Abstract
Centriole assembly plays an important role in centrosome duplication during the cell cycle and is a prerequisite for cilia formation during the differentiation of ciliated cells. In spite of numerous investigations, the molecular machinery that governs centriole/basal body formation remains enigmatic. Recent reports suggest that the ubiquitously expressed mammalian centrins, centrin2p and centrin3p, could be involved in the centriole duplication process. To better understand the specific functions of these proteins, we performed a systematic search for novel mammalian centrins. We isolated a cDNA and the corresponding gene coding for a novel murine centrin, centrin4p, which is more closely related to centrin2p. Like centrin2p, centrin4p accumulates to centrioles and procentrioles when ectopically expressed in HeLa cells. However, centrin4p possesses two splice variants that do not localize to centrioles, suggesting a posttranscriptional regulation mechanism. We also observed that centrin4p does not share the same centriolar targeting properties with centrin2p and 3p, indicating that these proteins could recognize different centriolar partners. Centrin4 mRNA possesses a restricted expression profile and is only detected in brain, kidney, lung, and ovary. In brain, centrin4p is exclusively expressed in ependymal and choroidal ciliated cells where it is localized to basal bodies. Together, our present data suggest that centrin4p could be more specifically involved in basal bodies assembly or in a subsequent step of ciliogenesis.
INTRODUCTION
In most animal cells, microtubules are nucleated and organized by a central organelle, the centrosome (for review, see Bornens, 2002). The centrosome is typically composed of a pair of centrioles, each associated with a cloud of pericentriolar material. The precise duplication of this organelle once during each cell cycle is essential for the establishment of the mitotic spindle and successful cell division. Although numerous microscopic studies have shown that centrosome reproduction is mainly characterized by the semiconservative centriole duplication (Kuriyama and Borisy, 1981; Vorobjev and Chentsov Yu, 1982; Kochanski and Borisy, 1990), the molecular machinery that governs centrosome duplication during the cell cycle is still unknown. It is one of the most fascinating cell biology questions because it has long been proposed that abnormal centrosome duplication could be at the origin of chromosome instability and progression to a cancerous phenotype. This hypothesis has been recently emphasized by the discovery of supernumerary abnormal centrosomes in different human tumor cells (Lingle et al., 1998; Pihan et al., 1998; Brinkley, 2001).
Beside the templated semiconservative duplication of centrioles during the cell cycle, centrioles can also self-assemble in absence of preexisting centrioles. In particular, during the differentiation of ciliated cells from various epithelia, hundreds of centrioles/basal bodies are assembled before migrating and anchoring to the apical plasma membrane where they trigger cilia formation (for review, see Dirksen, 1991). The molecular mechanisms that govern centriologenesis in these cells also remain unknown.
Some progresses in our understanding of centrosome duplication mechanisms came from genetic studies performed in unicellular organisms. In Saccharomyces cerevisiae, the CDC31 and KAR1 gene products were shown to be necessary for the initiation of the spindle pole body (SPB, the functional homolog of the animal centrosome) duplication, because cdc31 and kar1 mutants are both characterized by a large bud, a G2 DNA content, and a single unduplicated SPB (Baum et al., 1986; Rose and Fink, 1987). Both proteins are specifically localized to the half-bridge of the SPB on which the satellite (the precursor of the new SPB) assembles (Spang et al., 1993, 1995). A direct interaction between Kar1p and Cdc31p has been described and Kar1p seems to be required for the correct localization of Cdc31p, which could then interact with a downstream uncharacterized effector and initiate SPB duplication (Biggins and Rose, 1994; Vallen et al., 1994; Spang et al., 1995).
Genetic approaches in the green algae Chlamydomonas reinhardtii allowed to identify several mutants in the centriole/basal body duplication cycle. In particular, the variable flagella number mutant vfl2 is partially defective in templated centriole assembly and in their subsequent segregation during the cell cycle (Taillon et al., 1992; Marshall et al., 2001). Interestingly, the VFL2 gene encodes a small protein called centrin (Crcentrin), which is a homolog to Cdc31p, both proteins belonging to the EF-hand superfamily of calcium-binding proteins (Taillon et al., 1992).
Three centrin proteins (centrin1p, 2p, and 3p) have been identified so far in mammals (Lee and Huang, 1993; Errabolu et al., 1994; Middendorp et al., 1997). Sequence comparison revealed that centrin3p is close to ScCdc31p, whereas centrin1p and 2p are closer to Crcentrin (Middendorp et al., 1997). In contrast to centrin1p, which is mostly expressed in male germ cells, centrin2p and 3p were shown to be ubiquitously expressed, and both proteins are localized in the distal lumen of centrioles and in the procentriole bud (Paoletti et al., 1996; Hart et al., 1999). Their potential implication in the centrosome duplication process during the cell cycle was investigated in different cellular systems. Ectopic expression of both human recombinant proteins in Xenopus laevis embryos induced undercleavage of injected blastomeres and overexpression of Hscentrin3p was shown to impair centrosome duplication in this system (Paoletti et al., 1996; Middendorp et al., 2000). Moreover, it was recently shown that inactivation of centrin2p expression in HeLa cells by RNA interference results in the inhibition of centriole duplication and subsequent defects in the cell cycle progression (Salisbury et al., 2002). Together, these results obtained in different species, from yeasts to mammals, suggest a key role for centrin proteins in the centrosome duplication process.
In the present work, we report the identification and characterization of a novel, tissue-specific mammalian centrin that we called centrin4p.
MATERIALS AND METHODS
RNA Isolation
Total RNA was isolated by homogenizing dissected organs in TRIzol reagent according to manufacturer's recommendations (Invitrogen, Cergy-Pontoise, France). After a precipitation in isopropanol, RNA samples were resuspended in diethyl pyrocarbonate-treated water and quantified by UV spectrophotometry.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
For RT-PCR analysis of centrin2 or centrin4 mRNA expression, first-strand cDNA synthesis was performed with 1 μg of the different RNA samples by using 10 pmol of an oligo(dT)-adaptor primer (Invitrogen). Amplification reactions were performed using specific primers for centrin2 or centrin4 (Table 1). PCR conditions were as follows: one cycle (94°C, 2 min); 37 cycles (94°C, 30 s; 55°C, 45 s; 72°C, 1 min), and one cycle (72°C, 5 min). Final products obtained from kidney RNA sample were cloned and sequenced to confirm the specificity of the PCR amplifications.
Table 1.
Primers used in this study
| Primers for 5′ RACE | |
| 1st strand synthesis | CTGTAATTCATCTTCGGTTAA |
| I PCR | TATTTAGTGATATGCTTCCCGTA |
| II PCR | GCCGAATTCTTTCACTCATTTTTACGCTCATGAT |
| Primers for 3′ RACE | |
| I PCR | GCCGGATCCATGAGCGTAAAAATGAGTGAAAAAGATGA |
| II PCR 5′ primer | GCCGAATTCTTATTAGCTGCTTGGTGCCTTCCA |
| 3′ primer | GAGGGATCCGGCCACGCGTCGACTAGTAC |
| Primers for genomic cloning | |
| I PCR 5′ primer | GGATAACTGAATTGTCATTCTAGCT |
| 3′ primer | TCAGGTATTACATCATTGAACTGCT |
| II PCR 5′ primer | GCCCTGCAGACCCATGGTATGCTTATTGCA |
| 3′ primer | GCCGGATCCACCTCTTGGAAGGCACCAAGCAGCT |
| Primers for RT-PCR analysis | |
| Centrin4 5′ primer | GCCCTGCAGTCATGGCATCCAGCCAGCGCATAACT |
| 3′ primer | GCCGGATCCACCTCTTGGAAGGCACCAAGCAGCT |
| Centrin2 5′ primer | GCCGAATTCTTAACAACCGGTGGCAGAGTCCGTT |
| 3′ primer | GCCGGATCCAAATCTAGTCACATGTGCTTGCAGT |
Restriction site sequences introduced to facilitate subsequent cloning are in italic.
3′ and 5′ Rapid Amplification of cDNA Ends (RACE)
For 3′ RACE of centrin4 mRNA, first-strand cDNA synthesis was performed with 1 μg of brain (hemisphere) total RNA by using 10 pmol of an oligo(dT)-adaptor primer (Invitrogen). As a control for subsequent amplifications, the same reaction was also performed in absence of the reverse transcriptase. Two rounds of amplification were done with 10 pmol of each primer (Table 1). The final amplification product, which corresponds to the 3′-untranslated region (3′-UTR) of centrin4 mRNA, was cloned in a Bluescript SK+ vector, sequenced, and used to prepare the cRNA probe for in situ hybridization.
5′ RACE was carried out essentially as 3′ RACE, except that the first-strand cDNA synthesis was performed using 10 pmol of a specific primer for centrin4 (Table 1). After oligo(dC) tailing of the cDNA according to manufacturer's recommendations (Invitrogen), two successive rounds of amplification were performed using primers listed in Table 1.
