Abstract
Many annelids, including the earthworm Lumbricus terrestris, have giant cooperative respiratory proteins (molecular masses greater than 3.5 million Da) freely dissolved in the blood, rather than packaged in cells. These complexes, termed either erythrocruorins or hemoglobins, are assembled from many copies of both hemoglobin subunits and nonhemoglobin or “linker” subunits. In this paper, we present the crystal structure of Lumbricus erythrocruorin at 5.5-Å resolution, which reveals a remarkable hierarchical organization of 144 oxygen-binding hemoglobin subunits and 36 nonhemoglobin linker subunits. The hemoglobin chains arrange in novel dodecameric substructures. Twelve trimeric linker complexes project triple-stranded helical coiled-coil “spokes” toward the center of the complex; interdigitation of these spokes appears crucial for stabilization. The resulting complex of linker chains forms a scaffold on which twelve hemoglobin dodecamers assemble. This structure specifies the unique, self-limited assemblage of a highly cooperative single molecule.
Assembly of protein subunits into large complexes is an important mechanism used to attain greater efficiency and regulatory control of biological processes. Annelid erythrocruorins pose key problems in the design of such large macromolecular assemblages. The extracellular nature and giant size of these molecules have made them ideal systems for a number of seminal investigations into protein structure. Lumbricus erythrocruorin was the first protein ever reported to be crystallized, in 1840 (1), one of the first subjects of Svedberg's early ultracentrifugation experiments (2), and an early molecular subject of electron microscopic analysis (3).
Lumbricus erythrocruorin displays a double-layered hexagonal shape with overall molecular D6 symmetry, as revealed by both crystallographic analysis (4) and electron microscopy (5). Although its exact stoichiometry has been the subject of debate (6, 7), there is agreement that the giant complex comprises multiple copies of four different types of hemoglobin chains and four different types of nonhemoglobin “linker” chains. Because there are at least 180 polypeptide chains in the whole molecule, Lumbricus erythrocruorin has been proposed to be arranged in a hierarchy of symmetry (8). Befitting their large size, annelid erythrocruorins are highly cooperative, with binding properties that depend on the interplay of divalent cations and protons (9–11).
We present in this paper the crystal structure of Lumbricus erythrocruorin at 5.5-Å resolution. Our analysis reveals a remarkable arrangement of 180 subunits organized in an intricate hierarchical arrangement of symmetries and quasi-symmetries. This observed subunit arrangement dictates the self-limited assembly and underlies cooperative function.
Experimental Procedures
Crystallization.
Crystals of Lumbricus erythrocruorin were grown in the CO-liganded state directly from the blood of earthworms that had been concentrated by a factor of 2. The orthorhombic crystals grew from 1.8 M phosphate at pH 7.4 as described previously (12). The triclinic crystals used in this analysis were grown at 4°C by vapor diffusion against 0.2 M sodium sulfate/20% glycerol/10% polyethylene glycol 4000/0.1 M Tris, pH 8.5.
Data Collection.
Diffraction data from the high-salt crystal form were collected on x-ray film at Stanford Synchrotron Radiation Laboratory as described (4). Data collection from the triclinic crystals required crosslinking in 0.1% glutaraldehyde for 2 min before freezing in the nitrogen stream. Diffraction data were collected on a MarResearch 30-cm Image Plate system and processed with the denzo/hkl package (13). Crystal parameters and data statistics are given in Table 1.
Table 1.
Crystallographic data
| Crystal form | a, Å | b, Å | c, Å | α,° | β,° | γ,° | Space group | Z | dmin, Å | Measured reflections | Unique reflections | Completeness, % | Rmerge, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Triclinic, low-salt | 188.1 | 265.7 | 446.4 | 89.4 | 97.6 | 92.3 | P1 | 2 | 7.25 | 319,403 | 115,298 | 95.1 | 12.7 |
| Orthorhombic, high-salt | 502.1 | 297.8 | 350.1 | 90.0 | 90.0 | 90.0 | C2221 | 1/2 | 5.5 | 256,872 | 75,535 | 85.4 | 12.9 |
Z is the number of molecules per asymmetric unit.
