Abstract
The majority of Rab proteins are posttranslationally modified with two geranylgeranyl lipid moieties that enable their stable association with membranes. In this study, we present evidence to demonstrate that there is a specific lipid requirement for Rab protein localization and function. Substitution of different prenyl anchors on Rab GTPases does not lead to correct function. In the case of YPT1 and SEC4, two essential Rab genes in Saccharomyces cerevisiae, alternative lipid tails cannot support life when present as the sole source of YPT1 and SEC4. Furthermore, our data suggest that double geranyl-geranyl groups are required for Rab proteins to correctly localize to their characteristic organelle membrane. We have identified a factor, Yip1p that specifically binds the di-geranylgeranylated Rab and does not interact with mono-prenylated Rab proteins. This is the first demonstration that the double prenylation modification of Rab proteins is an important feature in the function of this small GTPase family and adds specific prenylation to the already known determinants of Rab localization.
INTRODUCTION
Rab proteins are GTPase superfamily members that regulate membrane trafficking through the secretory and endocytic pathways. The Rab proteins represent the numerically largest subgroup of the Ras superfamily, and it is thought that each stage of membrane transport through both constitutive cellular pathways and in differentiated cells with specialized organelles is associated with one or more Rab proteins (Pfeffer, 2001). The mechanism by which Rab proteins act to regulate membrane-trafficking events is not fully understood. A plethora of different effector proteins have been identified connecting the activated Rab proteins to a variety of cellular events such as cytoskeletal dynamics, phosphatidylinositol signaling events, protein kinase-mediated signal transduction, and the establishment of cell cycle-linked spatial cues (Ren et al., 1996; Finger et al., 1998; Christoforidis et al., 1999; Nielsen et al., 1999). Given the complexity of the exocytic and endocytic trafficking pathways, it makes sense for cells to link Rab protein activation to a variety of different outcomes depending on the requirements of the various organelles involved.
Recently, a consensus seems to be emerging that one commonality of Rab protein function is to participate in the tethering of a vesicle or membrane transport carrier (Pfeffer, 1999). Tethering refers to the process by which the membrane bound transport carriers dock onto the acceptor compartment. Tethering is the prelude to, and initiator of the cascade of events that terminate in a soluble N-ethylmaleimide-sensitive factor attachment protein receptor-mediated membrane fusion event. Even tethering may be controlled by Rab proteins through diverse mechanisms; for example, the tethering of constitutive post-Golgi vesicles at the plasma membrane is a transient event and very different from the rapid, signal-mediated fusion of synaptic post-Golgi vesicles that may exist in the tethered state for a prolonged time period (Wang et al., 1997). These examples, in turn, differ in their requirements from the fusion of post-Golgi vesicles with the plasma membrane of budding yeast, where tethering events must be spatially regulated and are coordinated with cell cycle progression.
Ras GTPases function as regulatory switches where the GDP-bound is the ground or “off” state and GTP-bound is the activated state (Vetter and Wittinghofer, 2001). It is still unclear whether Rab proteins function as binary switches in a similar manner to Ras with a single GTP turnover event for each round of transport. An alternative modality is suggested by analogy to the Rho family GTPase CDC42 where it is the rate of cycling, rather than the lifetime of the activated state, that is important for initiation of downstream events (Rybin et al., 1996; Lin et al., 1997).
One characteristic that Rab proteins do share with other members of the Ras superfamily is that these proteins are posttranslationally modified by the covalent attachment of isoprenoids on cysteine residues at the C terminus (Seabra, 1998). For the Ras, Rho, and Rab families, there are three major types of isoprenylation reactions mediated by three prenyl transferases that are conserved from yeast to human (for review, see Liang et al., 2002). The cysteine-containing motifs at the C terminus dictate the type of isoprenylation received by the small GTPase. A CAAX box where C is cysteine, A is aliphatic residue, and X is A, C, E, M, S, or V such as in H-Ras, K-Ras, and yeast Ras1p and Ras2p, is modified by farnesylation (C15 isoprenoid) by farensyl transferase (FTase). When X is leucine or a hydrophobic residue, typically found in Rho proteins such as CDC42, this is as substrate for geranylgeranyl transferase I (GGTase I), which attaches a C20 isoprenoid moiety. Rab proteins fall into a special category. The majority of them contain two cysteine residues at the C terminus in one of the following sequences: CXC, CC, CCX, CCXX, or CCXXX. The cysteine residues are subject to isoprenylation with two geranylgeranyl moieties catalyzed by geranylgeranyl transferase II (GGTase II). All the prenyltansferase enzymes consist of two subunits, however, in the case of GGTaseII, there is a third subunit, Rab escort protein (REP), which does not participate in the catalytic reaction but serves as a chaperone to introduce the prenyltransferase to its Rab protein substrate (Desnoyoyers et al., 1996).
Ras, Rho, and Rab superfamily members can be found in both membrane-associated and cytosolic pools. It is clear that prenylation is a necessary modification for the protein to be present in the membrane-bound pool, Ras superfamily members with mutations in their C-terminal cysteines that cannot be prenylated are soluble and nonfunctional (Walworth et al., 1989). Such experiments have propagated the view that the sole function of prenylation is to confer hydrophobic character onto a cytosolic protein, giving the recipient protein the physical ability to make a stable attachment with a lipid bilayer. In this study, we have focused on the question of what role, if any, is played by the particular type of lipid modification. Using Saccharomyces cerevisiae as a model system, we have examined the effect of different lipid modifications on Rab protein localization and function.
MATERIALS AND METHODS
Yeast Strains and Media
The S. cerevisiae strains used in these studies are listed in Table 1. All yeast strains were manipulated as described by Guthrie and Fink (2002). Yeast expressing various green fluorescent protein (GFP)-Rab proteins (both wild-type and prenylation mutants) were created by transforming the appropriate plasmids (Table 1) into NY605. Yeast strains were streaked on selection media plates and incubated at 30°C. Liquid media cultures were grown at room temperature. A single colony from each strain was inoculated into 5 ml of selective medium and grown to stationary phase. For fluorescence microscopy, selective media were inoculated with an aliquot of the stationary culture and grown to early to mid-log phase. Cells were then incubated for 5 min with 5 μg/ml Hoechst for nuclear visualization before image capture. For Triton X-114 partition experiments, an aliquot of the stationary phase was inoculated into 50 ml of selective media, and the cells were grown to mid-log phase. Turbidity measurements were made using a Thermo Spectronic Genesys (Rochester, NY) 10UV spectrophotometer at 600 nm.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY85 | MATa/α ura3-52/ura3-52 leu2-3, 112/leu2-3, 112 his3Δ200/his3Δ200 YPT1/YPT1ΔHIS3 | Brennwald laboratory |
| BY86 | MATa/α ura3-52/ura3-52 leu2-3, 112/leu2-3, 112 his3Δ200/his3Δ200 SEC4/SEC4ΔHIS3 | Brennwald laboratory |
| AG6 | LEU2::SEC4(L7,HV)YPSEC4Δ::HIS3 his3Δ200 ura3-52 | Brennwald laboratory |
| BY24 | MATa/α ura3-52/ura3-52 leu2-3, 112/leu2-3, 112 | Brennwald laboratory |
| NY605 | MATaura3-52 leu2-3, 112 | Novick laboratory |
| RCY1507 | MATα ura3-52 leu2-3, 112 his3Δ200 SEC4ΔHIS3 [YCP50 SEC4] | This study |
| RCY1510 | ura3-52 leu2-3, 112 his3Δ200 YPT1ΔHIS3 [pRS316 (pRC1762) YPT1] | This study |
| RCY 1530 | MATa/α ura3-52/ura3-52 leu2-3, 112/leu2-3, 112 YIP1/YIP1ΔKANR [YCP50 (pRC1245) YIP1] | This study |
| RCY1610 | MATaura3-52 leu2-3, 112 YIP1ΔKANR [YCP50 (pRC1245) YIP1] | This study |
| RCY1764 | MATaura3-52 leu2-3, 112 YIP1ΔKANR [pRS315 yip1-4] | This study |
| RCY1760 | MATaura3-52 leu2-3, 112 YIP1ΔKANR [pRS315 YIP1] | This study |
| Y190 | MATagal4Δ gal80Δ trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL10 → LacZ, LYS2::GAL10 → HIS3 cyhR | Elledge laboratory |
Plasmid Constructs
Plasmid constructs and oligonucleotides are listed in Tables 2 and 3. Yeast Rab genes were cloned under the control of an endogenous promoter and terminator with yeast-enhanced GFP (GenBank accession no. U73901) fused in frame at the N terminus by polymerase chain reaction (PCR) and cloned into the CEN LEU2 vector pRS315 or pRS316 to generate plasmids containing GFP-tagged genes. Plasmids with wild-type Rab genes were used as templates to generate the GFP-tagged C-terminal prenylation variants. The prenylation mutants with a terminal CTIM sequence were cloned into pRS315 or pRS316 by overlap PCR by placing the C terminus of RHO3 containing the sequence CTIM and terminator in place of the two terminal cysteines by using the forward primer MC25 and reverse primers with overlap sequence to MC25 for SEC4 (MC27), YPT7 (MC26), VPS21 (MC31), YPT1 (MC32), and YPT6 (MC30). The Rab prenylation variants with a terminal CIIL sequence were cloned into pRS315 or pRS316 by overlap PCR by using specific forward and reverse primers with nucleotide sequence coding for CIIL and the endogenous terminators of each Rab. Primers MC54 and MC55 were used for cloning SEC4, MC58, and MC59 for YPT7, MC60 and MC61 for YPT6, and MC64 and MC65 for YPT1. For SEC4C214S, YPT1C205S, SEC4ΔCC, and YPT1ΔCC, a similar approach was used with primers MC66, MC67, MC68, MC69, MC70, MC71, MC72, and MC73. VPS21CIIL was cloned with primer MC75, which anneals to the C terminus of VPS21 and overlaps with MC74, a primer-encoding CIIL sequence and the terminator of SEC4. Untagged wild-type SEC4 and YPT1 were constructed with genomic PCR by using primers YFSEC4, YRSEC4, YFYPT1, and YRYPT1 and cloned into pRS315 or pRS316. Untagged prenylation variants of SEC4 and YPT1 were cloned into pRS315 and pRS316 by overlap PCR by using YFSEC4 and YFYPT1 and primers as described above for the GFP-tagged variants. The primers RNC200 and RNC201 were used with genomic DNA template to clone full-length GDP-dissociation inhibitor 1 (GDI1) into the vectors YEP24 and YCP50. SEC4, SEC4CTIM, SEC4CIIL, SEC4C214S, and SEC4ΔCC were cloned into pAS2–1 to create two-hybrid “bait” plasmids. SEC4CTIM and SEC4C214S were cloned using primers NS1 with YRRHO3, and NS2 with RNC264, respectively, and subcloned in-frame into pAS2-1 vector. SEC4CIIL was cloned by genomic PCR with primers MC56 and MC57 and subcloned into pAS2–1. Other Y2H constructs used have been described previously (Calero et al., 2002). Sec7p was tagged with Discosoma red fluorescent protein (DsRed)T4 (Bevis and Glick, 2002) at the C terminus with the linker sequence GGPGG and subcloned into pRS316 with the endogenous promoter and 572 bp from the ADH1 3′ region to create pRC2240. A human open reading frame (ORF) encoding a protein with homology to Yip1p was reconstructed by alignment of accession numbers AA171435, AA373289, H83008, N73033, R88629, T71419, and W17013. The ORF was cloned by coupled reverse transcription-PCR by using the reverse transcription primer 5′-GGCCACGCGTCGACTAGTAC(T)17 and gene-specific PCR primers 5′-CTGGATCCTCGCAATGTCAGGCTTTGAAAACTTAAACACGG and 5′-GATGCGCGTCTCGAGTCAAAAGACGGAAATCAGGGCAAAGAC. Then 250 ng of human skeletal muscle poly(A)+ RNA (BD Biosciences Clontech, Palo Alto, CA) was reverse transcribed with Superscript II according to the manufacturer's protocol. Purified cDNA was used as a template in PCR reactions to amplify human YIP1. The sequence of the human ORF is identical to the previously reported YIP1A (Tang et al., 2001). Oligonucleotides used in this study were from by Integrated DNA Technologies and Sigma Genosys (The Woodlands, TX). DNA sequencing was performed by the Cornell Biotechnology Facility by using dye terminator chemistry on an ABI 373 sequencer (Applied Biosystems, Foster City, CA).