PCR Amplification of Genomic DNA
Centrin4 gene (CETN4) cloning was carried out by a PCR amplification approach on genomic DNA prepared from C57B16/DBA2 mouse tail (Miller et al., 1988). The first amplification was performed with 100 ng of genomic DNA and 10 pmol of primers listed in Table 1. The PCR conditions were as follows: one cycle (94°C, 2 min); 35 cycles (94°C, 30 s; 55°C, 45 s; 72°C, 2 min), and one cycle (72°C, 5 min). The second round of amplification was performed using 0.1% of the first PCR amplification products under the following conditions: one cycle (94°C, 2 min); 35 cycles (94°C, 30 s; 62°C, 45 s; 72°C, 2 min) and one cycle (72°C, 5 min). The final amplification product was cloned in a Bluescript SK+ vector and sequenced.
Northern Blot Analysis
To analyze the expression profile of centrin4 mRNA, 20 μg of total RNA from different tissues were separated on a denaturing agarose gel containing formaldehyde (Sambrook et al., 1989). Samples were transferred overnight to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech) using 50 mM NaPi, pH 7.2 as a transfer buffer, and visualized by methylene blue coloration. Prehybridization was performed in 1 M NaPi pH 7.2, 7% SDS, 1 mM EDTA at 68°C for one hour. A cDNA fragment corresponding to 3′ UTR of centrin4 mRNA (see above) was 32P-labeled using the rediprime II kit (Amersham Pharmacia Biotech) and purified using a Probe-Quant G-50 column (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, UK). The purified probe (106 cpm/ml) was added to fresh hybridization buffer and incubation was performed overnight at 65°C. After three washes in 40 mM NaPi buffer containing 1% SDS, membranes were subjected to an autoradiography analysis.
In Situ Hybridization
Adult mouse brains were collected and immediately frozen in isopentane. Coronal sections (15 μm in thickness) were performed on a cryostat, collected on SuperFrost Plus glass slides, and stored at —80°C before use. Brain sections were postfixed for 15 min with 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS), rinsed three times in PBS, and then acetylated with 0.25% acetic anhydride/0.1 M triethanolamine for 10 min. After three washes in PBS, sections were dehydrated in ethanol and air-dried.
One microgram of a Bluescript plasmid containing the 3′-UTR of centrin4 mRNA was linearized with BamHI or EcoRI to generate sense or antisense cRNA probes. In vitro transcription was performed using the Promega kit and T7 or T3 RNA polymerase in the presence of 35S-UTP (<1000 Ci/mmol; Amersham Biosciences UK). Hybridization was performed in a humid chamber overnight at 48°C by using 106 cpm/slide of 35S-labeled RNA probe in 20 mM Tris-HCl pH 7.4, 5 mM EDTA, 0.3 M NaCl, 10 mM phosphate buffer, 10 mM dithiothreitol (DTT), 50% formamide, 10% dextran sulfate, 1× Denhardt's solution, and 0.5 mg/ml yeast total RNA (Sigma Chemical, Poole, Dorset, United Kingdom). Sections were washed in 5× SSC, 1.6 mg/ml DTT at 42°C for 30 min, 2× SSC, 50% formamide, 12.5 mg/ml DTT at 60°C for 20 min and treated with 20 μg/ml RNAse A (Roche Diagnostics, Mannheim, Germany) for 30 min at 37°C. After two washes in 2× SSC and 0.1× SSC, 15 min each at 37°C, sections were dehydrated in ethanol and air-dried. Sections were first apposed to hyperfilms (β-max; Amersham Biosciences UK) for 5 d and then dipped in photographic emulsion (NTB-2; Kodak, Tokyo, Japan) and exposed for 2 to 3 weeks. After development, sections were counterstained with cresyl violet.
DNA Constructs and Cell Transfection
The different Hscentrin2p and Mmcentrin4p constructs were generated by a PCR approach and subcloned in the mammalian expression vector pcDNA3 (Invitrogen) in fusion with an NH2-terminal myc epitope tag or in the pGFP-C1 vector (BD Biosciences Clontech, Palo Alto, CA). The Nter4, Cter4, and III-4 domains of centrin4p correspond to amino acids 1–100, 89–168, and 1–138, respectively. The Nter2 and Cter2 domains of Hscentrin2p correspond to amino acids 1–104 and 93–172. The sequence corresponding to amino acids 232–272 of Kar1p (Spang et al., 1995) was inserted in pGFP-N1 vector (BD Biosciences Clontech) downstream of an initiating methionine.
Mouse L929 or human HeLa cells were grown as monolayers in DMEM containing 10% (vol/vol) inactivated fetal calf serum (Invitrogen) at 37°C in 5% CO2. For transient expression experiments, 5 × 106 cells were transfected by electroporation with 50 μg of plasmid DNA at 300 V and 960 μF by using a gene pulser (Bio-Rad, Hercules, CA). HeLa cell synchronization by a double thymidine block was performed as described by Stein et al. (1994).
Antibodies
Rabbit anti-centrin4p antiserum was generated against the following synthetic peptide conjugated to KLH: K-A-A-K-V-E-L-N-D-T-Q-K-Q-E-C. GT335 is a monoclonal antibody (mAb) directed against glutamylated tubulin, which stain axonemes of ciliated cells (Wolff et al., 1992; Tournier et al., 1998). CC310 is a mAb raised against ciliary cortices of quail oviduct epithelium, which stain striated rootlets in ciliated epithelia (Klotz et al., 1986; Peraldi-Roux et al., 1991). Rabbit anti-γ-tubulin antiserum has been described previously (Moudjou et al., 1996). Commercial monoclonal antibodies were anti-myc (9E10; Santa Cruz Biotechnology, Tebu, France) and anti-green fluorescent protein (GFP) (clones 7.1 and 13.1; Roche Diagnostics).
Immunofluorescence
Transfected cells growing on coverslips were briefly extracted with 0.5% NP-40 in PHEM buffer (45 mM PIPES, 45 mM HEPES, pH 6.9, 10 mM EGTA, 5 mM MgCl2) for 20 s and fixed in PBS containing 2% paraformaldehyde for 10 min at room temperature. Cells were subsequently permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 100 mM glycine in PBS for 15 min. Coverslips were then preincubated with 3% bovine serum albumin in PBS containing 0.1% Tween 20 for 30 min and incubated 1 h with primary antibodies in the same blocking buffer. After three washes with 0.1% Tween 20 in PBS, coverslips were incubated 1 h with rhodamine-conjugated secondary antibodies (Jackson Immunore-search Laboratories, West Grove, PA). After three new washes, coverslips were finally mounted with AF1 antifadent mountant solution (Citifluor; City University, London, England).
Frozen brain sections (15 μm in thickness) were air-dried for 5 min at room temperature and fixed in methanol at —20°C for 6 min. Brain sections were then blocked with 3% bovine serum albumin in PBS containing 0.1% Tween 20 for 2 h and incubated overnight at room temperature with primary antibodies. After three washes with 0.1% Tween 20 in PBS, sections were incubated 1 h with secondary antibodies. Nuclei were stained using Hoechst 33528 at 5 μg/ml for 5 min. After three new washes, brain sections were finally mounted with AF1 antifadent mountant solution.
Quantification
The relative quantification of the accumulation of the different GFP-centrin constructs to centrioles was performed as follows. HeLa cells were transfected with the different GFP-centrin constructs in the same conditions and 24 h later were briefly extracted with 0.5% NP-40 in PHEM buffer for 20 s before fixation with 2% paraformaldehyde in PBS for 10 min at room temperature. Then 10 sequential Z-axis 12-bit images were collected in 0.2-μm steps by using a DMIRBE microscope (Leica, Wetzlar, Germany) equipped with a piezoelectric device, which allowed to cover the entire centriolar signal in each transfected cell. For each Z-series, the maximal intensity signal to centriole was determined using MetaMorph software (Universal Imaging, Downingtown, PA) and the immediately surrounding background was subtracted. We checked that preextraction and paraformaldehyde fixation did not affect the GFP signal to centriole. Data are presented as the mean values obtained in 30 transfected cells in each condition for one given experiment. Results are representative of three independent experiments.
Coimmunoprecipitations
HeLa cells (5 × 106 cells) were cotransfected with 10 μg of an NH2-terminal myc epitope-tagged Mmcentrin4p or Hscentrin2p construct and 40 μg of a Kar1p peptide-GFP plasmid. Twenty-four hours later, cells were lysed in one-dimensional (1-D) buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM DTT, 0.5% NP-40) containing 2 mM CaCl2 or 2 mM EGTA. Cellular debris were removed by a centrifugation at 10,000 × g for 10 min at 4°C, and supernatants were then incubated with 5 μg of anti-GFP monoclonal antibodies coupled to protein G-Sepharose 4 Fast Flow beads (Amersham Biosciences UK) for 1 h at 4°C. After four washes in 1-D buffer containing 2 mM CaCl2 or 2 mM EGTA, immunoprecipitates were solubilized in SDS-PAGE sample buffer and processed for immunoblot analysis.
Calcium Binding and Electrophoretic Shifts
HeLa cells (5 × 106 cells) transfected with the different GFP-centrin constructs were lysed in three-dimensional buffer (0.5% NP-40, 0.5% deoxycholate, 0.05% SDS) for 10 min at 4°C and centrifuged at 10,000 × g for 10 min at 4°C. Supernatants were incubated with 5 μg of anti-GFP monoclonal antibodies coupled to protein G-Sepharose 4 Fast Flow beads (Amersham Biosciences UK) for 1 h at 4°C under mild agitation. After four washes in 1-D buffer, immunoprecipitates were solubilized in SDS-PAGE sample buffer. One-dimensional SDS-PAGE (12% acrylamide) and protein transfer on a nitrocellulose membrane were performed using standard protocols (Sambrook et al., 1989). After transfer, the membrane was washed in 10 mM imidazole-HCl pH 6.8, 60 mM KCl, 5 mM MgCl2 for 90 min and then incubated in the same buffer containing 20 μCi of 45Ca for 20 min (Maruyama et al., 1984). After three washes in distilled water for 2 min each, the membrane was processed for an autoradiography analysis.
