Abstract
Mitochondria undergo cycles of fusion and fission crucial for organelle homeostasis. Fission is regulated partially by recruitment of the large GTPase Dnm1p to the outer mitochondrial membrane. Using three-dimensional time-lapse fluorescence imaging of Saccharomyces cerevisiae cells, we found that Dnm1p-EGFP appears and disappears at “hot spots” along mitochondrial tubes. It forms patches that convert rapidly into different shapes regardless of whether mitochondrial fission ensues or not. Moreover, the thickness of the mitochondrial matrix displays frequent temporal fluctuations apparently unrelated to fission or to recruitment of Dnm1p-EGFP. These results suggest that mitochondrial fission requires coordination of at least two distinct processes.
INTRODUCTION
Mitochondria are branched, tubular structures surrounded by a double membrane. They continually undergo fission and fusion, both within single tubes and at branch points (Nunnari et al., 1997). In cells of the budding yeast Saccharomyces cerevisiae, the mitochondria form a highly interconnected reticulum distributed beneath the cell cortex (Hoffmann and Avers, 1973; Stevens and White, 1979; Nunnari et al., 1997). Regulation of this dynamic behavior is important for maintaining organelle structure and inheritance during growth, mating, and sporulation (Yaffe, 1999). Genetic studies have shown that several GTPases participate in mitochondrial dynamics. In particular, the large GTPase Dnm1p is important for regulated fission of the outer membrane (Otsuga et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999). Dnm1p thus provides a marker for mitochondrial fission in living cells.
Dnm1p and its higher eukaryotic homolog, Drp1 (Smirnova et al., 1998), show strong sequence and structural similarities to dynamin, another large GTPase known to regulate scission of the neck connecting budding clathrin coated pits with the donor membrane (for review, see Sever et al., 2000). In yeast and other cells, a fraction of Dnm1p/Drp1 is cytosolic, whereas the rest manifests by fluorescence microscopy as punctuate clusters on the cytosolic surface of mitochondria (Shin et al., 1997; Imoto et al., 1998; Otsuga et al., 1998; Smirnova et al., 1998). Immunoelectron microscopy shows colocalization of Dnm1p with constrictions in the organelle presumed to be sites of fission (Bleazard et al., 1999; Sesaki and Jensen, 1999). This view is strengthened by observations based on two-dimensional time-lapse fluorescence microscopy experiments in yeast, worms, or human cells expressing Dnm1p or Drp1 fused to enhanced green fluorescent protein (EGFP), showing that mitochondria divide at sites containing Dnm1p or Drp1 clusters (Labrousse et al., 1999; Mozdy et al., 2000; Smirnova et al., 2001; Shaw and Nunnari, 2002). Curiously, there are significantly more Dnm1p or Drp1 clusters than fission sites. This observation has led to the suggestion that the clusters represent sites of past and/or future scission events. Furthermore, time-lapse fluorescence microscopy of cells from Caenorhabditis elegans shows persistent spots before fission, suggesting that Drp1 recruitment to the mitochondrial membrane is not the rate-limiting step for division (Labrousse et al., 1999).
Static fluorescent images of mitochondria obtained from muscle cells in C. elegans show constrictions along the mitochondrial tubes, detected with fluorescent markers for both the matrix and the outer membrane (Labrousse et al., 1999). The number of these constrictions is significantly larger than the number of positions on a mitochondrion at which EGFP-Drp1p can be detected. The possibility that mitochondrial constriction and fission are distinct but linked processes is also suggested by the properties of certain mutations in the GTPase domain of Drp1p, which prevent mitochondrial outer membrane fission and at the same time lead to an uneven and exaggerated expansion of matrix at various portions along mitochondrial tubes. Outer membrane bridges link these expansions.
Although little is known about the mechanism of Dnm1p function, current views are strongly influenced by our understanding of dynamin's role in membrane tubulation and pinching of clathrin-coated vesicles from the donor membrane (for recent reviews, see Sever et al., 2000), by a number of genetic and cellular studies involving Dnm1p and Drp1p (for review, see Shaw and Nunnari, 2002), by the similarity of the in vitro assembly properties of Drp1 and dynamin, and finally by the close sequence and domain structure resemblance between Dnm1p/Drp1 and dynamin (Gammie et al., 1995; van der Bliek, 1999). Thus, a cyclic model for the involvement of Dnm1p/Drp1 in mitochondrial division has been proposed (for review, see Shaw and Nunnari, 2002): Dnm1p-GDP is first recruited by interaction with its partners such as Fis1p to the cytoplasmic face of the mitochondrial membrane; it then undergoes nucleotide exchange so that Dnm1p/Drp1-GTP can induce mitochondrial fission. The cycle ends with hydrolysis of GTP, disassembly of the cluster of Dnm1p/Drp1 and associated proteins from the outer surface of the mitochondria, and return of Dnm1p/Drp1 to the cytosol. It should be noted, however, that a mitochondrial tubule has an average diameter of ∼500 nm, significantly larger than the ∼10-nm diameter of the clathrin-coated vesicle neck around which dynamin binds. It is hard to imagine that Dnm1p makes a coherent collar around the tube before constriction.
