Abstract
Many eukaryotic proteins are anchored to the cell surface via glycosylphosphatidylinositol (GPI), which is posttranslationally attached to the carboxyl-terminus by GPI transamidase. The mammalian GPI transamidase is a complex of at least four subunits, GPI8, GAA1, PIG-S, and PIG-T. Here, we report Chinese hamster ovary cells representing a new complementation group of GPI-anchored protein-deficient mutants, class U. The class U cells accumulated mature and immature GPI and did not have in vitro GPI transamidase activity. We cloned the gene responsible, termed PIG-U, that encoded a 435-amino-acid hydrophobic protein. The GPI transamidase complex affinity-purified from cells expressing epitope-tagged-GPI8 contained PIG-U and four other known components. Cells lacking PIG-U formed complexes of the four other components normally but had no ability to cleave the GPI attachment signal peptide. Saccharomyces cerevisiae Cdc91p, with 28% amino acid identity to PIG-U, partially restored GPI-anchored proteins on the surface of class U cells. PIG-U and Cdc91p have a functionally important short region with similarity to a region conserved in long-chain fatty acid elongases. Taken together, PIG-U and the yeast orthologue Cdc91p are the fifth component of GPI transamidase that may be involved in the recognition of either the GPI attachment signal or the lipid portion of GPI.
INTRODUCTION
Various eukaryotic proteins are anchored to the cell surface membrane by glycosylphosphatidylinositol (GPI). GPI is synthesized by stepwise additions of sugars and other components to phosphatidylinositol in the endoplasmic reticulum (ER). A common backbone of GPI is conserved in yeast, protozoa, plants, and mammals. Precursor proteins destined to be GPI-anchored have two signal sequences: the N-terminal signal for translocation across the ER membrane and the C-terminal signal for attachment of the GPI anchor. When the precursor proteins are translocated into the lumen of the ER, the GPI transamidase recognizes the GPI attachment signal sequence, cleaves it, and generates an enzyme-substrate protein intermediate linked by a thioester bond. An amino group of the terminal ethanolamine of the GPI backbone attacks the thioester in the intermediate to complete the GPI anchoring.
The GPI transamidases of humans and Saccharomyces cerevisiae are complexes of at least four proteins, GAA1, GPI8, PIG-S, and PIG-T in humans and Gaa1p, Gpi8p, Gpi17p, and Gpi16p in yeast (Fraering et al., 2001; Ohishi et al., 2001). PIG-S and Gpi17p, and PIG-T and Gpi16p are orthologous to each other, respectively (Fraering et al., 2001; Ohishi et al., 2001). All the proteins are essential for GPI transamidase as shown by their mutant cells (Yu et al., 1997; Ohishi et al., 2000, 2001). GPI8/Gpi8p are most likely catalytic subunits because they have homology to the cysteine proteases of the C13 family (Benghezal et al., 1996; Meyer et al., 2000; Ohishi et al., 2000) and GPI8 associates with substrate proteins (Spurway et al., 2001; Vidugiriene et al., 2001). Thus, mutations in the cysteine and histidine residues of the putative active sites render GPI8/Gpi8p nonfunctional (Meyer et al., 2000; Ohishi et al., 2000). The recombinant GPI8 protein of protozoa, Trypanosoma brucei cleaved a synthetic peptide acetyl-S-V-L-N-aminomethyl-coumarine (Kang et al., 2002). PIG-T/Gpi16p are critical for the formation of the enzyme complex because the stable expressions of GPI8/Gpi8p are dependent upon PIG-T/Gpi16p (Fraering et al., 2001; Ohishi et al., 2001) and the expression of GAA1 is dependent on PIG-T (Ohishi et al., 2001). The roles of GAA1 and PIG-S/Gpi17p have yet to be clarified.
Aerolysin is a cytolytic toxin secreted by the Gram-negative bacterium Aeromonas hydrophila (Buckley, 1999). Aerolysin, secreted as proaerolysin, binds to GPI-anchored proteins on target cells, such as Thy-1, contactin, and erythrocyte aerolysin receptor, becomes active upon proteolysis by the cell-surface protease and lyses the cell by forming pores (Abrami et al., 2000). Mutant cells defective in GPI biosynthesis are resistant to aerolysin because of a lack of receptors (Abrami et al., 2001). We have been using aerolysin as a tool to isolate mutant cells defective in the biosynthesis of GPI-anchored proteins. Here, we report the isolation of new GPI transamidase mutant cells, termed class U cells, and the cloning of the gene responsible, PIG-U. We demonstrate that PIG-U is the fifth subunit of the GPI transamidase complex and that S. cerevisiae Cdc91p is the orthologue of PIG-U.
MATERIALS AND METHODS
Isolation of Aerolysin-resistant Mutants of CHO Cells
The generation of CHO(wt) cells, a parental CHO cell for the isolation of aerolysin-resistant mutants, was described previously (Hong et al., 2002). Briefly, we stably transfected PIG-L, DPM2, SL15, and PIG-A cDNAs (to avoid the isolation of known GPI-deficient mutants) into CHO-KI IIIB2A cells that stably expressed CD59 and DAF as marker GPI-anchored proteins (Nakamura et al., 1997). CHO(wt) cells were cultured in Ham's F-12 medium containing 10% FCS, 600 μg/ml G418, 200 μg/ml hygromycin, and 5 μg/ml puromycin to ensure the maintenance of the cDNAs. For mutagenesis, CHO(wt) cells (1 × 107 cells in a 15-cm dish) were treated with 400 μg/ml ethyl-methyl-sulfonate (Sigma, St. Louis, MO) for 48 h and cultured for 4 more days. They were then treated with 1 nM proaerolysin (Protox Biotech, Victoria, Canada) for 2 d, washed, and cultured for 2 d. Surviving cells were retreated with 5 nM proaerolysin for 1 d and cloned by limiting dilution.