For electrophoretic shifts analyses, transfected cells were lysed in 1-D buffer containing 2 mM CaCl2 or 2 mM EGTA. After a centrifugation at 10,000 × g for 10 min, supernatants were processed for immunoblot analyses with 2 mM CaCl2 or 2 mM EGTA in gels and 0.1 mM CaCl2 or 2 mM EGTA in running buffers.
RESULTS
Identification and Sequence Analysis of Murine Centrin4p
To better understand the specific roles of centrin proteins in the centriole/basal body assembly processes, we performed a systematic search for novel mammalian centrins in expressed sequence tags databases. We identified two mouse cDNA clones (accession nos. AA286656 and AI595077) derived from a 5-d postcoitum whole fetus and a mammary gland library, respectively, which contain almost identical sequences potentially coding for a novel centrin. Sequencing revealed that both cDNA clones contained only a partial open reading frame (ORF) (our unpublished data). 5′ and 3′ RACE were performed to determine the full sequence of the transcription unit (see MATERIALS AND METHODS). The corresponding mRNA (accession no. AF362367), which is 1073 nucleotides in length, contains an ORF of 507 bases with an ATG-initiating methionine flanked by a partial Kozak sequence containing a purine in position —3 and a guanine in position +4 (Figure 1A) (Kozak, 1986). The single-length 5′-UTR of 87 nucleotides has a high C + G (73.6%) content and the 3′-UTR of 464 nucleotides in length contains a perfectly conserved polyadenylation signal (AATAAA) 13 bases upstream of the poly(A) addition site (Figure 1A). The open reading frame encodes a novel 168 amino acid protein with a calculated molecular mass of 19,200 Da, that we propose to name centrin4p. Like other centrin proteins, centrin4p has an acidic isoelectric point (pI = 4.74) and contains four potential EF-hand calcium-binding domains, which consist of a central calcium-binding loop flanked on both sides by a α-helix (Figure 2A). Comparison of the four calcium-binding loop sequences with the consensus sequence predicted that only the fourth EF-hand calcium-binding domain of centrin4p could be functional (Figure 2B; see below) (Marsden et al., 1990).
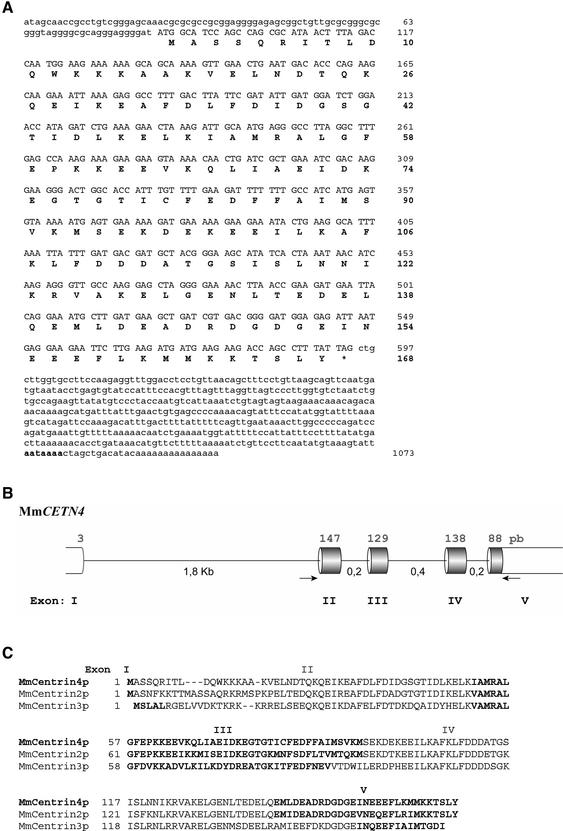
Figure 1.
Centrin4 cDNA sequence and structure of the corresponding gene. (A) Nucleotide sequence of the murine centrin4 cDNA with the deducted amino acid sequence (accession no. AF362367). The cDNA contains a coding sequence of 507 nucleotides corresponding to a polypeptide of 168 amino acids. Uppercase letters correspond to the coding sequence and lowercase letters to 5′- and 3′-untranslated regions. The polyadenylation site is indicated in bold. (B) Schematic representation of the intron/exon organization of murine centrin4 gene (MmCETN4). Upper numbers correspond to the length of coding regions and lower numbers to the length of introns. Primers used to amplify the coding region are indicated (arrows). (C) Alignment of amino acid sequences of murine centrin2p, 3p, and 4p. The contribution of the different exons is indicated by the alternation between bold and normal sequences. In contrast to centrin3, centrin4 and centrin2 genes share exactly the same exon/intron junctions (centrin1 gene is intronless).
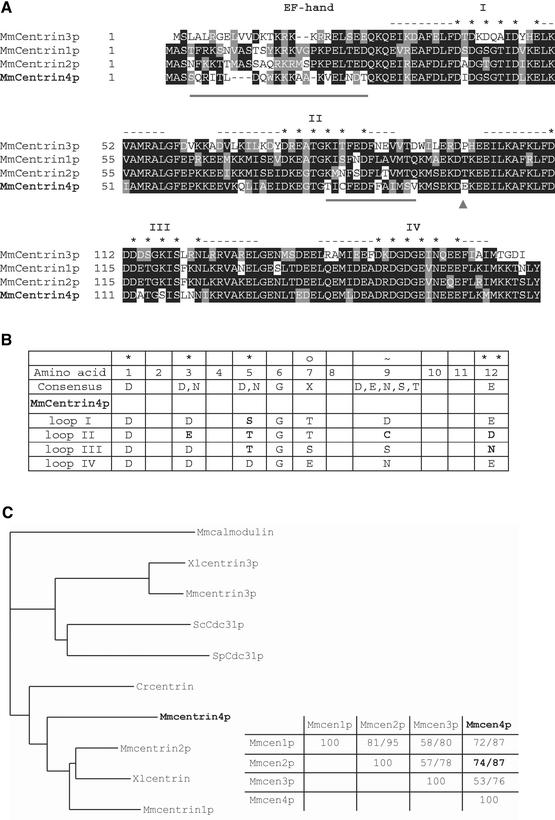
Figure 2.
Centrin4p is more closely related to centrin2p. (A) Alignment of amino acid sequences of murine centrin1p, 2p, 3p, and 4p. Identical amino acids are boxed in black and conservative changes are boxed in gray. The position of the four EF-hand calcium-binding domains is indicated by dashes (α-helix) and stars (calcium-binding loop). Centrin4p and 2p are closely related and differ mostly in two amino acid subdomains that are underlined. (B) Comparison of the four calcium-binding loops of centrin4p with the consensus sequence. Amino acids divergent from the consensus sequence are in bold. Note that only the fourth EF-hand of centrin4p could be functional. (C) Left, phylogenetic tree of centrin proteins identified in different species. Branch lengths are proportional to the evolutionary distances calculated. Centrin4p is closer to the subgroup of centrin1p/2p and Crcentrin than to the subgroup of centrin3p and yeasts centrins. Cr, C. reinhardtii; Mm, Mus musculus; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; and Xl: Xenopus laevis. Right, percentages of identity/similarity between murine centrins showing that centrin4p (Mmcen4p) is closely related to centrin2p (Mmcen2p).
Amino acid sequence comparison with other centrin proteins revealed that centrin4p is closer to the subgroup of centrin2p/centrin1p and Crcentrin than the subgroup containing centrin3p and yeast centrins (Figure 2C). In particular, Mmcentrin4p and Mmcentrin2p that are closely related share 74% identity and 87% similarity and differ mostly in two short amino acid domains (underlined in Figure 2A). The first domain corresponds to the amino terminal region of Mmcentrin4p that is the most divergent region between all known centrins and in part differentiates centrin proteins from the related EF-hand calcium-binding protein calmodulin (Figure 2A). The second specific amino acid domain of Mmcentrin4p is located between the second and third EF-hand calcium-binding domains, a region corresponding to the central helix of calmodulin (Figure 2A). Structural studies of calmodulin have revealed that the first two and the last two EF-hands form two globular functional units separated by a flexible central helix (for review, see Nelson and Chazin, 1998). The ability of calmodulin to recognize different target proteins is a consequence of the flexibility of this central helix, which is able to bend at the amino acid Ser81, thereby allowing the two globular domains to wrap around target peptides (O'Neil and DeGrado, 1990). This amino acid was also shown to be a substrate of casein kinase II in vitro (consensus phosphorylation site, S/T-X-X-E) (Quadroni et al., 1994). We noticed that the subgroup of centrin2p/centrin1p and Crcentrin have a serine or a threonine at the equivalent position (and share a potential casein kinase II phosphorylation site), whereas the subgroup of centrin3p and yeast centrins have a proline residue (Figure 2A, arrowhead). Interestingly, centrin4p contains a negatively charged amino acid at the equivalent position, which could confer conformational specificities (Figure 2A, arrowhead).