Does constriction of the inner and outer membranes occur before Dnm1p/Drp1 recruitment and follow by subsequent mitochondria fission? Or are these processes driven by Dnm1p/Drp1, either through mechanochemical forces resulting from conformational changes in Dnm1p/Drp1 or through lipid remodeling after recruitment of lipid-modifying enzymes by activated Dnm1p/Drp1? We have reinvestigated the possibility that constriction and fission are distinct with improved temporal and spatial resolution by simultaneous use of time-lapse fluorescence imaging and three-dimensional (3D) image reconstruction in living yeast cells. We have monitored the appearance of mitochondria by following the distribution of a red fluorescent protein targeted to the mitochondrial matrix (mito-RFP), representing the behavior of mitochondrial matrix and its surrounding membranes (Labrousse et al., 1999). Use of mito-RFP as a marker representing the overall mitochondrial geometry is also based on the extensive evidence obtained by electron microscopy showing a tight physical association of matrix with the surrounding membranes. The behavior of Dnm1p-EGFP, a chimera of Dnm1p fused at its carboxy terminus with EGFP. Dnm1p-EGFP is fully functional, because it is known to rescue the null phenotype of dnm1Δ cells (Sesaki and Jensen, 1999; Cerveny et al., 2001). We find abundant temporal and spatial fluctuations in the thickness of the matrix, most of them unrelated to fission events. We have confirmed that Dnm1p-EGFP assembles in discrete patches along mitochondrial tubes and branches, and we have found that the patches have different shapes, such as rings wrapping around a mitochondrion or clusters located on one side of a mitochondrial tube or branch. The majority of these clusters and rings undergo constant changes in their shape and size, sometimes disappearing completely, even though the underlying mitochondrion has not engaged in a fission event. These results favor the view that mitochondrial constriction and fission require distinct sets of molecular components, and that Dnm1p functions primarily or exclusively in the latter process.
MATERIALS AND METHODS
Yeast Media, Strains, and Plasmids
Yeast S. cerevisiae strains were grown in complete medium (YPD; 1% yeast extract, 2% peptone, and 2% glucose) or in synthetic complete medium (SC; 2% glucose, 0.67% yeast nitrogen base without amino acids, supplemented with amino acids and nutrients; Sherman, 1991). The wild-type (MAT a ura3 leu2 his3 trp1) and mutant strain dnm1Δ (Mat a ura3 leu2 dnm1::his3) and the plasmids pRS414-Dnm1p-EGFP, pVT100U-ADH/PreFoATPase (mito-EGFP), and pRS426ADH/PreFoATPase (mito-RFP) (subunit 9 of the F0 ATPase from Neurospora crassa fused to EGFP or RFP, respectively) were a generous gift of J. Shaw (University of Utah, Salt Lake City, UT). Yeast transformations were performed as described (Ito et al., 1983).
Time-Lapse Three-Dimensional Microscopy
Sample Handling. Yeast cells were cultured overnight to an optical density of 0.5–1. Cells were centrifuged at 2500 rpm for 5 min and suspended at 37°C in media containing melted 0.6% low-melting agarose (FMC Bioproducts, Rockland, ME). Then 200 μl of this suspension was placed on a 25-mm coverslip inside an open perfusion chamber (20/20 Technology, Wilmington, NC) and was kept at room temperature. On solidification of the agarose, the sample was overlaid with 300 μl of SC medium (without uracil, and also without tryptophan when looking at wild-type cells), and the perfusion chamber was transferred to the temperature controller (20/20 Technology) and kept at 30°C on the microscope stage. The imaging protocols were started after a stabilization period of 10–15 min and no effects were detected on cell growth, budding, and mitochondria morphology when monitored for at least 2 h.
Data Acquisition. Images were acquired with a fully motorized wide-field epifluorescence microscope (Axiovert 200 M; Carl Zeiss, Thornwood, NY) under control of SlideBook (Intelligent Imaging Innovations, Denver, CO). The microscope was equipped with phase contrast optics, motorized filter turret, and lens holder and a 63× lens (Pan Apochromat, numerical aperture 1.4; Carl Zeiss). 3D image stacks were recorded by sequential acquisition of views recorded every 70–300 ms along the z-axis by varying the position of the lens holder. A step size of 0.15 μm was used for single time point acquisitions or 0.5 μm to acquire time-lapse series. Due to the inherent over-sampling along the z-axis, little difference was observed in the data acquired by both modes. Samples were illuminated with a 175-W Xe lamp source (Sutter Instruments, Novato, CA) optically coupled to the microscope with a liquid guide. A 1 O.D. neutral density filter (Chroma Technology, Brattleboro, VT) was used to reduce photobleaching and photodamage effects.
Fluorescence recovery after photobleaching (FRAP) experiments were performed with a wavelength-tunable near-diffraction limited collimated laser beam (Micropoint, Photonics Instruments, St. Charles, IL) generated by a nitrogen pulse laser (VSL-337ND-S; Laser Science, Franklin, MA). The wavelength of the laser beam was tuned with a coumarine-based dye to specifically photobleach EGFP but not RFP. The laser spot was positioned on the image under computer control (Slidebook; Intelligent Imaging), and bleaching was achieved with minimal intensities and with exposures of <1 s to minimize possible phototoxic effects.
Data Processing. The images were restored in three dimensions by constrained iterative deconvolution (Agard et al., 1989) with Slide-book, by using experimentally determined point-spread functions corresponding to 170-nm beads labeled with fluorescent dyes (Molecular Probes, Eugene, OR) compatible with the fluorescein isothiocyanate and Cy3 filter sets. 3D surface rendering of the restored images was obtained with Volocity (Improvision, Lexington, MA). Five to eight iterations were used to deconvolve the EGFP images, whereas 10 to 12 cycles were used for the RFP images, respectively. Fluorescence intensities and volumes were determined using 3D segmentation (Slidebook) of the data before 3D rendering with Volocity. Likewise, size measurements (Slidebook) were done in the images before 3D rendering. After photobleaching, the mobile fraction and kinetics of exchange between a given mitochondria-associated Dn1mp-EGFP patch and the soluble cytosolic pool were determined as a function of time by calculating the ratio of its fluorescent signal before and after photobleaching determined in three dimensions.