Table 2.
Plasmids used in this study
| Plasmid name | Construct | Source |
|---|---|---|
| pRC651 | GFP-SEC4 pRS315 CEN LEU2 | This study |
| pRC2098 | GFP-SEC4 pRS316 CEN URA3 | This study |
| pRC1822 | GFP-SEC4CTIM pRS315 CEN LEU2 | This study |
| pRC1268 | GFP-SEC4CTIM pRS316 CEN URA3 | This study |
| pRC1286 | SEC4CTIM pRS426 2μ URA3 | This study |
| pRC1842 | GFP-SEC4CIIL pRS315 CEN LEU2 | This study |
| pRC1552 | GFP-SEC4CIIL pRS316 CEN URA3 | This study |
| pRC1860 | GFP-SEC4C214S pRS315 CEN LEU2 | This study |
| pRC1861 | GFP-SEC4C214S pRS316 CEN URA3 | This study |
| pRC1862 | GFP-SEC4ΔCC pRS315 CEN LEU2 | This study |
| pRC1863 | GFP-SEC4ΔCC pRS316 CEN URA3 | This study |
| pRC1820 | SEC4 pRS315 CEN LEU2 | This study |
| pNB139 | SEC4 YCP50 CEN URA3 | Novick lab |
| pRC1292 | SEC4CTIM pRS316 CEN URA3 | This study |
| pRC1824 | SEC4CTIM pRS315 CEN LEU2 | This study |
| pRC1743 | SEC4CIIL pRS315 CEN LEU2 | This study |
| pRC1728 | SEC4CIIL pRS316 CEN URA3 | This study |
| pRC1856 | SEC4C214S pRS315 CEN LEU2 | This study |
| pRC1857 | SEC4C214S pRS316 CEN URA3 | This study |
| pRC1858 | SEC4ΔCC pRS315 CEN LEU2 | This study |
| pRC1859 | SEC4ΔCC pRS316 CEN URA3 | This study |
| pRC2100B | GFP-YPT1 pRS315 CEN LEU2 | This study |
| pRC1840 | GFP-YPT1CTIM pRS315 CEN LEU2 | This study |
| pRC1752 | GFP-YPT1CIIL pRS315 CEN LEU2 | This study |
| pRC1840A | GFP-YPT1CTIM pRS316 CEN URA3 | This study |
| pRC1735 | YPT1 pRS315 CEN LEU2 | This study |
| pRC1762 | YPT1 pRS316 CEN URA3 | This study |
| pRC1829A | YPT1CTIM pRS315 CEN LEU2 | This study |
| pRC1828 | YPT1CTIM pRS316 CEN URA3 | This study |
| pRC1730 | YPT1CIIL pRS315 CEN LEU2 | This study |
| pRC1888A | GFP-YPT1C205S pRS315 CEN LEU2 | This study |
| pRC1889A | YPT1C205S pRS315 CEN LEU2 | This study |
| pRC1887A | GFP-YPT1ΔCC pRS315 CEN LEU2 | This study |
| pRC1884A | YPT1ΔCC pRS315 CEN LEU2 | This study |
| pRC1243 | GFP-YPT7 pRS316 CEN URA3 | This study |
| pRC1272 | GFP-YPT7CTIM pRS316 CEN URA3 | This study |
| pRC1560 | GFP-YPT7CIIL pRS316 CEN URA3 | This study |
| pRC650 | GFP-YPT6 pRS315 CEN LEU2 | This study |
| pRC1556 | GFP-YPT6CIIL pRS316 CEN URA3 | This study |
| pRC1544 | GFP-YPT6CTIM pRS316 CEN URA3 | This study |
| pRC680 | GFP-VPS21 pRS306 INT URA3 | This study |
| pRC1541 | GFP-VPS21CTIM pRS316 CEN URA3 | This study |
| pRC1964 | GFP-VPS21CIIL pRS315 CEN LEU2 | This study |
| pRC1462 | SEC4CTIM pAS2-1 | This study |
| pRC1798 | SEC4CIIL pAS2-1 | This study |
| pRC575 | YPT1 pACT2 | This study |
| pRC579 | SEC4 pACT2 | This study |
| pRC188 | YIP1 pAS1-CYH2 | This study |
| pRC2170 | PYIP1 → HsYIP1A pRS315 CEN LEU2 | This study |
| pRC1992 | yip1E70K pRS315 CEN LEU2 | This study |
| pRC1838b | YIP1 pRS315 CEN LEU2 | This study |
| pRC219 | pACT2 HsYIP1A | This study |
| pRC1803 | Rab8 pAS2-1 | This study |
| pRC1801 | Rab13 pAS2-1 | This study |
| pRC1802 | Rab13 pAS2-1 | This study |
| pRC787 | Rab1a pAS2-1 | This study |
| pRC763 | Rab5a pAS2-1 | This study |
| pRC2240 | Sec7p-T4DsRed pRS316 CEN URA3 | This study |
Table 3.
Oligonucleotides used in this study
| Primer name | Sequence 5′ to 3′ |
|---|---|
| MC25 | TGTACCATTATGTAATATAATAAG |
| MC26 | CTTATTATATTACATAATGGTACAAGAATTATTTTCTCCATCTAG |
| MC27 | CTTATTATATTACATAATGGTACAATTTGATTTAGAACTGTT |
| MC30 | CTTATTATATTACATAATGGTACAAGCGCTTTGTTCCTGCTC |
| MC31 | CTTATTATATTACATAATGGTACAAGCACTGTTTGCGCTGGT |
| MC32 | CTTATTATATTACATAATGGTACAGCCCCCACCGGTGTTGGTTAA |
| YRRH03 | TACCGGGCCCCCCCTCGAGGTCGACGCGTAAATCGTAACCATAGTAAG |
| YFSEC4 | TACCGGGCCCCCCCTCGAGGTCGACTTAGAACGAAATAAAAGTGCT |
| MC54 | AAATCAAATTGTATTATTTTGTGAAGAAAAGAAGATTTTTGCTTC |
| MC55 | TTCTTCACAAAATAATACAATTTGATTTAGAACTGTTTCC |
| MC56 | CATGCCATGGCATCAGGCTTGAGAACTGTTTC |
| MC57 | ATACTCGAGGCTTCTTTTCTTCACAAAATAA |
| MC58 | AATAATTCTTGTATTATTTTGTGAGCTGTACTACGTCGACCTCGA |
| MC59 | CAAAATAATACAAGAATTATTTTCTCCATCTAGGCGAATATT |
| MC60 | CAAAATAATACAAGAATTATTTTCTCCATCTAGGCGAATATT |
| MC61 | CAAAATAATACAAGCGCTTTGTTCCTGCTCCTCTGCTGTAGA |
| MC74 | TCTTCTTTTCTTCACAAAATAATGCATGCACTGTTTGCGCTGGT |
| MC75 | CATTATTTTGTGAAGAAAAGAAGATTTTTGC |
| YRSEC4 | ACCGGGCCCCCCCTCGAGGTCGACAAGTAGTTGAATAGTGGATTC |
| MC70 | GGTGGGGGCTGACTGCAGGCCTCTACCTTGCAGACCCATATAATA |
| MC71 | GAGGCCTGCAGTCAGCCCCCACCGGTGTTGGTTAA |
| MC72 | GGGGGCTCTTGTTGACTCGAGGCCTCTACCTTGCAGACCCATATA |
| MC73 | GGCCTCGAGTCAACAAGAGCCCCCGCCCCCACCGGTGTTGGTTAA |
| YFYPT1 | CGGCCGCTCTAGAACTAGTGGATCCTATATTACTTTGTGGAGATTT |
| YRYPT1 | TACCGGGCCCCCCCTCGAGGTCGACTAAACAAGAGAGATTGGGAAGGAA |
| MC66 | ATTGCTCTTGACTCGAGGTGAACTGGAATTA |
| MC67 | GTTCACCTCGAGTCAAGAGCAATTTGATTTAGAACTGTTTCCG |
| MC68 | ATTCAAATTGACTGCAGGTGAACTGGAATTAC |
| MC69 | CAGTTCACCTGCAGTCAATTTGATTTAGAACTGTTTCCGCT |
| MC64 | GCTTGTTCACAAAATAATACAGCCCCCACCGGTGTTGGTTAA |
| MC65 | GGTGGGGGCTGTATTATTTTGTGAACAAGCGCGCCTCTAC |
| NS1 | ATGGATCCTGTCAGGCTTGAGAACTGTTTCTG |
| NS2 | TTATCTCGAGTCAACAGCAATTTGATTTAGAACTG |
| RNC262 | TAGGATCCGATTGATATTCTTTTTGTTATTCGGAC |
| RNC263 | ATATACTCGAGTTCAGACAAAAATTACCATGAGG |
| RNC264 | ATACTCGAGTCAAGAGCAATTTGATTTAGAACTGTTTCC |
| RNC200 | TTGGAGCCACTATCGACTACGCGATCATGGCGACCAGGCCGTGGGAAGCTTC |
| RNC201 | TGATGCCGGCCACGATGCGTCCGGCGTAAGACTGACAGTTATACCCAAG |
| YFY1P1 | CGGCCGCTCTAGAACTAGTGGATCCCGTATCTCGTTAGTACTTGTT |
| YRYIP1 | TCACACAGGAAACAGCTATGACCATGAAGCTTGACCTTAGAGTACAGACGATG |
| KAH7 | ATAAACCTCCATTACTCGAGGAAATTGGAATAAATTTCG |
| KAH8 | CCTCGAGTAATGGAGGTTTATGTGGATATCCCTTAGTTGAAAGAG |
| RNC222 | TACCGGGCCCCCCCTCGAGGTCGACAGAATTCGGCATGCCGGTAGAGGTGTGGTC |
| CC16 | CTCGAGAGGGACTAATAGTTGT |
| CC22 | AACTATTAGTCCCTCTCGAGATGTCAGGCTTTGAAAACTTAAACA |
| MC1 | CGGGATCCCATCCAGCATGAATCCCGAA |
| MC2 | CCGCTCGAGGTTTAGCAGCAACCTCCACC |
| MC3 | ATGGATCCCAGCTAGTCGAGGCGCAACA |
| MC4 | ATACTCGAGGTTTAGTTACTACAACACTGATTCCT |
| RNC256 | ATGAATTCGCCAAAGCCTACGACCA |
| RNC257 | ATGGATCCTCAGCCCAGGGAGCAC |
| RNC258 | ATGAATTCGCGAAGACCTACGATTACCTG |
| RNC259 | ATGGATCCTCACAGAAGAACACATCGGAAAAAG |
| RNC263 | CAGAAACAACTCTGGCGCATC |
| RNC228 | CTATGGAACTGCCTCGGTGA |
| S1YIP1 | GCTACAAATTGGACGGGAAGTACTGCAAGACAACTATTAGTCCCTCTCGAGCGTACGCTGCAGGTCGAC |
| S2YIP1 | GTTCAGAAAAACATATATACAAATATCGCCCCTAAGCCAATTCCCTTCAATCGATGAATTCGAGCTCG |
Creation of yip1-4 Thermosensitive Allele
YIP1 gene deletion was carried out using the KANR module (Wach et al., 1994) as a selectable marker and the oligonucleotides S1YIP1 and S2YIP1 to precisely eliminate the YIP1 ORF in the diploid yeast strain BY24. The mutant allele of YIP1 was generated by a standard plasmid shuffling procedure (Sikorski et al., 1991). Briefly, RCY1610 was transformed with plasmid pRS315 containing the mutant yip1-4 gene created with primers KAH7 and KAH8 and genomic DNA template. Transformants were selected on synthetic media lacking leucine followed by colony purification on fluoroorotic acid (5-FOA)-containing media. 5-FOA–resistant colonies were tested for temperature-sensitive growth on rich media.
