the ligand-binding site of the other, providing an explanation for the inhibitory effects of ligands (Fig. 5A). However, despite the dramatic change in behavior, the tertiary structure of the FM subunit is almost identical to that of the wild-type protein (20) (root mean square deviation (rmsd) 1.24 Å for all atoms); the only appreciable shift is in the 80s loop, a region known to vary among FKBP structures (8) (Fig. 5B). The two monomers themselves are virtually superimposable (rmsd 0.82 Å for all atoms), although minor differences exist for some side chains, including Met-36.
Figure 5.
X-ray crystal structure of the FM homodimeric complex. (A) Overview of the complex in ribbon and space-filling formats. Monomer 1 [in the Protein Data Bank (PDB) file] is shown in red and monomer 2, in green. The Met-36 side chains are colored yellow. (B) Overlay of FM monomer 1 (green) with the structure of wild-type FKBP, PDB code 1FKF (20) (red), showing the minor 80s loop movement and change in disposition of Phe-36 (white) compared with Met-36 (yellow). The FK506 ligand in the wild-type structure is shown in magenta. (C) The dimerization interface. Side chains of monomer 1 are in gray and those of monomer 2 in green. Omitted from the foreground for clarity is Asp-37. (D) Quality of interaction between monomer side chains. van der Waals radii are shown for the side chains of Met-36 of monomer 1 and of Ile-90 of each protein; the hydrogen bond (2.94 Å) between Asp-37 Oδ2 and Gly-89 N is also shown. Ribbon figures were generated by using sybyl (Tripos Associates, St. Louis); all others were generated by using MidasPlus (25).
The interface between the two proteins is characterized by a remarkably extensive and complementary set of contacts suggestive of a bona fide protein–protein interaction rather than an artificial pairing. The key contacts are between the Pro-88–Ile-90 portion of the 80s loop of one protein and the hydrophobic binding pocket of the other (Fig. 5C). Pro-88 binds in a hydrophobic pocket formed by the side chains of Tyr-26 and Phe-46, and the Cα of Gly-89 packs tightly against the Phe-99 side chain and Gly-28. Further hydrophobic contacts are made by Ile-90, which directly contacts the Ile-90 side chain of the partner protein as well as Met-36 (Fig. 5D). Outside this hydrophobic core, there are several good electrostatic interactions, including a hydrogen bond between the Gly-89 main chain and Asp-37 (2.94 Å), and numerous water-bridged contacts. Thus the interaction strikingly resembles natural high-affinity protein–protein interfaces (21).
Although the two Met-36 side chains are close in space (5.9 Å), they are not in contact. However, they do contribute to the important binding pocket for the Ile-90 side chain of the partner protein (Fig. 5D), and modeling indicates that the presence of Phe-36 might preclude this contact (Fig. 5B). This result suggests that the F36M substitution may promote dimerization by subtly reshaping this hydrophobic pocket to relieve an inherent steric hindrance to intermolecular association.
Use of Reiterated FM Domains to Conditionally Aggregate Intracellular Proteins.
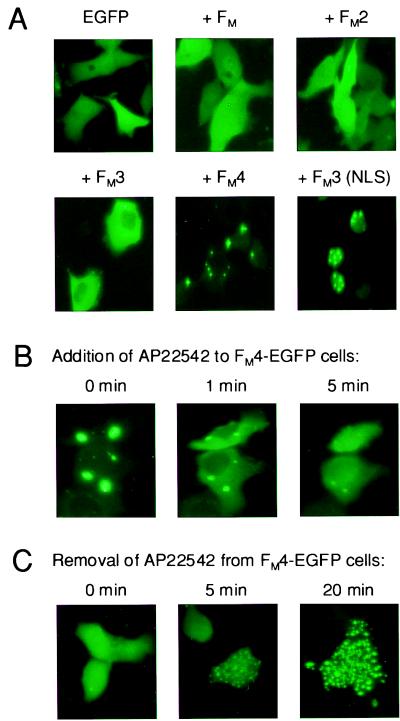
The biochemical and biophysical demonstration of FM self-dimerization suggests many uses as a protein–protein interaction tool. To test whether FM could be used to conditionally aggregate proteins inside cells, we engineered a series of fusion proteins with EGFP and tested their intracellular aggregation properties by transient expression in HT1080 cells followed by fluorescence analysis (Fig. 6). Proteins containing a single or two copies of the domain were evenly dispersed through the cytoplasm of transfected cells (Fig. 6A) similarly to unmodified EGFP. Addition of a third domain altered this appearance: several large fluorescent features were visible in most cells, presumably corresponding to aggregated fusion protein. Addition of a fourth domain dramatically increased the apparent level of aggregation: fluorescence was almost exclusively confined to a few (typically two to six) large, highly fluorescent sites. A protein with six FM domains behaved similarly (data not shown). We were also able to cause apparent aggregation specifically in the nucleus by adding a nuclear localization sequence to the fusion protein (Fig. 6A Lower Right). These observations indicate that multimerized FM domains can be used to aggregate heterologous proteins within cellular compartments, and that the degree of aggregation can be dictated by varying the number of domains fused. In the cytoplasm, maximal aggregation appears to occur with four FM domains, which leads to a dramatic sequestration into a few massive aggregates.
