Abstract
Tpr is a 267-kDa protein forming coiled coil-dominated homodimers that locate at the nucleoplasmic side of the nuclear pore complex (NPC). The proteins that tether Tpr to this location are unknown. Moreover, the question whether Tpr itself might act as a scaffold onto which other NPC components need to be assembled has not been answered to date. To assess Tpr's role as an architectural element of the NPC, we have studied the sequential disassembly and reassembly of NPCs in mitotic cells, paralleled by studies of cells depleted of Tpr as a result of posttranscriptional tpr gene silencing by RNA interference (RNAi). NPC assembly and recruitment of several nucleoporins, including Nup50, Nup93, Nup96, Nup98, Nup107, and Nup153, in anaphase/early telophase is shown to precede NPC association of Tpr in late telophase. In accordance, cellular depletion of Tpr by RNAi does not forestall binding of these nucleoporins to the NPC. In a search for proteins that moor Tpr to the NPC, we have combined the RNAi approach with affinity-chromatography and yeast two-hybrid interaction studies, leading to the identification of nucleoporin Nup153 as the binding partner for Tpr. The specificity of this interaction is demonstrated by its sensitivity to Tpr amino acid substitution mutations that abolish Tpr's ability to adhere to the NPC and affect the direct binding of Tpr to Nup153. Accordingly, cellular depletion of Nup153 by RNAi is shown to result in mislocalization of Tpr to the nuclear interior. Nup153 deficiency also causes mislocalization of Nup50 but has no direct effect on NPC localization of the other nucleoporins studied in this investigation. In summary, these results render Tpr a protein only peripherally attached to the NPC that does not act as an essential scaffold for other nucleoporins.
INTRODUCTION
The nuclear pore complex (NPC) serves as the gateway for the exchange of cellular material between cytoplasm and nucleus in eukaryotes. Its core structure of eightfold-radial symmetry consists of central globular subunits flanked by a ring-like structure at both the NPC's cytoplasmic (outer) and nucleoplasmic (inner) side. Both of these coaxial rings are attachment sites for fibrils that are also arranged in an eight-fold symmetrical pattern but are of distinctive shape and protein composition (for reviews, see Adam, 2001; Fahrenkrog et al., 2001; Rout and Aitchison, 2001; Vasu and Forbes, 2001). Fibrils emanating from the outer ring appear as short tufts of ∼35–50 nm in length that have free distal ends. In contrast, the eight rectilinear fibrils of ∼60–80 nm that are attached to the inner ring have been reported to be laterally interconnected at their distal ends by another ring-like structure, sometimes described as the “terminal ring.” Together, terminal ring and fibrils proximal to the NPC proper are considered to represent a structural and functional entity, called the nuclear “fishtrap” or “basket” (Ris, 1989, 1991; Jarnik and Aebi, 1991; Goldberg and Allen, 1992).
NPC proteins (nucleoporins) located at the nuclear side of the vertebrate NPC comprise Nup153 (Sukegawa and Blobel, 1993), Nup98 (Powers et al., 1995; Radu et al., 1995), Nup 50 (Fan et al., 1997; Guan et al., 2000; Smitherman et al., 2000), the components of the Nup160-Nup133-Nup96-Nup107 subcomplex (Radu et al., 1994; Fontoura et al., 1999; Belgareh et al., 2001; Vasu et al., 2001), the Nup93-Nup188-Nup205 subcomplex (Grandi et al., 1997; Miller et al., 2000), and a 267-kDa protein termed Tpr (Byrd et al., 1994; Cordes et al., 1997; Zimowska et al., 1997). However, because different antibodies and immunolabeling procedures have been used in different investigations, resulting in partly inconsistent findings, the exact localization of most of these proteins remains unknown. Although some have been attributed to the basket, it is uncertain which of them are in fact components of the inner coaxial ring, the associated fibrils, or the terminal ring.
Furthermore, in certain cell types additional fibers can be found emanating from the terminal ring region and projecting further into the nuclear interior. In amphibian oocytes this conspicuous fibrous material appears to form hollow cylinders several hundred nanometers in length that occasionally connect the NPCs with the cortex of amplified nucleoli (e.g., Ris and Malecki, 1993; Cordes et al., 1993; Arlucea et al., 1998). In the stage VI oocytes of the South African clawed frog, Xenopus laevis, at least two proteins, Nup153 and Tpr, reside not only in proximity to the NPC but are found associated with these distal fibers as well (Cordes et al., 1993, 1997). These filamentous structures might be specific for oocytes, known to stockpile individual proteins in large quantities and various forms. In fact, it has been considered possible that they might represent a common stockpile of Tpr, Nup153, and perhaps even some other nucleoporins, later to be used during embryogenesis (Hase et al., 2001).
The functions of Tpr are controversially discussed; propositions include roles in intranuclear and nucleocytoplasmic transport and as a scaffolding element of the nuclear interior and the NPC (e.g., Fontoura et al., 2001; Frosst et al., 2002, Shibata et al., 2002; Zimowska and Paddy, 2002). Although different metazoan species have been shown to contain only one Tpr ortholog, two probable homologues exist in both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Kuznetsov et al., 2002); in the budding yeast, these paralogs are called Mlp1 and Mlp2 (Strambio-de-Castilia et al., 1999; Kosova et al., 2000). Although overproduction of Mlp1 has been shown to cause nuclear accumulation of poly[A]+ RNA (Kosova et al., 2000), disruption of both Mlp genes is not lethal and does not notably affect any type of nucleocytoplasmic transport (Strambio-de-Castilia et al., 1999; Kosova et al., 2000). However, null mutants show increased sensitivity toward UV-induced DNA damage (Kosova et al., 2000), an impairment in DNA double-strand break repair, and alterations in the spatial organization of telomeric heterochromatin (Galy et al., 2000).
Tpr and Mlps are proteins forming coiled-coil dominated homodimers of extended rod-like shape (Kosova et al., 2000; Hase et al., 2001). Homodimerization occurs via the large N-terminal domain, which also includes a short sequence segment found to be essential for mediating binding of recombinant versions of Tpr to the NPC of transfected cells (Bangs et al., 1998; Cordes et al., 1998, Hase et al., 2001). However, binding partners responsible for positioning Tpr at the nuclear periphery have remained elusive. One mammalian nucleoporin, Nup98, has been described as an interaction partner of Tpr (Fontoura et al., 2001), but whether Nup98 serves as an NPC-attachment site for Tpr or whether Tpr itself provides an anchor site for Nup98 remained unanswered.
Amphibian NPCs reassembled from Xenopus egg extracts that had been immuno-depleted of Nup153 have been reported to be devoid not only of Nup153 but also of all other presumptive basket proteins investigated in that study, namely Nup93, Nup98, and Tpr (Walther et al., 2001). Accordingly, the authors suggested that Nup153 might be required for the formation or the structural integrity of a large part of the nuclear basket. However, whether Nup153, which seemingly lacks a yeast homolog, might serve as a direct binding partner for any of these proteins was not determined.
In yeast again, several nucleoporins have been found associated with either Mlp1 or Mlp2. Synthetic lethality screens had pointed at interactions between Nup145p-C, the probable yeast homolog of Nup96, and both Mlp1 and Mlp2 (Galy et al., 2000). On the other hand, in vivo cross-linking followed by affinity-purification of Nic96p, the yeast homolog of Nup93, resulted in cofractionation of Mlp2 (Kosova et al., 2000). Finally, in a yeast two-hybrid screen a cDNA clone encoding Mlp2 had been isolated by using Nup60p as bait, a mammalian homolog of which is not immediately apparent (Feuerbach et al., 2002). However, whether these proteins interact directly with Mlps, or whether binding is mediated by additional linker proteins, remained uncertain.
In the present study, we have identified nucleoporin Nup153 as the direct binding partner that links Tpr to the NPC in mammals. In addition, we have addressed the question whether Tpr itself might function as a central architectural element of the NPC and as a scaffold for other nucleoporins.
MATERIALS AND METHODS
Antibodies
Rabbit and guinea-pig antibodies against amino acids (aa) 2063–2084 of human Tpr (hTpr) and aa 2285–2314 of hNup358/RanBP2 (Cordes et al., 1996, 1997), rabbit antibodies against aa 1–14 and aa 604–615 of hTpr (Kuznetsov et al., 2002), mAb (mAb) 203-37 against hTpr (Cordes et al., 1997; Hase et al., 2001), mAb PF190x7A8 against an epitope located between aa 439 and 611 of rat Nup153 (Cordes et al., 1993), guinea-pig antibodies against aa 21–36 and 391–404 of hNup153 (Ferrando-May et al., 2001) and against murine p62 (Cordes et al., 1991), and mAb 9E10 for human c-Myc (ATCC CRL 1729) have been described. MAb R27 against mammalian lamin A and mAb X223 against B-type lamins (Höger et al., 1991) were kindly provided by G. Krohne. Novel guinea-pig and rabbit antibodies were raised against synthetic peptides corresponding to (i) aa 1–21 and (ii) aa 126–147 of hNup 50 (accession no. NM_007172) and (iii) aa 1743–1763 of hNup196 (AAL56659). Additional guinea-pig antibodies were raised against (iv) aa 350–369, (v) 371–389, and (vi) 586–606 of hNup93 (BAA07680); (vii) aa 206–219 and (viii) 596–618 of hNup98; (ix) aa 33–51 of hNup 107 (NM_020401); (x) aa 53–67 and (xi) 1784–1803 of the partial hNup 205 sequence (D86978); and (xii) aa 993-1009 and (xiii) 1437–1458 (C at position 1447 exchanged for amino butyric acid) of hNup196. Peptide synthesis and coupling to KLH (Cordes et al., 1997) via an additional cysteine residue at the peptide's N-terminus (ii, viii, ix, xii) or C-terminus (i, iii, iv, v, vi, vii, x, xi, xiii) was done at Peptide Specialty Laboratories (Heidelberg, Germany). Affinity purification of Igs on immobilized peptides followed standard procedures (Cordes et al., 1997). Additional guinea-pig antibodies were raised against bacterially expressed His-tagged aa 8–203 of hNup93. Species-specific secondary antibodies coupled to fluorochromes and horseradish peroxidase (HRP) were from Jackson ImmunoResearch (West Grove, PA) and DAKO (Glostrup, Denmark).
Expression Vectors and cDNA Cloning
pRC/CMV expression vectors myc.hTpr.1-775/SV40-NLS, myc.hTpr.1-775/SV40-NLS (L458P, M489P) and myc.hTpr.1-775/SV40-NLS (L458D, M489D) have been described (Cordes et al., 1998; Hase et al., 2001); in all constructs, silent point mutations (G/T) were introduced by in vitro mutagenesis at positions corresponding to nt 94, 97, and 100 of the human cDNA sequence (accession no. U69668). Yeast two hybrid expression vectors encoding segments of Tpr (pAS/pACT-hTpr.1-398, hTpr.76/172–651, hTpr.76/172-651[L458P/M489P; L458D/M489D], hTpr.72/305-775, hTpr.774-1370, and hTpr.1178-1640) have been described (Hase et al., 2001). Novel two-hybrid constructs based on vectors pAS2–1 and pACT-2 (Clontech, Palo Alto, CA) are summarized in Table 1; aa sequences are deduced from ORFs downstream of the vector's NcoI site. DNA segments encoding nucleoporins were amplified by PCR using Pfu polymerase (Stratagene, La Jolla, CA) or subcloned from the following constructs kindly provided by B. Burke (pCMV HA-hNup153; Bastos et al., 1996), J. Borrow (pCDNA3 Flag-hNup196), B. Fontoura (pAlter-MAX Myc-rNup197; Fontoura et al., 1999), F. Müller-Pillasch (pSport1 hNup107, NM_020401), T. Nagase (hNup93, KIAA 0095; hNup205, KIAA 0225), and the German Human Genome Project (pT7T3D-Pac hNup50, IMAGE clone 1352973). Novel derivatives of the bacterial expression vector pGEX6P1 (Amersham Pharmacia Biotech, Uppsala, Sweden) encoding hTpr aa 172–651 with and without aa substitutions L458D and M489D, and aa 228–611 of hNup153 are described in Table 2.