We next used the sequence of the two specific domains of centrin4p to search for homologs in other species. We identified numerous EST in some mammalian species (rat, cow, pig, and human) that allowed to obtain the full open reading frame of rat (accession no. BG662613) and cow centrin4p (accession nos. AV591122 and BE667637) (our unpublished data). On the other hand, we were unable to identify EST for centrin4p in nonmammalian species. Finally, we identified in databases a genomic clone containing the full sequence of the mouse centrin4 gene (CETN4, accession no. AL645982). We used mouse tail genomic DNA to amplify a part of the gene encompassing the coding exons (see MATERIALS AND METHODS). Sequencing revealed two nucleotide differences with the cDNA in the coding region but no differences at the amino acid level (our unpublished data). The centrin4 gene, located on chromosome 3, contains five exons and four introns with a large intron positioned just after the initiating methionine-coding triplet (Figure 1B). Comparison with the other murine centrin genes showed that in contrast to CETN3, CETN4, and CETN2 genes share exactly the same intron/exon junctions (centrin1 gene is intronless and was proposed to arise from a retrotransposition of centrin2 mRNA, (Hart et al., 1999) (Figure 1C). This observation confirms the phylogenetic relationship between centrin4p and centrin2p and suggests that the respective genes arise from a duplication of a common ancestor. Because they are closely related, we compared the properties of centrin4p and centrin2p.
Tissue-specific Expression of Centrin4 mRNA
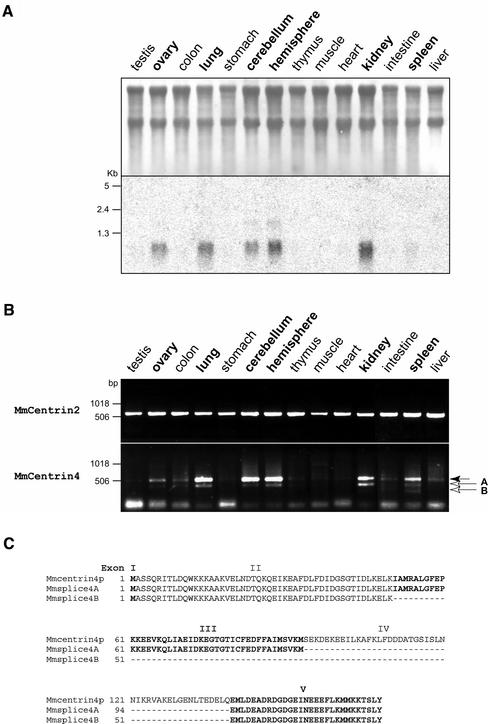
Given the relationship between CETN4 and CETN2 genes, we first wanted to determine whether centrin4 mRNA is also ubiquitously expressed. Northern blot analysis of total RNA from various adult mouse tissues with a specific 32P-labeled 3′-UTR probe revealed that centrin4 mRNA possesses a tissue-specific expression profile. Centrin4 mRNA is highly expressed in the brain (cerebellum and cerebral hemispheres), lung, kidney, and ovary (Figure 3A). A very low signal was also detected in spleen, whereas centrin4 mRNA was undetectable in other tissues (testis, colon, stomach, thymus, skeletal muscle, heart, intestine, and liver). This result was further confirmed by a RT-PCR analysis, which gave qualitatively the same expression profile (Figure 3B). Surprisingly, using primers designed to amplify the full coding sequence of centrin4p, we obtained one major band at the expected size and one or two lower bands in the same tissues (Figure 3B, arrows in A and B). The different bands were then subcloned and sequenced. As expected, the major band corresponds to the full coding sequence of centrin4p, indicating that the PCR amplification was specific. Sequence analysis revealed that bands A and B correspond to the same coding sequence with an internal deletion of different sizes (our unpublished data). More importantly, these internal deletions correspond exactly to a deletion of the sequence of exon IV (band A) or to a deletion of the sequence of exons III and IV (band B) (Figure 3C). The two corresponding mRNAs are thus most probably transcribed from the same gene but correspond to differential splicing variants of centrin4 mRNA. The corresponding proteins possess a deletion of the third EF-hand or of the second and third EF-hands and were called Mmsplice4A and Mmsplice4B, respectively (Figure 3C). Finally, using primers designed to amplify the full coding sequence of centrin2p and centrin3p, only one band was detected at the expected size in all tissues tested (Figure 3B; our unpublished data). Sequence analysis confirmed that the PCR products correspond to the centrin2p or centrin3p coding sequence (our unpublished data).
Figure 3.
Centrin4 mRNA expression profile and identification of two splice variants. (A) Northern blot analysis showing that centrin4 mRNA is highly expressed in the brain (cerebellum and hemispheres), lung, kidney, and ovary. (B) RT-PCR analysis of murine centrin4 or centrin2 expression in the same tissues by using primers designed to amplify the full coding sequences. Only one band corresponding to the full coding sequence of centrin2p is amplified in all tissues tested (top). In contrast, centrin4 specific primers allowed to amplify one major band corresponding to the full coding sequence and two minor bands (bands A and B) that encode splice variants of centrin4p (bottom). (C) Alignment of amino acid sequences of centrin4p and its two splice variants splice4A and 4B. Splice4A and 4B differ from centrin4p by a deletion of the amino acid sequence corresponding to exon IV or exon III and IV, respectively.
Together, our results show that in contrast to centrin2 and centrin3, centrin4 mRNA has a tissue-specific expression profile and possesses at least two splice variants.
Ectopically Expressed Centrin4p Localizes to Centrioles
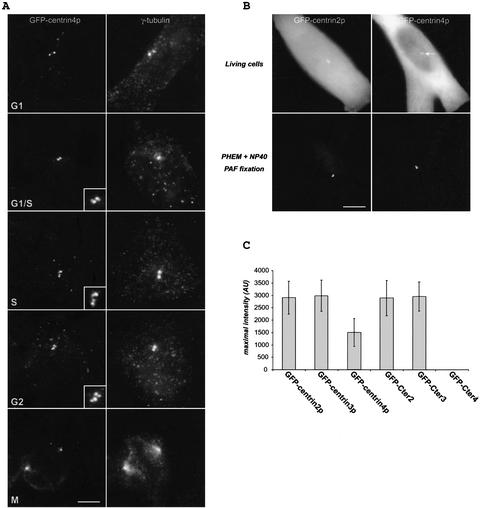
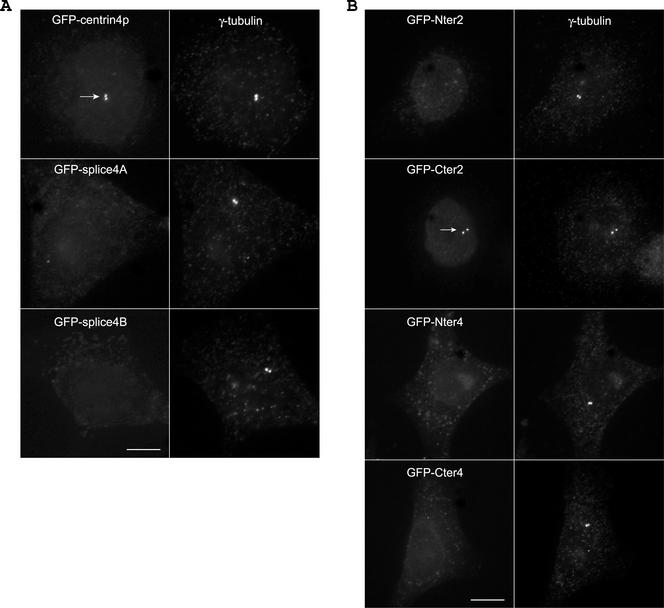
Previous work showed that mammalian centrin2p and centrin3p are present in the distal lumen of centrioles and in the procentriole bud (Paoletti et al., 1996; Middendorp et al., 1997). In a first step to examine its subcellular localization, centrin4p was ectopically expressed as a green fluorescent protein (GFP) fusion protein. As the same results were obtained in different cell lines, we illustrate the localization of centrin4p in HeLa cells. In asynchronous fixed HeLa cells, one or two pairs of dots located near the nucleus were observed (Figure 4A). Double immunostaining with anti-γ-tubulin antibodies confirmed that these dots correspond to centrioles (Figure 4A, G1). However, in a given pair, the two dots frequently differed in their intensity (Figure 4A, insets). To follow the modification of the dot staining during the centrosome duplication cycle, cells were synchronized at the G1/S transition by a double thymidine block and then allowed to progress in S, G2, and M phases. During the progression in the cell cycle, the intensity of the two dots in a given pair became progressively comparable, whereas the distance separating the two dots increased (compare Figure 4A, G1/S and G2). These observations strongly suggest that ectopically expressed centrin4p accumulates to procentrioles during their elongation and is most probably located at their distal part, as previously demonstrated for centrin2p (Paoletti et al., 1996). However, comparison of the localization of GFP-centrin2p and GFP-centrin4p in living cells gave a striking difference (Figure 4B). Both proteins were in a large part diffusely located in the cytoplasm but, in contrast to GFP-centrin2p, the accumulation of GFP-centrin4p to centrioles was more hardly detected (Figure 4B, arrow). This difference does not depend upon their respective stability because we checked that both proteins were expressed to a very similar level (our unpublished data). Relative quantification (see MATERIALS AND METHODS) revealed that the accumulation of GFP-centrin4p to centrioles is in average 2 or 3 times lower than GFP-centrin2p (or GFP-centrin3p) (Figure 4C).