RESULTS
Temporal Fluctuations in Thickness of Mitochondrial Matrix
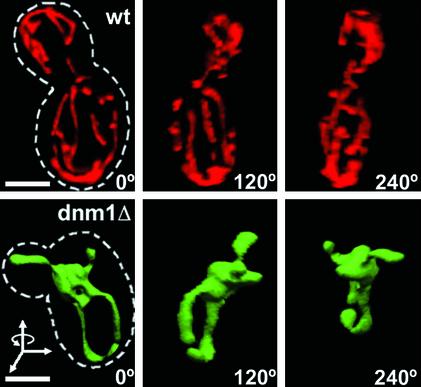
In S. cerevisiae, mitochondria are organized as an open, branched, tubular network located immediately under the cell cortex. To validate our imaging procedures, we first reproduced previous observations by using wild-type living yeast cells (wt) in which mitochondrial matrix had been labeled by expressing red fluorescent protein targeted to the mitochondrial matrix by the mitochondrial leader sequence of the F0 ATPase from N. crassa (mito-RFP) (Figure 1, top, and Movie 1 in Supplementary Materials). Mito-RFP marks only the matrix, but we assume that both inner and outer membranes follow its outline (except, of course, for cristae, which are bellow the resolutions of light microscopy). The views in Figure 1 correspond to different angular projections of the same 3D surface rendering (Volocity) of a single 3D image stack, restored by constrained iterative deconvolution (Slidebook). The stack represents 15 sequential optical sections acquired within 3 s by wide-field epifluorescence microscopy. In contrast to the extended reticulum observed in wild-type cells, haploid cells lacking Dnm1p (dnm1Δ) tagged with mito-EGFP presented the expected poorly resolved mitochondrial network corresponding to a structure mostly bundled to one side of the nucleus of the mother cell (Figure 1, bottom, and Movies 2–4 in Supplementary Materials) (Otsuga et al., 1998; Sesaki and Jensen, 1999).
Figure 1.
Three-dimensional imaging of mitochondria in living yeast cells. The mitochondrial matrix of S. cerevisae cells expressing (wt, top) or lacking (dnm1Δ, bottom) Dnm1p were labeled with mito-RFP or mito-EGFP, respectively. 3D image stacks were obtained by collecting 15 consecutive optical sections (step size 0.15 μm, 63×, 1.4 numerical aperture objective lens) followed by image restoration with eight cycles of constrained iterative deconvolution. The views correspond to 3D surface renderings of the image stacks rotated 120° around the y-axis. The cell contour (stippled line) is shown. Bar, 2 μm.
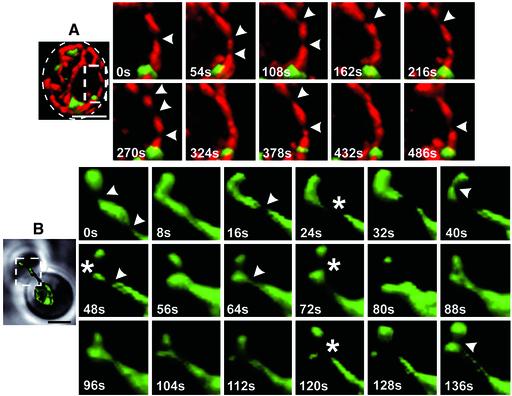
In accordance with the appearance of the elongated mitochondria in muscle cells of C. elegans, the overall thickness of the mito-RFP signal remained relatively constant along the yeast mitochondria, except for a thinning out observed in the rendered images at defined positions, referred to as “constrictions.” Time-lapse imaging coupled to 3D rendering of cells expressing wt Dnm1p showed frequent, rapid, and uncoordinated temporal fluctuations in the thickness of the mito-RFP signal, at locations mostly unrelated to sites of mitochondrial fission (Figure 2A) or of Dnm1p assembly (see below). Similar constrictions were also observed in dnm1Δ cells, notably in the smaller daughter cell generated during budding, where it was relatively easy to distinguish significant morphological changes in the shape of the inherited mitochondrion (Figure 2B, and Movie 5 in Supplementary Materials). The images also showed that before cytokinesis there were a number of transient mitochondrial matrix separation events at locations close to the neck between the mother and daughter cell; these separations were followed by local reattachment. Of note, in those cases with a large separation it would be hard to distinguish between reversible thinning or complete mitochondrial fission followed by fusion because we monitored the mitochondrial geometry with mito-RFP rather than with an outer membrane marker.
Figure 2.
Thickness of the mitochondria matrix fluctuates as a function of time. Time-lapse combined with 3D surface rendering acquired from selected areas of yeast cells (stippled rectangles) expressing mito-RFP and Dnm1p-EGFP (A) or mito-EGFP (B) in the presence or absence of Dnm1p, respectively. Transient constrictions of the mitochondria matrix (arrowheads) were observed regardless of Dnm1p expression. Example of extensive mitochondrial matrix constriction or fission occurring in the absence of Dnm1p is shown (stars). The area selected for visualization in B comprises the mitochondria compartment in the budding daughter cell and the connecting mitochondrial tubule in the mother cell. Each of the dual color image stacks for A required 24 s of acquisition, whereas the single color stacks for B required 3 s. Bar, 2 μm.