Table 1.
Yeast two-hybrid expression vector constructs
| pAS/pACT-hNup153.1-224: Both constructs encode spacer-free fusions between Gal4 domains and hNup153 aa1-244, followed by vector-derived C-terminal residues ARPAAKLIPGEFLMIYDFYY(pAS-end1), or AREJYES (pACT-end1). |
| pAS/pACT-hNup153.228-439: MGS, hNup153 aa228-439, plus pAS-end1, or pACT-end1. |
| pAS/pACT-hNup153.337-611: MGS, hNup153 aa337-611, plus pAS-end1, or pACT-end1. |
| pAS/pACT-hNup153.228-611: MEAGS, hNup153 aa228-611, plus pAS-end1, or pACT-end1. |
| pAS/pACT-rNup98.1-339: MEAEFPGIRI, rNup98 aa1-339, plus pAS-end1, and MEAPGIRI, rNup98 aa1-339, plus pACT-end1. |
| pAS/pACT-rNup98.316-844: rNup98 aa316-844, plus REIRRPAAKLIPGEFLMIYDFYY (pAS-end2), or REIYES (pACT-end2). |
| pAS-rNup98.2-844: MEAEFPGIRIGAFMST, rNup98 aa2-844, plus pAS-end2, and MEAPGIRIGAFMST, rNup98 aa2-844, plus pACT-end2. |
| pACT-rNup196.316-883: rNup196 aa316-883, plus VRSQEEAESHRGSVERSMNRRY. |
| pAS/pACT-hNup196.864-1261: MEAEFPGIRIPGS (for pAS2-1) or MEAPGIRIPGS (for pACT2), hNup196 aa864-1216, plus LTVDLGRSMNRRY. |
| pAS/pACT-rNup196.1158-1599: rNup196 aa1158-1599, plus RRPAAKLIPGEFLMIYDFYY for pAS2-1, and rNup196 aa1158-1599, plus RIR, and pACT-end1. |
| pAS/pACT-hNup93.1-312: MRIP, hNup93 aa1-312, plus pAS-end1, and MEAPGIP, hNup93 aa1-312, plus pACT-end1. |
| pAS/pACT-hNup93.226-603: hNup93 aa226-603, plus VRSQEEAESHR, and GSVDLQPS for pAS2-1, and GSEFELERSMNRRY for pACT2. |
| pAS/pACT-hNup93.226-819: hNup93 aa226-819. |
| pAS/pACT-hNup107.1-146: MEAEFPGIRI, hNup107 aa1-146, plus pAS-end1, and MEAPGIRI, hNup107 aa1-146, plus pACT-end1. |
| pACT-hNup107.101-606: hNup107 aa101-606, plus AKLIPGEFLMIYDFYY. |
| pAS/pACT-hNup107.101-769: hNup107 aa101-769, plus RGSVDLQPS for pAS2-1, and SSRDL for pACT2. |
| pAS-hNup107.101-925: hNup107 aa101-925. |
| pAS/pACT-hNup205.1-718: hNup205 aa1-718, plus S, and TCSQANSGRISYDL (pAS-end3), and RDL for pACT2. |
| pAS-hNup205.350-1907: ME, hNup205 aa350-1907, plus pAS-end3. |
| pAS/pACT-hNup205.1-1907: hNup205 aa1-1907, plus pAS-end3, and DL for pACT2. |
| pACT-hNup205.350-718: hNup205 aa350-718, plus RDL. |
| pAS-hNup205.1-1254: hNup205 aa1-1254, plus PS. |
| pAS-hNup205.350-1254: hNup205 aa350-1254, plus PS. |
| pAS/pACT-hNup50.1-126: MRIR, hNup50 aa1-126, plus pAS-end1, and MEAPGIRI, hNup50 aa1-126, plus pACT-end1. |
| pACT-hNup50.29-208: hNup50 aa29-208, plus RGSVDLQPS for pAS2-1, and SSRDL for pACT2. |
| pAS-hNup50.29-469: hNup50 aa29-469. |
| pACT-hNup160.1-301: MEAPGIP, hNup160 aa1-301, and pACT-end1. |
| pAS-hNup160.238-1255: hNup160 aa238-1255, plus YGPKKKRKVTGDPS, and pAS-end3. |
| pAS-hNup160.238-764: hNup160 aa238-765, plus AYGPKKKRKVTGDPS, and pAS-end3. |
Table 2.
Bacterial expression vector constructs
| pGEX.hTpr.76/172-651: Encodes an N-terminal GST-tag, followed by LEVLFQGP (Pre Scission recognition site, cleavage site between residues Q and G), spacer LGSMADTFLEH, hTpr aa1-76, hTpr aa172-651, plus TGDPNSSGRIVTD. |
| pGEX.hTpr.76/172-651DD and PP: As pGEX.hTpr.76/172-651 but with aa substitution mutations L458D and M489D or L458P and M489P within the hTpr aa sequence segment. |
| pGEX.hNup153.228-611: N-terminal GST-tag, followed by LEVLFQGP, spacer LGS, hNup153 aa228-611, plus SSGRIVTD. |
Immunofluorescence Microscopy, Posttranscriptional Gene Silencing, and Cell Synchronization
Culture conditions for HeLa cells (ATCC CCL-2) of low and high passage numbers and immunofluorescence (IF) microscopy of cells were as described (Kuznetsov et al., 2002). Confocal microscopy and image processing was performed with an LSM 510 and the release 2.3 software (Zeiss, Oberkochen, Germany). Tpr siRNA duplexes and transfection conditions used for posttranscriptional silencing of the tpr gene have been described (Elbashir et al., 2001; Kuznetsov et al., 2002). In brief, HeLa cells were split 1 d before transfection using Oligofectamine reagent (Invitrogen, Groningen, Netherlands), and transfected at 15–30% cell density; the degree of density depending on whether cells were cotransfected with expression vectors later. SiRNA duplexes against Tpr, lamin A, and Nup153 (Harborth et al., 2001) were applied at 15–25 pM/cm2 of culture dish surface and at 15–50 pM/cm2 for controls with single-stranded sense and anti-sense RNAs. Immunoblotting and IF microscopy was performed with cells harvested 24, 48, 72, 96, and 120 h after transfection with Tpr and lamin A siRNAs; Nup153 siRNA-treated cells were analyzed 72–120 h posttransfection. Additional transfection of Tpr siRNA-treated cells with mammalian expression vectors using Fu-Gene 6 Transfection Reagent (Roche, Mannheim, Germany) was performed 3 or 4 d after initial transfection with siRNAs; cells were fixed 14–18 h later. For cell cycle phase synchronization of Tpr siRNA-treated cells, 2.5 mM thymidine was added to the culture medium 3 d after the initial transfection with siRNAs. After an incubation of 24–26 h, the medium was replaced with fresh thymidine-free medium. Cells were analyzed at different time points 0–28 h later. In some experiments, a second 24-h thymidine block was performed 12 h after release from the first.
In Vivo Protein Cross-linking and Immunoprecipitation
HeLa cells grown to near confluency in 8-cm culture dishes were washed briefly with hand-warm PBS; subsequent steps were with ice-cold solutions. Cells were permeabilized with 0.5% Triton X-100 in PBS for 3 min, briefly washed and then covered with PBS. 3,3′-dithio-bis[sulfosuccinimidyl proprionate] (DTSSP; Pierce, Rockford, IL) dissolved in PBS was added at a final concentration of 0.2 mM. Cross-linking was performed on ice for 90 min, with cells remaining adherent to the culture dish. After further washes with PBS, cells were quenched with 0.5 M Tris, pH 7.5, for 30 min and then scraped off from the culture dish into 10 ml RIPA-buffer (40 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with Complete Protease Inhibitor Cocktail (Roche), and homogenized. Lysates were cleared by centrifugation at 20,000 × g and 2°C for 15 min. The resulting supernatants were first incubated with mAb 203-37 against hTpr at 2°C for 12 h, then supplemented with 50 μl bedvolume of Protein A Sepharose (Sigma-Aldrich, Stockholm, Sweden), and again incubated at 2°C for 4 h. Beads were sedimented by mild centrifugation, washed three times in RIPA-buffer, resuspended in SDS protein sample buffer containing 1.4 M β-mercaptoethanol, and boiled for 5 min, resulting in cleavage of cross-links. SDS-PAGE and immunoblotting were as described (Kuznetsov et al., 2002).
Bacterial Expression and Purification of Recombinant Polypeptides
Synthesis of GST-tagged Tpr polypeptides in Escherichia coli BL21-CodonPlus (DE3)-RIL (Stratagene) was induced in cultures of logarithmic growth phase by addition of 0.1 mM IPTG. Cells were grown further at 30°C for 3 h, harvested, and incubated in lysis buffer I (PBS, 0.1% Triton X-100, 0.1 mg/ml lysozyme, and Complete Protease Inhibitors) at 2°C for 30 min. After brief sonication, lysates were cleared by centrifugation. In contrast, synthesis of GST-tagged Nup153 polypeptides was induced in cultures in early stationary growth phase. After further incubation at 30°C for 3 h, cells were incubated in lysis buffer II (PBS, 0.1% Triton X-100, 10 mM MgCl2, 0.1 mg/ml lysozyme, Complete Protease Inhibitors, and 10 U/ml bovine pancreas deoxyribonuclease I; Pharmacia) at 2°C for 3 h. The resulting lysate was supplemented with 2 mM ATP, incubated at 37°C for 10 min, and centrifuged at 20000 × g and 4°C for 20 min. Cleared lysates were incubated with glutathione Sepharose 4B (Pharmacia) at 2°C for 2–4 h. The slurry containing GST-Tpr fusion proteins was washed with PBS containing 0.1% Triton X-100; bound proteins were eluted with 10 mM reduced glutathione in 100 mM Tris, pH 8.0, containing 120 mM NaCl, 5 mM DTT, and 0.1% Triton X-100, and stored at 4°C or dialyzed against PBS for further use. GST-Nup153 beads were washed with PBS first and then with PreScission buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 8.0) and then incubated with PreScission Protease (30 U/ml; Pharmacia) at 2°C for 12–16 h. Nup153 polypeptides of which GST tags had been proteolytically removed were eluted in PreScission buffer and stored at 4°C or dialyzed against PBS for further use.
