Abstract
The bullous pemphigoid antigen 1 (BP230) and desmoplakin (DP) are members of the plakin protein family of cytolinkers. Despite their homology, their COOH termini selectively bind distinct intermediate filaments (IFs). We studied sequences within their COOH termini required for their interaction with the epidermal keratins K5/K14, the simple epithelial keratins K8/K18, and type III IF vimentin by yeast three-hybrid, cell transfection, and overlay assays. The results indicate that BP230 interacts with K5/K14 but not with K8/K18 or vimentin via a region encompassing both the B and C subdomains and the COOH extremity, including a COOH-terminal eight-amino-acid stretch. In contrast, the C subdomain with the COOH-terminal extremity of DP interacts with K5/K14 and K8/K18, and its linker region is able to associate with K8/K18 and vimentin. Furthermore, the potential of DP to interact with IF proteins in yeast seems to be regulated by phosphorylation of Ser 2849 within its COOH terminus. Strikingly, BP230 and DP interacted with cytokeratins only when both type I and type II keratins were present. The head and tail domains of K5/K14 keratins were dispensable for their interaction with BP230 or DP. On the basis of our findings, we postulate that (1) the binding specificity of plakins for various IF proteins depends on their linker region between the highly homologous B and C subdomains and their COOH extremity and (2) the association of DP and BP230 with both epidermal and simple keratins is critically affected by the tertiary structure induced by heterodimerization and involves recognition sites located primarily in the rod domain of these keratins.
INTRODUCTION
The plakins are a family of proteins involved in the organization of the cytoskeleton. They act as cytolinkers and/or scaffolding proteins, connecting the intermediate filaments (IFs) to other cytoskeletal networks and/or distinct sites at the plasma membrane (Ruhrberg and Watt, 1997; Fuchs and Yang, 1999; Leung et al., 2001b, 2002). This protein family includes the bullous pemphigoid antigen 1 (BPAG1, also called BP230), desmoplakin (DP), plectin (PL), envoplakin, and periplakin. The plakin proteins are predicted to form dimers with a central coiled-coil domain flanked by globular domains (Green et al., 1992; Ruhrberg and Watt, 1997).
The NH2-terminal domain of plakins determines their localization at specialized sites of the membrane or their association with the microfilament system. For instance, DP, with its unique NH2-terminal sequence, is associated with adhesion complexes called desmosomes (Stappenbeck et al., 1993; Kowalczyk et al., 1997; Smith and Fuchs, 1998), whereas the epithelial isoform of BP230 is a specific constituent of junctional anchoring structures called hemidesmosomes (Borradori and Sonnenberg, 1999; Hopkinson and Jones, 2000). Finally, PL, the most versatile cytolinker, is found in desmosomes, hemidesmosomes, and focal contacts (Steinbock and Wiche, 1999).
Plakins bind to the IF cytoskeleton by their COOH terminus (Ruhrberg and Watt, 1997). Specifically, transfection studies have demonstrated that the COOH-terminal domain of BP230 is coaligned with keratin IFs (Yang et al., 1996; Leung et al., 1999). In analogy, the DP tail is also codistributed with and can bind to various keratins and vimentin in vitro (Stappenbeck et al., 1993; Kouklis et al., 1994; Meng et al., 1997). The COOH terminus of PL has been shown to bind to keratins, desmin, neurofilaments, glial fibrillary acidic protein, and vimentin in vitro (Foisner et al., 1988; Wiche et al., 1993; Nikolic et al., 1996; Steinbock et al., 2000). Finally, gene-targeted elimination of BP230, DP, or PL in mice has demonstrated the critical role of these proteins for the integrity of the architecture of the cytoskeleton in stratified epithelia and striated muscles (Guo et al., 1995; Andrä et al., 1997; Gallicano et al., 1998, 2001).