Phasing.
The approach used to solve the phase problem was to use models based on a cryo-electron microscopic image (14) to phase data at very low resolution, followed by molecular averaging to extend phasing to higher resolution. This approach was successful in the low-salt triclinic crystals, although similar attempts in the high-salt orthorhombic form were not. In retrospect, this outcome is probably attributable to poor quality of the very low (40–15 Å) diffraction data in the high-salt crystals resulting from low contrast between protein electron density and solvent electron density.
Starting orientations in the triclinic unit cell were determined by self-rotation functions calculated with the program glrf (15) that revealed similar orientations for the two molecules. Approximate body centering was evident from analysis of the diffraction data corresponding to spacings greater than 35 Å, providing an initial estimate of the vector between molecules. The orientation and positions for the two molecules were improved by systematic searches. Calculated phases were improved by 12 cycles of 24-fold molecular averaging at 26-Å resolution with the rave package of programs (16). Phases then were extended by small increases in resolution, using eight cycles of 24-fold averaging in each of 48 resolution steps to 7.25 Å. Improvement of the noncrystallographic matrices and the mask envelope was required at several steps.
The 7.25-Å electron density map from this process then was used to phase the diffraction data in the orthorhombic form. Self-rotation function calculations on the orthorhombic crystal form previously had revealed the molecular orientation in which a molecular dyad was coincident with the crystallographic twofold along the a axis (4). Electron microscopic analysis of sectioned crystals indicated that the molecular center was between 5 and 10 Å from x = 1/4a.¶ Placement of the density corresponding to one-half of a molecule from the triclinic structure showed a best fit when the molecular center was 5 Å from x = 1/4a (R = 47.3%; correlation coefficient = 0.272). Multiple crystal averaging between the 24 molecular asymmetric units in the triclinic cell and 6 molecular asymmetric units in the orthorhombic cell led to a improved map in which myoglobin-folded subunits were easily discerned. This map allowed construction of an improved mask for the 1/12th subunit and subsequent sixfold averaging in the orthorhombic form to 5.5 Å.
The initial 5.5-Å map was quite clear for hemoglobin dodecamers, but less so for the linker complex. To investigate the structure of the linker complex, a mask that covered all 18 linker subunits (per half molecule) and extended to the molecular center was used to perform proper sixfold averaging within the linker region, while using a separate dodecamer mask. This map revealed the long coiled-coil region that had been lost because of the earlier masking and allowed construction of an improved mask for a 1/12th subunit. After averaging with this new mask, the additional local 3-fold symmetry within the dodecamer region was incorporated allowing 18-fold averaging between hemoglobin subunits and 6-fold averaging within the linker region. Eight cycles of averaging in this way led to a final R factor of 21.9% and a correlation coefficient of 0.896 between the observed structure factors and those calculated from this map.
Results and Discussion
Structure Determination.
Two different crystal forms were used in our analysis of Lumbricus erythrocruorin, an orthorhombic form that grew from high-salt solutions and a triclinic form grown in low-salt. Initial phasing at 26-Å resolution in the low-salt crystal form was based on a model constructed from a cryo-electron microscopic image reconstruction (14). Phases then were extended from 26 Å to 7.25 Å by 24-fold molecular averaging between the two molecules in the asymmetric unit, with the resulting electron density used to phase the data from the orthorhombic crystal form. Molecular averaging between these two crystal forms followed, and phasing was extended to 5.5 Å in the orthorhombic crystal form.