Protein-Protein Interactions
For interaction studies between purified GST-Tpr and tag-free Nup153 polypeptides, the recombinant proteins in PBS were mixed in different molar ratios, supplemented with 5 mM MgCl2, and incubated by rotation at 2°C for 12 h. After addition of glutathione Sepharose 4B (30 μl/ml), the suspension was incubated further at 2°C for 1 h. The beads were washed three times with PBS and boiled in SDS protein sample buffer. For pull-down studies using mammalian cell extracts, purified GST or GST-Tpr polypeptides in PBS were first reimmobilized on glutathione Sepharose 4B by incubation at 2°C for 4 h. In parallel, human PLC cells (ATCC CRL 8024) grown to near-confluency were homogenized in PBS containing 500 mM NaCl, 0.5% Triton X-100, and Complete Protease Inhibitors. The homogenate was cleared by 20,000 × g centrifugation at 4°C for 20 min. The supernatant was diluted 1:5 in PBS without NaCl, resulting in a final concentration of 100 mM NaCl, supplemented with 1 mM MgCl2, and incubated with the GST or GST-Tpr beads at 2°C for 4 h. Beads were first washed with 10 vol PBS and then with 10 vol PBS containing 1 M MgCl2. Bound proteins were eluted stepwise with 10 and 20 mM reduced glutathione in 100 mM Tris-HCl, pH 8.0, containing 120 mM NaCl and 5 mM DTT. For RNase treatment of cell extracts before affinity-chromatography on GST and GST-Tpr beads, extracts were incubated with 2 mg/ml RNase A (Qiagen, Hilden, Germany) at 4°C for 60 min; efficiency of mRNA and tRNA digestion was controlled by gel electrophoresis. PBS used for washings of beads was supplemented with 1 mg/ml RNase A as well. Proteins from all wash and elution steps were precipitated with 80% methanol at –20°C, boiled in sample buffer, and separated by SDS-PAGE. Image analysis of protein gels stained with Coomassie Brilliant Blue R250 and quantification of protein band intensities was by using the Fujifilm Science Lab 99 Image Gauge software version 3.2 (Fuji Photo Film, Tokyo, Japan).
Yeast Two-Hybrid Assays
S. cerevisiae strains Y190 (Mat a) and Y187 (Mat α; both from Clontech) were used for all assays. Cell cultures, transformation procedures, filter lift assays, and control experiments with empty two-hybrid vectors were as described earlier (Hase et al., 2001). Liquid β-galactosidase activity assays using the fluorescent substrate o-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma) were performed according to the Yeast Protocols Handbook PT3024–1 from Clontech.
RESULTS
NPC Assembly and Recruitment of Nuclear Basket Proteins at the End of Mitosis Does Not Require the Presence of Tpr
NPC disassembly in prophase leads to the release of Tpr that then forms a soluble pool of homodimers in the mitotic cell (Byrd et al., 1994; Hase et al., 2001; our unpublished results). Reassociation of Tpr to the reassembling NPC in telophase is preceded by incorporation of several nucleoporins of the cytoplasmic side (RanBP2/Nup358, Can/Nup214) and the central NPC channel (p62, Pom121), but also by that of at least one presumptive protein of the nuclear basket, namely Nup153 (Byrd et al., 1994; Bodoor et al., 1999; Haraguchi et al., 2000). The sequel by which other basket nucleoporins are recruited back to the NPC and whether their presence might be essential for reincorporation of Tpr was not known. Further, it could not be excluded that Tpr itself might act as a scaffold for other basket proteins and that their recruitment would follow that of Tpr. It was similarly unknown whether NPC disassembly during prophase occurs in a sequential manner too and whether some nucleoporins are released after or before Tpr.
Because such knowledge was expected to provide insight into Tpr's role in NPC architecture, we raised antibodies against the human orthologues of Nup50, Nup93, Nup96, Nup98, Nup107, and Nup205. Raised in different species, some of these antibodies, and others available for Nup153 and Tpr, allowed to study mitotic HeLa cells by double-labeling IF microscopy.
In prophase, complete release of Nup98 is well in advance of Tpr dissociation (Figure 1), indicative that this protein is not required for tethering Tpr to the NPC. Similarly, release of Nup50 appears to begin earlier than that of Tpr, whereas Nup96 parallels the dissociation of Tpr or follows it shortly afterwards. Nup107 again is found to be released late in prophase and still be present at remnants of the nuclear envelope (NE) already negative for Tpr. Different from the other nucleoporins, disassociation of Nup153 appears to occur in a stepwise manner: a first pool is rapidly detached from the NE early during prophase (unpublished data), whereas the remaining pool of NPC-attached Nup153 disappears only later, concomittant to the release of Nup96 and Tpr (Figure 1; see also DISCUSSION). The disassembly of Nup93 during prophase could not be studied by IF microscopy, despite having raised Nup93 antibodies against different parts of the protein: When applying standard immunolabeling procedures, no Nup93 or only traces thereof were detectable at the NPCs of interphase and prophase cells. Instead, visualization of the NPC-bound form of Nup93 required detergent-extraction of cells before fixation (see below; also Grandi et al., 1997). However, this procedure resulted in near-quantitative extraction of Nup93 and several other nucleoporins from cells in prophase till anaphase. In nonextracted cells, accessibility of NPC-associated Nup93 was noted only in cells in telophase (see below). Similarly, the antibodies we have raised against Nup205 so far did not yet allow the detection of the NPC-bound protein by standard IF microscopy.
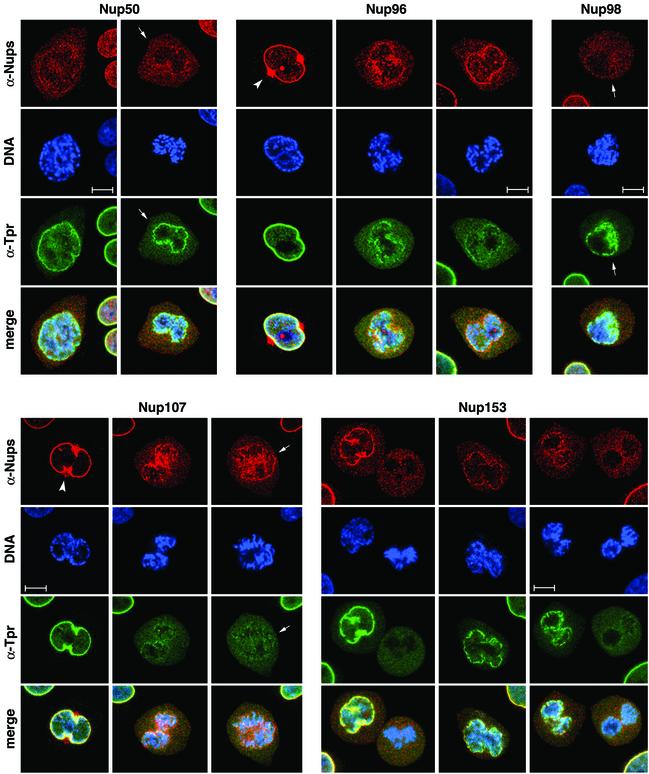
Figure 1.
Orderly disassembly of presumptive basket nucleoporins and Tpr in prophase. HeLa cells were fixed with formaldehyde and double-labeled with mouse or rabbit antibodies against Tpr (green staining), together with rabbit or guinea-pig antibodies against aa 126–147 of hNup50, aa 1743–1763 of hNup196 corresponding to the C-terminus of the proteolytic Nup96 product, aa 596–618 of hNup98, aa 33–51 of hNup107, and aa 21–36 of hNup153 (red staining). DNA staining with TO-PRO-3 is shown in blue. Dissociation of Tpr from the nuclear periphery of prophase cells is preceded by early loss of Nup50 and Nup98, but occurs before late disassembly of Nup107. Examples of cells in mid and late prophase/early prometaphase are labeled with arrows. Arrowheads demark a striking redistribution of Nup96 and Nup107 to the proximity of the centrosomes in cells early in prophase. Bars, 10 μm; for all micrographs.
Tpr is recruited back to the nucleus only late in telophase when chromosomes are mostly enclosed by a continuous NE (Figure 2; Bodoor et al., 1999) and nuclear import activity has been recovered (Haraguchi et al., 2000). In striking contrast, all the other presumptive basket nucleoporins are already present at the periphery of the newly segregated chromatids in anaphase, only preceded by even earlier association of Nup107 to kinetochores in metaphase (unpublished data; but see Belgareh et al., 2001) and of Nup96 to other structures proximal to prometaphase and metaphase chromatids (unpublished data). In early telophase, all these nucleoporins are largely incorporated into the newly formed NE. At this time point, Tpr is still found dispersed throughout the cytoplasm.
Figure 2.
Sequential recruitment of presumptive basket nucleoporins and Tpr into the reassembling NPC at the end of mitosis. HeLa cells were fixed with formaldehyde and double-labeled with antibodies against Tpr, Nup50, Nup96, Nup98, Nup107, and Nup153 as in Figure 1. Nup93 was labeled with antibodies against the recombinant N-terminus of hNup93 (B) and aa 350–369 of hNup93 (C). (A and B) Nucleoporins are recruited back to the periphery of the segregated chromatids in anaphase (A), whereas Tpr remains homogeneously dispersed throughout the cytoplasm until late in telophase (B). (C) Nucleoporins such as Nup93 are stably linked to the nuclear periphery in telophase cells and withstand detergent extraction of cells before fixation. Tpr, in contrast, is near-quantitatively extracted from such cells (arrows in left panel). Tpr is reincorporated into nuclei only late in telophase/early in G1, paralleling the decondensation of chromatin (arrowheads in right panel). Bar, 10 μm; for all micrographs.
Taken together these observations indicated that Tpr might not act as an anchoring site for other basket proteins at the NPC; on the contrary, most of these nucleoporins had to be rated as potential candidates through which Tpr itself might be tethered to the nuclear periphery.
Posttranscriptional tpr Gene Silencing Leads to Cellular Depletion of Protein Tpr, Which Does Not Prevent Other Basket Nucleoporins from Binding to the NPC
Similar conclusions as outlined above can be drawn from the analysis of HeLa cells in which tpr synthesis has been suppressed by RNA interference (RNAi).
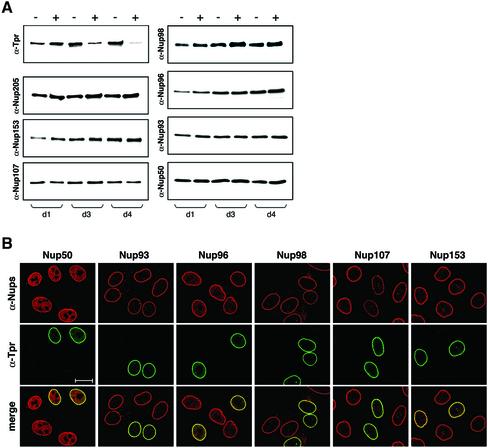
Use of small interfering RNA duplexes (siRNAs; Elbashir et al., 2001) is known to allow posttranscriptional silencing of the tpr gene, resulting in cellular depletion of the Tpr protein (Kuznetsov et al., 2002) without reducing the cellular content of other presumptive basket nucleoporins (Figure 3A). Some variability in the degree of the RNAi response can be noted between different established cell lines and also between their different laboratory substrains. For HeLa cells, the most striking RNAi effects can be seen in the original strain of low passage number (ATCC CCL-2) and in one of several laboratory HeLa substrains of high passage number (Chen, 1988). In these, near-complete loss of Tpr staining at the nuclear periphery of transfected cells can be noted ∼3.5 d after the initial transfection with siRNAs. From then on, Tpr-deficient cells still remain viable for at least a few more days. This behavior closely resembles that of HeLa cells in which we have silenced the nonessential lamin A gene by use of siRNAs (Harborth et al., 2001; our unpublished results). In our RNAi experiments, a small percentage of cells was generally found to have remained untransfected in siRNA-treated cultures, thus providing an internal subpool of Tpr-positive reference cells.