IFs consist of filaments 10-nm thick found ubiquitously in multicellular eukaryotes. All IFs have the intrinsic ability to self-assemble into filaments. Keratins are the main group of IF proteins (Fuchs and Weber, 1994; Parry and Steinert, 1999). The epithelial type I keratins form heterodimeric proteins with their type II partners and together assemble into IFs. Cytokeratins are expressed differentially in most epithelial cells, depending on development and differentiation stages. K5/K14 is the major pair in stratified squamous epithelia, whereas the K8/K18 pair is expressed in simple epithelia. Unlike keratins, type III IF proteins, such as vimentin, are homopolymer-forming. Nevertheless, they also appear to form heteropolymers with IF proteins of the same or other classes, such as nestin (Steinert et al., 1999). The ability to form such “mixed” IF networks probably enables vimentin to exert additional cell type–specific functions (Herrmann and Aebi, 2000).
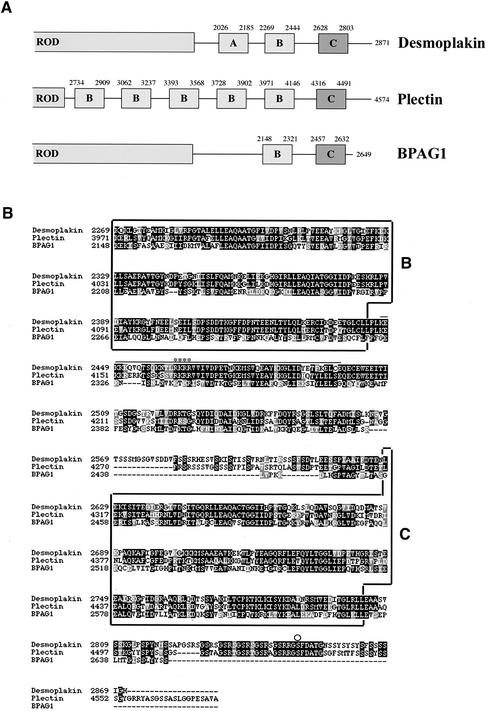
Thus far, attempts to identify the binding site(s) for IFs on DP, PL, and BP230 have not delivered a unifying picture, despite the presence in their COOH extremity of homologous domains comprising a series of 38-residue repeats (Wiche et al., 1991; Green et al., 1992). These repetitive elements are organized into subdomains, denoted A, B, and C in DP (Figure 1A). PL has five B subdomains, whereas BP230 and DP have only one (Green et al., 1992). Despite the complexity of the COOH extremity of PL, its binding to various IFs appears to rely primarily on a small stretch of basic residues located in the linker between the fifth B subdomain (B5) and the C subdomain of the protein (Figure 1B) (Nikolic et al., 1996). In contrast, the interaction of DP to IFs appears to be complex. Although a region of DP encompassing the rod domain, the A and B subdomains, and the downstream basic residues homologous to the B5 subdomain of PL seemed to be sufficient for the coalignment of DP with vimentin, the C subdomain was indispensable but not sufficient for its codistribution with K5/K14 and K8/K18 IFs (Stappenbeck et al., 1993). Moreover, DP interacts with the head domain of the single epidermal keratins K1 and K5 but only with heterodimeric K8/K18 independently of the head (Kouklis et al., 1994; Meng et al., 1997). Notably, recent crystallographic studies have shown that the B and C subdomains of DP have globular structure and exhibit a conserved basic groove, the features of which seem to be suitable for an interaction with the rod of vimentin (Choi et al., 2002).
Figure 1.
(A) Schematic of the COOH-terminal half of DP, PL, and BP230 showing the predicted subdomains within their COOH-terminal tail. (B) Sequence alignment of the COOH termini of human DP I, epithelial PL, and BP230 (GenBank accession numbers P15924, G02520, and Q03001, respectively). Alignment was performed with the CLUSTAL-W program. Black boxes indicate identical amino acids; gray boxes denote amino acid similarity. The bar indicates the stretch of amino acid residues of PL involved in binding to vimentin (Nikolic et al., 1996). The asterisks denote residues crucial for the interaction between PL and vimentin. The circle denotes Ser 2849 in DP. The subdomains B and C are de-lineated by boxes.