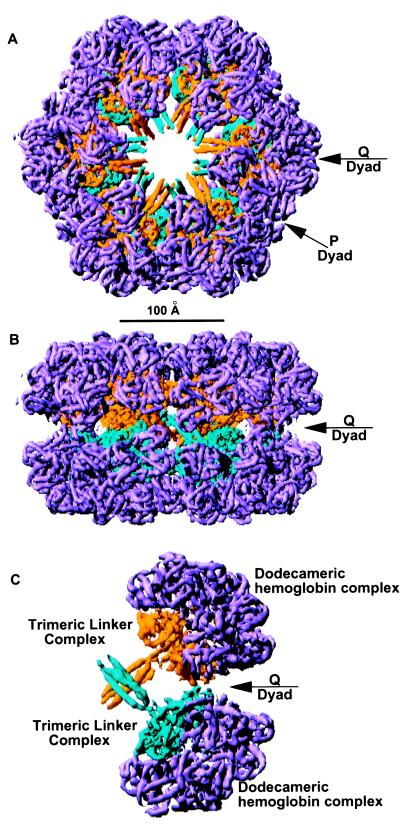
The overall electron density corresponding to one molecule is shown in Fig. 1. The map quality is excellent and unambiguously reveals all 144 hemoglobin subunits in the complex. The dimensions of the whole molecule are approximately 285 Å by 260 Å by 180 Å. The hemoglobin subunits occupy the surface of the complex (magenta portions in Fig. 1), whereas the linker chains occupy the interior of the molecule. This observed arrangement is consistent with a central role played by linker chains in the assembly of the entire complex (7, 17). The fundamental molecular unit, 1/12th of the whole molecule, comprises a dodecamer of hemoglobin subunits and a mushroom-shaped trimeric linker complex (Fig. 1C).
Figure 1.
Lumbricus erythrocruorin. The 5.5-Å electron density of Lumbricus erythrocruorin is represented at the 1σ level. Density corresponding to the hemoglobin chains is depicted magenta, the linker chains associated with the top of the molecule are shown in yellow, while those associated with the bottom linker subunits are shown in blue. (The scale bar refers to A and B.) (A) Whole molecule viewed down the molecular sixfold axis. (B) Whole molecule viewed along the P dyad, with sixfold axis vertical. (C) Pair of 1/12th subunits in the same orientation as in B. This, and subsequent figures, were produced with ribbons (32).
Dodecameric Hemoglobin Assemblage.
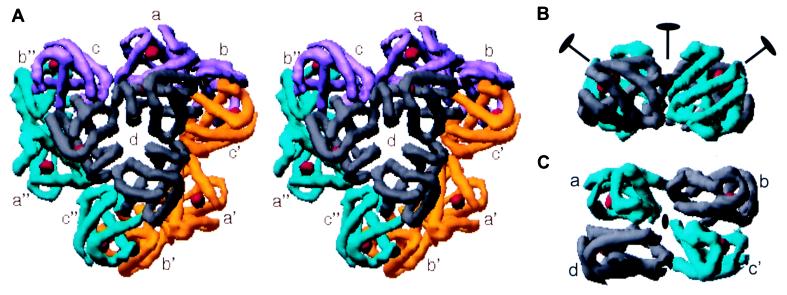
All hemoglobin subunits in a dodecamer have the classic myoglobin fold, the highly helical structures that can be traced easily in the electron density map. (The myoglobin fold is composed of seven or eight α-helices, with helices designated A, B, . . . H, that are connected by corners AB, BC, . . . GH.) Although the four types of hemoglobin chains, present in equal proportions, have at most 44% pairwise sequence identity (18), these subunits are indistinguishable in the map (correlation coefficients > 0.8). The dodecameric hemoglobin complex is assembled in a highly symmetrical arrangement. Three tetramers are related by a local threefold axis of symmetry (Fig. 2A). Subunits within each tetramer are related by a set of quasi-dyad axes arranged in a manner reminiscent of a molluscan tetrameric hemoglobin (19). One quasi-dyad axis, inclined by 56° from the threefold axis, applies to the entire tetramer. Two subordinate quasi-dyad axes, which intersect the tetramer dyad at angles of 54°, apply only to dimeric pairs of hemoglobin subunits (Fig. 2 B and C). Thus, the familiar D2 symmetry of mammalian tetrameric hemoglobin is not found here. The interfaces within these dimers, which are the most extensive in the assemblage, feature contacts between the heme-encapsulating E and F helices similar to those observed in cooperative dimers from both molluscs (20) and echinoderms (21). These dimeric contacts presumably also contribute to the observed cooperative oxygen binding in annelid erythrocruorins, but other contacts in this hierarchically symmetric dodecamer are required to explain the high level of cooperativity. A fit of oxygen-binding data from Lumbricus erythrocruorin at maximum cooperativity (n = 7.8) suggested up to 12 interacting heme groups (11).