Other Cells
A GPI(-).O cell line that was deficient in the PIG-O gene (Hong et al., 2000) was isolated from the CHO(wt) cells (Hong et al., 2002). CHO-K1 and class L CHO mutant (M2S2; Nakamura et al., 1997) cells were cultured in Ham's F-12 medium containing 10% FCS.
Flow-Cytometric Analysis
Cells were stained with anti-CD59 (5H8) plus FITC-conjugated antimouse IgG and biotinylated anti-DAF (IA10) plus PE-conjugated streptavidin (Biomeda, Foster City, CA) and analyzed in a FACScan (Becton Dickinson, Mountain View, CA). Cells were also stained with FLAER, an Alexa488-conjugated proaerolysin (Protox Biotech), dissolved in PBS.
Cell Viability Assay
Cells were incubated with various concentrations of proaerolysin for 3 h at 37°C. The viable cells were assessed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) as previously described (Hong et al., 2002).
Expression Cloning of Rat PIG-U
We used a rat C6 glioma cDNA library plasmid made with a mammalian expression vector, pME, bearing the polyoma virus origin of replication (Nakamura et al., 1997). We transfected 1.44 × 108 CHOPA16.1 cells (see RESULTS for characterization) with 480 μg each of the library plasmid and pcDNA-PyT(ori-) plasmid for the expression of the polyoma large T by electroporation at 360 V and 960 μF. Two days later, transfected cells were stained with biotinylated anti-CD59 plus PE-streptavidin and sorted with a FACS-Vantage (Becton Dickinson). Plasmids were recovered from the sorted CD59-positive cells and transfected again with pcDNA-PyT(ori-) into the mutant cells. CD59-positive cells were sorted again. Colonies of Escherichia coli transformed with the isolated plasmids were individually transferred into the wells of 96-well plates. Plasmids that could restore CD59 expression on the CHOPA16.1 cells were identified and sequenced.
Plasmid Construction
We amplified human PIG-U cDNA from a fibroblast cDNA library with primers having an XhoI or MluI site, 5′-ATACGCCTCGAGCCACCATGGCGGCTCCCTTGGTCCTGGTG (forward) and 5′-TAGTTCTAGAACGCGTCTTGAGCACGAGCATGGCCTCTGT (reverse). To add epitope tags at the carboxyl-terminus of human PIG-U, the amplified PCR fragment was cut with XhoI and MluI and cloned into XhoI- and MluI-cut pME-3HSV and pMEEB-Pig-n-GST-FLAG to generate pME-hPIG-U-3HSV and pMEEB-hPIG-U-GST-FLAG, in which triple HSV tags and tandem GST and FLAG tags, respectively, were linked to the C terminus of PIG-U.
To clone a cDNA of Chinese hamster PIG-U, we made subpools (20,000 clones each) of a CHO cell cDNA library (a gift from Drs. Osamu Kuge and Masahiro Nishijima, Institute of Infectious Diseases, Tokyo, Japan; Kuge et al., 2001) and screened these by PCR with primers, rU2F (5′-TTCTCTATCTCCTCCAGCGGCAGTACA, forward) and rU2R (5′-AATGAACATGAAGAAGATGGGGTGCTC, reverse) designed from the rat PIG-U cDNA sequence. From a subpool showing positive bands by PCR, we amplified the 5′ fragment of hamster PIG-U with an upstream vector primer and rU2R. We also amplified the 3′ fragment with rU2F and a downstream vector primer and sequenced the amplified bands. The fulllength hamster PIG-U was amplified with primers having a restriction enzyme site, 5′-ATACGCTCTCGAGCCACCATGGCGGCTCCCTTGGCCCTTGTG (forward) and 5′-TGCCAGCCAACGCGTCTTGAGCACAAGCATAGCCTCTGTGCC (reverse). Amplified PCR fragments were cut with XhoI and MluI and cloned into XhoI- and MluI-cut pME-3HSV to generate pME-CHOPIG-U-3HSV.
The expression plasmids for HA-PIG-S (PIG-S tagged with triple HA at the N-terminus), Myc-PIG-T (PIG-T tagged with hexad Myc at the C terminus), and GST-GPI8 (GPI8 tagged with GST at the C terminus) were the same as in our previous reports (Ohishi et al., 2000, 2001). HSV-GAA1 (GAA1 tagged with triple HSV at the N-terminus) was prepared by transferring human GAA1 cDNA cut from pMEPyori-FLAG-hGAA1 with SalI and NotI (Ohishi et al., 2000) into SalI- and NotI-cut pME bearing the HSV tag sequence.
To clone S. cerevisiae CDC91 DNA, we designed two primers bearing an XhoI or MluI site, 5′-AAAGGACTCGAGCCATGGATTCCACACTTAAGGTAG (forward) and 5′-TGGAGGACGCGTAATTTGTGTTACCTTCAATTTG (reverse) based on the CDC91 sequence and amplified the coding region of CDC91 by PCR from an S. cerevisiae genomic DNA library. The amplified product was cut with XhoI and MluI and cloned into XhoI- and MluI-cut pME-FLAG to generate pME-CDC91-FLAG, a plasmid for the expression of Cdc91p with a FLAG tag at the C terminus. The sequence of CDC91 was confirmed by sequencing the plasmid.