Figure 4.
Centrin4p localizes to centrioles and procentrioles. (A) Ectopically expressed GFP-centrin4p in HeLa cells localizes to centrioles and growing procentrioles during the cell cycle, as identified by a γ-tubulin staining. Bar, 5 μm; insets, twofold magnification. (B) Top, in contrast to centrin2p, the accumulation of centrin4p to centrioles is more hardly detected over a cytoplasmic background in living cells (arrow). Bottom, cells were briefly extracted with 0.1% NP-40 before paraformaldehyde fixation. Accumulation of centrin4p to both centrioles is clearly visible. Bar, 5 μm. (C) Quantification of the accumulation to centrioles of centrins 2p, 3p, and 4p and of their corresponding COOH-terminal domains (Cter2, 3, and 4) (see text).
To further investigate the centriolar targeting of centrin4p, we next examined the intracellular localization of its two splice variants. In contrast to centrin4p, splice4A and splice4B were never detected at the centrioles, as identified by a γ-tubulin staining (Figure 5A). This suggests that the third EF-hand of centrin4p is at least necessary for its centriolar targeting. In an attempt to define a minimal amino acid domain necessary for its centriolar localization, we generated different constructs of centrin4p containing either the two first or the two last EF-hand calcium-binding domains (Nter4 and Cter4 domains, respectively) or lacking only the last EF-hand calcium-binding domain (III-4 domain) (Figure 6B). Transient expression in HeLa cells showed that all of these truncated forms were unable to accumulate to centrioles, suggesting that the four EF-hand calcium-binding domains of centrin4p are essential for its proper localization (Figure 5B; our unpublished data). Surprisingly, using equivalent constructs of Hscentrin2p, we observed that the COOH-terminal domain (Cter2) of centrin2p accumulates to centrioles, whereas the NH2-terminal domain (Nter2) was diffusely located in the cytoplasm (Figure 5B). The same result was also obtained with the two truncated forms Cter3 and Nter3 of Hscentrin3p (our unpublished data). Moreover, relative quantification showed that the COOH-terminal domain Cter2 (or Cter3) accumulates to centrioles to a similar level as the full-length protein, suggesting that this domain is necessary and sufficient for the proper localization of centrin2p (or centrin3p) (Figure 4C). Together, these results show that ectopically expressed centrin4p, but not its two splice variants, is able to accumulate to centrioles and procentrioles. However, the centriolar anchoring mechanism seems to differ between centrin4p and centrin2p (or centrin3p) because the four EF-hand calcium-binding domains of centrin4p are essential for its centriolar localization, whereas only the two last EF-hand domains of centrin2p (or centrin3p) are needed for its proper localization. One possibility to explain the different behavior between centrin4p and centrin2p (or centrin3p) could be that they did not recognize the same centriolar partners (see below).
Figure 5.
Splice variants are unable to localize to centrioles. (A) In contrast to centrin4p, its two splice variants, splice4A and splice4B, are unable to localize to centrioles, as identified by a γ-tubulin staining. Note that transfected cells were easily identified as they keep a low cytoplasmic background even after a brief detergent extraction. (B) Whereas the NH2 and COOH-terminal domains of Mmcentrin4p (Nter4 and Cter4) are unable to localize to centrioles, the COOH-terminal domain of Hscentrin2p (Cter2) is necessary and sufficient for its proper localization (arrow). Bar, 5 μm.
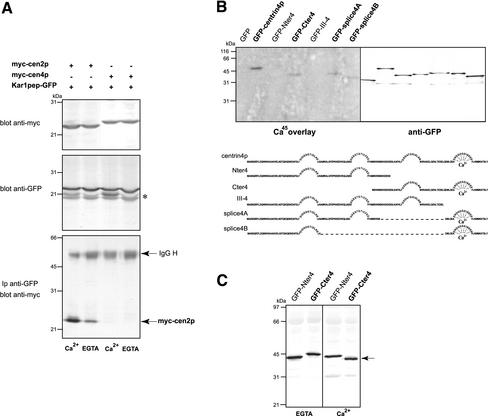
Figure 6.
Centrin4p is a Ca2+-binding protein that does not recognize Kar1 peptide. (A) HeLa cells were cotransfected with a myc-tagged Mmcentrin4p (myc-cen4p) or Hscentrin2p (myc-cen2p) construct and a Kar1p peptide-GFP (Kar1pep-GFP) plasmid. Top, Western blot analysis of centrin2p and centrin4p expression showing that both proteins were expressed to a very similar level (25 μg of total protein extract per lane). Middle, Western blot analysis of Kar1p peptide expression. Lower bands correspond most probably to degradation products (asterisk). Bottom, In contrast to centrin4p, centrin2p coimmunoprecipitates with Kar1p peptide in a calcium-regulated manner. (B) GFP-tagged centrin4p and different truncated forms were transiently expressed in HeLa cells and immunoprecipitated using anti-GFP antibodies. After SDS-PAGE and transfer on nitrocellulose membrane, proteins were incubated with 45Ca (left) or revealed with anti-GFP antibodies (right). Note that centrin4p and only truncated forms containing its fourth EF-hand domain bind calcium, suggesting that only this domain is functional. (C) NH2- and COOH-terminal domains (Nter4 and Cter4) of centrin4p were submitted to SDS-PAGE in presence of 2 mM Ca2+ or 2 mM EGTA in gels and samples. Only the Cter4 domain exhibits a Ca2+-dependent electrophoretic mobility shift, suggesting that it effectively binds calcium, in agreement with 45Ca binding results.
Centrin4p Does Not Bind to Kar1p Peptide
Despite numerous investigations, the centriolar partners of mammalian centrins are still unknown. In contrast, in S. cerevisiae, the centrin protein Cdc31p anchors to the SPB through its interaction with Kar1p (see INTRODUCTION). The Cdc31p-binding peptide in Kar1p was previously characterized and was shown to be recognized by Hscentrin2p in vitro in a calcium-regulated manner (Spang et al., 1995; Geier et al., 1996). To investigate whether Kar1p peptide is a target of centrin4p, NH2-terminal myc epitope-tagged Mmcentrin4p or Hscentrin2p were cotransfected in HeLa cells with Kar1p peptide in fusion with GFP (Figure 6A). The interactions were tested by examining whether centrin proteins were coimmunoprecipitated with Kar1p peptide in presence (Ca2+) or absence of calcium (EGTA). We observed that centrin2p effectively binds to Kar1p peptide and that this interaction is calcium dependent (Figure 6A, bottom). In contrast, no interaction between centrin4p and Kar1p peptide could be detected independently of the presence of calcium. As centrin proteins were expressed to a similar level (Figure 6A, top), this suggests that they do not recognize the same target peptide and that they could have different centriolar partners.
Centrin4p Is a Ca2+-binding Protein
As the four potential EF-hand calcium-binding domains of centrin4p seem to be essential for its proper localization to centrioles, we wanted to determine which domain(s) can bind a calcium ion. GFP-tagged centrin4p, its two splice variants, and the different truncated forms were ectopically expressed in HeLa cells and immunoprecipitated using anti-GFP monoclonal antibodies. Proteins were then separated by gel electrophoresis, transferred on a nitrocellulose membrane, and either revealed with anti-GFP antibodies or incubated with 45Ca (Figure 6B). Autoradiography revealed that the full-length centrin4p binds calcium (Figure 6B, lane 2). A radioactive signal was also obtained with the COOH-terminal domain (Cter4) and the two splice variants (Splice4A and 4B) of centrin4p that share only the fourth EF-hand calcium-binding domain. However, the signals obtained were weaker than with centrin4p and could reflect that deletions of calcium-binding domains modify the conformational stability of these truncated forms. In contrast, no signal was obtained with the NH2-terminal domain (Nter4) or with the truncated form (III-4 domain) lacking only the last EF-hand calcium-binding domain. Together, these results strongly suggest that only the fourth EF-hand calciumbinding domain of centrin4p is functional, in agreement with sequence analyses (Figure 2B). We further investigated electrophoretic mobility of centrin4p (our unpublished data) and of its COOH- and NH2-terminal domains (Cter4 and Nter4) in presence or absence of calcium, because it is well known that calmodulin shows a Ca2+-dependent migration behavior in SDS-PAGE (Figure 6C). We observed that centrin4p (our unpublished data) and its COOH-terminal domain migrate with a lower apparent molecular weight in presence (Ca2+) than in absence of calcium (EGTA), suggesting that the Ca2+-bound form could adopt a more compact conformation (Figure 6C). In contrast, the migration behavior of the NH2-terminal domain was unaffected in both conditions (Figure 6C). These results are in good agreement with our observation that centrin4p binds calcium only through its fourth EF-hand calcium-binding domain.