Figure 2.
Hemoglobin dodecamer complex. (A) Stereo view oriented down local threefold with labels indicating the tentative identification of subunits. Density corresponding to d chains is depicted in gray, while each of the three trimers is shown in a unique color. The course of the myoglobin fold of each subunit is evident in this map, as are the heme positions that appear as isolated red spheres of density. Note how each subunit is involved in an extensive interaction using its E and F helices. (B) Tetrameric hemoglobin complex corresponding to abc′d with tetrameric quasi-dyad (vertical) and dimeric quasi-dyads indicated by ovals with lines. (C) Tetramer abc′d viewed down the tetrameric quasi-dyad.
Although the different types of hemoglobin subunits (termed a, b, c, and d) cannot be distinguished by density features, probable assignments can be made from the known covalent linkages. Disulfide bridges link subunits a, b, and c into a trimer. Cysteines near their amino termini link subunits a and c, whereas cysteines at the GH corners link subunits a and b (18). Given the arrangement of subunits in the dodecamer (Fig. 2), two possible assignments meet these constraints. The more probable of these assignments is indicated by the labels in Fig. 2. This assignment places the monomeric d subunits closest to the threefold axis, which is consistent with results from an electron microscopic investigation of erythrocruorin molecules reconstituted in the absence of d subunits (22). The abc trimers are then extended in an almost linear array that links two adjacent tetramers. Subunits labeled a and b are abutted with density apparent for the disulfide bridge between them. Subunits a and c have their amino-terminal regions near each other, although density for this disulfide bridge is not clear in the map. An alternative assignment, possible because of symmetry within the tetramers, would have abcd, respectively, in the places shown in Fig. 2 for c′da′b. Lack of density at either putative disulfide bridge and inconsistency with the electron microscopic results make this a less likely alternative.
Linker Scaffold Complex.
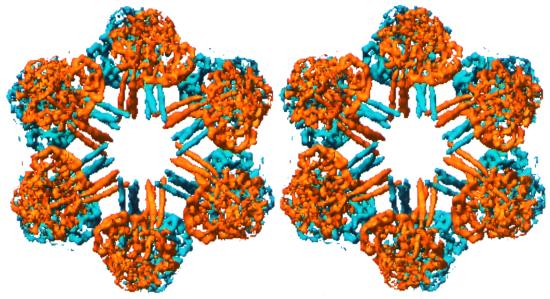
A most striking and unexpected aspect of this structure is the interdigitation of 12 triple-stranded coiled-coil helices near the center of the complex, as illustrated in Fig. 3. Emanating from each 1/12th unit are three 45-Å long rods consistent with coiled-coil α-helices. Analysis of the four known linker sequences (ref. 23 and A. F. Riggs, personal communication), both manually and by using the program multicoil (24), indicates the potential of 50-residue segments near the amino terminus to form triple-stranded coiled-coils. Moreover, there are no cysteine residues in this segment of any linker sequence from which disulfide bridges could form to preclude coiled-coil formation, whereas there are a number of cysteine residues in the carboxyl-terminal region. The simplest explanation for our observed electron density is that there are three linker chains within a 1/12th unit, each of which contributes one long amino-terminal helix to the observed coiled-coil. Twelve such trimeric linker complexes then are associated in a whole molecule, with each projecting a triple-stranded coiled-coil “spoke” toward the molecular center. The extensive interdigitation of the six spokes from the upper half with those of the lower half (Fig. 3) provides an explanation for the absence of half-molecules among observed subassemblies in sedimentation data of annelid erythrocruorins (8).