Analysis of the GPI Transamidase Complex
CHOPA16.1 cells (2 × 107) were transfected with 5 μg each of HA-PIG-S, Myc-PIG-T, GST-GPI8, and HSV-GAA1 cDNAs plus 5 μg of FLAG-PIG-U or empty pME vector. Two days after transfection, the cells were dissolved in 1% NP40/20 mM Tris-HCl, pH 7.4/150 mM NaCl/1 mM EDTA/protease inhibitors. HA-PIG-S was immunoprecipitated with 1 μg of anti-HA (Roche Molecular Biochemicals, Mannheim, Germany) antibody plus protein G beads (Amersham Pharmacia Biotech, Piscataway, NJ). The precipitate was divided into five aliquots and Western blotted with anti-HA, anti-Myc (Oncogene Research Products, Boston, MA), anti-GST (Clontech, Palo Alto, CA), anti-HSV (Novagen, Darmstadt, Germany), and anti-FLAG M2 (Sigma) antibodies. The bands that reacted with the antibodies were visualized by horseradish peroxidase (HRP)-conjugated protein G (Bio-Rad, Hercules, CA) and chemiluminescence (Dupont, Wilmington, DE).
Other Methods
In vivo mannose labeling and TLC were performed as previously described (Hirose et al., 1992). The in vitro GPI transamidase assay originally developed by Kodukula et al. (1991) was performed as previously described (Ohishi et al., 2000).
The GPI transamidase complex was isolated by a two-step affinity purification procedure from GPI8-deficient K562 cells expressing FLAG- and GST-tagged GPI8 as previously described (Ohishi et al., 2001). To enhance the denaturation of PIG-U, 6 M urea was included in the SDS-PAGE sample buffer. The N-terminal sequence of PIG-U was determined with a G1005A Hewlett-Packard Protein Sequencing System using Coomassie Blue–stained proteins eluted from a gel after SDS-PAGE.
Site-directed mutants of PIG-U were generated using an oligonucleotide-directed mutagenesis method. RT-PCR was performed using total RNA, random primers for reverse transcription and forward (5′-ATACGCTCTCGAGCCACCATGGCGGCTCCCTTGGCCCTTGTG for PIG-U and 5′-CCTTTCCCCTGCCAGGAGGTCCTATGGCC for DPM3) and reverse (5′-TGCCAGCCAACGCGTCTTGAGCACAAGCATAGCCTCTGTGCC for PIG-U and 5′-GCCCCCTGCGGGCTAAATCTGCTCGCGCC for DPM3) primers for PCR.
RESULTS
Isolation of New Mutant Class U Cells Defective in GPI Transamidase
We obtained a group of mutant CHO cells, CHOPA9.1, 12.1, and 16.1, that were resistant to aerolysin. They were deficient in the surface expression of CD59 and DAF, GPI-anchored proteins (Figure 1, C–E). CHOPA16.1 cells were nearly completely deficient in CD59 and had ∼1% of the normal level of DAF (panel E), whereas CHOPA9.1 and 12.1 cells had heterogeneous expressions of CD59 and DAF (panels C and D). The reasons for the heterogeneous expression of GPI-anchored proteins on CHOPA9.1 and 12.1 cells were unclear but this phenotype was maintained after repeated limiting dilution.
Figure 1.
Expression of GPI-anchored proteins on three class U mutant CHO cells. Cells were stained for CD59 and DAF and analyzed by flow cytometry. (A) Control staining of the parental cell, CHO(wt) with nonrelevant first antibodies; (B) CHO(wt); (C) CHOPA9.1; (D) CHOPA12.1; (E) CHOPA16.1 cells.
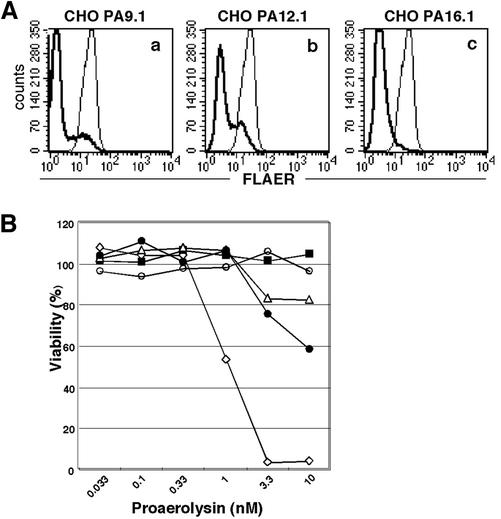
The binding of aerolysin to these cells was markedly decreased and roughly correlated with the profile of GPI-anchored protein expression, i.e., CHOPA9.1 and 12.1 cells containing CD59- and DAF-positive cells had a population that bound aerolysin significantly (Figure 2A). These mutant cells were clearly more resistant to aerolysin than the wild-type cells (Figure 2B). CHOPA16.1 cells, like the GPI-deficient PIG-O mutant CHO cells, GPI(-).O, were resistant to 10 nM aerolysin (open circles and closed squares), whereas some of the CHOPA9.1 and 12.1 cells, presumably those expressing CD59 and DAF, were killed at 3.3 nM aerolysin (triangles and closed circles).
Figure 2.
Aerolysin-binding and -sensitivity of class U mutant CHO cells. (A) Binding of fluorescence-tagged proaerolysin (FLAER) on class U cells. Thick lines, CHOPA9.1 (a); CHOPA12.1 (b); CHOPA16.1 (c) cells. Thin lines, CHO(wt) cells. (B) Sensitivity to aerolysin. The cell viability after a 3-h incubation with increasing concentrations of proaerolysin was measured by an MTT assay. Diamonds, CHO(wt); ▪, GPI(-). O cells defective in the PIG-O gene; triangles, CHOPA9.1; •, CHOPA12.1; ○, CHOPA16.1.