Figure 6.
Rapid, reversible aggregation of cytoplasmic EGFP fusion proteins containing FM domains. (A) Extent of aggregation depends on FM copy number. EGFP was transiently expressed in HT1080 cells as a fusion to 0–4 copies of FM, as indicated, and intracellular fluorescence was observed 20 h after transfection. The final panel shows nuclear aggregation of an EGFP-FM3 fusion protein expressed with a nuclear localization sequence (NLS). (B) Rapid dissolution of FM4-EGFP aggregates upon addition of ligand AP22542. At time 0, 2 μM AP22542 was added, and fluorescence was observed at the indicated time points. The same field of cells is shown in all three panels. (C) Rapid reaccumulation of FM4-EGFP aggregates after removal of ligand. Cells exposed to 2 μM AP22542 for 2 h were rapidly washed three times at time 0 to remove the compound. Each panel shows a different field of cells.
To assess whether such aggregates could be disrupted by ligand, we expressed the cytoplasmic EGFP fusion protein with four copies of FM and monitored intracellular fluorescence after addition of AP22542 (Fig. 6B). Within 1 min of adding ligand, substantial dispersion of fluorescence could be observed throughout the cytoplasm, concomitant with a decrease in the fluorescence of the aggregates. Within 5 min, the aggregates had almost completely disappeared. To determine the reversibility of this process, we exposed the same cells to AP22542 for several hours and then washed away the ligand (Fig. 6C). Within 5 min, most cells showed a striking punctate fluorescence, with several hundred aggregates apparently forming. By 20 min these strengthened in intensity and began to coalesce, and eventually reformed the large mega-aggregates seen in untreated cells (not shown). These data show that the dynamic monomer–dimer equilibrium observed in vitro can be exploited in vivo to obtain extremely rapid control over the oligomerization state of fusion proteins.
Discussion
We have discovered that a single point mutation converts the normally monomeric protein FKBP into a discrete dimer, both in vitro and when expressed in mammalian cells. Although the affinity is weak (Kd ≈ 30 μM), the interaction appears highly specific, because minimal interactions with the wild-type or F36V proteins can be detected (Fig. 1). Dimerization is abolished by adding ligand, and the kinetics of FM association and dissociation are extremely rapid—on the order of minutes (Fig. 6 B and C), presumably reflecting the efficient cellular penetrance of the ligands, the rapid dynamic equilibrium of the monomer–dimer transition, and the high ligand affinity. These properties allow the use of reiterated copies of FM as conditional aggregation domains (CADs), to drive high-avidity interactions between proteins of interest inside cells that can be abolished at will by adding cell-permeant ligands.
Potential Applications.
The FM-based reverse dimerization system represents a complementary technology to dimerizer approaches for controlling protein–protein associations, and it suggests two broad types of application. First, the system should be generally useful for rapidly turning off processes that are activated by oligomerization. We demonstrated the use here as an off switch for transcription (Fig. 2). FM fusions could be used analogously to create inhibitable alleles of any signaling protein that is activated by protein–protein associations. The extent of association in the absence of ligand could be controlled by choice of the number of FM domains; or fusions to a single domain could be used to uniquely drive dimerization, albeit with low affinity. We showed here that FM-driven interactions can be achieved in the nucleus or cytoplasm by appropriate use of targeting sequences (Fig. 6A). Fusion proteins could also be targeted to the plasma membrane by appending myristoylation motifs (3, 22).
A second general use for the system is to rapidly turn on processes that are inactivated by oligomerization. We showed here that massive aggregates of EGFP-FM fusion proteins can be rapidly created and dissolved inside cells (Fig. 6 B and C). Such aggregation can be used to sequester an FM fusion partner in an inactive form, allowing rapid activation by adding ligand (data not shown), analogously to the use of steroid receptor fusions (23). An extension of this approach is to use aggregation to conditionally target a protein to a particular cellular compartment. We have found that adding a secretion signal sequence to FM fusion proteins results in accumulation of aggregated protein exclusively in the endoplasmic reticulum (ER), suggesting a way to bring protein secretion under small molecule control (24). A protein of interest is fused to multiple FM domains, leading to aggregation and consequently retention in the ER. Addition of ligand rapidly dissolves the aggregates and releases the protein into the secretory pathway. The FM domains are then removed from the protein by virtue of a cleavage site for the trans-Golgi protease furin that is interposed between them and the protein of interest. This system is much faster than transcriptional regulatory systems for controlling protein production; we have used regulated secretion of insulin to transiently correct glucose levels in a mouse model of hyperglycemia (24).
Structural Considerations.