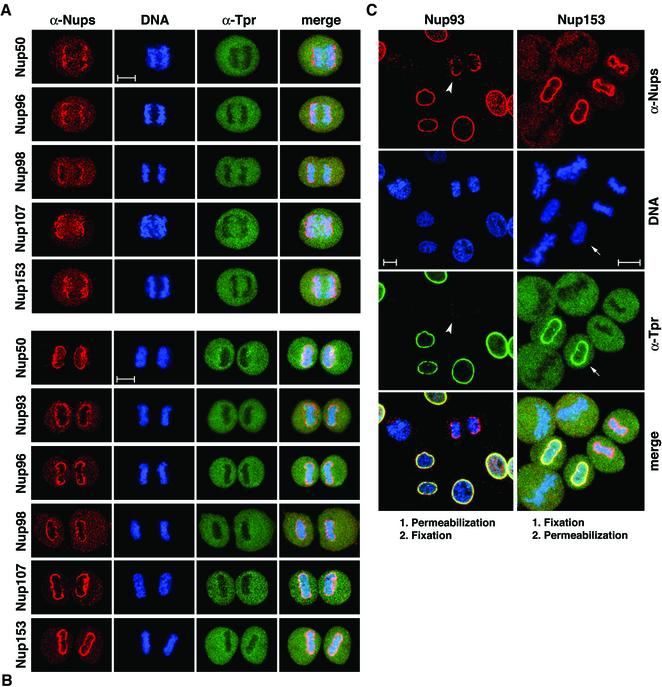
Figure 3.
Posttranscriptional tpr gene silencing leads to cellular depletion of protein Tpr which neither alters NPC distribution nor prevents synthesis and NPC-association of other presumptive basket nucleoporins. (A) Immunoblotting of total cell proteins from control (–) and siRNA-treated (+) HeLa cells harvested 1, 3, and 4 d (d) posttransfection. Proteins from corresponding pairs of control and RNAi cells were loaded in near-equal amounts. Filters with identical loadings prepared in parallel were immunolabeled with rabbit antibodies against the recombinant C-terminus of Tpr, mAb PF190x7A8 against Nup153, and guinea-pig antibodies against aa 1784–1803 of hNup205, aa 33–51 of hNup107, 206–219 of hNup98, aa 993-1009 of hNup196 (Nup96), and aa 371–389 of hNup93. Note that the total amount of Tpr in a siRNA-treated cell population reaches a minimum after three to four days; the residual amount of Tpr mainly reflects a subpopulation of nontransfected cells (Kuznetsov et al., 2002). In contrast, no reduction is seen for the other nucleoporins. (B) IF microscopy of HeLa cells 3 or 4 d after transfection with Tpr siRNA duplexes. Cells were fixed with formaldehyde and double-labeled with antibodies against Tpr and the other nucleoporins as in Figure 1. Cells labeled with antibodies against Nup93 were permeabilized with detergent before fixation. Note that in populations transfected with siRNA duplexes, most cells show only traces of Tpr staining at the nuclear periphery; the usually bright nuclear rim staining is visible only in a minor subpopulation of cells that have remained untransfected. Tpr deficiency, however, neither inhibits the recruitment of other nucleoporins to the nuclear periphery nor affects the normal distribution of NPCs within the NE. Bar, 20 μm.
Here we show that Tpr deficiency neither alters the NPC distribution within the plane of the NE nor prevents NPC association of other known nucleoporins in late mitosis and interphase (Figure 3B). Moreover, using antibodies specific for both lamin A and B-type lamins (Höger et al., 1991), lamina staining in both Tpr-deficient and control nuclei was found to be indistinguishable (unpublished data).
In both synchronized and asynchronous cell populations, no differences in NPC labeling for Nup96 and Nup358 were observed between Tpr-deficient and control nuclei. Likewise, Nup93 staining at the NE of control and Tpr-deficient cells that had been detergent-extracted before fixation was indistinguishable as well. Similarly, in most Tpr-deficient nuclei Nup153 labeling also remained unchanged; only few nuclei showed a minor reduction of Nup153 staining intensity at the NE. Also no reduction was noted for Nup50 and Nup98; in fact, in some Tpr-deficient nuclei NPC labeling with these antibodies appeared slightly more intense than in control cells. A striking difference between Tpr-deficient and control nuclei was only noted for Nup107; in the absence of Tpr, Nup107 labeling at the nuclear periphery was generally far more intense (Figure 3B).
In summary, these results indicate that Tpr does not act as a central scaffold for these presumptive nuclear basket proteins.
In Vivo Cross-linking Confirms Proximity between Presumptive Basket Proteins and Tpr
Several vertebrate nucleoporins form stable subcomplexes (e.g., Kita et al., 1993; Fornerod et al., 1997, Bastos et al., 1997), some of which also remain associated during mitosis (Grandi et al., 1997; Matsuoka et al., 1999; Miller et al., 2000; Belgareh et al., 2001; Vasu et al., 2001). In contrast, the mitotic form of Tpr does not remain bound to other nucleoporins, and extraction conditions required for releasing NPC-bound Tpr from interphase nuclei result in the release of soluble Tpr homodimers (our unpublished results; also Shah et al., 1998; Hase et al., 2001). Therefore, in order to stabilize the interaction between Tpr and its NPC binding partner(s), and thus allow coimmunoprecipitation of the proteins, HeLa cells were treated with DTSSP, a homo-bifunctional and thiol-cleavable cross-linker with a short spacer arm of 1.2 nm. After immunoprecipitation of the cross-link products with Tpr antibodies, the cross-linkers were cleaved and proteins analyzed by immunoblotting (Figure 4).
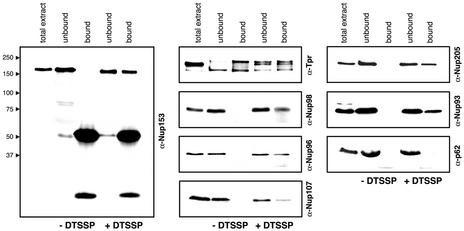
Figure 4.
Chemical cross-linking reveals physical proximity between presumptive basket nucleoporins and Tpr. HeLa cells were treated with or without thiol-cleavable cross-linker DTSSP and then solubilized with ionic detergents. The resulting extracts were immunoprecipitated with Tpr antibodies, and bound vs. unbound proteins analyzed by immunoblotting. Gels were loaded with the same percentage of each fraction. The left lanes show total cell extracts as reference. Relative masses of marker proteins in kDa are given at the left margin. The disulfide bond of DTSSP was reduced by treatment with β-mercaptoethanol before SDS-PAGE. Note that nucleoporins located at the nuclear side of the NPC coprecipitate with Tpr, whereas nucleoporin p62 located within the central pore channel is not.
Although p62, a component of the central pore channel did not become cross-linked to Tpr, we could coimmunoprecipitate all but one of the presumptive basket proteins, pointing at close spatial relationships to Tpr. Only the distribution of Nup50 remained uncertain, because of technical problems that were a consequence of cross-linker treatment, preventing unambiguous evaluation of the Nup50 results (unpublished data).
However, despite variations in DTSSP concentration and length of incubation, these and other cross-link approaches did not allow to distinguish unequivocally between nucleoporins that were bound directly to Tpr and those located only in close proximity.
Amino Acid Substitution Mutations That Abrogate NPC Binding of Recombinant Tpr Directly Affect the Interaction between Tpr and Another Nucleoporin
Recently we have shown that certain Tpr amino acid (aa) substitution mutations can abolish NPC binding of recombinant Tpr polypeptides in transfected cultured cells in which the natural tpr gene is expressed in parallel (Hase et al., 2001). To use these mutants in another approach aiming at identifying Tpr's NPC binding partner, we first had to verify that these mutations indeed abolish the interaction between Tpr and other nucleoporins. Though considered unlikely (Hase et al., 2001), we could not exclude definitively that the NPC binding observed in such transfected cells merely reflected the result of potential homo-oligomeric interactions between recombinant and wild-type Tpr homodimers and that such interactions might be sensitive for the mutations we had introduced.
To clarify this issue, we made use of our finding that cultured cells depleted of wild-type Tpr by RNAi can be readily posttransfected with expression vectors encoding a broad range of different proteins and that synthesis of these proteins is not impaired (our unpublished results). However, to allow synthesis of recombinant Tpr variants in the presence of siRNAs that target the wild-type Tpr mRNA, concomitant degradation of the recombinant mRNA had to be prevented. To this end, we introduced silent point mutations into the nucleotide sequence encompassing the start codon (Figure 5A), thus preventing “RNAi initiation” by siRNA duplexes that efficiently target the corresponding wild-type sequence (Elbashir et al., 2001). Second, all original 5′-UTR sequences of the Tpr mRNA upstream of these mutations were exchanged for Tpr-unrelated sequences to prevent potential “transitive RNAi effects” known to occur in some nonmammalian species (e.g., Nishikura, 2001; Hutvágner and Zamore, 2002).
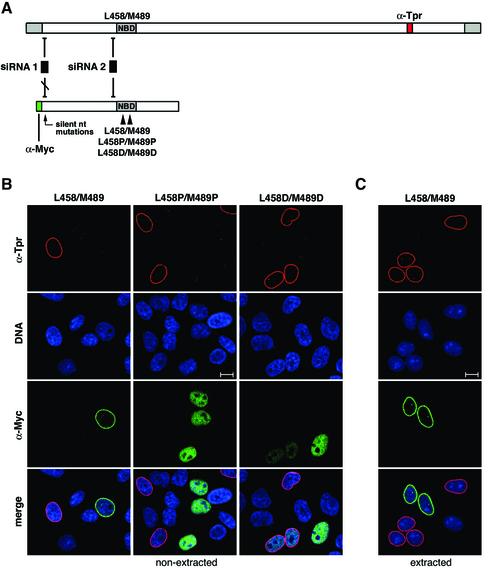
Figure 5.
NPC binding of recombinant Tpr in cells silenced for wild-type tpr expression can be abolished by aa substitution mutations. (A) Silent point mutations introduced into the nt sequence encompassing the start codon of Tpr cDNAs protect from being targeted by siRNA duplexes that mediate wild-type mRNA degradation: Schematic presentation of the wild-type Tpr mRNA and one recombinant Tpr mRNA variant, here encoding aa 1–774 fused to an N-terminal Myc-tag (green box) and a C-terminal NLS. Relative positions of siRNA duplexes, both complementary to the wild-type sequence, are represented as boxes 1 and 2. Duplex 2 triggers degradation of both the wild-type and the recombinant mRNA. In contrast, duplex 1 affects only the wild-type mRNA since silent nt exchanges that prevent the RNAi initiation step were introduced into the recombinant sequence. In addition, 5′-UTR sequences (light gray box) identical to the wild-type mRNA and upstream of the silent mutations have been removed. Arrowheads demark relative positions of substitution mutations at L458 and M489 located within the NPC binding domain of Tpr (NBD). The red box stands for the Tpr sequence encoding the epitope recognized by α-Tpr aa 2063–2084. (B) Expression of Myc-tagged Tpr 1–774 and corresponding mutants with double proline or aspartic acid substitutions at aa 458 and aa 489 in HeLa cells transfected with Tpr siRNA duplex no.1 and consequently depleted of endogenous wild-type Tpr. Normal levels of wild-type Tpr in nontransfected cells shown as reference were detected with rabbit antibodies against Tpr aa 2063–2084 (red); recombinant Tpr was stained with mAb 9E10 against the N-terminal Myc-tag (green), and DNA with TO-PRO-3. Note that in wild-type Tpr-deficient cells only recombinant Tpr polypeptides with an intact NBD associate with the NE. In contrast, double substitution mutations at aa 458 and aa 489 abolish NPC binding and cause intranuclear accumulation. Confocal micrographs are of cells fixed with FA before permeabilization with detergents. (C) Expression of Myc-tagged Tpr 1–774 in wild-type Tpr-deficient cells. Before FA-fixation and labeling with antibodies, cells were extracted with Triton X-100. Bars, 10 μm.