Figure 3.
Stereo view of complex of 36 linker chains viewed along molecular sixfold axis. The density corresponding to the 18 linker chains associated with the top half of the molecule is depicted in yellow, whereas the bottom 18 linker chains are shown in blue. Note the extensive interdigitation of the triple-stranded coiled-coils near the molecular center.
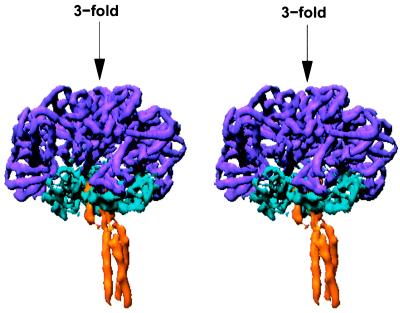
The linker chain spokes are connected to their globular head portions in a disjointed, asymmetric manner (Fig. 4). The 45-Å long coiled-coil spoke density is offset from a second, 20-Å long coiled-coil within the head portion of the linker complex in a 1/12th unit. Density within the linker head portion, including the 20-Å coiled-coil core at its center, is related by a quasi-triad axis of symmetry that essentially is coincident (within 2°) with the nearly perfect threefold axis that relates hemoglobin subunits within the dodecamer complex. The quasi-triad axis that relates the three α-helices in the coiled-coil spoke is inclined by 9° and displaced by 7 Å from the quasi-triad axis relating the rest of the 1/12th subunit. The density is clear for interhelical connections for two of the helix pairs uniting the 45-Å and 20-Å coiled-coils, whereas that for the third connection, possibly in an extended conformation, is unclear. Because of this disjointed connectedness, the three linker chains can be distinguished from one another. The quasi-triad superposition among linker heads has correlation coefficients (between 0.55 and 0.6) that are lower than those between hemoglobin subunits, which may reflect real differences between three linker chains of distinct sequences.
Figure 4.
Stereo view illustrating local symmetry within a 1/12th subunit. The density corresponding to the dodecameric hemoglobin complex is shown in magenta, the density corresponding to both the long and short triple-stranded coiled-coils is shown in yellow, and the remainder of the linker density is depicted in blue. The approximate local threefold that relates the hemoglobin subunits and the globular portion of the linker complex is vertical. Note the sharp break in symmetry axis between the short and long coiled-coils.
Apart from the central coiled-coil, the density in the linker head portion has no helix-like rods and the chains cannot be traced. Linker chains, however, are known from sequence analyses to contain a segment of approximately 40 residues that is highly homologous with the cysteine-rich low-density lipoprotein (LDL) receptor domain (23). This segment follows immediately after the amino-terminal segment that we now identify with the α-helical coiled-coils. Details of how this domain is involved in the assembly of Lumbricus erythrocruorin will require higher resolution data, but the density features are compatible with a possible location near the carboxyl terminus of the 20-Å coiled-coil, based on the known crystal structure of an LDL receptor domain (25). Reasonable boundaries also can be defined for the density associated with the remaining portion of the linker chains, which must correspond to the ≈125-residue disulfide-bridged carboxyl-terminal domain that can be imputed from the sequences. The carboxyl-terminal region of each chain, which includes both the LDL-related domain and the carboxyl-terminal domain, forms contacts with hemoglobin subunits b and c, and more tenuously with a. This interface within the 1/12th unit necessarily involves a mismatch between the symmetric dodecameric hemoglobin shell and the sequence-distinctive linker chains in the complex.
Symmetrical Arrangement of Lumbricus Erythrocruorin.