The discrete quaternary structural change elicited by the F36M mutation is remarkable and, to our knowledge, unprecedented. The crystal structure suggests that, rather than being “actively” caused by the Met-36 substitution, dimerization is actually a latent property of FKBP that is sterically prevented by the wild-type Phe-36 side chain. More mutagenesis and structural work will likely be required to confirm this model. However, the striking and previously unappreciated self-complementarity of the FKBP binding site raises the question of whether FKBP self-association may occur naturally. No known natural FKBP sequence has methionine at position 36 (6), and our data (Figs. 1C and 3), along with a wealth of experience with dimerizer systems, show that the wild-type and F36V proteins do not self-interact. However, the property of dimerization may not be restricted to FM: we have preliminary evidence that another mutation at a different residue in the binding site (W59V; see Fig. 5C) also provokes dimerization (A. Golden, V.M.R. and T.C., unpublished results). Together with the crystal structure, this suggests that a subset of FKBP variants may be prone to self-interaction, and that some of these may exist naturally. Although the physiological significance is unclear, the structure represents another example of the extraordinary versatility of the FKBP binding site, which can accommodate small molecules, linear peptides, and discontinuous protein epitopes of highly diverse structure (8, 9).
The crystal structure also provides a rational framework for designing conditional aggregation domains with improved properties. One attractive goal would be to increase the affinity of the interaction to reduce the number of FM domains required to obtain complete aggregation, or to allow discrete dimerization to be driven with high affinity by fusion with single copies of FM. It may also be possible to engineer directionality into the interaction, for example by inserting a steric “bump” on one protein and a compensating “hole” on the other, to produce a system for ligand-reversible heterodimerization of proteins.
Acknowledgments
T.C. and D.N.W. dedicate this work to the memory of Cara Marks. We thank Lauren Stevenson for her early work on FM, Tom Phillips for E. coli fermentations, Xiaode Lu for help with crystallization, and Surinder Narula and David Dalgarno for graphics assistance. D.N.W. thanks the Biotechnology and Biological Sciences Research Council of the U.K. for a grant (JEI09469) to purchase a Beckman XL-I analytical ultracentrifuge. FM ligands and FM-containing constructs are being distributed to the academic community for research purposes and can be requested through our website at www.ariad.com.
Abbreviations
- FKBP
12-kDa human FK506-binding protein
- FM
FKBP mutant with Phe-36 replaced by Met
- EGFP
enhanced green fluorescent protein
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1EYM).
See commentary on page 6921.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100101997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100101997
References
- 1.Klemm J D, Schreiber S L, Crabtree G R. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 2.Spencer D M. Trends Genet. 1996;12:181–187. doi: 10.1016/0168-9525(96)10013-5. [DOI] [PubMed] [Google Scholar]
- 3.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 4.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 5.Rivera V M, Clackson T, Natesan S, Pollock R, Amara J F, Keenan T, Magari S R, Phillips T, Courage N L, Cerasoli F, Jr, et al. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 6.Kay J E. Biochem J. 1996;314:361–385. [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber S L. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- 8.Clardy J. Proc Natl Acad Sci USA. 1995;92:56–61. doi: 10.1073/pnas.92.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse M, Chen Y G, Massague J, Kuriyan J. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 10.Amara J F, Clackson T, Rivera V M, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage N L, Holt D A, Gilman M. Proc Natl Acad Sci USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clackson T, Yang W, Rozamus L W, Hatada M, Amara J F, Rollins C T, Stevenson L F, Magari S R, Wood S A, Courage N L, et al. Proc Natl Acad Sci USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Rozamus L W, Narula S, Rollins C T, Yuan R, Andrade L J, Ram M K, Phillips T B, van Schravendijk M R, Dalgarno D C, et al. J Med Chem. 2000;43:1135–1142. doi: 10.1021/jm9904396. [DOI] [PubMed] [Google Scholar]
- 13.Pollock R, Rivera V M. Methods Enzymol. 1999;306:263–281. doi: 10.1016/s0076-6879(99)06017-6. [DOI] [PubMed] [Google Scholar]
- 14.Rivera V M. Methods. 1998;14:421–429. doi: 10.1006/meth.1998.0596. [DOI] [PubMed] [Google Scholar]
- 15.Attar R M, Gilman M Z. Mol Cell Biol. 1992;12:2432–2443. doi: 10.1128/mcb.12.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomerantz J L, Sharp P A, Pabo C O. Science. 1995;267:93–96. doi: 10.1126/science.7809612. [DOI] [PubMed] [Google Scholar]
- 17.Yang T T, Cheng L, Kain S R. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ackers G K. In: The Proteins. Neurath H, Hill R L, editors. Vol. 1. New York: Academic; 1975. pp. 1–94. [Google Scholar]
- 19.Stevens F J. Biochemistry. 1986;25:981–993. doi: 10.1021/bi00353a006. [DOI] [PubMed] [Google Scholar]
- 20.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 21.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 22.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picard D, Salser S J, Yamamoto K R. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 24.Rivera V M, Wang X, Wardwell S, Courage N L, Volchuk A, Keenan T, Holt D A, Gilman M, Orci L, Cerasoli F, et al. Science. 2000;287:826–830. doi: 10.1126/science.287.5454.826. [DOI] [PubMed] [Google Scholar]
- 25.Ferrin T E, Huang C L, Jarvis L E, Langridge R. J Mol Graph. 1988;6:13. [Google Scholar]