In the Adult Mouse Brain, Centrin4 mRNA Is Specifically Expressed in Ciliated Cells
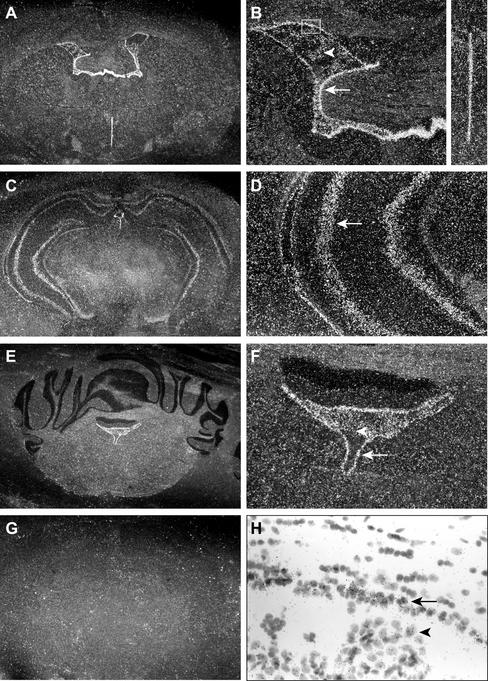
Centrin2p and centrin3p that seem to be involved in the regulation of centriole duplication in proliferating cells are both ubiquitously expressed in adult mouse tissues (see INTRODUCTION). In contrast, we showed by Northern blot analyses that centrin4 mRNA has a restricted expression profile with an unexpected high expression in the adult brain (cerebellum and hemispheres), a tissue containing mostly quiescent cells. To better understand the potential function(s) of centrin4p, we examined its distribution in this particular tissue by in situ hybridization with a specific 35S-labeled cRNA probe corresponding to the 3′-UTR of centrin4 mRNA. A strong expression of centrin4 mRNA was detected in cells lining the different ventricular cavities (i.e., lateral ventricles, third and fourth ventricles) (Figure 7, A–F). Centrin4 mRNA was also detected in the choroid plexus epithelium inside the lateral and fourth ventricles (Figure 7, B and F, arrowheads) and to a weaker level in the hippocampus (Ammon's horn and dentate gyrus) (Figure 7, C and D). Finally, no signal was detected in other brain areas. This restricted expression profile was very informative because it is known that ependymal cells lining the ventricles as well as cells from the plexus choroid have numerous cilia on their surface, which drive the transport of cerebrospinal fluid (Roth et al., 1985). Moreover, the presence of a primary nonmotile cilium in some neuronal cells, in particular hippocampal cells, has been described previously (Popov and Tsyganova, 1996; Handel et al., 1999). Together, these results suggest that centrin4 mRNA is specifically expressed, at least in the brain, in cells bearing motile cilia and to a lower extent in some cells with a primary nonmotile cilium.
Figure 7.
Centrin4 mRNA is expressed in ciliated cells in the brain. In situ hybridization views of centrin4 mRNA distribution in successive coronal sections of adult mouse brain. The orientation is antero-posterior from top to bottom. Dark field images (A–G), white field image (H). B, D, and F correspond to fourfold magnification of A, C, and E, respectively. (A and B) Centrin4 mRNA is expressed in ependymal cells lining lateral (arrow) and third ventricles as well as in choroidal cells (arrowhead). (C and D) Centrin4 mRNA is detected in hippocampal cells (arrow). (E and F) Centrin4 mRNA is also expressed in ependymal cells (arrow) lining the fourth ventricle as well as in choroidal cells (arrowhead) in a cerebellum section. (G) Control hybridization with the corresponding sense probe on a brain section equivalent to A. (H) White field image of an area corresponding to the white box in B. Radioactive signals are clearly associated with ependymal cells (arrow) and choroidal cells (arrowhead).
Centrin4p Is Localized to Basal Bodies in Ciliated Cells
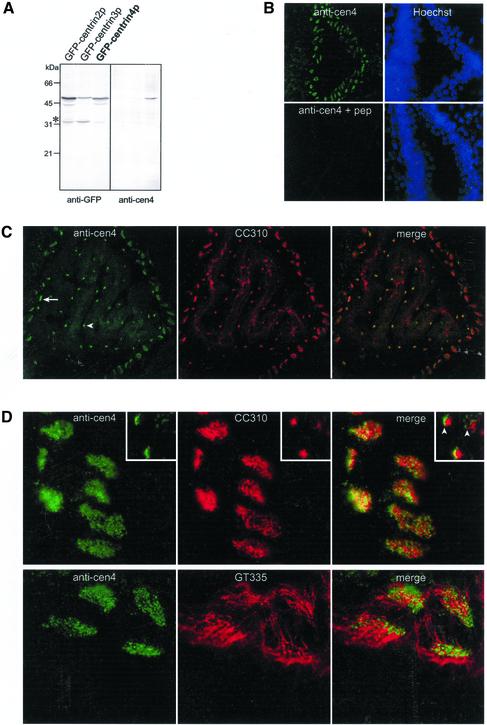
To confirm the expression of centrin4p in ciliated cells, we generated a rabbit polyclonal antiserum directed against a specific amino-terminal peptide (see MATERIALS AND METHODS). We first examined its specificity by an immunoblot analysis. Mouse L929 cells were transiently transfected with GFP-tagged centrin2p, centrin3p, or centrin4p and processed for immunoblotting with anti-centrin4p or anti-GFP antibodies (Figure 8A). We observed that the anti-centrin4p antiserum recognized one polypeptide corresponding to GFP-centrin4p but did not cross-react with GFP-centrin2p or GFP-centrin3p. Moreover, no other protein was detected in cell mouse extracts (Figure 8A). We next tested the specificity of this antiserum by immunofluorescence on mouse brain slices. The anti-centrin4p antiserum yielded a strong signal in cells lining the ventricles (see below) that was totally abolished by a preincubation with the corresponding peptide (Figure 8B). Finally, we were unable to detect endogenous centrin4p by a Western blot analysis on mouse brain extracts probably due to the fact that this protein is expressed in a very small fraction of cells. Together, these results show that our antiserum specifically recognizes centrin4p.
Figure 8.
Centrin4p localizes to basal bodies in ciliated cells. (A) Western blot analysis of the specificity of the anti-centrin4p antiserum. Mouse L929 cells were transiently transfected with GFP-tagged centrin2p, centrin3p, or centrin4p and processed for immunoblotting with anti-centrin4p or anti-GFP antibodies (25 μg of total protein extract per lane). Note that only GFP-centrin4p is recognized by anti-centrin4p antiserum. Lower bands correspond most probably to degradation products (asterisk). (B) Mouse brain sections were labeled with anti-centrin4p antiserum (1:500) preincubated or not with the corresponding peptide (5 μg/μl serum). Nuclei were visualized by a DAPI staining. A centrin4p staining was detected in cells lining the lateral ventricle and was totally abolished by a preincubation with the antigenic peptide. (C) Double-labeling experiments with anti-centrin4p antiserum and a mAb that recognizes ciliary rootlets (CC310). Centrin4p is specifically expressed in ependymal (arrow) and choroidal (arrowhead) ciliated cells. (D) Top, centrin4p and CC310 staining in ependymal cells do not colocalize. Insets, on a lateral view of choroidal cells, centrin4p dot staining is detected near the apical surface (arrowheads). Bottom, double labeling experiments with anti-centrin4p antiserum and a mAb that stains cilia axonemes (GT335). Centrin4p staining is restricted at the base of cilia.
Using this antiserum, centrin4p was exclusively detected in the brain in cells lining the different ventricles and in the choroid plexus, in good agreement with our in situ hybridization results (Figure 8C, left). However, we were unable to detect centrin4p in hippocampal cells, possibly as a consequence of a lower expression level in these cells, which possess only a monocilium. To unambiguously identify cells expressing centrin4p, we performed a double-immunostaining with a mAb (CC310) that specifically recognizes ciliary rootlets in ependymal and choroidal cells (Peraldi-Roux et al., 1991). This experiment confirmed that centrin4p is exclusively expressed in ciliated cells (Figure 8C). Closer examination revealed a punctuate staining of centrin4p that is less abundant in choroidal than in ependymal cells (Figure 8D, top, insets correspond to choroidal cells). This dot staining does not colocalize with the ciliary rootlet staining in ependymal and choroidal cells (Figure 8D, top). Moreover, on a lateral view of choroidal cells, centrin4p staining is clearly localized in a more apical position than the ciliary rootlet staining (arrowheads). Together, these observations strongly suggest that centrin4p is localized to basal bodies, in agreement with our observations that centrin4p accumulates to centrioles when ectopically expressed in different cell lines. Finally, we performed a double-immunostaining using GT335, a mAb directed against glutamylated tubulins, which stain axonemes of ciliated cells (Wolff et al., 1992; Tournier et al., 1998). Centrin4p staining was restricted at the base of cilia and no colocalization was detected in cilia axonemes (Figure 8D, bottom). Together, our results show that centrin4p is specifically expressed in ciliated cells in the brain and strongly suggest that it is concentrated to basal bodies at the origin of cilia axonemes.
DISCUSSION
In the present study, we isolated a novel mammalian centrin that we called centrin4p. Examination of its amino acid sequence revealed that centrin4p is closely related to the previously identified centrin2p. Moreover, we observed that centrin4 and centrin2 genes share exactly the same intron/exon organization, suggesting that both genes arise from the duplication of a common ancestor. However, in contrast to centrin2p, centrin4p has a restricted tissue expression profile and possesses two splice variants. We observed that ectopically expressed centrin4p accumulates to centrioles and procentrioles, as described previously for centrin2p, but with different molecular properties (see below, Paoletti et al., 1996). In contrast to centrin4p, its two splice variants were never found associated with centrioles. We do not investigate at the present time the regulation of the splicing of centrin4 mRNA, but our observations raise the possibility that centrin4p activity could be regulated by a posttranscriptional regulation mechanism.