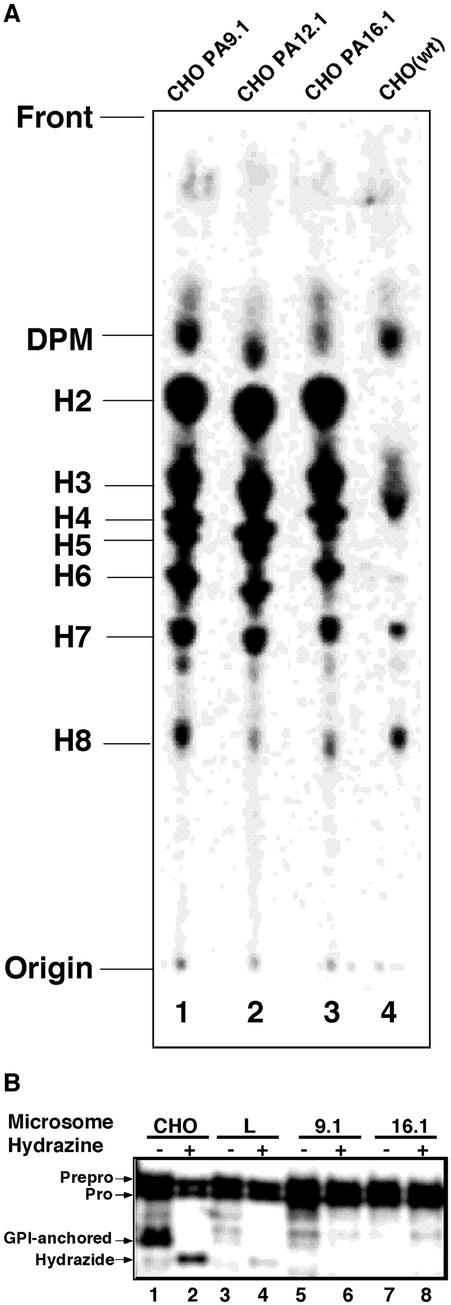
To determine the bases of the defective surface expressions of GPI-anchored proteins on these cells, we analyzed the biosynthesis of GPI by metabolically radiolabeling the cells in a medium containing [3H]mannose. All three had a similar, but obviously abnormal, profile of GPI (Figure 3A, lanes 1–3 vs. lane 4). The three mutants showed a typical profile for cells defective in GPI transamidase, i.e., they generated mature forms of GPI, H7, and H8, and accumulated large amounts of intermediates, such as H2 to H6.
Figure 3.
(A) GPI biosynthesis in class U cells. Lipids from cells labeled with [3H]mannose were separated by TLC in a solvent, CHCl3:MeOH:H2O = 10:10:3. Lane 1, CHOPA9.1; lane 2, CHOPA12.1; lane 3, CHOPA16.1; lane 4, CHO(wt) cells. The origin, front, and identities of the mannolipids are on the left. DPM, dolichol-phosphate-mannose; H2–H6, GPI intermediates; H7 and H8, mature GPI (Hirose et al., 1992). (B) Class U cells do not have GPI transamidase activity in a cell-free system. Mini-PLAP mRNA was translated using rabbit reticulocyte lysates and microsomes from the indicated cells in the absence (-) or presence (+) of hydrazine. Lanes 1 and 2, CHO(wt); lanes 3 and 4, class L CHO cells defective in the PIG-L gene; lanes 5 and 6, CHOPA9.1; lanes 7 and 8, CHOPA16.1. The identities of the forms of mini-PLAP according to Kodukula et al. (1991) are shown on the left.
We then measured the GPI transamidase activities in the microsomes of CHOPA9.1 and 16.1 as well as wild-type cells. When the mRNA of mini-PLAP, a model GPI-anchored protein, was translated in the presence of wild-type microsomes, GPI-anchored mini-PLAP proteins were generated (Figure 3B, lane 1). The addition of hydrazine, which cleaves a thioester bond formed between GPI transamidase and mini-PLAP within the enzyme-substrate intermediate, resulted in the formation of the hydrazide-form instead of the GPI-anchored form (lane 2) as described previously (Maxwell et al., 1995). Microsomes of class L CHO mutant cells that were deficient in the second step of GPI biosynthesis did not generate the GPI-anchored form (lane 3). The hydrazide-form appeared in the presence of hydrazine, indicating that mini-PLAP was processed to the formation of the enzyme-substrate intermediate (lane 4). The microsomes of two class U mutant CHOPA9.1 and 16.1 cells generated only a small amount of and almost no GPI-anchored form (lanes 5 and 7). They also did not generate the hydrazideform in the presence of hydrazine (lanes 6 and 8), indicating that these cells were not able to make the intermediate state. This phenotype is common among mutant cells defective in components of the GPI transamidase complex.
Transfection of cDNAs of four components of GPI transamidase, GPI8, GAA1, PIG-S, and PIG-T, did not restore the surface expressions of GPI-anchored proteins on the three mutants (unpublished data). Therefore, the three mutants represent the fifth gene involved in the attachment of the GPI anchor. We grouped these mutants into a new class, class U, and termed the gene responsible PIG-U.
Expression Cloning and Characterization of PIG-U
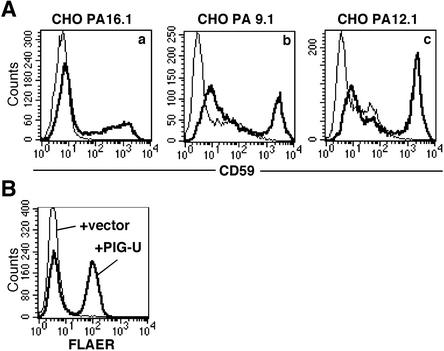
To obtain PIG-U cDNA, we transfected a rat cDNA expression library into CHOPA16.1 cells, collected CD59-positive cells with a cell sorter, and rescued the plasmids. One plasmid containing a 1.6-kb cDNA restored the surface expression of CD59 (Figure 4Aa) and DAF (unpublished data) on CHOPA16.1 cells after transfection. It also restored the binding of aerolysin (Figure 4B). The same cDNA complemented CHOPA12.1 and 9.1 cells as well (Figure 4A, b and c).
Figure 4.