Triton X-114 Partition Experiments
Triton X-114 (Roche Diagnostics, Indianapolis, IN) was purified by precondensation as described previously (Bordier, 1981). Then 5 OD units of yeast strains were harvested and washed in 1 ml of TAZ buffer. Postnuclear supernatants were generated by two sequential centrifugation steps for 5 min at 500 × g. Then 500 μl of phosphate-buffered saline (PBS) containing 2% Triton X-114 with protease inhibitors (1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 10 μg pepstatin A) was added to the post-nuclear supernatants. The samples were incubated for 20 min at 4°C to solubilize membrane proteins. To separate the detergent-enriched and the -soluble phases, samples were incubated for 3 min at 30°C followed by low-speed centrifugation at room temperature. This cycle was repeated two times with the detergent-enriched and -soluble phases individually. The detergent phase was washed twice with PBS containing 0.05% Triton X-114 and the soluble phase with 2% Triton X-114. Samples were then incubated with an equal volume of 10% trichloroacetic acid on ice for 30 min followed by a centrifugation at 4°C for 15 min. The protein pellets were washed twice with 300 μl of cold acetone and resuspended in 15 μl of SDS sample buffer. The samples were then analyzed by SDS-PAGE and Western blot. Affinity-purified α-GFP polyclonal antibody (gift from Pam Silver, Harvard University, Cambridge, MA; Seedorf et al., 1999) and alkaline phosphatase-conjugated anti-rabbit secondary antibody (Bio-Rad, Hercules, CA) were used to detect the GFP-tagged Rab protein. Snc1/2p, an integral membrane protein, was detected with anti-Snc1/2p antisera (gift from P. Brennwald, Cell and Developmental Biology, University of North Carolina, Chapel Hill, NC).
Microscopy
For direct fluorescence microscopy, yeast strains were grown to mid-log phase in selection media. For visualization of the nuclei, the samples were incubated with 5 μg/ml Hoechst 33258 (Molecular Probes, Eugene, OR) for 5 min. For immunofluorescence microscopy, cells were grown to early log phase in YPD or selection media. A 2× fixative (2× PBS, 4% glucose, 40 mM EGTA, and 7.4% formaldehyde) was added to an equal volume of medium containing 3 OD units of cells and incubated for 20 min at room temperature. Cells were then collected by centrifugation, resuspended in 5 ml of 1× fixative, and incubated for a further 1 h. The cells were washed twice in 2 ml of spheroplasting buffer (100 mM KPi pH 7.5 and 1.2 M sorbitol) and then incubated in spheroplasting buffer containing 0.2% 2-mercaptoethanol and 0.08 mg/ml zymolyase for 30 min at 37°C with gentle mixing. Then 20 μl of the cell suspension was placed on individual wells of a polylysine-coated printed microscope slides (Carlson Scientific, Peotone, IL) for 10 min. The cells were then washed three times with PBS/bovine serum albumin (BSA) (1 mg/ml BSA) and permeabilized for 5 min with 0.1% SDS. After washing five times in PBS/BSA, cells were blocked for 30 min in PBS/BSA. Monoclonal 1.2.3 antibody was used to detect Sec4p (gift from P. Brennwald). Alexa 488-labeled anti-mouse secondary antibody (Molecular Probes) was used at a dilution of 1:250. Cells were examined with an Eclipse E600 (Nikon, Tokyo, Japan) equipped with a 60× objective and 1.5× optovar. A Spot-RT monochrome charge-coupled device camera (Diagnostic Instruments, Stirling Heights, MI) with software version 3.5 was used for image capture. All images shown are representative images from small budded cells in logarithmic phase growth.
Yeast Two-Hybrid (Y2H) Experiments
ORF sequences were subcloned into pAS1-CYH2 or pAS2–1 for “bait” and pACTII for “fish” constructs and transformed into the yeast strain Y190, which contains the reporter genes lacZ and HIS3 downstream of the binding sequences for Gal4 (Bai and Elledge, 1996). Double transformants were plated on selective media and incubated for 2–3 d at 30°C before processing for β-galactosidase activity as described previously (Calero et al., 2002).
RESULTS
Functionality of Lipid Tail Mutants
In our functionality studies, we initially focused on the Rab GTPases YPT1 and SEC4, because these genes are unique and essential for viability at all temperatures and conditions. To study the effects of prenylation on Rab protein function, we cloned the Rab genes into centromeric, single copy plasmids under the control of endogenous promoter and terminator elements with four variants at the C terminus that would result in different, defined prenylation outcomes. The double cysteine motif of the Rab proteins was replaced with two different CAAX boxes. The first CAAX box contained a CTIM sequence derived from Rho3p that would make the protein a substrate of FTase (see INTRODUCTION) and result in addition of a single farnesyl group. The second CAAX box contained the sequence CIIL at the C terminus, making the protein a substrate for either GGTaseI or GGTaseII and resulting in a single geranylgeranyl group. The CIIL sequence was chosen to ensure the protein would be geranylgeranylated and not a substrate of FTase. We also created a mono-geranylated protein by removing one cysteine from the double cysteine motif (ypt1C205S and sec4C214S). Such proteins are mono-geranylated by GGTaseII exclusively (Pereira-Leal et al., 2001). Finally, the two cysteines were deleted creating ypt1ΔCC and sec4ΔCC, rendering the proteins unable to be prenylated.
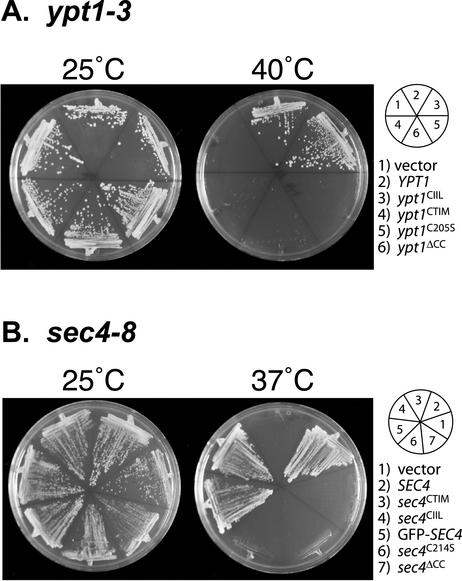
We began our studies by investigating whether the prenylation variants could complement the thermosensitive alleles sec4-8 and ypt1-3. The proteins encoded by these alleles are temperature sensitive, resulting in a complete loss of function at 37 and 40°C, respectively. Each prenylation mutant on CEN vectors was transformed into the sec4-8 temperature-sensitive strain or the ypt1-3 temperature sensitive strain. Transformants were streaked at both permissive and restrictive-temperatures and growth was assessed 2 to 3 d later. YPT1 and ypt1CIIL but not ypt1CTIM, ypt1C205S, or ypt1ΔCC could complement ypt1-3 at restrictive temperature (Figure 1A). Similarly, in the case of sec4-8 cells, only SEC4, sec4CIIL, and GFP-SEC4 could rescue the temperature growth defect but not sec4CTIM, sec4C214S, and sec4ΔCC (Figure 1B).
Figure 1.
Suppression of sec4 and ypt1 temperature-sensitive strains indicate that correct lipid modification is required for full function. The indicated constructs were transformed into ypt1ts (A) and sec4ts (B) strains. These strains contain the temperature-sensitive alleles ypt1-3 and sec4-8 that grow at 25°C but not at 40°C or at 37°C, respectively. Transformants were then streaked on selection media and incubated at the indicated temperatures for 2–3 d.
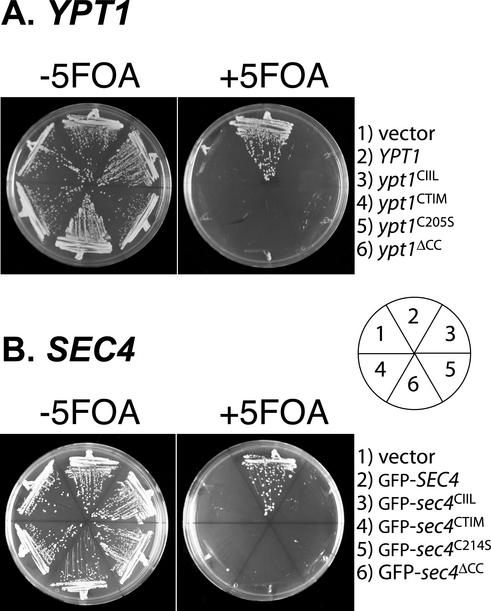
These experiments suggest that Rab proteins have a degree of dependence on their prenylation status for full function. However, it remained possible that the C-terminal variants that were unable to suppress the temperature-sensitive phenotype could in fact function at lower temperatures, a possibility that would not be revealed by suppression analysis. To address this issue, we investigated whether any of the prenylation variants were capable of function as the only copy of the Rab gene in the cell. For these experiments, we transformed the LEU2 CEN plasmids containing the wild-type and prenylation mutants of YPT1 and SEC4 into a SEC4Δ::HIS3 strain or a YPT1Δ::HIS3 strain containing a CEN URA3 plasmid with either SEC4 or YPT1 as the sole source of wild-type SEC4 or YPT1. Transformants were streaked on 5-FOA–containing media to select for loss of the URA3 plasmid containing wild-type SEC4 or YPT1. In this way, we could assess whether the mutants were able to supply the essential function of SEC4 and YPT1 genes. In Figure 2, we show the results of these experiments. Only the wild-type Rab ORF (YPT1 or SEC4) could function as the sole source of these essential genes. None of the singly prenylated or unprenylated mutants can act as the sole source of the Rab protein, indicating that di-geranylgeranylation of SEC4 and YPT1 is critical for function. The results we obtain for YPT1 differ from those reported by Gallwitz and colleagues who find that mono-prenylated Ypt1p is fully functional as the sole copy (Molenaar et al., 1988). It is possible that strain differences and protein expression levels differ between these two sets of experiments and can account for the difference in results.
Figure 2.