These sequence alterations allowed expressing recombinant versions of Tpr despite on-going silencing of the wild-type tpr gene. Recombinant polypeptides possessing an intact NPC binding domain (NBD; Hase et al., 2001) were found to bind to the NPC even though the wild-type protein was no longer present (Figure 5B). In contrast, Tpr polypeptides with aa substitution mutations L458P and M489P, or with L458D and M489D, bound neither to the NPCs of control cells nor to those of wild-type Tpr-depleted cells. Instead, these mutants accumulated within the nuclear interior (Figure 5B) from where they could be quantitatively extracted by brief detergent-permeabilization of cells (unpublished data; but see also Hase et al., 2001). However, polypeptides with an intact NBD remained stably bound to the NPC despite detergent-extraction of cells (Figure 5C).
The NPC Binding Domain of Tpr Attracts Nup153
Having established that certain Tpr aa substitution mutations abolish NPC binding, we searched for nucleoporins whose interaction with Tpr would depend on sequence integrity of Tpr's NBD
To this end, recombinant GST and GST-Tpr fusion proteins with and without aa substitution mutations L458D and M489D were immobilized on glutathione Sepharose beads and used for affinity chromatography of human cell extracts. Bound and unbound nucleoporins were detected by immunoblotting. Only Nup153 was found to bind specifically to the intact NBD of Tpr (Figure 6). RNase treatment of cell extracts before affinity chromatography did not affect this interaction (unpublished data; see MATERIALS AND METHODS), indicative that RNA molecules possibly bound to Nup153 (Dimaano et al., 2001) are not likely to be involved in the binding of Tpr to Nup153. Moreover, this interaction clearly depended on sequence integrity of Tpr's NBD because only traces of Nup153 were found associated with the Tpr mutant. GST alone did not bind Nup153.
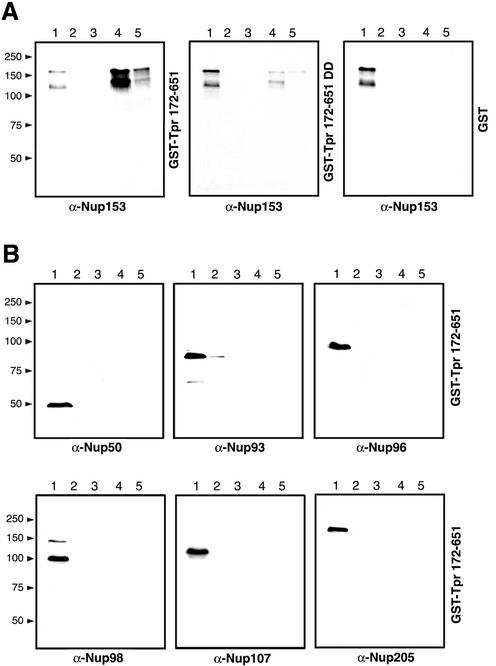
Figure 6.
Sequence integrity of Tpr's NPC binding domain mediates specific binding to Nup153. GST fusion proteins encompassing the NBD of wild-type Tpr (GST-Tpr 172–651), the corresponding mutant with aspartic acid substitutions at aa 458 and aa 489 (GST-Tpr 172–651 DD) and GST alone were immobilized on glutathione Sepharose beads. After incubation with Hela cell extracts, bound and unbound proteins were analyzed by immunoblotting. Unbound material representing the initial column run-through was loaded in lane 1, material from column washes with PBS in lane 2, and material eluted with 1 M MgCl2 in PBS in lane 3. Final elutions with 10 mM and 20 mM reduced glutathione are shown in lane 4 and 5. (A) Immunoblots with mAb PF190x7A8 against Nup 153. The second immunoreactive polypeptide of lower molecular mass is considered being a product of Nup153 degradation during extract incubation. (B) Immunoblots with antibodies against aa 1–21 and aa 126–147 of hNup50, aa 371–389 of hNup93, aa 1743–1763 of hNup196 (Nup96), aa 206–219 of hNup98, aa 33–51 of hNup107, and aa 1784–1803 of hNup205. Here only the results with GST-Tpr 172–651 are shown. Note that salt-resistant protein-protein interactions only occur between Nup153 and the intact NBD of Tpr, but not between the NBD and other nucleoporins.
Yeast Two-hybrid Analysis Confirms a Specific Interaction between Tpr and Nup153 That Depends on Sequence Integrity of Tpr's NPC Binding Domain
As an independent approach to study interactions between Tpr and other nucleoporins, we made use of the yeast two-hybrid methodology (Fields and Song, 1989). To this end we constructed two-hybrid expression vectors encoding the full-length sequences and segments of presumptive basket nucleoporins. These were then studied for their ability to engage in two-hybrid interactions with the intact or mutated forms of Tpr's NBD.
Only coexpression of Nup153 with Tpr polypeptides containing an intact NBD caused lacZ reporter gene activation as determined by β-galactosidase filter lift assays (Figure 7A). Remarkably, Tpr amino acid substitutions L458P and M489P that abolish Tpr's ability to bind to the NPC but do not inhibit Tpr homodimerization (Hase et al., 2001) near-completely abolished binding to Nup153 (Figure 7B).
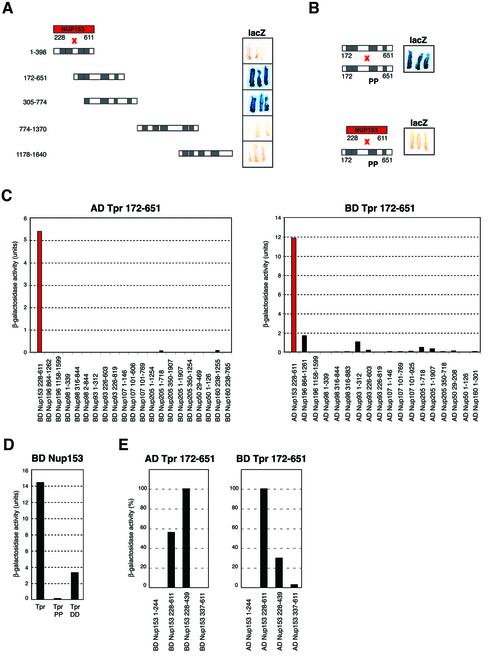
Figure 7.
Yeast two-hybrid analysis reveals specific interaction between Tpr's NPC binding domain and a segment of the N-terminal domain of Nup153 in vivo. (A and B) Schemes of Tpr and Nup153 fragments, and representative results of β-galactosidase filter lift assays. Here, interactions between Nup153 polypeptide 228–611 fused to yeast GAL4-BD and Tpr segments fused to GAL4-AD are shown. (A) LacZ reporter gene activation only occurs upon coexpression of Nup153 and those Tpr segments that contain the region known to be essential for NPC binding (∼ aa 450–550; Hase et al., 2001). Filters shown were prepared in parallel, with settings for color reproduction being identical. (B) Tpr aa substitution mutations that do not impair homodimerization but inhibit binding to the NPC, abolish the interaction with Nup153. (C) Quantification of two-hybrid interactions between Tpr and nucleoporins by liquid β-galactosidase assays. Sequences of Nup 50, Nup93, Nup 96, Nup 98, Nup107, Nup153, and Nup205 were fused to GAL4-BD, and Tpr aa 172–651 to GAL4-AD, and vice versa. The vertical scale represents units of β-galactosidase activity. Each value represents the average of enzyme activity measured for three independent yeast clones; a minor background of β-galactosidase activity measured for cells coexpressing GAL4-AD and GAL4-BD alone was subtracted from each data set. (D) Quantification of two-hybrid interactions between Nup153 and Tpr's intact NBD, Tpr 172–651 DD, or Tpr 172–651 PP. (E) Quantification of two-hybrid interactions between Tpr's intact NBD and different segments of Nup153.
The results of liquid β-galactosidase assays (Figure 7C) that allow to assess the relative strength of two-hybrid interactions (Estojak et al., 1995) further emphasized the specific binding between Nup153 and Tpr. Fused to the activation domain (AD) of GAL4, the Tpr NBD did not interact with any of the nucleoporins except for Nup153 (Figure 7C, left diagram). Similarly, in the reciprocal combination in which the GAL4 DNA binding domain (BD) was fused to Tpr, high levels of β-galactosidase activity were only noted when Tpr was coexpressed with Nup153 (Figure 7C, right diagram). Such β-galactosidase activity was significantly reduced by Tpr amino acid mutations L458D and M489D and was near-quantitatively suppressed by the L458P and M489P mutations (Figure 7D).
Tpr's NBD was found to interact not only with Nup153 aa 228–611 but also with a shorter Nup153 segment comprising aa 228–439 (Figure 7E). In contrast, no or only traces of β-galactosidase activity were detected in cells coexpressing Tpr and Nup153 segments comprising aa 1–244 and aa 337–611.
Specific Binding between Purified Polypeptides Reveals Direct Interaction between Nup153 and the NPC Binding Domain of Tpr
Yeast two-hybrid interactions and affinity chromatography of cell extracts indicated that Nup153 might be the direct binding partner of Tpr at the NPC. To rule out the possibility that another, yet undiscovered protein acts as a linker between both proteins, we investigated whether purified Tpr and Nup153 polypeptides that had been synthesized in bacteria were capable of direct interaction. To this end, a molar surplus of tag-free Nup153 polypeptide 228–611 was incubated with smaller amounts of GST-Tpr fusion protein 172–651 with and without aa substitutions L458D and M489D. As a further control, Nup153 was incubated with GST alone. The L458P/M489P mutant, however, could not be used because expression in bacteria went along with enhanced degradation, preventing purification of sufficient amounts of the full-sized polypeptide.
After incubation in PBS supplemented with 5 mM MgCl2, potential complexes were bound to glutathione Sepharose via GST, eluted, and analyzed by SDS-PAGE and Coomassie staining (Figure 8A) and immunoblotting (Figure 8B). Nup153 polypeptides and GST-Tpr were bound to glutathione in seemingly stoichiometric amounts. Image analysis of protein gels from two different experiments, and quantification of the amount of Coomassie dye bound to the Tpr and Nup153 bands revealed an average Tpr:Nup153 staining ratio of 2.1:1. In contrast, the relative amount of Nup153 bound to the Tpr aa substitution mutant was significantly reduced; the average ratio of Tpr:Nup153 staining being 9.1:1. Once again, GST alone did not bind Nup153. Interestingly, no or only dramatically reduced binding between Tpr and Nup153 was observed in buffers lacking MgCl2 (unpublished data).