PIG-U restores the surface expresssion of GPI-anchored proteins and aerolysin binding to class U cells. Class U cells were transfected with PIG-U cDNA or a mock vector, and 2 days later were stained with an anti-CD59 antibody or incubated with 5 nM FLAER. Thick lines, PIG-U-transfected; thin lines, mock vectortransfected. (A) CD59 expression on CHOPA16.1 (a), 9.1 (b), and 12.1 (c) cells. (B) Binding of FLAER to CHOPA16.1 cells.
The rat PIG-U cDNA encoded 435 amino acids (DDBJ/GenBank/EMBL accession number AB086841; Figure 5A). On the basis of the sequence homology, we cloned Chinese hamster and human PIG-U with 98 and 97% amino acid identities, respectively, to rat PIG-U (Accession numbers AB086843 and AB086842, respectively). Hamster and human PIG-U cDNAs restored the expression of GPI-anchored proteins on CHOPA16.1 cells (unpublished data). In the databases, we found PIG-U homologues of S. cerevisiae (Figure 5A), Schizosaccharomyces pombe (Accession number O13883), and Drosophila melanogaster (Accession number AAF52689) with 28, 30, and 39% amino acid identities to rat PIG-U, respectively. S. cerevisiae homologue corresponded to CDC91 encoding ORF YLR459w.
Figure 5.
Characterization of PIG-U proteins. (A) Amino acid sequences of rat, Chinese hamster, and human PIG-U and S. cerevisiae Cdc91p. Two highly conserved regions are underlined. Black and gray squares are identical and similar amino acids, respectively. (B) Hydropathy plot (Kyte and Doolittle, 1982) of human PIG-U.
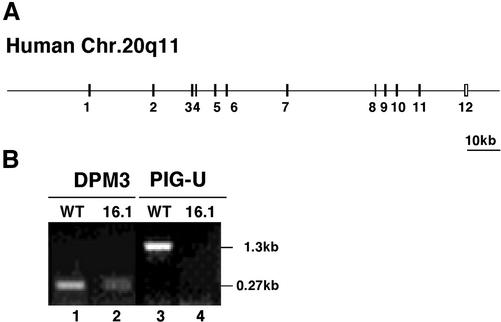
PIG-U was a highly hydrophobic protein with nine putative transmembrane domains as predicted by the TMAP (Persson and Argos, 1994) programs (Figure 5B). The human PIG-U gene consists of 12 exons spanning ∼110 kb in chromosome 20q11 (GenBank accession number AL118520; Figure 6A).
Figure 6.
Genomic structure of human PIG-U and RT-PCR analysis of PIG-U mRNA. (A) Exon-intron organization of human PIG-U. (B) RT-PCR analysis of CHO(wt) and CHOPA16.1 cells for PIG-U and DPM3 mRNAs. Lanes 1 and 2, DPM3; lanes 3 and 4, PIG-U; lanes 1 and 3, CHO(wt); lanes 2 and 4, CHOPA16.1.
RT-PCR analysis demonstrated that CHOPA16.1 cells had no detectable PIG-U mRNA but expressed a normal size mRNA of the ubiquitously expressed DPM3, a subunit of dolichol-phosphate-mannose synthase (Figure 6B; Maeda et al., 2000). This result, taken together with the result of a flow-cytometric analysis showing that CHOPA16.1 cells expressed almost no CD59 and only 1% of the normal level of DAF (Figure 1) indicates that CHOPA16.1 was a nearly null mutant of PIG-U.
PIG-U Is a Subunit of the GPI Transamidase Complex
In a previous study we did not find a clear band of PIG-U when we analyzed an isolated GPI transamidase by SDS-PAGE and silver staining but saw faint smears at around the 80- and 120-kDa positions (see Figure 1 in Ohishi et al., 2001). Because PIG-U might have an anomalous mobility in SDS-PAGE because of its highly hydrophobic characteristics, we reanalyzed a similar preparation of GPI transamidase containing epitope-tagged GPI8 after adding 6 M urea to enhance the denaturing conditions (Figure 7A). A distinct but broad band appeared at the 38-kDa position in addition to the four bands corresponding to the known components (lane 2). We determined the N-terminal sequence of the 38-kDa band to be AAPLVLV, which corresponded to residues 2–8 of the predicted human PIG-U (Figure 5A). Therefore, the human GPI transamidase complex consists of five subunits, GPI8, GAA1, PIG-S, PIG-T, and PIG-U.
Figure 7.
PIG-U is the fifth subunit of the GPI transamidase complex. (A) Two-step purification of the GPI transamidase complex. Class K cells stably expressing GST-FLAG-tagged GPI8 (lane 2) were solubilized in a buffer with 1% NP40. GST-FLAG-tagged GPI8 was purified with anti-FLAG beads followed by glutathione beads. The identities of the bands and the determined N-terminal sequence of PIG-U protein (AAPLVLV) are shown on the right. Lane 1, size markers. (B) Formation of a stable complex of PIG-S, PIG-T, GAA1, and GPI8 in the absence of PIG-U. A mixture of HA-PIG-S, Myc-PIG-T, HSV-GAA1, and GST-GPI8 was transfected into CHOPA16.1 cells with or without FLAG-PIG-U cDNA. Complexes were precipitated with anti-HA to collect HA-PIG-S, and the other coprecipitated components were assessed by Western blotting using antitag antibodies. Lanes 1, 3, 5, 7, and 9, cells with PIG-U; lanes 2, 4, 6, 8, and 10, cells without PIG-U; lanes 1 and 2, anti-HA; lanes 3 and 4, anti-Myc; lanes 5 and 6, anti-HSV; lanes 7 and 8, anti-GST; lanes 9 and 10, anti-FLAG.