Finally, the ability of the BP230 tail to bind to vimentin and various IF proteins in the nervous system, in which neuronal isoforms of BP230 are expressed, is debated (Yang et al., 1996; Leung et al., 1999). New evidence has uncovered the existence of at least four structurally distinct isoforms encoded by the BPAG1/dystonin gene, BPAG1-e/BP230, BPAG1-n, BPAG1-a, and BPAG1-b, of which the latter harbors a region similar to the A subdomain of DP, but it remains to be established whether it can interact with IFs (Brown et al., 1995; Yang et al., 1996; Dowling et al., 1997; Leung et al., 2001a).
The identification of the sequences conferring binding specificities is the key for interpreting the interactions of IFs with the cell membrane and might increase our understanding of plakin- and IF-related human diseases. Hence, in this study, we investigated the ability of the tail of BP230 and DP to bind to the specific IF keratins K5/K14, K8/K18, and vimentin and characterized the sequences required and sufficient for their interaction with these IFs by using the yeast-three-hybrid system, cell transfection studies, and biochemical assays.
MATERIALS AND METHODS
Cell Culture, Transfection, and Immunofluorescence Microscopy Studies
The African monkey kidney cell line COS-7 and rat kangaroo PtK2 cells were cultured in DMEM (Invitrogen, Basel, Switzerland) supplemented with 10% bovine FCS, 100 U/ml penicillin, and 100 U/ml streptomycin. Cells from an immortalized keratinocyte cell line, derived from a patient with junctional epidermolysis bullosa that stably expresses a full-length integrin β4 subunit transgene cDNA (PA-JEB/β4 keratinocytes) (Sterk et al., 2000) were cultured in keratinocyte-SFM medium or in HAMF12:DMEM (1:3) (Invitrogen). The cells were grown at 37°C in a humidified, 5% CO2 atmosphere.
For transfection, cells were plated at 40–60% confluence on glass coverslips in six-well tissue culture plates. The PA-JEB/β4 keratinocytes and PtK2 cells were transiently transfected with 1.5 and 4 μg, respectively, of plasmid DNA using 2 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's procedure. COS-7 cells were transfected using the DEAE-dextran method. Two days after transfection, cells were processed for confocal microscopy as previously described (Favre et al., 2001) and viewed under a Zeiss LSM410 confocal inverted laser scanning microscope (Zeiss, Oberkochen, Germany). The following immunoreagents were used: rabbit polyclonal anti–green fluorescent protein (GFP) (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal antibody (mAb) anti-K14 (Sigma, St. Louis, MO), mouse mAb anti-K8 Cam5.2 (Becton Dickinson, Mountain View, CA), mouse mAb antivimentin (Immunotech, Marseille, France), Alexa-488 conjugated goat anti-rabbit IgG (Molecular Probes Inc., Eugene, OR), and Texas Red-labeled sheep anti-mouse IgG (Amersham Biosciences, Arlington Heights, IL).
Western Blot Analysis
The following antibodies were used: the rabbit polyclonal anti-GFP (Santa-Cruz Biotechnology), an affinity-purified rabbit polyclonal NW 6 directed against the carboxyl-terminus of DP (Angst et al., 1990), the human mAb 5E that recognizes the carboxyl-terminus of BP230 (Hashimoto et al., 1993), the mouse mAb and rabbit polyclonal antibody anti–Gal4-AD or –Gal4-BD (Santa Cruz), peroxidase-coupled anti-rabbit IgG (Bio-Rad, Hercules, CA), or anti–human IgG (Jackson ImmunoResearch, West Grove, PA).
Lysates of transfected COS-7 cells were prepared in SDS sample buffer and heated for 5 min at 95°C. Protein extracts were analyzed by SDS-PAGE followed by Western blotting as previously described (Favre et al., 2001).
Immunoblot analysis of transfected cell extracts showed that the expression levels of the various recombinant proteins of BP230 and DP were comparable. The apparent molecular weight was uniformly lower than that predicted on the basis of the cDNA as previously described (Skaria et al., 2000) (not shown).