Examination of the subcellular localization of centrin4p expressed in different cell lines showed that this protein shares with other mammalian centrins the ability to accumulate to centrioles but with different properties. In fact, centrin4p accumulates to centrioles at a lower level than centrin2p or centrin3p. Moreover, we demonstrated that the COOH-terminal domain (constituted of the two last EF-hand calcium-binding domains) of centrin2p and centrin3p is necessary and sufficient for their localization to centrioles, whereas both NH2 and COOH-terminal domains of centrin4p are required for its proper localization. Based on the relationship between centrin proteins and calmodulin, centrin4p could have a target interaction mechanism more similar to calmodulin than other centrins, in spite of the fact that only its fourth EF-hand calcium-binding domain seems to be functional. In fact, in vitro structural studies have shown that calmodulin binds both ends of its target peptides through their interaction with its NH2 and COOH-terminal domains (for review, see Zhang and Yuan, 1998). On the other hand, the observation that only half of centrin2p or centrin3p is sufficient for their anchoring to centrioles was totally unexpected. However, it could be related to mutational studies of the yeast centrin, suggesting that the COOH-terminal domain of Cdc31p mediates its interaction with Kar1p and possibly its correct localization to the SPB (Ivanovska and Rose, 2001). Moreover, recent structural analyses of C. reinhardtii centrin showed that its interaction with Kar1p peptide in vitro is primarily mediated by its COOH-terminal domain (Veeraraghavan et al., 2002). Thus, the target interaction mechanism of centrin2p and centrin3p could be different from that of calmodulin and centrin4p and involve mostly their COOH-terminal domain. Interestingly, one centrin-related protein was recently discovered in Dictyostelium discoideum, which possesses only two EF-hands in its COOH-terminal domain, suggesting that this protein could share the same target interaction mechanism with centrins 2p/3p (Daunderer et al., 2001).
In contrast to centrin2 and centrin3 mRNA, which are ubiquitously expressed, we observed that centrin4 mRNA is highly expressed in the brain, lung, kidney, and ovary and is expressed to a very low level in the spleen. Interestingly, this expression profile is very similar to that of Polaris, the product of the Tg737 gene, which is expressed in brain, kidney, lung, and in the reproductive tracts (oviduct and testis) (Taulman et al., 2001). Moreover, in brain, Polaris shares the same cellular distribution with centrin4p and is specifically expressed in multiciliated ependymal and choroidal cells where it is localized to basal bodies and cilia axonemes (Taulman et al., 2001). In the other tissues, Polaris was detected in the ciliated lung epithelium, in sperm, and in kidney cells harboring a monocilium where it is also localized to basal bodies and cilia or flagella axonemes. Thus, similar tissue expression profiles as well as similar cellular and subcellular localizations in the brain raise the possibility that centrin4p could be expressed in other ciliated epithelia bearing motile cilia (possibly ciliated epithelia from lung and oviduct). Centrin4p could be also expressed in different cells harboring a primary immotile cilium. Interestingly, presence of cells harboring a monocilium in kidney, spleen, and ovary has been described previously (Abdel-Bari and Sorenson, 1965; Motta et al., 1971). Further experiments will be performed to investigate this hypothesis. We also noticed similarities in the expression pattern of centrin4 and that of HFH-4 mRNA, which encodes a forkhead transcription factor involved in the ciliogenesis process of lung, oviduct, choroid plexus, and ependyma epithelia (Blatt et al., 1999; Brody et al., 2000). The target genes regulated by this transcription factor are still unknown, but a DNA consensus sequence specifically recognized by HFH-4 was identified (Lim et al., 1997). Interestingly, we found in the CETN4 gene sequence a putative HFH-4 binding site localized upstream of the transcription initiation site (our unpublished data). Thus, it will be of interest to investigate whether CETN4 gene expression is under control of this transcription factor.
We observed that centrin4p is specifically expressed in ciliated cells in the adult mouse brain where it is localized to basal bodies at the origin of cilia axonemes, in agreement with its ability to accumulate to centrioles when ectopically expressed in different cell lines. Examination of the expression profiles of centrin2p and centrin3p in the brain showed that both proteins are also expressed in ciliated cells lining the different ventricles where they are localized to basal bodies (Gavet et al., unpublished data). However, both proteins are broadly expressed in the brain and are found associated to the centrosome in neuronal and glial cells (Gavet et al., unpublished data). Thus, the more restricted expression profile of centrin4p versus centrins 2p/3p in the brain is a first indication that centrin4p could be more specifically involved in the differentiation process of some ciliated cells. In agreement with this hypothesis, we observed that centrin4p expression correlates with the apparition of CC310 immunoreactivity in ependymal cells during brain development (Gavet et al., unpublished data). The ciliogenesis process in multiciliated cells of vertebrate tissues has been morphologically described (for review, see Dirksen, 1991). The first cytoplasmic structures involved in centriologenesis are fibrogranular aggregates that consist of clouds of filamentous material associated with electron-dense granules. Immature centrioles occur around these electron-dense granules and then mature and migrate to the apical membrane where they trigger axonemal microtubule polymerization and cilia formation. Interestingly, during the differentiation of nasal epithelial ciliated cells in culture, centrin2p and centrin3p were found associated with fibrogranular aggregates and elongating procentrioles, suggesting a role of these proteins in an early step of centriologenesis (Laoukili et al., 2000). Because centrin2p and centrin3p were detected in ependymal and choroidal ciliated cells and as we observed that centrin4p expression in these cells correlated with their differentiation, one could imagine that centrins 2p/3p could be involved in an early step of centriologenesis, whereas centrin4p could be involved later in basal body maturation or in cilia formation and associated functions. In agreement with this hypothesis, it was shown that the incubation of permeabilized nasal epithelial ciliated cells with an anti-centrin antibody that recognizes both centrin1p and 2p inhibits ciliary beating, suggesting that some centrin proteins, possibly centrin4p, could be involved in cilia associated functions (Laoukili et al., 2000). However, at the present time, we cannot exclude that centrin4p could be also involved in an early step of de novo centriole assembly.
It will be difficult to investigate centrin4p function(s) in primary cultures of ciliated cells. In fact, the differentiation process of ciliated cells in vitro takes many weeks and limits the utilization of classical approaches like the overexpression of dominant negative forms or the inhibition of expression by RNA interference. Thus, the best way to investigate whether centrin4p is effectively involved in the differentiation process of ciliated cells and in which step of ciliogenesis it could be required will be to generate CETN4 null mice. Interestingly, inactivation of the HFH-4 gene as well as partial loss of Polaris functions in Tg737orpk (hypomorphic allele) mutant mice leads to a hydrocephalus phenotype as a consequence of the absence or reduced number of cilia in ependymal cells (Brody et al., 2000; Taulman et al., 2001). Detailed analyses of the morphology of ciliated cells in these mutant mice allowed to identify in which step of ciliogenesis the corresponding proteins are required. Thus, Polaris seems to be involved in ciliary assembly, whereas HFH-4 seems to control the expression of proteins required for centriole migration and/or docking to the apical membrane (Brody et al., 2000; Pazour et al., 2000).
To date, vertebrate organisms seem to express two ubiquitous centrins (centrin2p and centrin3p) and two more specific “ciliary” centrins (centrin1p and centrin4p). Due to the absence of analyses of centrin1 mRNA expression by in situ hybridization and the lack of anti-centrin antibodies that fully discriminate between centrin1p and centrin2p, the expression profile of centrin1p and its relation with that of centrin4p is still unclear. Centrin1 mRNA was clearly detected in testis by a Northern blot analysis, in contrast to centrin4 mRNA (Hart et al., 1999; this work). Centrin1 expression was also detected by RT-PCR in retina photoreceptor cells, which possess a very specialized primary nonmotile cilium, the connecting cilium (Wolfrum and Salisbury, 1998). Interestingly, in these cells, centrin1p was shown to interact with the heterotrimeric G protein transducin of the visual transduction cascade (Pulvermuller et al., 2002). However, the expression profile of centrin1 mRNA in the epithelial cells of respiratory tracts seems to be more confusing. In fact, centrin1 mRNA was not detected in lung by a Northern blot analysis, in contrast to centrin4 mRNA (Hart et al., 1999; this work). On the other hand, centrin1 expression was detected by RT-PCR in human nasal epithelial ciliated cells but not in human tracheal epithelial ciliated cells (LeDizet et al., 1998; Laoukili et al., 2000). Thus, it will be interesting to examine whether ciliated cells of the different epithelia of respiratory tracts, which harbor multiple motile cilia, can express both centrin1p and centrin4p or whether the expression patterns of these proteins are mutually exclusive.
Acknowledgments
We thank O. Smrzka, Y. Abraham, and O. Cases for stimulating discussions. We thank F. Tournier, V. Marthiens, and V. Doye for critical reading of this manuscript. This work was supported by the Center National pour la Recherche Scientifique and the Institut Curie.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-11-0709. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0709.