We next tested whether the four other components could form a complex in the absence of PIG-U. For this, we transfected cDNAs of differentially tagged PIG-S, PIG-T, GAA1, and GPI8 into CHOPA16.1, a nearly null mutant of PIG-U, with an empty vector or FLAG-tagged PIG-U cDNA. When HA-tagged PIG-S was immunoprecipitated from detergent extracts of these cells, the amounts of PIG-S and three other coprecipitated components were similar in the absence or presence of PIG-U (Figure 7B). Therefore, PIG-S, PIG-T, GAA1, and GPI8 did form a stable complex in the absence of PIG-U. When FLAG-tagged PIG-U was immunoprecipitated, all four other components were coprecipitated (unpublished data), further confirming that PIG-U is a subunit of the GPI transamidase complex.
Yeast Cdc91p Partially Restored the Expression of GPI-anchored Proteins on Class U Cells
As shown in Figure 5A, S. cerevisiae Cdc91p is a structural homologue of PIG-U. To test whether Cdc91p is functionally homologous to PIG-U, we stably transfected CHOPA16.1 cells with a CDC91 expression plasmid. Cells transfected with CDC91 partially restored the surface expression of CD59 and DAF (Figure 8). Therefore, yeast Cdc91p is a functional homologue of PIG-U.
Figure 8.
S. cerevisiae Cdc91p partially restores the expression of GPI-anchored proteins on class U cells. CHOPA16.1 cells were stably transfected with PIG-U, CDC91, or a mock vector and stained with anti-CD59 and anti-DAF antibodies. Left panel, PIG-U-transfected; middle panel, CDC91-transfected; right panel, mock vectortransfected cells.
Functionally Important Regions in PIG-U
Mammalian PIG-U and S. cerevisiae Cdc91p share two highly conserved short regions (Figure 5A, underlined regions). Using a BLAST program, we searched for nearly exact matches with these short regions (National Center for Biotechnology Information, Bethesda, MD). With 17 amino acids corresponding to amino acids 239–255 of Cdc91p, we hit corresponding sequences of the human PIG-U (amino acids 273–289) and the S. pombe PIG-U/CDC91 homologue with expectation values of 0.10 and 5e-05, respectively. Under these conditions, we hit 17 amino acid sequences in mouse and rat fatty acid elongase 1 (Accession numbers NM_134255 and NM_134382), their human homologue (XP_113474) and fish Scophthalmus maxims polyunsaturated long-chain fatty acid elongase (AAL69984; Oh et al., 1997), all with an expectation value of 0.017 (Figure 9A). A similar analysis with 17 amino acids in human PIG-U (amino acids 273–289) hit the corresponding Cdc91p and S. pombe sequences with expectation values of 0.10 and 0.017, respectively, but did not hit any fatty acid elongases. Similar searches with the other conserved sequence (amino acids 380–392 in PIG-U) did not make any significant hits.
Figure 9.
Functionally important short sequence in PIG-U. (A) Alignment of similar short sequences of PIG-U (residues 273–289), Cdc91p (residues 239–255) and long-chain fatty acid elongases. (B) Flow-cytometric analysis of a function of the F274L/W275L mutant of hamster PIG-U. CHOPA16.1 cells were transfected with wild-type PIG-U (thin line), the mutant PIG-U (thick line) and a mock vector (broken line), and 2 days later, stained for CD59. (C and D) Normal expression and incorporation into the GPI transamidase complex of the F274L/W275L mutant of PIG-U. The HSV-tagged mutant and wild-type PIG-U cDNAs were transfected into CHOPA16.1 cells together with HA-PIG-S, Myc-PIG-T, FLAG-GAA1, GST-GPI8, and HSV-ALDH (as a control to assess transfection efficiency). To measure the expression level, the mutant and wild-type PIG-U, and ALDH were immunoprecipitated by anti-HSV beads and analyzed by Western blotting with anti-HSV antibody (C). To assess incorporation of the mutant PIG-U into the GPI transamidase complex, HA-PIG-S was immunoprecipitated with anti-HA antibody and coprecipitated four other proteins were determined by Western blotting with antitag antibodies (D). Lane 1, wild-type PIG-U; lane 2, F274L/W275L PIG-U.
To determine whether the region spanning amino acids 273–289 is functionally important for PIG-U, we generated site-directed mutants in which the conserved aromatic amino acids were changed to leucines. Among them, the F274L/W275L mutant was expressed at a level comparable to that of the wild-type PIG-U (Figure 9C) and was incorporated normally into the protein complex (Figure 9D). Two other mutants were not expressed well and hence were not informative. The F274L/W275L mutant had no activity in restoring the surface expression of CD59 on CHOPA16.1 cells (Figure 9B). This indicates that this conserved region is important for a specific function of PIG-U.
DISCUSSION
In this study, we found that PIG-U is the fifth component of GPI transamidase. We isolated three new mutant CHO cells, termed class U cells, that were defective in GPI transamidase. Using one of the mutant cells for expression cloning, we cloned PIG-U cDNA that complemented all three mutant cells. PIG-U encodes a hydrophobic protein of 435 amino acids. The isolated GPI transamidase complex contained PIG-U, indicating that PIG-U is a subunit of the enzyme complex. We also found that S. cerevisiae Cdc91p, a hydrophobic protein of 394 amino acids, had 28% amino acid identity with PIG-U, and partially restored the expression of GPI-anchored proteins on class U CHO cells. Therefore, Cdc91p is the orthologue to PIG-U.
Characteristics of the GPI Transamidase Complex
The present study and previous reports (Hamburger et al., 1995; Benghezal et al., 1996; Yu et al., 1997; Hiroi et al., 1998; Ohishi et al., 2000, 2001; Fraering et al., 2001) indicate that human and S. cerevisiae GPI transamidases are very similar, both consisting of five proteins. Human GAA1, GPI8, PIG-S, PIG-T, and PIG-U are orthologous with S. cerevisiae Gaa1p, Gpi8p, Gpi17p, Gpi16p, and Cdc91p, respectively, having 25, 44, 23, 30, and 28% amino acid identity, respectively. The human GPI transamidase affinity-purified by taking advantage of epitope tags on GPI8 contained only these five proteins at stoichiometric levels. Whether the five components are sufficient for the attachment of GPI to proteins is, however, yet to be determined because an assay for GPI transamidase with the solubilized enzyme has not been established.