The association of protomers (1/12th units) related by the molecular D6 symmetry in Lumbricus erythrocruorin involves three unique interfaces. There are lateral contacts within each ring between adjacent protomers related by the sixfold axis, and protomers within one ring are related to those in the apposed ring by dyad axes of two kinds, designated P and Q (Fig. 1). The assemblage is specified by hemoglobin:hemoglobin and linker-head:linker-head contacts as well as the central interdigitation of linker spokes. Lateral contacts between sixfold-related adjacent protomers are composed entirely from hemoglobin contacts. The inclination of local threefold axes by approximately 45° from the sixfold axis, as first identified by cryo-electron microscopic reconstructions (14), is such that subunits a and b from each protomer interact with a" and c", respectively, from an adjacent one (see Fig. 2A). The Q-dyad interface involves a two-helix to two-helix contact between related linker spokes (Fig. 1C), crossing at 90°, and intimate contacts between carboxyl-terminal domains of the Q-related third linker chains, but no contacts between hemoglobin subunits. The intriguing break in local symmetry between the linker chain spokes and globular heads is essential for this arrangement. The P-dyad interface is less extensive. It involves the third helices of apposed spokes interacting with one another and also with one of the other two helices (Fig. 3) and somewhat distant contacts of hemoglobin subunits b′ with one another and of each with the b subunits of sixfold-related protomers in opposite rings. There are no P contacts between linker-head domains.
The assemblage of 180 subunits of Lumbricus erythrocruorin into the molecule that we observe involves an intricate hierarchy of symmetries and quasi-symmetries among at least seven structurally distinct subunits. Despite the lack of atomic detail at this resolution, one can nevertheless begin to consider how this structural hierarchy impacts on aspects of macromolecular assembly and biological function. Giant molecules are needed in extracellular respiratory proteins for vascular retention and for adequate oxygen capacity at a manageable osmotic pressure, but this size demand poses issues for a spontaneous assembly into a unique, self-limited molecule with suitable genetic economy. Separation of the oxygen-binding function of erythrocruorin from much of the assembly specification is an important mechanistic ingredient in this case and is served by the surprising involvement of heterotrimeric coiled-coils. Coiled-coil structures are found to play key roles in assembly of transcription factors (26, 27), membrane fusion (28–30), and cytoskeletal assembly (31); here, it is an intricate set of interactions between coiled-coils that are exploited. The structure gives witness to the need for distinctive linker chains to specify the assemblage. What roles there might be for four kinds of linker chains is less clear. The existence of stable four-protomer dissociation products (8) may offer a clue. Barring rearrangements, such would be unexpected from D6 symmetry, but an alteration of two kinds of 1/12th units (perhaps containing L1:L2:L3 and L1:L2:L4) could generate quasi-dyad P axes and only true D3 symmetry. What underlies the requirement for distinct hemoglobin sequences also is unclear in light of the very similar structures they achieve. It may be that the plethora of interactions in the hierarchy of the assembly, including cooperative interfaces within the dodecamer, contacts with the linker chains, and interprotomer contacts in the assemblage, require the specialization afforded by four kinds of hemoglobin subunits.
Acknowledgments
We thank W. Love and E. Lattman for advice and encouragement in the early stages of this project, R. Craig for assistance with the electron microscopy of crystal sections, and A. Riggs for many helpful discussions and for sharing the linker sequences before publication. This work was supported by National Institutes of Health Grant DK43323 to W.E.R. and by an American Heart Association Established Investigator Award to W.E.R.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Strand, K., Craig, R. W. & Royer, W. E., Jr. (1993) Biophys. J. 64, A63 (abstr.).