Different Shapes of Dnm1p-EGFP Assemblies
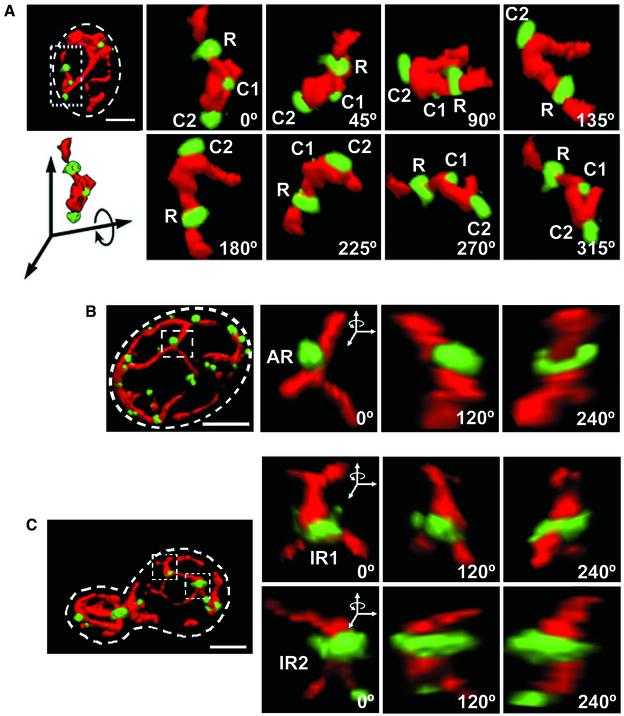
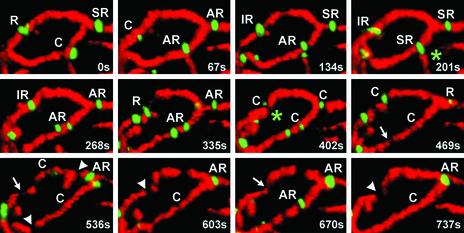
Using cells simultaneously expressing mito-RFP and a chimera of Dnm1p fused at its carboxy terminus with EGFP (Dnm1p-EGFP) we confirmed that Dnm1p-EGFP assembled at unique locations, appearing as bright patches, along the mitochondrial outer surface (Figure 3, and Movie 6 in Supplementary Materials). Most of the patches could be described as either clusters (C) or rings (R). Clusters were ragged and of variable size and could be found on the side of a tube or branch point (Figure 3A, and Movie 7 in Supplementary Materials). Rings were more homogeneous in size and wrapped around a single mitochondrial tube (Figure 3A), a mitochondrial branch (Figure 3C, top, IR1), or two adjacent tubes (Figure 3C, bottom, IR2). Approximately 50% of the rings presented an asymmetric radial distribution of the Dnm1p-EGFP signal (Figure 3B, AR, and Movie 8 in Supplementary Materials). Some rings completely wrapped around the mitochondrion, whereas others surrounded only part of it (semi-ring) (Figure 7).
Figure 3.
Dnm1p-EGFP assemblies adopt different shapes. 3D renditions of yeast cells expressing Dnm1p-EGFP and mito-RFP visualized from different viewpoints rotated along the indicated axis. Selected areas (stippled rectangles and squares) were selected to highlight Dnm1p-EGFP assemblies of different shapes position on different locations along the mitochondria. (A) Dnm1p-EGFP arranged in the form of a regular ring surrounding a mitochondria tubule (R) or as a cluster on one side of the mitochondria tubule (C1 and C2). (B) Dnm1p-EGFP arranged as an asymmetric ring (AR) with the majority of the molecules located toward one side of the ring. (C) Dnm1p-EGFP arranged as an irregular ring around a mitochondrial branch (IR1, top) or surrounding two juxtaposed mitochondrial tubules (IR2, bottom). Bar, 2 μm.
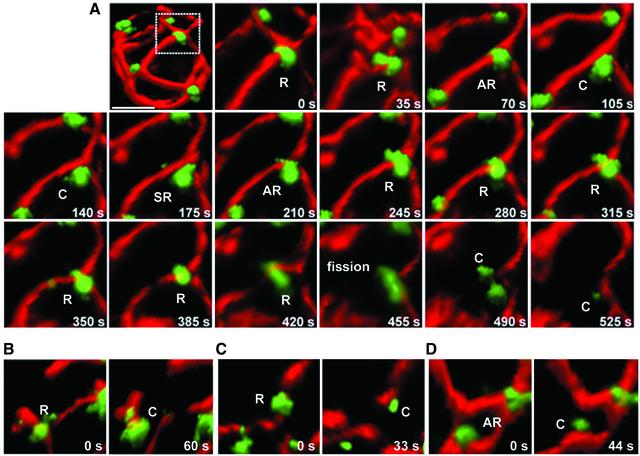
Figure 7.
Relation between mitochondrial fission and Dnm1p assemblies. (A) Time-lapse 3D renditions were acquired every 35 s from different yeast cells to follow the coupling between mitochondria fission events and the appearance of Dnm1p-EGFP assemblies adjacent to the fission event (monitored with mito-RFP). (B–D) These images illustrate several examples acquired from different cells before and after fission occurring next to Dnm1p-EGFP rings. After fission and separation of the free mitochondrial ends, Dnm1p-EGFP remained associated with one but not the other end. Bar, 2 μm.