The sum of the molecular weights of the five components is ∼300 kDa. The GPI transamidase complex extracted from HeLa cells with digitonin sedimented at ∼17S corresponding to ∼460 kDa (Vainauskas et al., 2002). The size of the digitonin-extracted GPI transamidase of S. cerevisiae was assessed to be 430–650 kDa by blue native gel electrophoresis (Fraering et al., 2001). As discussed previously (Vainauskas et al., 2002), the large size of the digitonin-extracted GPI transamidase may be attributed to a number of possible reasons: a nonglobular shape of the complex, bound detergent, the presence of multiple copies of one or more subunits and/or the presence of unidentified subunits that may have been lost during the affinity-purification. A GPI transamidase assay applicable to the isolated enzyme complex should be established to solve this problem.
Function of PIG-U/Cdc91p
We did not detect the PIG-U transcript in CHOPA16.1 cells by RT-PCR (Figure 6B). CHOPA16.1 cells expressed only a trace amount of GPI-anchored proteins on the cell surface (Figure 1) and had no detectable GPI transamidase activity in the cell-free assay (Figure 3B). Therefore, we conclude that PIG-U is an essential component of GPI transamidase.
A lack of PIG-U did not affect the formation of a complex of the four other components (Figure 7B). In the cell-free assay, microsomes of CHOPA16.1 cells, which should contain protein complexes consisting of the four other components, did not generate the carbonyl-intermediate between mini-PLAP and GPI8 (Figure 3B). If the protein complex formed without PIG-U had an otherwise normal structure, the result of the cell-free assay would suggest that PIG-U is involved in an event preceding the cleavage of the GPI attachment signal sequence in the precursor protein, such as recognition of the GPI attachment signal or presentation of the precursor proteins to the catalytic site of GPI8. It is possible, however, that PIG-U does not contribute to the recognition and/or presentation of the GPI attachment signal peptide if the protein complex formed with the four other components has a somewhat altered structure so that generation of the carbonyl-intermediate was impaired.
The other possible function of PIG-U/Cdc91p was suggested by analysis of sequence homology. The sequence of the 21-amino-acid region spanning residues 269–289 in PIG-U (PNIGLFWYFFAEMFEHFSLFF) is highly similar to that of the corresponding region in Cdc91p (PNLGLWWYFFIEMFDTFIPFF) spanning residues 235–255, having 14 identical (bold) and 3 similar (underlined) amino acids. Nine of the 21 amino acids are aromatic in both PIG-U and Cdc91p. The F274L/W275L mutant was nonfunctional (Figure 9), indicating that this region, particularly one or both of these aromatic amino acids, is essential for the function of PIG-U.
A part of the sequence in Cdc91p (LWWYFFIEMFDTFIPFF) was similar to the sequence LWWYYFSKLIEFMDTFFF found in mammalian and fish proteins that structurally belong to a long-chain fatty acid elongase family (Tvrdik et al., 2000; Moon et al., 2001). Fatty acid elongase activities of these proteins have not been reported but CIG30 and SSC1, two human members in the same family, complemented S. cerevisiae mutants defective in the ELO2 and ELO3 genes, respectively (Tvrdik et al., 2000). ELO2 is involved in the elongation of the C16 acyl CoA to the C24 chain, whereas ELO3 is required for the generation of the C26 chain (Oh et al., 1997). The sequence found in the mammalian and fish putative long-chain fatty acid elongases belongs to a motif conserved in yeast and mammalian fatty acid elongases. It is, therefore, possible that the seventeen amino acid regions in PIG-U and Cdc91p are involved in the recognition of long-chain fatty acids in GPI. Further studies are necessary to determine whether PIG-U is involved in the recognition and/or presentation of the GPI attachment signal sequence and/or whether PIG-U is involved in the recognition of GPI.
Consistent with previous reports that GPI transamidase is essential for the growth of S. cerevisiae (Hamburger et al., 1995; Benghezal et al., 1996; Fraering et al., 2001; Ohishi et al., 2001), CDC91 is a gene essential for growth (Dolinski, K., Balakrishnan, R., Christie, K.R., Costanzo, M.C., Dwight, S.S., Engel, S.R., Fisk, D.G., Hong, E.L., Issel-Tarver, L., Sethuraman, A., Theesfeld, C.L., Binkley, G., Lane, C., Schroeder, M., Dong, S., Weng, S., Andrada, R., Botstein, D., and Cherry, J.M. Saccharomyces Genome Database; http://genome-www.stanford.edu/Saccharomyces/). The phenotype of cdc91 mutants has not been published but cdc mutants were isolated on the basis of a defect in the cell division cycle. It is likely that defective cell wall genesis caused by a lack of GPI-anchoring accounts for the cdc91 phenotype.
Acknowledgments
We thank Dr. Kisaburo Nagamune for discussion, Drs. Osamu Kuge and Masahiro Nishijima for the CHO cDNA library, Kohjiro Nakamura for the cell sorting, and Keiko Kinoshita and Fumiko Ishii for their technical assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-12-0794. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-12-0794.
References
- Abrami, L., Fivaz, M., Kobayashi, T., Kinoshita, T., Parton, R.G., and van der Goot, F.G. (2001). Cross-talk between caveolae and glycosylphosphatidylinositol-rich domains. J. Biol. Chem. 276, 30729–30736. [DOI] [PubMed] [Google Scholar]
- Abrami, L., Fivaz, M., and van der Goot, F.G. (2000). Adventures of a pore-forming toxin at the target cell surface. Trends Microbiol 8, 168–172. [DOI] [PubMed] [Google Scholar]
- Benghezal, M., Benachour, A., Rusconi, S., Aebi, M., and Conzelmann, A. (1996). Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15, 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Buckley, J.T. (1999). The channel-forming toxin aerolysin. In: The Comprehensive Sourcebook of Bacterial Protein Toxins, ed. J.E. Alouf and J.H. Freer, London: Academic Press, 362–372.