Only di-geranylgeranylated proteins can function as the sole cellular source of the essential Rab proteins Sec4p and Ypt1p. The indicated constructs were transformed into RCY1510 (A) or RCY1509 (B), disruption strains for YPT1 or SEC4 that contain a URA3 CEN plasmid with either YPT1 or SEC4. Transformants were then streaked on plates containing 5-FOA. Growth was assessed 2–3 d later. Only the wild-type copies of YPT1 or SEC4 are able to act as the only source of the Rab in the cell. None of the prenylation mutants are able to provide the essential function of the wild-type Rab gene. Note that GFP-SEC4 is functional when present as the sole cellular source of SEC4.
Singly Prenylated Rab Proteins Do Not Localize to the Correct Subcellular Membrane
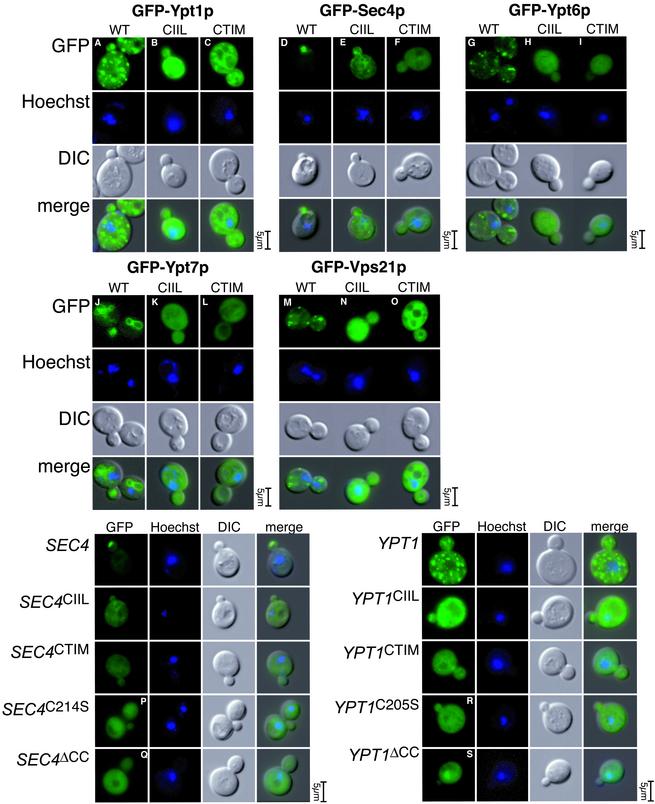
To examine the lack of function of the prenylation variants, we wished to determine the effect of these prenylation variants on localization. For this purpose, we created centromeric plasmids containing the GFP-tagged Rab proteins Sec4p, Ypt1p, Ypt6p, Vps21p, and Ypt7p with different C-terminal variants that would result in different types of prenylation. This group embodies a representative set of yeast Rab proteins: Ypt1p, Ypt6p, and Sec4p are involved at different stages of exocytosis; Vps21p is involved in endocytosis; and Ypt7p is involved in vacuolar transport (for review, see Lazar et al., 1997). In addition, all these Rab proteins have been well studied and their characteristic localization has been firmly established.
Centromeric vectors containing GFP-YPT1, GFP-ypt1CTIM, GFP-ypt1CIIL, GFP-SEC4, GFP-sec4CTIM, GFP-sec4CIIL, GFP-YPT6, GFP-ypt6CTIM, GFP-ypt6CIIL, GFP-YPT7, GFP-ypt7CTIM, GFP-ypt7CIIL, GFP-VPS21, GFP-vps21CTIM, and GFP-vps21CIIL were transformed into yeast, and their localization was examined by fluorescence microscopy (Figure 3). The wild-type GFP-Rab constructs localized to patterns identical to the wild-type untagged protein according to published results. For SEC4, an essential gene, GFP-SEC4 could function as the sole cellular source of SEC4 (as shown in Figure 2), demonstrating that physiological function of the Rab is unimpaired by the N-terminal GFP tag. GFP-YPT1 can also function as the sole cellular source of YPT1 (our unpublished data). GFP-Sec4p is localized to the bud tip as a bright fluorescent spot (Brennwald and Novick, 1993, #22; Figure 3D); GFP-Ypt1p (Figure 3A) and GFP-Ypt6p (Figure 3G) are localized to punctate structures representing yeast Golgi cisternae (Beranger et al., 1994); GFP-Ypt7p (Figure 3J) is localized to the vacuole (Haas et al., 1995; Figure 1D); and GFP-Vps21p (Figure 3M) is localized to distinct punctate endosomal structures (Singer-Kruger et al., 1995; Figure 1E).
Figure 3.
Rab lipid tail variants are unable to correctly localize in vivo. Wild-type and CAAX-containing variants of GFP-YPT1,-SEC4,-YPT6,-YPT7, and -VPS21 were cloned in expression plasmids at wild-type protein levels. These constructs were transformed into NY605 and the localization was assessed by fluorescence microscopy. The mutants containing a CAAX box with CIIL sequence (B, E, H, K, and N) should contain a single geranylgeranyl lipid group and mutants containing CTIM sequence as the CAAX box (C, F, I, L, and O) should contain a single farnesyl lipid group. These mutants do not localize to the typical wild-type compartment of their respective Rab (A, D, G, J, and M). In addition to the CAAX mutants, we cloned GFP-SEC4C214S, GFP-SEC4ΔCC, GFP-YPT1C205S, and GFP-YPT1ΔCC. The point mutants should contain a single geranylgeranyl lipid group, and the ΔCC mutants are unprenylated and should remain cytosolic. These constructs were transformed into cells and the localization was assessed by fluorescence microscopy. These mutants (O–R), similar to the CAAX-containing mutants (shown immediately above for direct comparison), do not localize to the typical wild-type compartment of Sec4p (bud tip) or Ypt1 (Golgi). Cells were incubated with Hoechst to visualize the nuclei.
In each case examined, the farnesylated (CTIM) or the mono-geranylgeranylated (CIIL) Rab showed marked mislocalization (Figure 3, B and C, E and F, H and I, K and L, and N and O). In some cases such as ypt1CIIL (B), ypt1CTIM (C), and sec4CIIL (E), the fluorescence pattern reflected reticular structures suggestive of endoplasmic reticulum. In the case of sec4CTIM (F), ypt6CTIM (F), ypt6CIIL (H), ypt7CTIM (L), ypt7CIIL (K), vps21CTIM (O), and vps21CIIL (N), the fluorescence seemed to be a rather nonspecific cytoplasmic signal. For each experiment, the Hoechst and differential interference images are included as reference points to indicate the cell cycle stage of the cells.
We continued our analysis of the localization of singly prenylated Rab proteins by studying the localization of GFP-sec4C214S and GFP-ypt1C205S (Figure 3, P–S). In principle, these mutants should result in equivalent mono-geranylgeranylation status to sec4CIIL or ypt1CIIL, the exclusive substrate of GGTaseII in the former case and of either GGTaseI or GGTaseII in the latter case. The suppression analysis (Figure 1), however, suggested that CIIL box-containing mutants, but not the single point mutants, were able to complement the temperature-sensitive alleles of sec4-8 or ypt1-3, suggesting that there are differences between these mutants. Because REP is the chaperone that presents the Rab protein to GGTaseII, it is thought that REP is responsible for mediating the very first membrane-targeting event in the existence of the Rab protein. If this is the case, and if the initial REP-mediated targeting is critical, perhaps the CAAX box variants we constructed are unable to localize correctly because these sequences are in vivo substrates for FTase or GGTaseI, but not REP/GGTaseII. As a control, we included the localization of GFP-sec4ΔCC and GFP-ypt1ΔCC, mutants that lack prenylation and are therefore soluble and cytoplasmic. The localization of GFP-sec4C214S (Figure 3P) and ypt1C205S (Figure 3R) reflected a nonspecific cytoplasmic signal far from the typical localization of the wild-type proteins. It may be that the singly prenylated Rab is unable to detach from REP and be delivered onto membranes and we did observe slower growth rates of cells expressing sec4C214S and ypt1C205S, which would agree with the suggestion that such mutants are acting as dominant blockers of REP-mediated prenylation in vivo. Interestingly, the localization of the partly functional sec4CIIL and ypt1CIIL is reticular and not cytoplasmic (Figure 3, B and E), which could explain the partial functionality, if sufficient Rab protein reached the correct location via indirect means.
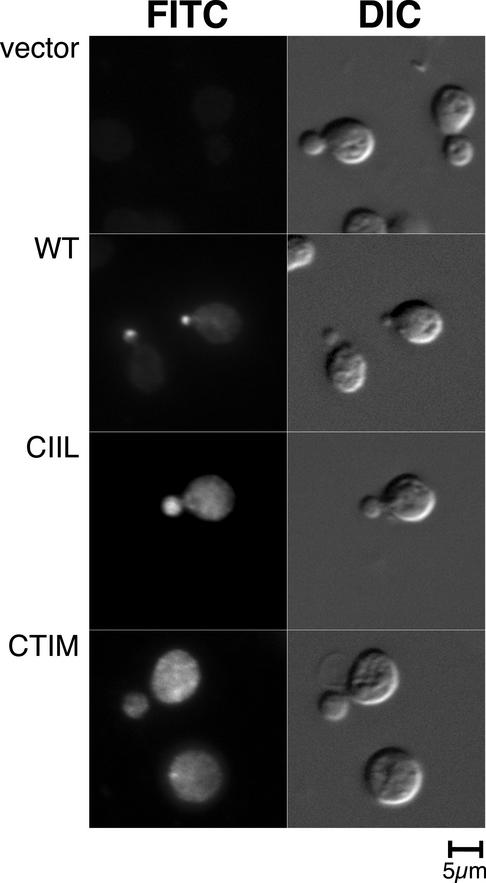
For Ypt1p, Sec4p, Ypt6p, Vps21p, and Ypt7p, the GFP-tagged protein gave a subcellular localization pattern that is identical to previously published reports from immunofluorescence experiments with untagged proteins. However, it remained formally possible that the GFP tag may have affected the prenylation mutants in a manner different to wild type. We therefore carried out immunofluorescence studies to determine the localization of untagged wild-type SEC4 in comparison with prenylation variants. Because the prenylation variants cannot support growth at single sole copy, we made use of a yeast strain, AG6 with a functional SEC4 gene that is antigenically silent to the anti-Sec4p monoclonal antibody (mAb) 1.2.3 (Brennwald and Novick, 1993). In this strain, SEC4 is deleted and a Sec4p chimera with Loop7 and the hypervariable C-terminal domain derived from Ypt1p covers Sec4p function. Because this construct has wild-type SEC4 function but is not recognized by the mAb 1.2.3, it could be used to examine the localization of the SEC4 variants by immunofluorescence. The only SEC4 constructs recognized by the antibody in this strain background should be the prenyl variants and controls that are expressed from episomal plasmids. The results of this experiment are shown in Figure 4. The plasmid-dependent nature of the immunofluorescence signal is demonstrated in Figure 4a, where the vector only control gave only background immunofluorescence. Neither sec4CIILp or sec4CTIMp localized in a manner similar to wild-type Sec4p (Figure 4b) where the signal is tightly restricted to the bud tip of small budded cells. The localization of the SEC4 mutants by indirect immunofluorescence was very similar to that of the GFP-SEC4 mutants by direct fluorescence, indicating that the GFP-tag is not responsible for alterations in the localization of the prenyl variants of Sec4p shown in Figure 3. Together, these experiments confirm that double prenylation is absolutely required for correct targeting of Rab GTPases.