Assembly and Disassembly of Dnm1p-EGFP on Mitochondrial “Hot Spots”
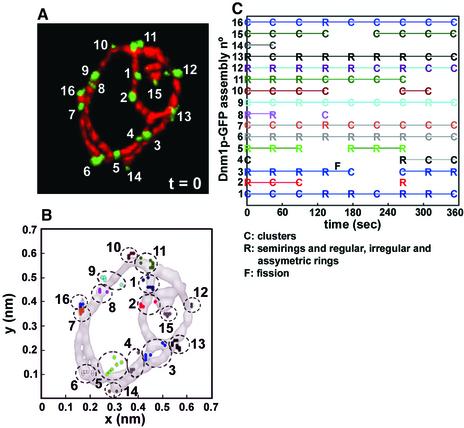
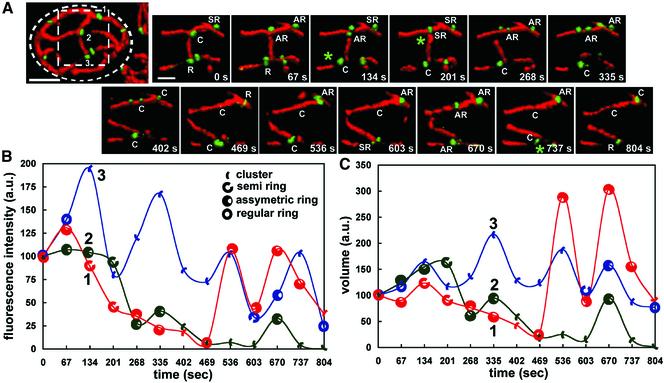
The temporal characteristics of shape, position and intensity of Dnm1p-EGFP assemblies were monitored over a period of 6 min by acquiring sets of nine sequential 3D image stacks as in the example shown for the individual cell in Figure 4A. Most assemblies remained centered at their original positions, displaying a limited lateral displacement of their centers of volume of <100 nm (presented as orthogonal projections in Figure 4B). About half of these Dnm1p-EGFP assemblies, of various shapes, disappeared and subsequently reappeared at essentially the same positions, defining what we call hot spots (Figure 4, B and C, e.g., spots 2–5, 8, 10, and 15). During the lifetime of a given Dnm1p-EGFP patch, we noticed clear changes in its geometry, without any particular order in the shape transition from clusters to rings and vice versa (Figure 5 A). In addition, there was no obvious correlation between changes in fluorescence intensity or volume of a given Dnm1p-EGFP assembly and its shape transition (Figure 5, B and C). These shape variations were not apparent when the images were visualized with more conventional two-dimensional (2D) wide-field epifluorescence microscopy or by inspection of the orthogonal 2D projections from restored 3D views (our unpublished data).
Figure 4.
Assembly and disassembly of Dnm1p-EGFP occurs on mitochondria hot spots. Consecutive nine timelapse 3D renditions from a yeast cell expressing Dnm1p-EGFP and mito-RFP were acquired every 44 s to monitor the temporal and spatial characteristics of Dnm1p-EGFP. (A) The image depicts the 3D rendition for the first time point (t = 0) and highlights the location of 16 different Dnm1p-EGFP assemblies whose presence, relative positions, and shape were monitored as a function of time. (B) Plots of the centers of mass presented as an orthogonal projection along the z-axis corresponding to the 16 Dnm1p-EGFP selected in A. The absolute position of the mass centers was determined in each time frame and is represented by a colored dot; each cluster (stippled circle) corresponds to a single Dnm1pEGFP assembly. (C) Schematic representation of the Dnm1p-EGFP assemblies high lighting variations in their shape and presence during the imaging period. Scoring of the assemblies as clusters (C) or rings (R) was done by visual inspection of the 3D renditions. For simplicity, all forms of rings were grouped together. About one-half of the Dnm1p-EGFP assemblies disappeared during the imaging period and subsequently reappeared at approximately the same position, irrespective of the original shape and of a mitochondria fission event (F).
Figure 5.
Shape transitions of Dnm1p-EGFP assemblies. Fourteen sequential 3D renditions were obtained every 67 s from a single yeast cell expressing Dnm1p-EGFP and mito-RFP, and three Dnm1p-EGFP spots (labeled 1–3) located within the broken square area were selected for analysis. (A) Images correspond to the time-lapse series of the selected area visualized from the same viewpoint. The shapes of the Dnm1p-EGFP assemblies, scored by inspection from this and other viewpoints (our unpublished data), indicated frequent changes in shape between clusters (C) and rings of variable appearance (R, AR, or SR); these changes occurred at sites unrelated to a mitochondria fission event. Mitochondria fissions (*) occurred adjacent to Dnm1p-EGFP rings (R, SR, and AR) but never clusters. (B and C) Time-dependent plots corresponding to the fluorescence intensities (proportional to the number of Dnm1p-EGFP molecules) and volumes (proportional to the spatial density of Dnm1p-EGFP) determined for the Dnm1p-EGFP assemblies 1–3; they were normalized to the values obtained at the beginning of the time series. The plots highlight the apparent lack of correlation between shape transitions, fluorescence intensity and relative volume. Bar, 2 μm.