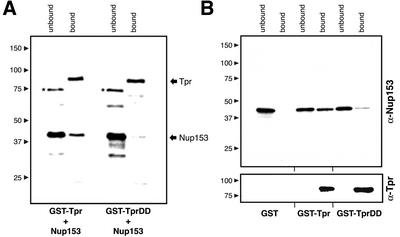
Figure 8.
Direct interaction of Nup153 and Tpr in vitro. (A) GST-Tpr 172-651 and GST-Tpr 172-651 DD were first incubated together with recombinant Nup 153 polypeptide 228–611 and then immobilized on glutathione Sepharose beads. Bound and unbound proteins were separated by SDS-PAGE and stained by Coomassie Blue. Arrows mark bands of Nup153 and Tpr, respectively. Asterisks mark one of several contaminating bacterial proteins not removed in the course of Nup153 purification. (B) Immunoblot identification of bound and unbound Nup153 proteins after incubation with same molar amounts of GST alone, GST-Tpr 172-651, and GST-Tpr 172-651 DD. The recombinant Tpr polypeptide was immunodetected with rabbit antibodies against Tpr aa 604–615. Lanes with bound proteins were loaded at eight times higher ratio of fraction than lanes with unbound material.
Posttranscriptional Nup153 Gene Silencing Leads to Mislocalization of Tpr and Nup50 to the Nuclear Interior but Does Not Prevent Other Nucleoporins from Binding to the NPC
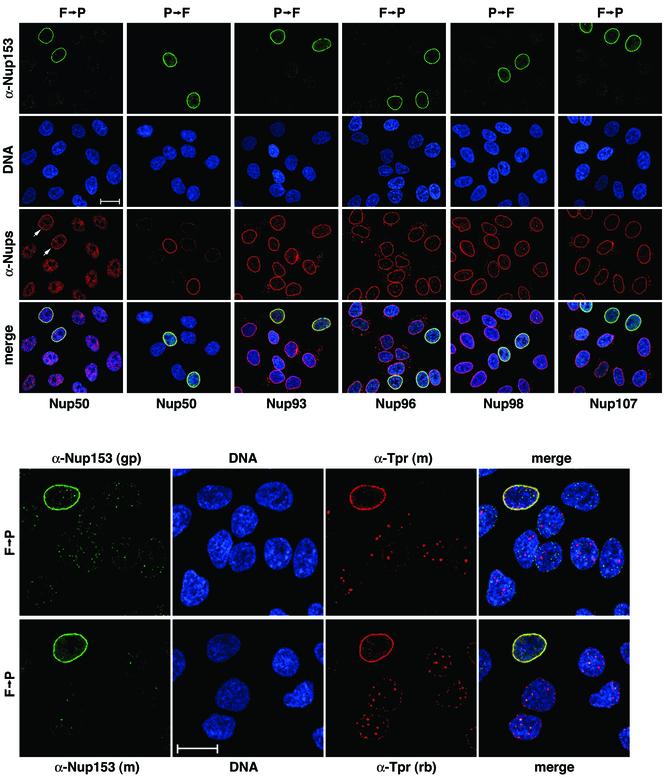
Recently, silencing of the Nup153 gene in HeLa cells has been reported to lead to cell growth arrest several days after transfection with Nup153 siRNAs (Harborth et al., 2001). Here we now wanted to investigate to which extent Nup153 deficiency in a cultured mammalian cell might affect NPC composition and distribution in general, and the localization of Tpr in particular.
Near-complete loss of Nup153 staining at the nuclear periphery was noted 3–4 d after the initial transfection of nonsynchronized HeLa cells with Nup153 siRNAs. NPC distribution within the plane of the NE of such Nup153-deficient cells was indistinguishable from that seen in control cells.
At 4 d posttransfection, bright staining for Nup96 and Nup107 at the nuclear periphery appeared largely unaltered in most Nup153-deficient cells (Figure 9). In fact, similar as in Tpr-deficient cells, intensity of Nup107 staining at the NE was often noted to be more intense than in neighboring control cells, perhaps reflecting facilitated Nup107 accessibility for antibodies in the absence of Nup153. Similarly, Nup93 and Nup98 remained stably bound to the Nup153-deficient NE, even after detergent extraction of cells before fixation (Figure 9). Staining intensity for these nucleoporins was found to diminish only later, concomittant to morphological and physiological alterations of the Nup153-deficient nuclei (unpublished data). Such late decrease of NE staining intensity was accompanied by the appearance of numerous variably-sized dot-like structures in the cytoplasm, reminiscent of the staining for annulate lamellae (AL) seen in certain cell lines (Cordes et al., 1996).
Figure 9.
Posttranscriptional Nup153 gene silencing leads to cellular depletion of protein Nup153 and nuclear redistribution of Tpr. IF microscopy of HeLa cells four days after transfection with Nup153 siRNA duplexes. Cells were either fixed with formaldehyde first, and then permeabilized with detergent (F→P), or permeabilized before fixation (P→F). Double-labeling was with mouse, rabbit, or guinea-pig antibodies as in Figure 1. In populations transfected with siRNA duplexes, most cells show only traces or no Nup153 staining at the nuclear periphery; the usually bright nuclear rim staining is visible only in a minor subpopulation of cells that have remained untransfected. Nup153 deficiency neither inhibits the recruitment of Nup93, Nup96, Nup98, and Nup107 to the nuclear periphery nor affects the normal distribution of NPCs within the NE, but strikingly impairs NPC-binding of Nup50 and Tpr. Normal NE-staining for Nup50 (arrows) is only seen in Nup153-positive cells; Nup50 released from the NE in Nup153-deficient cells often appears to add to the large pool of Nup50 generally found distributed throughout the nuclear interior. Such intranuclear forms of Nup50 can be near-quantitatively extracted by brief detergent treatment. Tpr molecules that have been prevented from binding to the NE in Nup153-deficient cells tend to form intranuclear aggregates that are detectable with rabbit antibodies (rb) against the C-terminal tail as well as with mAb (m) 203-37 against the rod domain of Tpr. Nup153 was labeled with either polyclonal guinea-pig antibodies (gp) against the N-terminal region, or with mAb PF190x7A8 against an epitope located between aa 439 and 611. Bars, 20 μm.
In contrast, the nuclear localization of Nup50 and Tpr was strikingly altered in Nup153-deficient cells already at 3 to 4 d posttransfection with siRNAs. Nup50 staining at the nuclear periphery was largely diminished; instead, the staining for Nup50 deep within the nuclear interior (see also Figure 3 and Guan et al., 2000) often appeared more intense than in control cells. This intranuclear pool of Nup50 could be extracted by brief detergent treatment of cells before fixation, resulting in near-complete loss of Nup50 staining from Nup153-depleted cells, but not affecting the Nup50 staining at the NE of control cells (Figure 9). Similarly, on-going depletion of Nup153 went along with a progressive decrease of Tpr staining at the NE. Although minor residual amounts of Tpr could still be observed at the nuclear periphery of some Nup153-deficient cells (see also DISCUSSION), a striking accumulation of Tpr in several detergent-resistant intranuclear aggregates was quite frequently observed 4 d after the initial transfection with siRNAs (Figure 9). In addition, loss of Nup153, and release of Tpr from the NPC, was also paralleled by a gradual decrease of the total cellular content of Tpr (unpublished data). Furthermore, in contrast to the other nucleoporins, Tpr and Nup50 in Nup153-depleted cells were never found to accumulate in AL-reminiscent cytoplasmic structures.
DISCUSSION
The mammalian tpr gene encodes one major isoform of ∼267 kDa, which locates near-exclusively at the NPC in most somatic cell types (Kuznetsov et al., 2002). In the present study, we have assessed Tpr's role as an architectural element of the NPC in HeLa cells. In addition, we have identified the NPC binding partner that links Tpr to the nuclear periphery.
Tpr and NPC Architecture
Tpr is shown to be only peripherally attached to the NPC and not to act as a scaffold onto which several other nucleoporins need to be assembled. In Tpr-deficient cells, binding of these nucleoporins to the NPC was found to be neither impaired in G1 phase nor in G2 when NPC numbers have doubled (Maul, 1977). These findings are in harmony with recent studies in which Tpr has been prevented from reincorporation into the NPC by injection of Tpr antibodies into mitotic cells, resulting in sequestration of Tpr into large, mostly cytoplasmic aggregates at or near the nuclear periphery in G1 cells (Frosst et al., 2002; Shibata et al., 2002; our unpublished results). NPC staining intensity for Nup153 and Nup98 in such injected cells has been reported to remain largely similar to that seen in noninjected controls (Frosst et al., 2002). Here we show that Tpr is also dispensable for NPC binding of Nup50, Nup93, Nup96, and Nup107. In fact, we noted that NPC staining for some of these presumptive basket proteins was even more intense in Tpr-deficient nuclei than in control nuclei. However, it remains to be clarified whether this reflects an actual increase in protein copy numbers at the NPC or whether it simply reflects better antibody accessibility to some nucleoporins when Tpr is absent.
Furthermore, NPC distribution within the NE remains unaffected by lack of Tpr, demonstrating that Tpr is not an essential anchoring element for the NPC as proposed earlier. Apart from Tpr, only Nup98 has been shown to be dispensable for correct NPC positioning in vertebrates (Wu et al., 2001). Here, we also observe that NPC positioning in Nup153-depleted cells is indistinguishable from that seen in control cells. NPCs, however, that lack at least four presumptive basket components at the same time, namely Nup93, Nup98, Nup153, and Tpr, have been shown to be mobile and form clusters within the plane of the NE (Walther et al., 2001), suggesting that several nucleoporins in concert might be involved in holding the NPCs in place.
Tpr and Nucleoporin Binding Partners at the NPC
Binding of Tpr to the NPC is mediated by only a short segment of its aminoterminal domain. Aa substitution mutations introduced into this NBD abolish Tpr's ability to bind to the NPC, rendering the protein soluble and resulting in its accumulation in the nuclear interior (Hase et al., 2001). None of the Tpr regions outside the NBD are sufficient to stably tether the protein to the nuclear periphery (Bangs et al., 1998; Cordes et al., 1998; Hase et al., 2001).
In the present study, we have searched for those proteins that specifically interact with the intact version of Tpr's NBD but disdain the mutated form. This objective was tackled by different experimental approaches in parallel, each of them of different probative force. Nevertheless, all approaches consistently lead to the identification of Nup153 as a specific binding partner for the NBD of Tpr. This finding does not rule out the possibility that still other nucleoporins might bind to other regions of Tpr and that such additional interactions might help stabilize Tpr's association with the NPC. It is equally well imaginable that once Tpr has been linked to the NE, this localization might be stabilized further by interactions with also other, non-NPC molecules located at the nuclear periphery. On their own, however, none of these additional interactions can be expected to suffice to stably and durably secure Tpr at the NPC. Cellular depletion of Nup153 by RNAi clearly caused a mislocalization of Tpr to the nuclear interior but at the same time had no apparent effect on the localization of several other nucleoporins.
A previous study has reported a direct binding between in vitro translated full-length Tpr and Nup98 in reticulocyte lysates; however, the region of Tpr involved in such binding was not determined (Fontoura et al., 2001). Because Nup98 does not bind to Tpr's NBD, it is unlikely that Nup98 is directly involved in the attachment of Tpr to the NPC. On the other hand, the possibility remains that other regions of Tpr might contain binding sites for Nup98. However, because staining for Nup98 at Tpr-deficient NPCs remains unaltered or sometimes is even slightly more pronounced than in control cells, the majority of binding sites for Nup98 at the NPC must be provided by proteins other than Tpr.