Figure 4.
Sec4p immunofluorescence of untagged constructs in an antigenically silent background. Untagged plasmid constructs containing SEC4, SEC4CTIM, SEC4CIIL, and control vector only were transformed into a strain with functional SEC4 gene that is antigenically silent to the anti-Sec4p antibody 1.2.3. The cells were grown to log phase and processed for immunofluorescence with the mAb 1.2.3. As expected, the control, vector only strain gave no signal demonstrating that the antibody only recognizes the episomal plasmid protein product (5a). The localization of wild-type Sec4p (5b) is very similar to the localization of GFP-Sec4p (3D, 4a). The CAAX box mutants (5, c and d) did not show to the typical localization of Sec4p, indicating that the mislocalization of the equivalent GFP-tagged constructs shown in Figures 3 and 4 are independent of the GFP-tag.
Rab C-Terminal Variants Are Modified by Prenylation
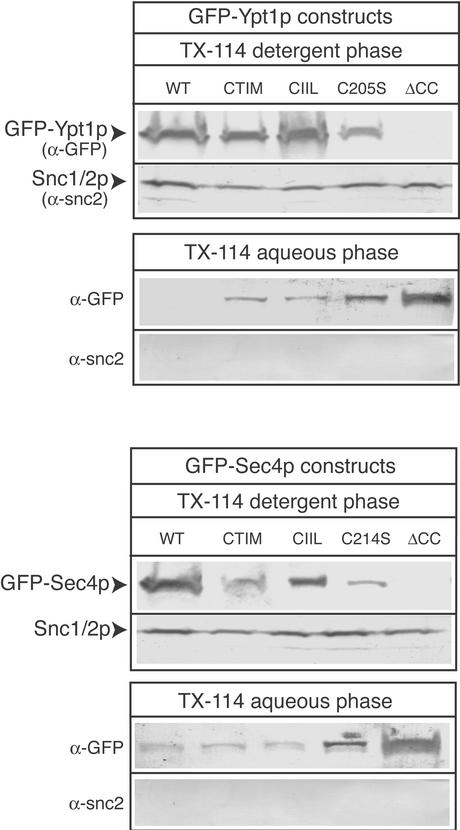
In our examination of the localization of singly prenylated Rab proteins by fluorescence microscopy, we found the mutant Rab proteins to be present both on particulate, endomembrane structures and also observed diffuse cytoplasmic signals. One reason for the observed cytoplasmic localization could be that the singly prenylated Rabs are complexed to chaperone-like proteins such as Gdi1p or Mrs6p, which enable the prenylated proteins to reside in the cytosol. Alternatively, our mutants could be unmodified by prenylation and therefore resident in the cytosol. To rule out the latter possibility, we determined whether our mutant constructs were being modified by prenylation by carrying out Triton X-114 partition experiments. Triton X-114 is a nonionic detergent with a cloud point at the physiological temperature of 30°C (Pryde, 1986). By incubating postnuclear supernatants with this detergent at 30°C followed by a low-speed centrifugation, it is possible to separate the detergent phase that contains membrane proteins and lipid modified proteins from the aqueous phase that contains cytosolic proteins. Triton X-114 phase partitioning was performed on lysates from yeast strains expressing GFP-Ypt1p, GFP-Ypt1CTIM, GFP-Ypt1CIIL, GFP-Ypt1C205S, GFP-Ypt1ΔCC, GFP-Sec4p, GFP-Sec4pCTIM, Sec4pCIIL, GFP-Sec4pC214S, and GFP-Sec4ΔCC. As a positive control, we probed the blots for the presence of an integral membrane protein, Snc2/1p. The results of this experiment are shown in Figure 5. All mutants except for the GFP-Ypt1pΔCC and GFP-Sec4pΔCC, partitioned in the detergent phase, which indicates that they are modified by prenylation. The expression levels of the constructs are all roughly equivalent (our unpublished data), indicating that the degree of partitioning into the detergent phase shown in Figure 5 probably reflects the fraction of lipid modified Rab protein. In the case of Sec4CTIMp and Sec4C214Sp, the fraction of lipid modified is less than for wild-type Sec4p, suggesting perhaps inefficient prenylation. However, even when the prenyl variants of Ypt1p and also Sec4CTIMp and Sec4C214Sp are overexpressed from a multi-copy plasmids, they cannot rescue function (our unpublished data), suggesting that it is not the overall amount of prenylated protein, but the type of modification that is the important factor. The result of this experiment gave us confidence that the prenyl Rab variants we created are indeed modified in the expected manner as predicted from the enzymology and in vivo action of the prenyltransferase enzymes.
Figure 5.
TX-114 extraction demonstrates hydrophobic modifications to Rab lipid tail variants. Triton X-114 fractionation generating a detergent-enriched and aqueous phase was performed as described under MATERIALS AND METHODS on cells expressing GFP-Ypt1p, -Ypt1pCTIM, -Ypt1pCIIL, -Ypt1pC205S, -Ypt1pΔCC, -Sec4p, -Sec4pCTIM, -Sec4pCIIL, -Sec4pC214S, and -Sec4pΔCC. The detergent-enriched phase was then subjected to trichoroacetic acid precipitation followed by SDS-PAGE electrophoresis and Western blotting to detect the GFP-fusion proteins. As a control, the fractions were probed for the transmembrane protein Snc1/2p. Relevant protein markers are indicated.
Yip1p Is Sensitive to Rab Protein Prenylation Status
Our results indicated that di-geranylgeranyl groups are required for correct targeting and function of Rab proteins. Mono-geranylgeranylation of Rab proteins, although conferring hydrophobic character sufficient to mediate membrane association, cannot substitute for the double geranylgeranylation. One possibility is that the functionality is related to the hydrophobicity of the lipid modification, with a gradient from farnesylation (C15) to double geranylgeranylation (two C20 moeities). An alternative explanation might be the existence of Rab-interacting proteins whose interaction is dependent on specific C-terminal prenylation and who play a role in mediating specific localization of Rab proteins. Currently, there are only seven known factors, Rab-GDI, Yip4p, Yip5p, Yif1p, Yip3p/Pra1p, the Rab3-specific GAP, and Rab3-GEF, that require prenylation for productive Rab protein interactions. Rab-GDI is a soluble protein whose recognition site consists of both the GDP-bound Rab and its prenylation moiety (for review, see Wu et al., 1996). It is conserved throughout evolution and its in vivo role is to remove Rab protein from the membrane and recycle the protein through the cytosol before delivering the protein back onto donor membranes. Yif1p, Yip4p, and Yip5p are members of an evolutionarily conserved YIP1-like membrane protein family (Matern et al., 2000; Calero et al., 2002). YIP1-related proteins seem to play roles in membrane transport, and it has been suggested that all of these family members interact with Rab proteins in a prenylation-dependent manner (Calero et al., 2002). PRA1 and its yeast homolog Yip3p/Pra1p are membrane proteins originally identified as Rab-interacting factors (Martincic et al., 1997; Calero et al., 2002), although it has since been demonstrated for Pra1p/Yip3p that the interaction solely relies on a prenyl moiety and simple addition of a CAAX box onto a soluble protein such as GFP is sufficient for interaction (Figueroa et al., 2001). The Rab3-specific GEP and -GEF are specific to mammalian cells where they regulate the activation and deactivation of the neuronal Rab3A (Fukui et al., 1997; Wada et al., 1997).
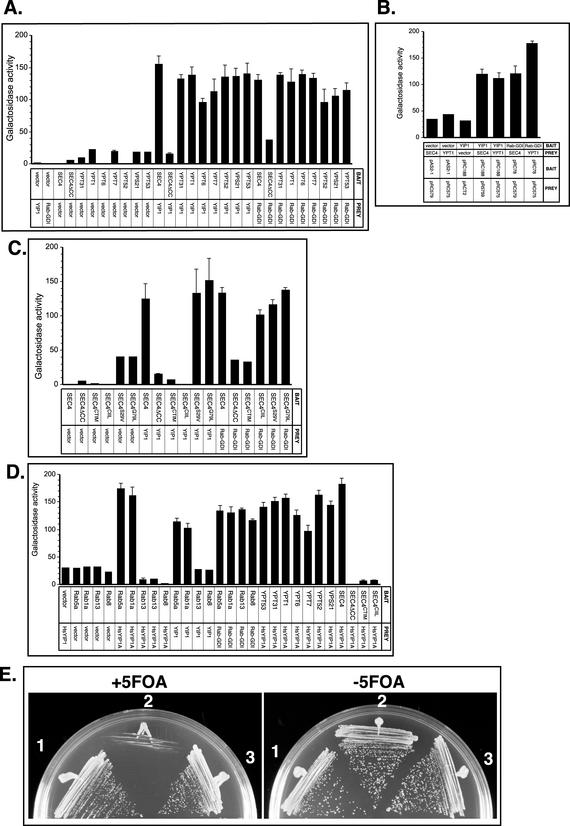
We examined the prenylation status of Rab interactions with Yip1p as a representative member of the YIP1 family. We started by testing the interactions of Yip1p and various Rab proteins and compared these interactions with those of Rab-GDI with Rab proteins. Interactions were monitored by Y2H assay. Pairs of constructs were transformed into the Y190 reporter strain and leu+trp+ transformants were analyzed by β-galactosidase assays. The results of these experiments are shown in Figure 6A where Rab proteins expressed as bait constructs are tested for interactions with Rab-GDI and YIP1 “prey” constructs. The interactions between Yip1p and Rab proteins were very similar to the interactions of Rab-GDI and Rab proteins (Figure 6A). Namely, all of the Rab proteins tested (Sec4p, Vps21p, Ypt1p, Ypt6p, Ypt7p, Ypt52p, Vps21p, and Ypt53p) are capable of interaction with Rab-GDI and Yip1p. However, Sec4ΔCCp does not interact with either Yip1p or Rab-GDI, confirming that the interactions are dependent upon C-terminal prenylation in this system. The requirement for geranylgeranylation of Rab proteins for productive interaction with Rab-GDI has been established previously (for review, see Pfeffer et al., 1995). No constructs showed autoactivation when partnered with vector only, no insert control plasmids.
Figure 6.