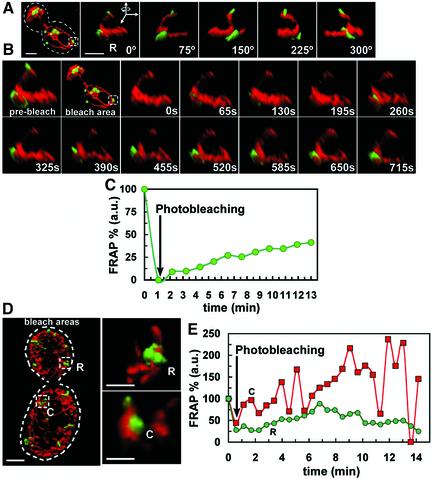
The extent of Dnm1p-EGFP exchange between the cytosolic and mitochondrial-bound pools was measured in selected rings and clusters following a FRAP protocol carried out with the aid of a diffraction-limited wavelength-tunable pulsed ablation laser. The implementation of this system allowed us to acquire a time-lapse 3D rendered series after the selective photobleaching of single Dnm1p-EGFP spots without affecting the fluorescence of Dnm1p-EGFP in the cytosol or the fluorescence of mito-RFP in the matrix immediately adjacent to the photobleached Dnm1p-EGFP assembly (Figure 6). This FRAP procedure did not seem to reduce the viability of the cells or to affect the dynamics of the Dnm1p-EGFP spots, because the oscillations in shape and intensity of the Dnm1p-EGFP assemblies were similar to those observed with nonbleached Dnm1p-EGFP spots. The examples presented in the figure show slow and steady recovery of up to 50% in the fluorescence signal of the photo-bleached Dnm1p-EGFP rings (Figure 6, B, C, and E, top). They also show that the recovery is uneven and occurs at random positions around the rings, reflecting the continuous exchange between cytosolic and assembled Dnm1p along different locations in its circumference, suggesting that this type of assembly does not correspond to a molecular coherent collar. The recovery of clusters was faster (Figure 6, D and E), often associated with broad oscillations in fluorescence intensity similar to those observed with non-bleached Dnm1p-EGFP spots (Figure 5).
Figure 6.
Exchange between the cytosolic and mitochondria-bound pools of Dnm1p-EGFP. FRAP experiments were carried out on single Dnm1p-EGFP assemblies imaged in yeast cells expressing Dnm1p-EGFP and mito-RFP. Photobleaching restricted to the EGFP but not the RFP signal was achieved in <1 s of illumination of the selected Dnm1p-EGFP assemblies by using a diffraction-limited laser whose wavelength was tuned using a coumarin-based fluorescent dye optimized for GFP photobleaching. 3D renditions were obtained at the indicated times. (A) General and close-up views of a Dnm1p-EGFP ring (R) selected for photobleaching visualized immediately before FRAP (see Movie 9 in Supplementary Materials). (B) Recovery of the fluorescent signal of the Dnm1p-EGFP assembly imaged in A after photobleaching. (C) Plot as a function of time corresponding to the recovery of fluorescent signal of the Dnm1p-EGFP assembly depicted in B normalized to its fluorescence intensity before photobleaching; the recovery reflects the incomplete exchange between the cytosolic and mitochondria-bound pools of Dnm1p-EGFP when organized as a ring. (D and E) Comparison between the relative recoveries of fluorescent signals determined for a Dnm1p-EGFP ring (R) and a cluster (C) after photobleaching. Fluorescence recovery for a ring was slower and less complete than for a cluster. Bar, 2 μm.
Dnm1p and Mitochondrial Fission
We obtained 20 independent data sets of two-color 3D image stacks, sequentially acquired every 35 s over periods ranging between 5 and 10 min, from cells expressing Dnm1p-EGFP. Fission occurred at sites containing Dnm1p-EGFP, often located at a branch point with at least one branch moving away after scission (Figure 7A). The spot immediately before fission was always ring shaped (Figure 7, A–D; see complete time-lapses in Supplementary Materials), and after fission it remained attached to one free mitochondrial end, but not to both; local fusion was rarely observed. After a period that could vary significantly (in the range of 30–90 s), the fluorescence intensity of the Dnm1p-EGFP ring decreased significantly, presumably reflecting its depolymerization and cycling to the cytosolic pool. Some apparent mitochondrial fission events occurred at locations on tubes that were devoid of any fluorescence signal of Dnm1p-EGFP (Figure 8, arrowheads). This type of mitochondrial separation was generally followed by immediate local fusion. Because the sensitivity limit for fluorescence detection at a single location is ∼20–30 Dnm1p-EFGP molecules, it is still possible that a very small number of Dnm1-EGFP were recruited but not imaged. We note, however, that similar mitochondrial constrictions and fissions were observed even in cells lacking Dnm1p (Figure 2B). Hence, we believe that these events reflect a Dnm1p-independent reversible thinning and separation of the matrix, still linked by an outer membrane neck, rather than complete mitochondrial scission.
Figure 8.
Mitochondrial constriction and fission in the absence of Dnm1p-EGFP assemblies. 3D image stacks of a wt cell coexpressing Dnm1p-EGFP and mito-RFP were captured every 67 s and surface rendered as mentioned previously. Examination of an area of this cell reveals that mitochondria matrix constriction (arrows) and fissions (arrowheads) do not necessarily require the presence of Dnm1p. During the time lapse it is also possible to observe Dnm1p-dependent fission events (see green stars). Rapid oscillations in the shape of several Dnm1p assemblies and formation of new ones occurred during the time lapse, without necessarily concluding in mitochondria fission.