References
- Abdel-Bari, W., and Sorenson, G.D. (1965). Ciliated cells in the spleen of adult rats. Anat. Rec. 152, 481–485. [DOI] [PubMed] [Google Scholar]
- Baum, P., Furlong, C., and Byers, B. (1986). Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 83, 5512–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and Rose, M.D. (1994). Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 125, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, E.N., Yan, X.H., Wuerffel, M.K., Hamilos, D.L., and Brody, S.L. (1999). Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am. J. Respir. Cell. Mol. Biol. 21, 168–176. [DOI] [PubMed] [Google Scholar]
- Bornens, M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34. [DOI] [PubMed] [Google Scholar]
- Brinkley, B.R. (2001). Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 11, 18–21. [DOI] [PubMed] [Google Scholar]
- Brody, S.L., Yan, X.H., Wuerffel, M.K., Song, S.K., and Shapiro, S.D. (2000). Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell. Mol. Biol. 23, 45–51. [DOI] [PubMed] [Google Scholar]
- Daunderer, C., Schliwa, M., and Graf, R. (2001). Dictyostelium centrin-related protein (DdCrp), the most divergent member of the centrin family, possesses only two EF hands and dissociates from the centrosome during mitosis. Eur. J. Cell Biol. 80, 621–630. [DOI] [PubMed] [Google Scholar]
- Dirksen, E.R. (1991). Centriole and basal body formation during ciliogenesis revisited. Biol Cell 72, 31–38. [DOI] [PubMed] [Google Scholar]
- Errabolu, R., Sanders, M.A., and Salisbury, J.L. (1994). Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 107, 9–16. [DOI] [PubMed] [Google Scholar]
- Geier, B.M., Wiech, H., and Schiebel, E. (1996). Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J. Biol. Chem. 271, 28366–28374. [DOI] [PubMed] [Google Scholar]
- Handel, M., Schulz, S., Stanarius, A., Schreff, M., Erdtmann-Vourliotis, M., Schmidt, H., Wolf, G., and Hollt, V. (1999). Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 89, 909–926. [DOI] [PubMed] [Google Scholar]
- Hart, P.E., Glantz, J.N., Orth, J.D., Poynter, G.M., and Salisbury, J.L. (1999). Testis-specific murine centrin, Cetn1: genomic characterization and evidence for retroposition of a gene encoding a centrosome protein. Genomics 60, 111–120. [DOI] [PubMed] [Google Scholar]
- Ivanovska, I., and Rose, M.D. (2001). Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics 157, 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz, C., Bordes, N., Laine, M.C., Sandoz, D., and Bornens, M. (1986). A protein of 175,000 daltons associated with striated rootlets in ciliated epithelia, as revealed by a monoclonal antibody. Cell Motil. Cytoskeleton 6, 56–67. [DOI] [PubMed] [Google Scholar]
- Kochanski, R.S., and Borisy, G.G. (1990). Mode of centriole duplication and distribution. J. Cell Biol. 110, 1599–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Kuriyama, R., and Borisy, G.G. (1981). Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 91, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili, J., Perret, E., Middendorp, S., Houcine, O., Guennou, C., Marano, F., Bornens, M., and Tournier, F. (2000). Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113, 1355–1364. [DOI] [PubMed] [Google Scholar]
- LeDizet, M., Beck, J.C., and Finkbeiner, W.E. (1998). Differential regulation of centrin genes during ciliogenesis in human tracheal epithelial cells. Am. J. Physiol. 275, L1145–L1156. [DOI] [PubMed] [Google Scholar]
- Lee, V.D., and Huang, B. (1993). Molecular cloning and centrosomal localization of human caltractin. Proc. Natl. Acad. Sci. USA 90, 11039–11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L., Zhou, H., and Costa, R.H. (1997). The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proc. Natl. Acad. Sci. USA 94, 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle, W.L., Lutz, W.H., Ingle, J.N., Maihle, N.J., and Salisbury, J.L. (1998). Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA 95, 2950–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden, B.J., Shaw, G.S., and Sykes, B.D. (1990). Calcium binding proteins. Elucidating the contributions to calcium affinity from an analysis of species variants and peptide fragments. Biochem. Cell Biol. 68, 587–601. [DOI] [PubMed] [Google Scholar]
- Marshall, W.F., Vucica, Y., and Rosenbaum, J.L. (2001). Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 11, 308–317. [DOI] [PubMed] [Google Scholar]
- Maruyama, K., Mikawa, T., and Ebashi, S. (1984). Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 95, 511–519. [DOI] [PubMed] [Google Scholar]
- Middendorp, S., Kuntziger, T., Abraham, Y., Holmes, S., Bordes, N., Paintrand, M., Paoletti, A., and Bornens, M. (2000). A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp, S., Paoletti, A., Schiebel, E., and Bornens, M. (1997). Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA 94, 9141–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.A., Dykes, D.D., and Polesky, H.F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta, P., Takeva, Z., and Palermo, D. (1971). On the presence of cilia in different cells of the mammalian ovary. Acta Anat. 78, 591–603. [DOI] [PubMed] [Google Scholar]
- Moudjou, M., Bordes, N., Paintrand, M., and Bornens, M. (1996). γ-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J. Cell Sci. 109, 875–887. [DOI] [PubMed] [Google Scholar]
- Nelson, M.R., and Chazin, W.J. (1998). Calmodulin and Signal Transduction, San Diego: Academic Press.
- O'Neil, K.T., and DeGrado, W.F. (1990). How calmodulin binds its targets: sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64. [DOI] [PubMed] [Google Scholar]
- Paoletti, A., Moudjou, M., Paintrand, M., Salisbury, J.L., and Bornens, M. (1996). Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., Vucica, Y., Seeley, E.S., Rosenbaum, J.L., Witman, G.B., and Cole, D.G. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraldi-Roux, S., Klotz, C., Nguyen-Thanh-Dao, B., and Gabrion, J. (1991). A common epitope is shared by ciliary rootlets and cell-cell adherens junctions in ciliated ependymal cells. J. Cell Sci. 99, 297–306. [DOI] [PubMed] [Google Scholar]
- Pihan, G.A., Purohit, A., Wallace, J., Knecht, H., Woda, B., Quesen-berry, P., and Doxsey, S.J. (1998). Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58, 3974–3985. [PubMed] [Google Scholar]
- Popov, V.I., and Tsyganova, V.G. (1996). Replication of centrioles and differentiation of neurons in hippocampal slice cultures and olfactory neuroepithelium in the rat. Neurosci. Lett. 203, 135–138. [DOI] [PubMed] [Google Scholar]
- Pulvermuller, A., Giessl, A., Heck, M., Wottrich, R., Schmitt, A., Ernst, O.P., Choe, H.W., Hofmann, K.P., and Wolfrum, U. (2002). Calcium-dependent assembly of centrin-G-protein complex in photoreceptor cells. Mol. Cell. Biol. 22, 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadroni, M., James, P., and Carafoli, E. (1994). Isolation of phosphorylated calmodulin from rat liver and identification of the in vivo phosphorylation sites. J. Biol. Chem. 269, 16116–16122. [PubMed] [Google Scholar]
- Rose, M.D., and Fink, G.R. (1987). KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48, 1047–1060. [DOI] [PubMed] [Google Scholar]
- Roth, Y., Kimhi, Y., Edery, H., Aharonson, E., and Priel, Z. (1985). Ciliary motility in brain ventricular system and trachea of hamsters. Brain Res. 330, 291–297. [DOI] [PubMed] [Google Scholar]
- Salisbury, J., Suino, K., Busby, R., and Springett, M. (2002). Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Spang, A., Courtney, I., Fackler, U., Matzner, M., and Schiebel, E. (1993). The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., Courtney, I., Grein, K., Matzner, M., and Schiebel, E. (1995). The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, G.S., Stein, J.L., Lian, J.B., Last, T.J., Owen, T., and McCabe, L. (1994) Cell Biology: A Laboratory Handbook, San Diego: Academic Press.
- Taillon, B.E., Adler, S.A., Suhan, J.P., and Jarvik, J.W. (1992). Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119, 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulman, P.D., Haycraft, C.J., Balkovetz, D.F., and Yoder, B.K. (2001). Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol. Biol. Cell 12, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier, F., Laoukili, J., Giuliani, I., Gendron, M.C., Guennou, C., and Marano, F. (1998). Ciliated differentiation of rabbit tracheal epithelial cells in vitro. Eur. J. Cell Biol. 77, 205–213. [DOI] [PubMed] [Google Scholar]
- Vallen, E.A., Ho, W., Winey, M., and Rose, M.D. (1994). Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics 137, 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraraghavan, S., Fagan, P.A., Hu, H., Lee, V., Harper, J.F., Huang, B., and Chazin, W.J. (2002). Structural independence of the two EF-hand domains of caltractin. J. Biol. Chem. 277, 28564–28571. [DOI] [PubMed] [Google Scholar]
- Vorobjev, I.A., and Chentsov Yu, S. (1982). Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93, 938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, A., de Nechaud, B., Chillet, D., Mazarguil, H., Desbruyeres, E., Audebert, S., Edde, B., Gros, F., and Denoulet, P. (1992). Distribution of glutamylated α and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59, 425–432. [PubMed] [Google Scholar]
- Wolfrum, U., and Salisbury, J.L. (1998). Expression of centrin isoforms in the mammalian retina. Exp. Cell Res. 242, 10–17. [DOI] [PubMed] [Google Scholar]
- Zhang, M., and Yuan, T. (1998). Molecular mechanisms of calmodulin's functional versatility. Biochem. Cell Biol. 76, 313–323. [DOI] [PubMed] [Google Scholar]