Prenylation status of Rab proteins is a critical determinant for interactions with Yip1p. (A) Yip1p has the ability to interact with several Rab proteins in yeast. Pairs of constructs were coexpressed in the reporter strain Y190, and β-galactosidase activity (arbitrary units) in the resulting transformants was measured as described in Calero et al. (2002). The β-galactosidase activity of 12 independent transformants was tested for each pair. The Rab protein bait constructs as indicated on the x-axis were tested against prey constructs of Yip1p, yeast Rab-GDI, or vector only controls. The plasmids pAS2–1 and pACTII were used for vector only bait and prey controls, respectively. Plasmid constructs are listed in Table 2. (B) Yip1p Y2H interactions with Rab proteins are preserved with bait/prey reversal. The Yip1p or Rab-GDI protein bait constructs were tested against prey constructs of Ypt1p, Sec4p, or vector only controls as indicated on the x-axis. Transformants were processed for β-galactosidase activity as described in Figure 7A. Both Yip1p and Rab-GDI bait constructs maintain Y2H interactions with prey constructs of Yptp1p and Sec4p. Background activity is observed with vector only controls cotransformed with each construct. (C) YIP1 interaction with Sec4p requires di-geranylgeranylation. Rab protein bait constructs expressing wild-type Sec4p, Sec4p with no C-terminal cysteines, CTIM, or CIIL lipid tail variants and the point mutations S29V and Q79L were tested against prey constructs of Yip1p, as indicated on the x-axis. Interactions with Rab-GDI and vector only controls are shown for comparison. Transformants were processed for β-galactosidase activity as described in Figure 7A. Both Yip1p and Rab-GDI will interact with wild-type Sec4p, Sec4S29Vp and Sec4Q79Lp. Neither Yip1p or Rab-GDI will interact with the farnesylated Sec4CTIMp or unprenylated Sec4ΔCCp, and only Rab-GDI but not Yip1p will interact with the mono-geranylgeranylated Sec4CIILp construct. Plasmid constructs are listed in Table 2. (D) Human YIP1A will not interact with human Rab proteins containing C-terminal CAAX motifs, although they will interact with highly homologous di-geranylgeranylated Rab proteins. Rab protein bait constructs as indicated on the x-axis were tested against prey constructs of human YIP1A (HsYIP1A). Interactions with Rab-GDI, yeast YIP1 and vector only controls are shown for comparison, also see Figure 6, A and B. Transformants were processed for β-galactosidase activity as described in Figure 7A. Note that yeast Yip1p and human YIP1A are promiscuous in their ability to interact with several Rab proteins, which include mammalian Rab1a, mammalian Rab5a, Sec4p, Ypt31p, Ypt1p, Ypt6p, Ypt7p, Vps21p, Ypt52p, and Ypt53p. Neither human or yeast YIP1 can interact with the mono-geranylgeranylated version of Sec4p or with the mammalian CAAX box containing Rab proteins Rab8 or Rab13, even though these are highly homologous to wild-type Sec4p. For comparison, interactions with Rab-GDI are shown, both Rab13 and Rab8 are fully capable of interaction with Rab-GDI. None of the constructs used showed any autoactivation with vector only cotransformations. Plasmid constructs are listed in Table 2. (E) Human homolog of Yip1p, HsYIP1A, can substitute for YIP1 function in budding yeast. Cells bearing their only copy of YIP1 on plasmid containing the counterselectable marker URA3 were tested for ability to grow on 5-FOA after transformation with the human ORF YIP1A. Colonies transformed with centromeric, single-copy vectors containing (1) YIP1, (2) no insert vector only, or (3) human YIP1A placed under the control of the endogenous YIP1 promoter and ADH1 terminator elements. Transformants were tested for growth on complete media at 30°C with and without 5-FOA to select against retention of the URA3 YIP1 plasmid. Both cells containing wild-type YIP1 or the human homolog YIP1A plasmid can survive the loss of the URA3 YIP1-containing plasmid on 5FOA, whereas transformants containing the no insert control plasmid are dead.
The ability of Yip1p to interact with both Ypt1p and Sec4p was confirmed by reversal of the bait and prey constructs in the Y2H system (Figure 6B). Both YIP1 and Rab-GDI show positive interactions with Sec4p and Ypt1p when expressed as bait constructs with Sec4p and Ypt1p expressed as prey constructs. No constructs showed autologous activation with vector only plasmids.
Having established that prenylation is necessary for both Yip1p interactions with Sec4p, we next tested whether the interactions would be conserved with our monoprenylated variants Sec4CTIMp and Sec4CIILp. Neither Yip1p nor Rab-GDI will interact with the farnesylated Sec4CTIMp, but, although Rab-GDI was able to interact with the mono-prenylated Rab protein Sec4CIILp, Yip1p was not (Figure 6C). These results are in agreement with previous studies showing Rab-GDI can interact with mono-geranylgeranylated Rab proteins (Soldati et al., 1993) and indicate that Yip1p has a specific recognition determinant for doubly prenylated Rab GTPases. In this experiment, we also included the Sec4p point mutations S29V and Q79L. These point mutations influence the conformation of Sec4p, the former toward the GDP-bound and the latter toward the GTP-bound due to its effect on the GTP hydrolysis rate. In the Y2H system, the Sec4S29Vp mutant, unlike the wild-type protein, is capable of productive interactions with its exchange factor, and the Sec4Q79Lp mutant interacts with its effector protein in a manner greatly stimulated over wild-type Sec4p, demonstrating that these point mutants retain their effects in this assay. These mutants are nontoxic when expressed with intact di-cysteine motifs, unlike the dominant negative point mutations, and we used them to examine whether the interactions between Yip1p and Sec4p are influenced by the nucleotide-binding confirmation. Both Sec4S29Vp and Sec4Q79Lp interacted with Yip1p in a manner identical to wild-type Sec4p (Figure 6C), suggesting no strong influence of nucleotide-dependent confirmation on Yip1p interaction with Sec4p. We also found a similar result for the interaction of Sec4p with Rab-GDI, in the Y2H assay, Rab-GDI cannot discriminate between Sec4p, Sec4S29Vp, and Sec4Q79Lp.
Our experiments (Figures 1, 2, 3) indicated that double prenylation is a requirement for proper functioning and localization of Rab GTPases; however, it is of note that there are several Rab proteins that contain CAAX boxes instead of the double cysteine motif and are therefore mono-prenylated (Wilson et al., 1998). Such Rab proteins are not present in yeast but are found in mammalian cells. We therefore investigated whether the human homolog of YIP1, YIP1A, will interact with these naturally monoprenylated Rab proteins. In Figure 6D, we demonstrate that human YIP1A does interact with human Rab5a and canine Rab1a but not with the CAAX box-containing Rab proteins Rab8 and Rab13. Human YIP1A will also interact with a number of yeast Rab proteins: Sec4p, Ypt1p, Vps21p, and Ypt6p. Neither human YIP1A, or yeast YIP1, will interact with the mono-prenylated SEC4 variants. Notably, although Rab8 and Rab13 were incapable of human YIP1A or yeast Yip1p interaction, they were fully able to interact with Rab-GDI.
The two-hybrid experiments in Figure 6D revealed that human YIP1A and yeast Yip1p show striking cross-species conservation of interactions, with human YIP1A capable of interactions with yeast Rab proteins, and yeast Yip1p with mammalian Rab proteins. These results prompted us to test whether this conservation of protein interactions was functionally significant. We asked whether human YIP1A could functionally replace its yeast homolog and act as the only cellular source of the otherwise essential YIP1 gene. For this experiment, we created a LEU2 CEN plasmid containing the human YIP1A ORF with the endogenous YIP1 promoter and 572 base pairs from the ADH1 3′ region to provide a generic yeast termination signal element. The HsYIP1A plasmid (pRC2170) was transformed into a YIP1Δ strain containing a URA3 CEN plasmid with Yip1p as the sole source of Yip1p. Transformants were streaked to 5-FOA plates to select for loss of the URA3 plasmid to assess whether human YIP1A could act as the only copy of Yip1 in the cell. In Figure 6E, we show that both yeast containing wild-type YIP1 and human YIP1A can survive equivalently on the 5-FOA–containing media, whereas a control plasmid with no insert cannot. The fact that human YIP1A can function similarly to yeast YIP1, an essential gene in S. cerevisiae underscores the conservation of Rab protein interactions presented in Figure 6D.
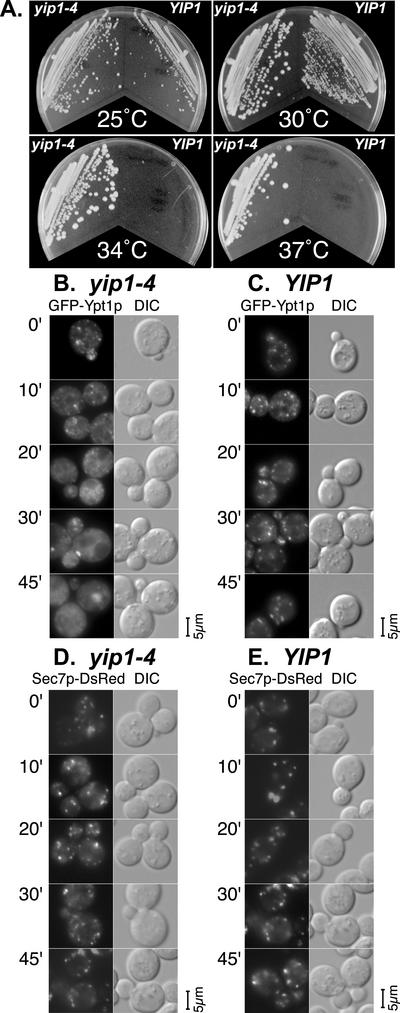
Our data indicate that Yip1p is factor that interacts with Rab proteins in a di-geranylgeranylation–dependent manner and that di-geranylgeranylation is critical for Rab protein function and localization. The exact role of Yip1p is not known and to get an insight into Yip1p function in vivo, we created a mutant allele of YIP1, yip1-4. yip1-4 contains a single point mutation E70K in the cytoplasmic domain. Yip1-4 cells are thermosensitive and do not grow on YPD at 34 or 37°C (Figure 7A). Using the temperature-sensitive allele yip1-4, we examined the effect of conditional loss of Yip1p function on the localization of Ypt1p in vivo. GFP-Ypt1p was transformed into yip1-4 and isogenic wild-type control cells. Cells were grown to log phase at 25°C before shifting them to the restrictive temperature of 37°C. Samples were taken after shift at times of 0, 10, 20, 30, and 45 min for visualization of GFP-Ypt1p location by fluorescence microscopy. The localization of GFP-Ypt1p in cells bearing the yip1-4 allele becomes diffuse and cytoplasmic after only 10 min shift to restrictive temperature (Figure 7B). By 30 min at restrictive temperature, the punctate structures characteristic of Golgi cisternae are very rare and by 45 min, GFP-Ypt1p in these cells is exclusively diffuse in appearance. Electron microscopy of the yip1-4 cells at restrictive temperature shows a predominant endoplasmic reticulum accumulation (our unpublished data), leading us to conclude that the diffuse GFP-Ypt1p localization is cytoplasmic and represents an increase in the soluble pool. In contrast, GFP-Ypt1p in the isogenic wild-type control remains in Golgi cisternae after shift to 37°C for the entire experiment (Figure 7C). These results show that loss of Yip1p function can influence the localization of the Rab protein Ypt1p. To confirm that the loss of Golgi Ypt1p localization was not due to a generalized disruption of Golgi structure caused by the yip1-4 allele, we also examined these cells with a Sec7p. Sec7p is an abundant peripheral membrane protein of the Golgi (Franzusoff et al., 1991), and, in a typical cell, Sec7p-DsRed labels cytoplasmic spots that correspond to individual Golgi cisternae (Preuss et al., 1992; Seron et al., 1998). With Sec7p-labeled Golgi in yip1-4 cells, there was no change in the apparent number or intensity of fluorescent puncta after a shift to the restrictive temperature (Figure 7D). These results indicate that the yip1 mutant allele does not cause a generalized disruption of Golgi structure and are consistent with a more selective action of this protein.
Figure 7.