- Fraering, P., Imhof, I., Meyer, U., Strub, J.M., van Dorsselaer, A., Vionnet, C., and Conzelmann, A. (2001). The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol. Biol. Cell 12, 3295–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger, D., Egerton, M., and Riezman, H. (1995). Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol. 129, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi, Y., Komuro, I., Chen, R., Hosoda, T., Mizuno, T., Kudoh, S., Georgescu, S.P., Medof, M.E., and Yazaki, Y. (1998). Molecular cloning of human homolog of yeast GAA1 which is required for attachment of glycosylphosphatidylinositols to proteins. FEBS Lett. 421, 252–258. [DOI] [PubMed] [Google Scholar]
- Hirose, S., Prince, G.M., Sevlever, D., Ravi, L., Rosenberry, T.L., Ueda, E., and Medof, M.E. (1992). Characterization of putative glycoinositol phospholipid anchor precursors in mammalian cells. Localization of phosphoethanolamine. J. Biol. Chem. 267, 16968–16974. [PubMed] [Google Scholar]
- Hong, Y., Maeda, Y., Watanabe, R., Inoue, N., Ohishi, K., and Kinoshita, T. (2000). Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275, 20911–20919. [DOI] [PubMed] [Google Scholar]
- Hong, Y., Ohishi, K., Inoue, N., Kang, J.Y., Shime, H., Horiguchi, Y., van der Goot, F.G., Sugimoto, N., and Kinoshita, T. (2002). Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicum α-toxin. EMBO J. 21, 5047–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, X., Szallies, A., Rawer, M., Echner, H., and Duszenko, M. (2002). GPI anchor transamidase of Trypanosoma brucei: in vitro assay of the recombinant protein and VSG anchor exchange. J. Cell Sci. 115, 2529–2539. [DOI] [PubMed] [Google Scholar]
- Kodukula, K., Micanovic, R., Gerber, L., Tamburrini, M., Brink, L., and Udenfriend, S. (1991). Biosynthesis of phosphatidylinositol glycan-anchored membrane proteins. J. Biol. Chem. 266, 4464–4470. [PubMed] [Google Scholar]
- Kuge, O., Yamakawa, Y., and Nishijima, M. (2001). Enhancement of transport-dependent decarboxylation of phosphatidylserine by S100B protein in permeabilized Chinese hamster ovary cells. J. Biol. Chem. 276, 23700–23706. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Maeda, Y., Tanaka, S., Hino, J., Kangawa, K., and Kinoshita, T. (2000). Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 19, 2475–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, S.E., Ramalingam, S., Gerber, L.D., Brink, L., and Udenfriend, S. (1995). An active carbonyl formed during glycosylphosphatidylinositol addition to a protein is evidence of catalysis by a transamidase. J. Biol. Chem. 270, 19576–19582. [DOI] [PubMed] [Google Scholar]
- Meyer, U., Benghezal, M., Imhof, I., and Conzelmann, A. (2000). Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry 39, 3461–3471. [DOI] [PubMed] [Google Scholar]
- Moon, Y.A., Shah, N.A., Mohapatra, S., Warrington, J.A., and Horton, J.D. (2001). Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 276, 45358–45366. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Inoue, N., Watanabe, R., Takahashi, M., Takeda, J., Stevens, V.L., and Kinoshita, T. (1997). Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem. 272, 15834–15840. [DOI] [PubMed] [Google Scholar]
- Oh, C.S., Toke, D.A., Mandala, S., and Martin, C.E. (1997). ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272, 17376–17384. [DOI] [PubMed] [Google Scholar]
- Ohishi, K., Inoue, N., and Kinoshita, T. (2001). PIG-S, and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1, and GPI8. EMBO J. 20, 4088–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi, K., Inoue, N., Maeda, Y., Takeda, J., Riezman, H., and Kinoshita, T. (2000). Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell 11, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B., and Argos, P. (1994). Prediction of transmembrane segments in proteins utilizing multiple sequence alignments. J. Mol. Biol. 237, 182–192. [DOI] [PubMed] [Google Scholar]
- Spurway, T.D., Dalley, J.A., High, S., and Bulleid, N.J. (2001). Early events in glycosylphosphatidylinositol anchor addition: substrate proteins associates with the transamidase subunit Gpi8p. J. Biol. Chem. 276, 15975–15982. [DOI] [PubMed] [Google Scholar]
- Tvrdik, P., Westerberg, R., Silve, S., Asadi, A., Jakobsson, A., Cannon, B., Loison, G., and Jacobsson, A. (2000). Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J. Cell Biol. 149, 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainauskas, S., Maeda, Y., Kurniawan, H., Kinoshita, T., and Menon, A.K. (2002). Structural requirements for the recruitment of gaa1 into a functional glycosylphosphatidylinositol transamidase complex. J. Biol. Chem. 277, 30535–30542. [DOI] [PubMed] [Google Scholar]
- Vidugiriene, J., Vainauskas, S., Johnson, A.E., and Menon, A.K. (2001). Endoplasmic reticulum proteins involved in glycosylphosphatidylinositol-anchor attachment: Photocrosslinking studies in a cell-free system. Eur. J. Biochem. 268, 2290–2300. [DOI] [PubMed] [Google Scholar]
- Yu, J., Nagarajan, S., Knez, J.J., Udenfriend, S., Chen, R., and Medof, M.E. (1997). The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc. Natl. Acad. Sci. USA 94, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]