DISCUSSION
Using a combination of three-dimensional fluorescence imaging with time-lapse acquisition methods, we have followed the behavior of mito-RFP and Dnm1p-EGFP in living yeast cells. We report three basic findings. 1) The thickness of the mitochondrial matrix marked with mito-RFP is uneven, with acute constrictions along its length. These constrictions appear and disappear at different sites, with no obvious spatial or temporal correlation. 2) Dnm1p-EGFP assembles in patches at specialized sites (hot spots) on the outer surface of the mitochondrion. We describe these patches as either clusters or rings of different geometries; their presence at a given position is transient, and their shapes can change from rings to clusters or back. Formation of these patches is not dependent on constriction of the inner membrane/matrix and does not, in general, lead to fission. 3) When mitochondria fission does occur, it is at the position of a Dnm1p ring.
Most mitochondrial fission models assume a direct coupling between the constriction of the matrix, inner and outer membrane with the fission of the inner and outer membranes (Shaw and Nunnari, 2002). This is due in part to the fact that the majority of previous studies used visualization imaging methods that did not allow the temporal dissection of morphological events separating constriction from fission. Variations in thickness along mitochondrial tubules have been noted before in images acquired by light or electron microscopy (Nunnari et al., 1997; Labrousse et al., 1999; Cerveny et al., 2001; Smirnova et al., 2001), but because of the static nature of the images, the significance of the constrictions could not be established.
By using 3D-time lapse imaging as depicted in Figure 2 we can now see clear examples of temporal fluctuations in matrix thickness, largely uncoupled from actual fission. It is striking that the majority of these fluctuations also occur at sites unrelated to the recruitment and assembly of Dnm1p on the mitochondrial outer surface. The proteins responsible for constriction are unknown. They are likely to be different than those involved in outer membrane scission, as expression in C. elegans of dominant mutants of Drp1 (including some that cannot hydrolyze GTP) results in significant accumulations of expansions and thinning of the matrix, without loss of outer membrane continuity (Labrousse et al., 1999). Consistent with the apparent absence of strict correlation between constriction and scission in cells expressing Dnm1p, we also noticed fluctuations in the thickness of the mitochondrial matrix in yeast cells lacking Dnm1p (Figure 2). Thus, in yeast cells, there seem to be at least two separable machineries, one responsible for mitochondrial constriction and another for mitochondrial scission.
The molecular basis for some or all of the non-Dnm1p constrictions remains to be determined. A possibility is that because mitochondria movement and morphology is dependent of myosin motors associated to actin cables (Simon et al., 1995; Suelmann and Fischer, 2000), tension along the mitochondrial tubules might stretch it out, resulting in local squeezing at points of least resistance followed by Dnm1p recruitment or activation.
By analogy with how we believe dynamin works during the pinching of the neck joining a budding vesicle to the membrane, it has been proposed that Dnm1p functions in a cycle of assembly, constriction, GTP hydrolysis, and disassembly (Shaw and Nunnari, 2002). In this model, cytosolic Dnm1p is recruited into a large spiral that wraps around the outer membrane of the mitochondrion, leading to constriction of the assembly and/or recruitment of effectors required to promote mitochondrial constriction and fission, followed by rapid disassembly and return to the cytosolic pool. This model predicts the sequential recruitment of cytosolic Dnm1p to fission sites. We find, however, that the Dnm1p assembles along the mitochondrial surface, only seldom in association with fission events, while adopting different, interconverting shapes. Fission occurs mostly at sites adjacent to ring-shaped Dnm1p assemblies that remain attached to one free end of the segmented mitochondrion and that often do not disappear.
Appearance and disappearance of the Drp1, the C. elegans ortholog of Dnm1p, has been examined by 2D wide-field fluorescence time-lapse imaging in muscle cells expressing EGFP-Drp1 (Labrousse et al., 1999). In that work, it was noticed that as with Dnm1p, Drp1 spots appeared and disappeared along the mitochondrial reticulum. Most of the appearing (assembling) spots were, however, located on tubes, whereas the disappearing (disassembling) ones mapped to free ends or tips. These observations led to the conclusion that appearance and disappearance represented the initial and final stages of the scission process regulated by Drp1. Our data indicate that in yeast cells constriction of the matrix and Dnm1p assembly are not tightly coupled and that only when the two processes coincide does ring formation lead to scission. Most of the assembly events we record are abortive, presumably because they do not coincide with suitable constrictions, and, as shown by the apparent fission and refusion of the matrix at positions where Dnm1p is absent, constriction can likewise occur without Dnm1p assembly.
Based primarily on the results of genetic studies in yeast, proteins such as the outer mitochondrial membrane protein Fis1p and the cytosolic protein Mdv1p are known not only to interact with Dnm1p but also to be required for fission (Tieu and Nunnari, 2000; Tieu et al., 2002). Exactly how they work is still unclear. It is thought that Fis1p acts as a Dnm1p receptor, somehow participating in the recruitment and regulation of the assembly of cytosolic Dnm1p; Mdv1p is thought to act at a later stage after Dnm1p assembly. Fis1p might also have a role in communicating the state of matrix and inner membrane constriction with the assembly state of Dnm1p. In this context, Mgm1p, an intermembrane space protein required for inner membrane remodeling events, also participates in the coupling of both inner and outer membrane fission, where Mgm1p functions in coordination with Dnm1p-dependent outer membrane fission to regulate inner membrane division (Wong et al., 2000).