(A) Cells bearing the yip1-4 allele are thermosensitive. yip1-4 mutant cells bearing the single point mutation E70K are thermosensitive with a restrictive temperature of 34°C on rich media. Growth of yip1-4 cells on YPD are compared with isogenic wild-type controls at the temperatures indicated. (B–E) yip1-4 mutant cells are defective in Golgi localization of the Rab protein Ypt1p at the restrictive temperature. The localization of GFP-Ypt1p was measured in yip1-4 mutant and wild-type cells following a shift to the restrictive temperature (37°C) for the time indicated. The cells were visualized by fluorescence microscopy. The left side of each panel shows GFP fluorescence, whereas the right side is differential interference contrast optics. The characteristic punctate Golgi distribution of GFP-Ypt1p becomes diffuse and cytosolic after only a 10-min shift to the restrictive temperature and becomes maximal by 30 min of shift. This is in contrast to another peripheral Golgi marker, Sec7p, which maintains characteristic Golgi puncta at restrictive temperatures in yip1-4 cells. (B) yip1-4 mutant cells (RCY1764) expressing GFP-Ypt1p after shift to restrictive temperature for the times indicated. (C) isogenic wild-type cells (RCY1768) expressing GFP-Ypt1p after shift to restrictive temperature for the times indicated. (B) yip1-4 mutant cells (RCY1764) expressing Sec7p-DsRed after shift to restrictive temperature for the times indicated. Sec7p-DsRed after shift to restrictive temperature for the times indicated.
DISCUSSION
Rabs are intrinsically cytosolic proteins, yet only function when in association with membranes. The posttranslational modification of prenylation is a prerequisite for membrane attachment. In addition to membrane association, proper targeting of Rab proteins is essential for their function in regulating membrane traffic; a characteristic feature of Rab proteins is their steady-state localization to the cytosolic surface of a particular subcellular membrane. Pioneering experiments examining the relationship of membrane attachment and function led to the idea that prenylation is a convenient method of giving proteins the physiochemical ability to stably attach to lipid bilayers, enabling regulation through reversible recruitment onto membranes (Leevers et al., 1994; Stokoe et al., 1994). In the case of Rab proteins, this idea was underscored by the dramatic finding that the lipid tail could be circumvented by giving the Rab proteins a transmembrane domain, provided that the transmembrane domain took them to the correct location (Ossig et al., 1995). Obviously, Rab-GDI–mediated recycling did not occur in this situation; however, this requirement could be eliminated by protein overexpression. In these experiments, the replacement of the lipid anchor with a transmembrane domain that targeted the Rab protein to the correct compartment in this experiment may have obscured any possible specific role played by the lipid anchor. The availability of genomic information, a better understanding of the enzymes involved in prenylation, and genetic models that can distinguish between different levels of function have allowed us to examine this question.
To explore the specific role of the lipid modification in Rab protein function, we asked whether a single lipid geranylgeranyl group could substitute for the two geranylgeranyl groups found on most Rab proteins, and, if so, could a shorter lipid group such as a farnesyl group substitute for the longer geranylgeranyl groups? We created Rab prenylation variants by replacing the double cysteine motif at their C terminus with CAAX boxes to study the localization and function of the singly prenylated Rab proteins. C-terminal CTIM or CIIL box versions of the essential Rab genes YPT1 and SEC4 were unable to function in vivo when expressed as the only copy in the cell. Although Rab-GDI plays a critical role in the membrane targeting and recycling of Rab proteins, it is also thought that the homologous protein REP can function in this manner. Because REP is the chaperone that presents the Rab protein to GGTaseII, it is thought that REP mediates the very first membrane-targeting event in the existence of the Rab protein. If this is the case, and if the REP-mediated targeting is critical, perhaps the CAAX box variants we constructed were unable to function correctly because these sequences are in vivo substrates for FTase and GGTaseI. To eliminate this possibility we created C-terminal variants that contained a single cysteine-to-serine point mutation of one of the residues that is prenylated by GGTaseII. It has been previously demonstrated that such mutants remain the substrates of a single round of prenylation by GGTaseII and so would exist as a complex with REP, which could then target them to membranes (Wilson et al., 1996). Such mutants would be singly geranylgeranylated exclusively by GGTaseII in combination with REP so eliminating any contribution from GGTaseI. Our finding that even singly geranylgeranylated YPT1 and SEC4 variants that are the substrates of GGTaseII cannot function as the only copy in the cell indicates that it is the specific double prenylation modification that is required for full function. We did, however, uncover differences in the mono-geranylgeranylated proteins that result from different prenyltransferase enzymes. ypt1CIIL and sec4CIIL, the substrates for either GGTaseI or GGTaseII, were able to suppress temperature-sensitive alleles ypt1-3 and sec4-8, while the exclusive GG-TaseII substrates ypt1C205S and sec4C214S were not. These data agree with previous studies demonstrating that Rab proteins mutated to GGTaseI substrate CAAX boxes can in fact support function, provided that sufficient Rab protein reaches the correct membrane (Soldati et al., 1993; Overmeyer et al., 2001). It is possible that the ypt1C205S and sec4C214S are not released from REP after a single round of prenylation, because REP has been reported to form a very tight, stable complex with mono-geranylgeranylated Rab protein (Thoma et al., 2001), and this could explain differences observed between the two set of mutants.
Using Ypt1p and Sec4p as examples, we also investigated whether the prenylation variants we created are indeed modified by asking if they could still partition into the detergent phase of a Triton X-114 partition. Each of the prenyl variants was able to partition into the detergent phase, in contrast to an unprenylated ΔCC mutant (Figure 5). These data suggest that the effects we observe in Rab protein functionality with these variants can be attributed to the alternative Rab prenylation. Although other lipid anchor sequences on Rab proteins receive lipid modifications, they do not lead to correct function.
Why do mono-prenylated Rab proteins fail to function? One possibility is that alternative lipid modifications fail to stably associate with membranes. This may well be the case for farnesylated proteins (C15 moiety). However, singly geranylgeranylated proteins, with their C20 lipid tails are more than two log(P) units more nonpolar than farnesyl groups (Black, 1992). Geranylgeranylation significantly enhances the bilayer partitioning ability of the modified protein. Although mono-geranylgeranylated proteins have the biophysical ability to stably associate with membranes, our data indicate that they are nonfunctional because they are unable to localize to the correct subcellular compartment. In each case examined, Sec4p, Ypt1p, Ypt6p, Ypt7p, and Vps21p, the monoprenylated variants did not localize in the same manner as their wild-type equivalents. Moreover, in the case of Sec4p, untagged prenyl variants examined by indirect immunofluorescence, gave similar results (Figure 4). These data suggest that for Rab proteins, lipid modification plays dual functions. It is required for both membrane association and localization or clustering; prenylation is necessary for the former, and di-geranylgeranylation is required for the latter.
How applicable are these results to Rab proteins in general? In this study, we have examined the functionality of two different Rab proteins and the localization of prenylation variants of five different Rab proteins to reach our conclusion that dual prenylation is specifically required for Rab protein function and localization. While preparing this article, we became aware of a similar study in mammalian cells that reached the same conclusions (Gomes et al., 2003). We therefore believe that our results show a common principle of Rab protein function, namely, a specific requirement for double prenylation. The original impetus for the experiments we report in this study was the desire to create prenylated peptide constructs of Rab hypervariable sequences to examine the possibility that such constructs might act as dominant inhibitors of endogenous Rab membrane recruitment. We expected that singly geranylgeranylated Rab proteins would be indistinguishable from wild type and were surprised by our results that mono-prenylated Rab proteins were nonfunctional. However, double prenylation is a characteristic hallmark of the majority of Rab GTPase family members, a family that is conserved in all eukaryotes. In fact, it would be surprising that a group of proteins would evolve this specialized dual prenylation modification and the machinery to produce it without a biological imperative.
We examined known Rab-interacting factors for the possible existence of protein entities that recognize the specialized dual prenylation of Rab proteins. We confined our list to factors conserved from yeast to human that are known to require an intact C-terminal cysteine motif for productive Rab protein interactions. The results of these experiments lead us to propose the YIP1 family of proteins as potential candidates through which the di-geranylgeranylation specificity is mediated. Yip1p was originally identified as a factor specific for Ypt1p and Ypt31p interaction (Yang et al., 1998). However, Sec4p is as homologous to Ypt1p and Ypt31p as either is to each other, and it has become appreciated recently that Yip1p is capable of pleiotropic Rab protein interactions (Matern et al., 2000; Calero et al., 2002), which we confirm in this study. Our data show that Yip1p can interact with the di-geranylgeranylated Rab proteins Ypt1p, Sec4p, Ypt31p, Vps21p, Ypt6p, Ypt7p, Ypt52p, and Ypt53p. Yip1p does not interact with mono-geranylated Sec4p proteins. It is also of note that several mammalian Rab proteins such as Rab8 contain CAAL motifs that are singly geranylgeranylated both by REP/GGTase II and by GGTaseI (Wilson et al., 1998). We would predict that such proteins may be insensitive to the impact of YIP1-like family members and demonstrated that such proteins are unable to interact with human YIP1A, although, as we have demonstrated for Sec4p mutants with CAAX boxes, Rab-GDI can still bind these monoprenylated Rab proteins. It should be noted that Sec4p is more homologous in primary sequence to either Rab8 or Rab13 (49.3 and 52.2% identity, respectively) than to Ypt1p (44.4% identity), its closest homolog in yeast. The fact that both human YIP1A and yeast Yip1p are capable of interactions with Sec4p, Rab1a, Ypt1p, and Ypt31p, all di-geranylgeranylated members of the same Rab subfamily (Pereira-Leal and Seabra, 2000), but not with mono-geranylgeranylated Rab8 or Rab13, leads us to conclude that it is the di-geranylgeranylation that is the critical factor for YIP1 interaction. The relevance of our findings showing cross-species protein interaction is reflected in our demonstration showing the conservation of YIP1 protein function. Human YIP1A can fully substitute for YIP1, an essential gene in yeast. Together with our data showing no interaction between Yip1p and mono-geranylgeranylated Sec4p variants (Figure 7C), these results show that di-geranylgeranylation is critical for interactions between Yip1p and Rab GTPases and additionally demonstrate that the interactions of Yip1p with Rab GTPases are well conserved in evolution. Due to our finding that the requirement of di-geranylgeranylation for Rab protein function correlates with specific Rab protein localization, we sought to examine whether Yip1p might play a role in Rab protein localization. Using the mutant allele, yip1-4, we demonstrate that loss of functional Yip1p has an impact on the localization of Ypt1p, shifting it from Golgi localization to a diffuse pool. These results demonstrate that Yip1p can impact Ypt1p localization in vivo. Together with our results showing loss of localization of the mono-prenylated Rab proteins, and the failure of such mutants to interact with Yip1p, these data suggest that Yip1p and other YIP1-family members are candidates for factors through which di-geranylgeranylated Rab proteins work to achieve correct membrane localization. It should be noted, however, that in this study we only tested known Rab-interacting factors, and there may be additional proteins present in the proteome that also specifically recognize digeranylgeranylated Rabs and aid in their correct localization. YIP1 is an essential gene, and yip1-4 cannot be suppressed by overexpression of other YIP1 family members in yeast (our unpublished data). These data are surprising considering that an ability to promiscuously associate with dual prenylated Rab proteins is the only known function for YIP1-family proteins and suggest either that Yip1p contains additional unique functions or that it interacts with, and is responsible for, an essential Rab protein. Four members of the YIP1-protein family and 11 Rab proteins have been identified in yeast. YIP1-family members associate both among themselves as well as with other proteins (Matern et al., 2000; Calero et al., 2001; Calero and Collins, 2002), and one possibility may be that a combinatorial assortment of YIP1 family complexes confer specificity toward different Rab proteins. In vivo, the accessibility of Yip1p to Rab proteins may be restricted by its localization and interacting partners.