Affinity purification of the yeast homolog of Nup93, Nic96p, from glutaraldehyde-treated yeast nuclei has been shown to result in copurification of one of the probable yeast homologues of Tpr, Mlp2p, together with several other nucleoporins (Kosova et al., 2000). The possibilities that Nic96p could serve as an NPC-attachment site for Mlp2p or that Mlp2p itself might provide an anchor site for Nic96p at the nuclear basket were proposed as equally likely alternatives. However, whether Mlp2p and Nic96p interact directly with each other or via other nucleoporins remained uncertain (Kosova et al., 2000). NPC binding of mammalian Nup93 in any case does not depend on the presence of Tpr. And because Nup93 does not interact with Tpr's NBD, a direct role for Nup93 in tethering Tpr to the mammalian NPC basket seems unlikely too. However, the consistent finding that Mlp2 and Nic96p as well as Tpr and Nup93 can be coenriched from nuclei treated with cross-linkers points at similarly close, though not necessarily direct spatial relationships in both lower and higher eukaryotes.
Again in yeast, potential NPC docking sites for Mlp proteins have also been searched for by performing synthetic lethality screens. This resulted in the identification of C-Nup145p, the probable homolog of mammalian Nup96. In cells lacking C-Nup145p, both Mlp1p and Mlp2p were found to be mislocalized to the nuclear interior (Galy et al., 2000). Although the experimental approach did not allow to argue for direct interactions between these proteins, it clearly pointed at an essential role of C-Nup145p in positioning the Mlps at the NPC. Although our present study did not reveal any direct interaction between the mammalian Nup96 homolog and Tpr's NBD (see also Fontoura et al., 2001), the possibility remains that also Nup96 might act as a chain link, perhaps via its interaction with Nup153 (Vasu et al., 2001), and might therefore be essential for indirectly tethering Tpr to the NPC. Our observation that Nup153-depletion of cells leads to mislocalization of Tpr, but has no direct and immediate effect on the localization of Nup96 at the nuclear periphery, supports this point of view.
Most recently, Mlp2p has also been identified in a yeast two-hybrid screen as a binding partner of yeast Nup60p; the latter having been used as the bait. Subsequent deletion of the Nup60 gene led to mislocalization of both Mlps to the nuclear interior, similar to what had been seen in cells negative for C-Nup145p. Additional results then indicated that Nup60p provides a link between C-Nup145p and Mlp proteins (Feuerbach et al., 2002). Supposing Nup60p might be the direct binding partner that tethers Mlp2p to the NPC, its function as an Mlp anchor site would correspond to that of Nup153 in Tpr binding. At first sight, however, Nup153 and Nup60p exhibit no apparent sequence homology. Most strikingly, the central Zn-finger domain present in Nup153 (Sukegawa and Blobel, 1993) does not have any equivalent in Nup60p. Because other candidates that might represent possible counterparts of Nup153 and Nup60p in the respective other species are not at hand either, their similar function in binding Tpr and Mlps might be considered an example of convergent evolution. Closer inspection of both protein sequences, however, reveals some interesting similarities between the N-terminal segment of Nup60p covering aa 1–300 and the Nup153 segment comprising aa 1–600, the latter including the binding sites for Tpr and other nucleoporins. Although overall sequence homology between both segments is rather poor, secondary structures are predicted to be very similar (Garnier et al., 1996; at http://npsa-pbil.ibcp.fr/NPSA). Furthermore, both segments are similarly charged (pI of 9.6 for Nup60p, 9.28 for hNup153), are rich in hydroxy amino acids (23.7% for Nup60p, 24.3% for hNup153), and include a few regions with similarly positioned proline residues. Although such crude sequence similarities can also be found in pairs of analogous proteins, the additional occurrence of short but seemingly conserved sequence segments within these and other regions of the two proteins might indicate a common ancestor. Moreover, both proteins share further common properties. For example, Nup153 interacts with Nup50 (Guan et al., 2000; Smitherman et al., 2000), and Nup60p represents a binding partner for Nup2p, the probable yeast homolog of Nup50. The binding site for Nup2p is located within the N-terminal half of Nup60p (aa 1–388; Denning et al., 2001), resembling the Nup50 binding region of Nup153, located between aa 337 and 611 (our unpublished results). Moreover, the Nup153 region (aa 228–439) that binds Tpr also contains the binding sites for the Nup160 complex including Nup96 (Vasu et al., 2001); this region is also essential for anchoring Nup153 to the NPC itself (Enarson et al., 1998). Because Nup50 and Tpr are dispensable for NPC binding of Nup153 (Smitherman et al., 2000; this study), it might be the Nup160 complex that provides the anchor site for Nup153. This scenario would then again resemble the situation in yeast where Nup60p provides a link between C-Nup145p and Mlps. Finally, Nup153 contains binding sites for nuclear transport factors, including importin beta and Ran-GTP (e.g., Saitoh et al., 1996; Moroianu et al., 1997; Shah et al., 1998; Nakielny et al., 1999), as does Nup60p for the corresponding yeast homologues Kap95p and Gsp1p-GTP (Denning et al., 2001). In view of these similarities, Nup153 and Nup60p might be considered blown-up respectively truncated versions of each other; the Zn-finger domain present in the one and missing in the other might have been acquired or lost later during evolution, possibly concomittant with the acquirement or loss of additional functions.
Wild-type Tpr in vivo and tag-free recombinant Tpr polypeptides in vitro have recently been shown to readily form homodimers; such homodimers have been proposed to represent the units that bind to the NPC (Hase et al., 2001; see also below). Here we have noted that the GST-tagged NBD of Tpr binds Nup153 in seemingly stoichiometric amounts. Although we do not exclude that the dimerization property of GST may contribute to the homodimerization of the GST-Tpr fusion protein, Tpr's NBD on its own is already well capable of homodimerizing (Hase et al., 2001). Binding of recombinant Tpr to Nup153 in an approximate 2:1 ratio would therefore be consistent with the conception of Tpr homodimers being bound to the NPC. In this context it might be interesting to note that the molar content of Nup153 in at least some mammalian cell lines can exceed that of Tpr ∼2.5-fold (our unpublished results). Assuming that one Nup153 monomer acts as the binding partner for one Tpr homodimer, only ∼20% of the cellular content of Nup153 would be required for NPC-binding of Tpr in such cells. In this case, the majority of Nup153 molecules would be free to fulfill other functions, including those that do not necessarily require all Nup153 polypeptides to be stable structural components of the NPC (e.g., Nakielny et al., 1999; Dimaano et al., 2001; Daigle et al., 2001). In fact, we consider it possible that HeLa cells contain at least two distinct pools of protein Nup153: a major more dynamic one (e.g., Nakielny et al., 1999; Daigle et al., 2001; our unpublished results) and a smaller pool that comprises those Nup153 molecules that represent intrinsic components of NPC structure (e.g., Walther et al., 2001; our unpublished results) and act as Tpr binding partners. Tpr itself, once bound to this NPC-architectural form of Nup153, might even have a mild stabilizing effect on such Nup153 positioning, thus providing one possible explanation for the mild reduction of Nup153-staining intensity occasionally noted at the NE of some Tpr-deficient cultured cells.
Because binding between recombinant Tpr and Nup153 was only observed in the presence of MgCl2, we finally also want to point at earlier electron microscopic studies that have reported on the necessity of bivalent cations to preserve nuclear basket integrity and the association of its filaments to the NPC proper (Jarnik and Aebi,1991; Arlucea et al., 1998).
Future Perspectives
It still remains to be determined in how far nuclear basket structure might have been altered upon depletion of Tpr. Standard transmission electron microscopy (TEM) of ultra-thin sections through plastic-embedded HeLa cells does not allow to identify this structure, let alone visualize any of its morphological characteristics. That is also why descriptions of NPC basket structure mainly build on scanning electron microscopic (SEM) investigations. These again have used manually isolated nuclei from amphibian and avian oocytes and from insect salivary gland cells, which allow rather easy access to the inner side of the NE (e.g., Ris, 1989; Jarnik and Aebi, 1991; Goldberg and Allen, 1992; Kiseleva et al., 1996; Goldberg et al., 1997). Similar SEM investigations of the inner side of the NPC in cultured somatic cells are technically far more demanding and have not been reported to date. However, once methodological improvements will allow to visualize the inner side of the NE in HeLa cells by SEM, it will be tempting to study the alterations in mammalian NPC structure after the depletion of Tpr and other nucleoporins by RNAi. Moreover, provided that expression of individual genes in amphibian oocytes can be similarly well suppressed by siRNAs as recently reported for genes transcribed during Xenopus embryogenesis (Zhou et al., 2002), Tpr-depleted Xenopus oocytes would be an obvious target for SEM investigations as well.
Furthermore, the exact locations of Tpr, Nup153, and other presumptive basket nucleoporins in space, relative to each other and to the NPC proper, remain largely unknown. In fact, several immunolocalization studies using different antibodies and immunolabeling procedures have produced conflicting results with regard to the localization of individual basket components, including Tpr. Our present study has now addressed the question as to the relative position of Tpr from a different angle. The finding that Tpr binds only to properly assembled NPCs and is dispensable for NPC binding of other nucleoporins, renders Tpr a protein only peripherally attached to the NPC proper. Provided that some of the other nucleoporins formerly envisioned to be basket components might instead be located closer to the NPC proper, as recently reported for human Nup107 (Belgareh et al., 2001) and the N-terminal domain of Xenopus Nup153 (Walther et al., 2001), the coiled-coil homodimer of Tpr (Hase et al., 2001) would now emerge as the most likely candidate for the central architectural element of the nuclear basket (see also Frosst et al., 2002). In this scenario, close spatial relationships between Tpr and other nucleoporins can still be easily envisioned, especially in view of their closely neighboring binding sites on the Nup153 molecule. However, taking Tpr's extended rod-like shape into consideration (Hase et al., 2001), close neighborhood of Tpr with other nucleoporins might be limited to a region encompassing Tpr's NBD. The availability of novel antibodies specific for individual domains not only of Tpr but also other nucleoporins will now allow follow-up on this idea and reinvestigate the locations of these NPC proteins by immunoelectron microscopy under identical experimental conditions. Moreover, further use of the RNAi execution machinery to deplete cultured cells also of presumptive basket components other than Tpr and Nup153 will provide additional insight into the spatial arrangements of these proteins at the inner side of the NPC.
Acknowledgments
We thank Dr. Hans-Richard Rackwitz at Peptide Specialty Laboratories Heidelberg for peptide syntheses; Maarten Fornerod for stimulating discussions; and Drs. Brian Burke, Julian Borrow, Beatriz Fontoura, Georg Krohne, Friederike Müller-Pillasch, and Takahiro Nagase for providing reagents. This investigation was supported by grants to V.C.C. from the Swedish Natural Research Council, the foundation Cancerföreningen i Stockholm, and the Marianne and Marcus Wallenberg Foundation.
DOI: 10.1091/mbc.E02-09-0620.
Abbreviations used: aa, amino acid; AD, activation domain; AL, annulate lamellae; BD, binding domain; FA, formaldehyde; IF, immunofluorescence microscopy; NE, nuclear envelope; NBD, nuclear pore complex binding domain; NPC, nuclear pore complex; nt, nucleotide; RNAi, RNA interference; siRNA, small interfering RNA.