Given uneven photorecovery of bleached Dnm1p-EGFP ring and the large diameter (300–700 nm) of a nonconstricted mitochondrial tube (much greater than the ∼10-nm diameter of a vesicle neck or of the rings generated by self assembled dynamin or Dnm1p/Drp1), it seems unlikely that Dnm1p and its homologs can assemble as molecular rings or spirals that wrap around an intact, unconstricted mitochondrion. We suggest that when constriction of the mitochondria reaches a diameter consistent with assembly of a complete molecular collar of Dnm1p/Drp1, the dynamin-like protein can then induce scission (Figure 9). Scission might result from mechanochemical forces produced by conformational changes dependent on the hydrolysis of GTP, or more likely from the recruitment of other proteins, yet to be determined that change the physicochemical properties of the outer membrane. It is clear, at least in yeast cells that Dnm1p cycles through different stages of spatial organization on the mitochondrion surface, without a strict correlation with scission. This behavior may represent a mechanism for sampling the physical state of the mitochondrial tube, so that when constriction occurs at a Dnm1p hot spot, scission can then ensue.
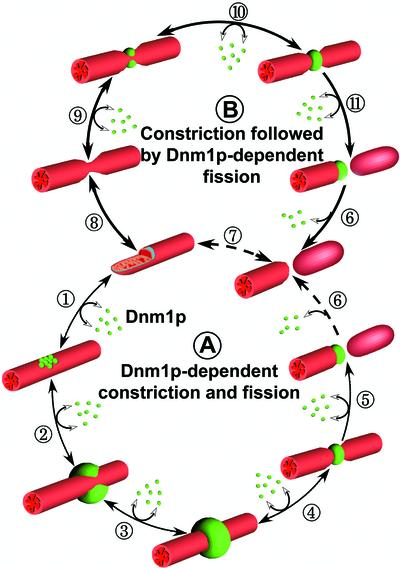
Figure 9.
Schematic representation for two models of mitochondrial fission. (A) Cytosolic Dnm1p (green spheres) is actively recruited to specific sites (hot spots) on the surface of the outer mitochondrial membrane where it first organizes as clusters (1) and continues growing to become a semi-ring (SR, 2) to end as a complete ring (3). These steps are reversible. Eventually, the Dnm1p ring induces deformation of the outer membrane coupled with inner membrane constriction (4) followed by scission (5). Dnm1p remains attached to one mitochondrial free end although full disassembly back to the cytosol is not required (6). (B) Inner and outer mitochondrial membranes can fluctuate in diameter irrespective of Dnm1p presence (7). If recruitment of cytosolic Dnm1p coincides with a constricted region that is followed by the rapid assembly of Dnm1p to form a ring (8) then scission (9) and dissociation occurs (10). Although with significantly less efficiency, mitochondria scission might also occur without Dnm1p (11).
Supplementary Material
Acknowledgments
We thank Drs. J. Shaw and J. Nunnari for generously supplying yeast strains and plasmids, and Stephen C. Harrison for reviewing the manuscript. We thank the Perkin Foundation for making available funds to help in purchasing the imaging equipment. This study was supported by National Institutes of Health grant GM-036548. A.L.M. was a Dorit Fellow.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-10-0657. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-10-0657.
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
References
- Agard, D.A., Hiaroka, Y., Shaw, P., and Sedat, J.W. (1989). Fluorescence microscopy in three dimensions. Methods Cell Biol. 30, 353–377. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell 12, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A.E., Kurihara, L.J., Vallee, R.B., and Rose, M.D. (1995). DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 130, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, H.P., and Avers, C.J. (1973). Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science 181, 749–751. [DOI] [PubMed] [Google Scholar]
- Imoto, M., Tachibana, I., and Urrutia, R. (1998). Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase Vps1p. J. Cell Sci. 111, 1341–1349. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, A.M., Zappaterra, M.D., Rube, D.A., and van der Bliek, A.M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari, J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B.R., Brisch, E., Thatcher, J.W., Hermann, G.J., Bleazard, W., and Shaw, J.M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000). Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic 1, 385–392. [DOI] [PubMed] [Google Scholar]
- Shaw, J.M., and Nunnari, J. (2002). Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12, 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Shin, H.W., Shinotsuka, C., Torii, S., Murakami, K., and Nakayama, K. (1997). Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J. Biochem. 122, 525–530. [DOI] [PubMed] [Google Scholar]
- Simon, V.R., Swayne, T.C., and Pon, L.A. (1995). Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J. Cell Biol. 130, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Griparic, L., Shurland, D.L., and van der Bliek, A.M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Shurland, D.L., Ryazantsev, S.N., and van der Bliek, A.M. (1998). A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, B.J., and White, J.G. (1979). Computer reconstruction of mitochondria from yeast. Methods Enzymol. 56, 718–728. [DOI] [PubMed] [Google Scholar]
- Suelmann, R., and Fischer, R. (2000). Mitochondrial movement and morphology depend on an intact actin cytoskeleton in Aspergillus nidulans. Cell Motil. Cytoskeleton 45, 42–50. [DOI] [PubMed] [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek, A.M. (1999). Functional diversity in the dynamin family. Trends Cell Biol. 9, 96–102. [DOI] [PubMed] [Google Scholar]
- Wong, E.D., Wagner, J.A., Gorsich, S.W., McCaffery, J.M., Shaw, J.M., and Nunnari, J. (2000). The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P. (1999). The machinery of mitochondrial inheritance and behavior. Science 283, 1493–1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.