In summary, our findings demonstrate a specific lipid requirement of double geranylgeranylation for the Rab GTPase class of proteins to function correctly and show that double geranylgeranyl groups are required for the Rab protein to localize to its characteristic organelle membrane. The exact mechanism by which the di-geranylgeranylated proteins act to achieve correct localization remains to be uncovered. Although different prenylation will affect the membrane-partitioning ability of the modified protein, isoprenylation may have an additional role and be recognized by another protein. Our data indicate the YIP1 family as possible effector candidates for the di-geranylgeranylated Rab proteins, although further work is needed to explore the biochemical basis and physiological relevance of the YIP1–Rab interactions.
Acknowledgments
Many thanks to Pat Brennwald for reagents and helpful discussions, to Miguel Seabra for generously providing mammalian Rab constructs, and to Gary Whittaker for help with microscopy, and together with Elysa Goldberg, for critical reading of the manuscript. K.A.H. is a participant of the 2002 Cornell Leadership Program for Veterinary Students; many thanks to the Program director Dr. D. McGregor for support and insights. Casey Kurtz, a Cornell undergraduate, provided valuable technical assistance. M.C. is the recipient of Army Predoctoral Fellowship DAMD17-00-1-0218. This work was supported in part by the National Institutes of Health grant GM-61221 (to C.B.), USDA Animal Health and Disease Research Program, American Heart Association grant 0030316T, and National Science Foundation grant MCB-0079045 (to R.C.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-11-0707. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0707.
Abbreviations used: 5-FOA, 5-fluorooroic acid; FTase, farnesyl transferase; GDI, GDP-dissociation inhibitor; GGTase I, geranylgeranyl transferase type I; GGTaseII, type II geranylgeranyl transferase; GFP, green fluorescent protein; mAb, monoclonal antibody; PCR, polymerase chain reaction; REP, Rab escort protein; Y2H, yeast two-hybrid.
References
- Bai, C., and Elledge, S.J. (1996). Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996, 331–347. [DOI] [PubMed] [Google Scholar]
- Beranger, F., Paterson, H., Powers, S., de, G.J., and Hancock, J.F. (1994). The effector domain of Rab6, plus a highly hydrophobic C terminus, is required for Golgi apparatus localization. Mol. Cell. Biol. 14, 744–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis, B.J., and Glick, B.S. (2002). Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20, 83–87. [DOI] [PubMed] [Google Scholar]
- Black, S.D. (1992). Development of hydrophobicity parameters for prenylated proteins. Biochem. Biophys. Res. Commun. 186, 1437–1442. [DOI] [PubMed] [Google Scholar]
- Bordier, C. (1981). Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256, 1604–1607. [PubMed] [Google Scholar]
- Brennwald, P., and Novick, P. (1993). Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature 362, 560–563. [DOI] [PubMed] [Google Scholar]
- Calero, M., and Collins, R.N. (2002). Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem. Biophys. Res. Commun. 290, 676–681. [DOI] [PubMed] [Google Scholar]
- Calero, M., Whittaker, G.R., and Collins, R.N. (2001). Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J. Biol. Chem. 276, 12100–12112. [DOI] [PubMed] [Google Scholar]
- Calero, M., Winand, N., and Collins, R.N. (2002). Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 515, 89–98. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., Miaczynska, M., Ashman, K., Wilm, M., Zhao, L., Yip, S.-C., Waterfield, M.D., Backer, J.M., and Zerial, M. (1999). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1, 249–252. [DOI] [PubMed] [Google Scholar]
- Desnoyoyers, L., Anant, J.S., and Seabra, M.C. (1996). Geranylgeranylation of Rab proteins. Biochem. Soc. Trans. 24, 699–703. [DOI] [PubMed] [Google Scholar]
- Figueroa, C., Taylor, J., and Vojtek, A.B. (2001). Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J. Biol. Chem. 276, 28219–28225. [DOI] [PubMed] [Google Scholar]
- Finger, F.P., Hughes, T.E., and Novick, P. (1998). Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell 92, 559–571. [DOI] [PubMed] [Google Scholar]
- Franzusoff, A., Redding, K., Crosby, J., Fuller, R.S., and Schekman, R. (1991). Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 112, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, K., Saaki, T., Imazumi, K., Matsuura, Y., Nakanishi, H., and Takai, Y. (1997). Isolation and characterization of GTPase activating protein specific for the Rab3 subfamily of small G proteins. J. Biol. Chem. 272, 4655–4658. [DOI] [PubMed] [Google Scholar]
- Gomes, A.Q., Ali, B.R., Ramalho, J.S., Godfrey, R.F., Barral, D.C., Hume, A.N., and Seabra, M.C. (2003). Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Guthrie, C., and Fink, G.R. (2002). Guide to Yeast Genetics and Molecular and Cell Biology. San Diego: Academic Press.
- Haas, A., Scheglmann, D., Lazar, T., Gallwitz, D., and Wickner, W. (1995). The GTPase Ypt7p of S. cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 14, 5258–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, T., Götte, M., and Gallwitz, D. (1997). Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem. Sci. 22, 468–472. [DOI] [PubMed] [Google Scholar]
- Leevers, S.J., Paterson, H.F., and Marshall, C.J. (1994). Requirement for ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 369, 411–414. [DOI] [PubMed] [Google Scholar]
- Liang, P.-H., Ko, T.-P., and Wang, A.H.-J. (2002). Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269, 3339–3354. [DOI] [PubMed] [Google Scholar]
- Lin, R., Bagrodia, S., Cerione, R., and Manor, D. (1997). A novel CDC42Hs mutant induces cellular transformation. Curr. Biol. 7, 794–797. [DOI] [PubMed] [Google Scholar]
- Martincic, I., Peralta, M.E., and Ngsee, J.K. (1997). Isolation and characterization of a dual prenylated Rab and VAMP2 receptor. J. Biol. Chem. 272, 26991–26998. [DOI] [PubMed] [Google Scholar]
- Matern, H., Yang, X., Andrulis, E., Sternglanz, R., Trepte, H.-H., and Gallwitz, D. (2000). A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 19, 4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar, C.M., Prange, R., and Gallwitz, D. (1988). A carboxyl-terminal cysteine residue is required for palmitic acid binding and biological activity of the ras-related yeast YPT1 protein. EMBO J. 7, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., Severin, F., Backer, J.M., Hyman, A.A., and Zerial, M. (1999). Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1, 376–382. [DOI] [PubMed] [Google Scholar]
- Ossig, R., Laufer, W., Schmitt, H.D., and Gallwitz, D. (1995). Functionality and specific membrane localization of transport GTPases carrying C-terminal membrane anchors of synaptobrevin-like proteins. EMBO J. 14, 3645–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeyer, J.H., Wilson, A.L., and Maltese, W.A. (2001). Membrane targeting of a Rab GTPase that fail to associate with Rab escort protein (REP) or Rab-GDI. J. Biol. Chem. 276, 20379–20386. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J.B., Hume, A.N., and Seabra, M.C. (2001). Prenylation of Rab GTPases: molecular mechanisms and involvement in genetic disease. FEBS Lett. 498, 197–200. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J.B., and Seabra, M.C. (2000). The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. (1999). Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1, E17–E22. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. (2001). Rab GTPases; specifying a deciphering organelle identity and function. Trends Cell Biol. 11, 487–491. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R., Dirac-Svejstrup, A.B., and Soldati, T. (1995). Rab GDP dissociation inhibitor: putting Rab GTPases in the right place. J. Biol. Chem. 270, 17057–17059. [DOI] [PubMed] [Google Scholar]
- Preuss, D., Mulholland, J., Franzusoff, A., Segev, N., and Botstein, D. (1992). Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell 3, 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde, J.G. (1986). Triton X-114: a detergent that has come in from the cold. Trends Biochem. Sci. 11, 160–163. [Google Scholar]
- Ren, M., Zeng, J., De Lemos-Chiarandini, C., Rosenfeld, M., Adesnik, M., and Sabatini, D.D. (1996). In its active form, the GTP-binding protein rab8 interacts with a stress-activated protein kinase. Proc. Natl. Acad. Sci. USA 93, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybin, V., Ullrich, O., Rubino, M., Alexandrov, K., Simon, I., Seabra, C., Goody, R., and Zerial, M. (1996). GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature 383, 266–269. [DOI] [PubMed] [Google Scholar]
- Seabra, M.C. (1998). Membrane association and targeting of prenylated Ras-like GTPases. Cell Signal. 10, 167–172. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., Damelin, M., Kahana, J., Taura, T., and Silver, P. (1999). Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19, 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron, K., et al. (1998). A yeast t-SNARE involved in endocytosis. Mol. Biol. Cell 9, 2873–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., Boeke, J.D., Zhao, H., and Arnold, F.H. (1991). In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast: optimization of DNA shuffling for high fidelity recombination. Methods Enzymol. 194, 302–318. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger, B., Stenmark, H., and Zerial, M. (1995). Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J. Cell Sci. 108, 3509–3521. [DOI] [PubMed] [Google Scholar]
- Soldati, T., Riederer, M.A., and Pfeffer, S.R. (1993). Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell 4, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe, D., Macdonald, S.G., Cadwallader, K., Symons, M., and Hancock, J.F. (1994). Activation of Raf as a result of recruitment to the plasma membrane. Science 264, 1463–1467. [DOI] [PubMed] [Google Scholar]
- Tang, B.L., Ong, Y.S., Huang, B., Wei, S., Wong, E.T., Qi, R., Horstman, H., and Hong, W. (2001). A membrane protein enriched in ER exit sites interacts with COPII. J. Biol. Chem. 276, 40008–40017. [DOI] [PubMed] [Google Scholar]
- Thoma, N.H., Niculae, A., Goddy, R.S., and Alexandrov, K. (2001). Double prenylation by RabGGase can proceed without dissociation of the mono-prenylated intermediate. J. Biol. Chem. 276, 48631–48636. [DOI] [PubMed] [Google Scholar]
- Vetter, I.R., and Wittinghofer, A. (2001). The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in S. cerevisiae. Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wada, M., Nakanishi, H., Satoh, A., Hirano, H., Obaishi, H., Matsuura, Y., and Takai, Y. (1997). Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J. Biol. Chem. 272, 3875–3878. [DOI] [PubMed] [Google Scholar]
- Walworth, N.C., Goud, B., Kabcenell, A.K., and Novick, P.J. (1989). Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 8, 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Okamoto, M., Schmitz, F., Hofmann, K., and Sudhof, T.C. (1997). Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388, 593–598. [DOI] [PubMed] [Google Scholar]
- Wilson, A.L., Erdman, R.A., and Maltese, W.A. (1996). Association of Rab1B with GDP-dissociation Inhibitor (GDI) is required for recycling but not initial membrane targeting of the Rab protein. J. Biol. Chem. 271, 10932–10940. [DOI] [PubMed] [Google Scholar]
- Wilson, A.L., R.A., E., Castellano, F., and Maltese, W.A. (1998). Prenylation of Rab8 GTPase by type I and type II geranylgeranyl transferases. Biochem J. 333, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.K., Zeng, K., Wilson, I.A., and Balch, W.E. (1996). Structural insights into the function of the Rab GDI superfamily. [Review]. Trends Biochem. Sci. 21, 472–476. [DOI] [PubMed] [Google Scholar]
- Yang, X., Matern, H.T., and Gallwitz, D. (1998). Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J. 17, 4954–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]