References
- Adam, S.A. (2001). The nuclear pore complex. Genome Biol. 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlucea, J., Andrade, R., Alonso, R., and Arechaga, J. (1998). The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J. Struct. Biol. 124, 51–58. [DOI] [PubMed] [Google Scholar]
- Bangs, P.L., Burke, B., Powers, C., Graig, R., Purohit, A., and Doxsey, S. (1998). Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell Biol. 143, 1801–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos, R., Lin, A., Enarson, M., and Burke, B. (1996). Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos, R., Ribas de Pouplana, L., Enarson, M., Bodoor, K., and Burke, B. (1997). Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J. Cell Biol. 137, 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N. et al. (2001). An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor, K., Shaikh, S., Salina, D., Raharjo, W.H., Bastos, R., Lohka, M., and Burke, B. (1999). Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112, 2253–2264. [DOI] [PubMed] [Google Scholar]
- Byrd, D.A. et al. (1994). Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. J. Cell Biol. 127, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.R. (1988). Re-evaluation of HeLa, HeLa S3, and HEp-2 karyotypes. Cytogenet. Cell. Genet. 48, 19–24. [DOI] [PubMed] [Google Scholar]
- Cordes, V.C., Waizenegger, I., and Krohne, G. (1991). Nuclear pore complex glycoprotein p62 of Xenopus laevis and mouse: cDNA cloning and identification of its glycosylated region. Eur. J. Cell Biol. 55, 31–47. [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., Köhler, A., Stuurman, N., van Driel, R., and Franke, W.W. (1993). Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol. 123, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., and Franke, W.W. (1996). Cytoplasmic annulate lamellae in cultured cells: composition, distribution, and mitotic behavior. Cell Tissue Res. 284, 177–191. [DOI] [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., Rackwitz, H.-R., and Franke, W.W. (1997). Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., Hase, M.E., and Müller, L. (1998). Molecular segments of protein Tpr that confer nuclear targeting and association with the nuclear pore complex. Exp. Cell Res. 245, 43–56. [DOI] [PubMed] [Google Scholar]
- Daigle, N., Beaudouin, J., Hartnell, L., Imreh, G., Hallberg, E., Lippincott-Schwartz, J., and Ellenberg, J. (2001). Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D., Mykytka, B., Allen, N.P., Huang, L., Burlingame, A., and Rexach, M. (2001). The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154, 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano, C., Ball, J.R., Prunuske, A.J., and Ullman, K.S. (2001). RNA association defines a functionally conserved domain in the nuclear pore protein Nup153. J. Biol. Chem. 276, 45349–45357. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Enarson, P., Enarson, M., Bastos, R., and Burke, B. (1998). Aminoterminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma 107, 228–236. [DOI] [PubMed] [Google Scholar]
- Estojak, J., Brent, R., and Golemis, E.A. (1995). Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15, 5820–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., Stoffler, D., and Aebi, U. (2001). Nuclear pore complex architecture and functional dynamics. Curr. Top. Microbiol. Immunol. 259, 95–117. [DOI] [PubMed] [Google Scholar]
- Fan, F., Liu, C.-P., Korobova, O., Heyting, C., Offenberg, H.H., Trump, G., and Arnheim, N. (1997). cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics 40, 444–453. [DOI] [PubMed] [Google Scholar]
- Ferrando-May, E., Cordes, V.C., Biller-Ckovric, I., Mirkovic, J., Görlich, D., and Nicotera, P. (2001). Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors, and modulation of nuclear permeability in apoptosis. Cell Death Differ. 8, 495–505. [DOI] [PubMed] [Google Scholar]
- Feuerbach, F. et al. (2002). Nuclear architecture, and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4, 214–221. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O.-K. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Fontoura, B.M.A., Blobel, G., and Matunis, M.J. (1999). A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144, 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M.A., Dales, S., Blobel, G., and Zhong, H. (2001). The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc. Natl. Acad. Sci. USA 98, 3208–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., van Deursen, J., van Baal, S., Reynolds, A., Davis, D., Murti, K.G., Fransen, J., and Grosveld, G. (1997). The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst, P., Guan, T., Subauste, C., Hahn, K., and Gerace, L. (2002). Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V., Olivo-Marin, J.-C., Scherthan, H., Doye, V., Rascalou, N., and Nehrbass, U. (2000). Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403, 108–112. [DOI] [PubMed] [Google Scholar]
- Garnier, J., Gibrat, J.-F., and Robson, B. (1996). GOR secondary structure prediction method version IV. Methods Enzymol. 266, 540–553. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.W., and Allen, T.D. (1992). High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 119, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M.W., Solovei, I., and Allen, T.D. (1997). Nuclear pore complex structure in birds. J. Struct. Biol. 119, 284–294. [PubMed] [Google Scholar]
- Grandi, P., Dang, T., Pante, N., Shevchenko, A., Mann, M., Forbes, D., and Hurt, E. (1997). Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell 8, 2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, T., Kehlenbach, R.H., Schirmer, E.C., Kehlenbach, A., Fan, F., Clurman, B.E., Arnheim, N., and Gerace, L. (2000). Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20, 5619–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, T., Koujin, T., Hayakawa, T., Kaneda, T., Tsutsumi, C., Imamoto, N., Akazawa, C., Sukegawa, J., Yoneda, Y., and Hiraoka, Y. (2000). Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113, 779–794. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Bechert, K., Tuschl, T., and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- Hase, M.E., Kuznetsov, N., and Cordes, V.C. (2001). Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol. Biol. Cell 12, 2433–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höger, T.H., Grund, C., Franke, W.W., and Krohne, G. (1991). Immunolocalization of lamins in the thick nuclear lamina of human synovial cells. Eur. J. Cell Biol. 54, 150–156. [PubMed] [Google Scholar]
- Hutvágner, G., and Zamore, P.D. (2002). RNAi: nature abhors a double-strand. Curr. Opin. Genet. Develop. 12, 225–232. [DOI] [PubMed] [Google Scholar]
- Jarnik, M., and Aebi, U. (1991). Towards a more complete 3 D-structure of the nuclear pore complex. J. Struct. Biol. 107, 291–308. [DOI] [PubMed] [Google Scholar]
- Kiseleva, E., Goldberg, M.W., Daneholt, B., and Allen, T.D. (1996). RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol. 260, 304–311. [DOI] [PubMed] [Google Scholar]
- Kita, K., Omata, S., and Horigome, T. (1993). Purification and characterization of a nuclear pore glycoprotein complex containing p62. J. Biochem. 113, 377–382. [DOI] [PubMed] [Google Scholar]
- Kosova, B., Pante, N., Rollenhagen, C., Podtelejnikov, A., Mann, M., Aebi, U., and Hurt, E. (2000). Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with Nic96p. J. Biol. Chem. 275, 343–350. [DOI] [PubMed] [Google Scholar]
- Kuznetsov, N.V., Sandblad, L., Hase, M.E., Hunziker, A., Hergt, M., and Cordes, V.C. (2002). The evolutionarily conserved single-copy gene for murine Tpr encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma 111, 236–255. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y., Takagi, M., Ban, T., Miyazaki, M., Yamamoto, T., Kondo, Y., and Yoneda, Y. (1999). Identification and characterization of nuclear pore subcomplexes in mitotic extract of human somatic cells. Biochem. Biophys. Res. Commun. 254, 417–423. [DOI] [PubMed] [Google Scholar]
- Maul, G. (1977). The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int. Rev. Cytol. Suppl. 6, 75–186. [PubMed] [Google Scholar]
- Miller, B.R., Powers, M., Park, M., Fischer, W., and Forbes, D.J. (2000). Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay. Mol. Biol. Cell 11, 3381–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu, J., Blobel, G., and Radu, A. (1997). RanGTP-mediated nuclear export of karyopherin alpha involves its interaction with the nucleoporin Nup153. Proc. Natl. Acad. Sci. USA 94, 9699–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny, S., Shaikh, S., Burke, B., and Dreyfuss, G. (1999). Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura, K. (2001). A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107, 415–418. [DOI] [PubMed] [Google Scholar]
- Powers, M.A., Macaulay, C., Masiarz, F.R., and Forbes, D.J. (1995). Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128, 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu, A., Blobel, G., and Wozniak, R.W. (1994). Nup107 is a novel nuclear pore complex protein that contains a leucine zipper. J. Biol. Chem. 269, 17600–17605. [PubMed] [Google Scholar]
- Radu, A., Moore, M.S., and Blobel, G. (1995). The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81, 215–222. [DOI] [PubMed] [Google Scholar]
- Ris, H. (1989). Three-dimensional imaging of cell ultrastructure with high resolution low voltage SEM. Inst. Phys. Conf. Ser. 98, 657–662. [Google Scholar]
- Ris, H. (1991). The three-dimensional structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull. 21, 54–56. [Google Scholar]
- Ris, H., and Malecki, M. (1993). High-resolution field emission scanning electron microscope imaging of internal cell structures after epon extraction from sections: a new approach to correlative ultrastructural and immunocytochemical studies. J. Struct. Biol. 111, 148–157. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., and Aitchison, J.D. (2001). The nuclear pore complex as a transport machine. J. Biol. Chem. 276, 16593–16596. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., Cooke, C.A., Burgess, W.H., Earnshaw, W.C., and Dasso, M. (1996). Direct and indirect association of the small GTPase ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevis egg extracts. Mol. Biol. Cell 7, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, S., Tugendreich, S., and Forbes, D. (1998). Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 141, 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, S., Matsuoka, Y., and Yoneda, Y. (2002). Nucleocytoplasmic transport of proteins, and poly(A)+ RNA in reconstituted Tpr-less nuclei in living mammalian cells. Genes Cells. 7, 421–434. [DOI] [PubMed] [Google Scholar]
- Smitherman, M., Lee, K., Swanger, J., Kapur, R., and Clurman, B.E. (2000). Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol. Cell. Biol. 20, 5631–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castilia, C., Blobel, G., and Rout, M.P. (1999). Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144, 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa, J., and Blobel, G. (1993). A nuclear pore complex protein that contains zinc finger binding motifs, binds DNA, and faces the nucleoplasm. Cell 72, 29–38. [DOI] [PubMed] [Google Scholar]
- Vasu, S., Shah, S., Orjalo, A., Park, M., Fischer, W.H., and Forbes, D.J. (2001). Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu, S.K., and Forbes, D.J. (2001). Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13, 363–375. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Fornerod, M., Pickersgill, H., Goldberg, M., Allen, T.D., and Mattaj, I.W. (2001). The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20, 5703–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Kasper, L.H., Mantcheva, R.T., Mantchev, G.T., Springett, M.J., and van Deursen, J.M.A. (2001). Disruption of the FG nucleoporin Nup98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA 98, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Ching, Y.-P., Kok, K.H., Kung, H., and Jin, D.-Y. (2002). Post-transcriptional suppression of gene expression in Xenopus embryos by small interfering RNA. Nucleic Acids Res. 30, 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska, G., Aris, J.P., and Paddy, M.R. (1997). A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J. Cell Sci. 110, 927–944. [DOI] [PubMed] [Google Scholar]
- Zimowska, G., and Paddy, M.R. (2002). Structures and dynamics of Drosophila Tpr inconsistent with a static, filamentous structure. Exp. Cell Res. 276, 223–232. [DOI] [PubMed] [Google Scholar]