References
- 1.McPherson A. Crystallization of Biological Macromolecules. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 2.Svedberg T, Ericksson-Quensel I B. J Am Chem Soc. 1933;55:2834–2841. [Google Scholar]
- 3.Roche J, Bessis M, Thiery J P. Biochim Biophys Acta. 1960;41:182–184. doi: 10.1016/0006-3002(60)90397-8. [DOI] [PubMed] [Google Scholar]
- 4.Royer W E, Jr, Hendrickson W A. J Biol Chem. 1988;263:13762–13765. [PubMed] [Google Scholar]
- 5.Boekema E J, van Heel M. Biochim Biophys Acta. 1989;957:370–379. doi: 10.1016/0167-4838(88)90228-2. [DOI] [PubMed] [Google Scholar]
- 6.Martin P D, Kuchumov A R, Green B N, Oliver R W-A, Braswell E H, Wall J W, Vinogradov S N. J Mol Biol. 1996;255:154–169. doi: 10.1006/jmbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Ownby D W, Riggs C K, Nolasco N J, Stoops J K, Riggs A F. J Biol Chem. 1996;271:30007–30021. doi: 10.1074/jbc.271.47.30007. [DOI] [PubMed] [Google Scholar]
- 8.Hendrickson W A, Royer W E., Jr Biophys J. 1986;49:177–189. doi: 10.1016/S0006-3495(86)83633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuneshige A, Imai K, Hori H, Tyuma I, Gotoh T. J Biochem (Tokyo) 1989;106:406–417. doi: 10.1093/oxfordjournals.jbchem.a122866. [DOI] [PubMed] [Google Scholar]
- 10.Santucci R, Chiancone E, Giardina B. J Mol Biol. 1984;179:713–727. doi: 10.1016/0022-2836(84)90163-3. [DOI] [PubMed] [Google Scholar]
- 11.Fushitani K, Imai K, Riggs A F. J Biol Chem. 1986;261:8414–8423. [PubMed] [Google Scholar]
- 12.Royer W E, Jr, Hendrickson W A, Love W E. J Mol Biol. 1987;197:149–153. doi: 10.1016/0022-2836(87)90618-8. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Schatz M, Orlova E V, Dube P, Jager J, vanHeel M. J Struct Biol. 1995;114:28–40. doi: 10.1006/jsbi.1995.1003. [DOI] [PubMed] [Google Scholar]
- 15.Tong L A, Rossmann M G. Acta Crystallogr A. 1990;46:783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- 16.Kleywegt G J, Jones T A. In: From First Map to Final Model. Bailey S, Hubbard R, Waller D A, editors. Warrington, U.K.: Science and Engineering Research Council Daresbury Lab.; 1994. pp. 59–66. [Google Scholar]
- 17.Kuchumov A R, Taveau J-C, Lamy J N, Wall J S, Weber R E, Vinogradov S N. J Mol Biol. 1999;289:1361–1374. doi: 10.1006/jmbi.1999.2825. [DOI] [PubMed] [Google Scholar]
- 18.Fushitani K, Matsuura M S A, Riggs A F. J Biol Chem. 1988;263:6502–6517. [PubMed] [Google Scholar]
- 19.Royer W E, Jr, Heard K S, Harrington D J, Chiancone E. J Mol Biol. 1995;253:168–186. doi: 10.1006/jmbi.1995.0543. [DOI] [PubMed] [Google Scholar]
- 20.Royer W E., Jr J Mol Biol. 1994;235:657–681. doi: 10.1006/jmbi.1994.1019. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell D T, Kitto G B, Hackert M L. J Mol Biol. 1995;251:421–431. doi: 10.1006/jmbi.1995.0445. [DOI] [PubMed] [Google Scholar]
- 22.de Haas F, Kuchumov A, Traveau J-C, Boisset N, Vinogradov S N, Lamy J N. Biochemistry. 1997;36:7330–7338. doi: 10.1021/bi970131l. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, Riggs A F. J Biol Chem. 1993;268:13548–13555. [PubMed] [Google Scholar]
- 24.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fass D, Blacklow S, Kim P S, Berger J M. Nature (London) 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 26.O'Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 27.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 28.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 29.Singh M, Berger B, Kim P S. J Mol Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton R B, Fasshauer D, Jahn R, Brunger A T. Nature (London) 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 31.Faix J, Steinmetz M, Boves H, Kammerer R, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerish G. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- 32.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]