Yeast Two- and Three-Hybrid Assays
Generation of pACT2-URA The cDNA sequence encoding LEU2 in the yeast vector pACT2 (Clontech, Palo Alto, CA) was replaced by a sequence encoding the gene URA3 as described below. First, a pACT2 vector deleted of its LEU2 sequence was generated by PCR with Pfu DNA polymerase (Promega, Madison, WI) using a 5′ primer that contained the ClaI restriction site (italicized) and nucleotides corresponding to sequence 3572–3598 of pACT2 (GCTTAAATCGATTCTCTTTTTTTATGATATTTGTAC) and a 3′ primer that also contained the ClaI restriction site and nucleotides corresponding to sequence 2443–2466 of pACT2 (GCTAATCGATGACATTAGAATGGTATATCCTTGA). Second, the URA3 fragment was generated by PCR using pGBDUC (James et al., 1996) as template, a 5′ primer containing the ClaI restriction site, and the initiation starting codon of the URA3 sequence (TCGAATCGATAATGTCGAAAGCTACATATAAG), and a 3′ primer that contained the ClaI restriction site and the stop codon (bold) of the URA3 sequence (CACTATCGATTTAGTTTTGCTGGCCGCATC). These two fragments were digested with ClaI and ligated. The resulting vector was sequenced and named pACT2-URA.
Yeast Strain, Transformation, and Interaction Analysis The Saccharomyces cerevisiae strain PJ69–4A (a gift from Dr. P. James, Department of Biomolecular Chemistry, University of Wisconsin, Madison, WI), which contains the genetic markers trp1–901, leu2–3112, ura3–52, his3–200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, and met2::GAL7-lacZ (James et al., 1996), was used for all assays. It contains two regulated and selectable reporter genes, HIS3 and ADE2, each driven by a different promoter under the control of GAL4 transcriptional factor. Cells were cotransformed with two or three defined constructs in pAS2–1, pACT2 (Clontech), or pACT2-URA according to the manufacturer's procedure. Double and triple transformants were selected on agar synthetic complete (SC) medium lacking leucine and tryptophan or leucine, tryptophan, and uracil, respectively (SC-LW or SC-LWUra). For each transformation, eight colonies were arrayed in 96-well microtiter plates and then transferred onto agar SC-LW(Ura) (positive control), SC-LW(Ura) without adenine, and SC-LW(Ura) without histidine and supplemented with 2 mM 3-amino 1,2,3-triazole. Growth was determined after 5 d of incubation at 30°C. Transactivation controls were systematically performed for each construct with the opposite vector without insert.
To ascertain that the transformed yeast expressed the various recombinant proteins, cell extracts were prepared by the trichloroacetic acid method (Clontech Protocol Handbook) and subjected to immunoblotting (see above) with antibodies directed to the GAL4-AD or GAL4-BD. The apparent molecular weight of the various recombinant proteins was uniformly lower than that predicted on the basis of the cDNA, whereas their expression levels was comparable (Skaria et al., 2000) (not shown).
Immunoprecipitation and Dephosphorylation Assay
Yeast protein extracts prepared as described above were diluted 1/20 in buffer A (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5% vol/vol Triton X-100, 2 mM EDTA) and used for immunoprecipitation using 150 μl of 10% wt/vol protein A Sepharose 4B (Amersham) and 3 μg of rabbit anti-Gal 4 BD. Immobilized immunocomplexes were then resuspended in calf intestine alkaline phosphatase (CIAP) buffer (Promega) and dephosphorylated by 1 U of CIAP for 30 min at 30°C. Immunocomplexes were then washed twice with ice-cold buffer A and solubilized in SDS sample buffer previously analyzed by SDS-PAGE and Western blot.
cDNA Constructs
The various plakin deletion mutants were cloned into the yeast Gal4 DNA-BD vector pAS2–1 (Clontech) and into the eukaryotic expression vector pEGFP-C3 (Clontech) and pcDNA3-myc. IF proteins were cloned into the yeast GAL4-AD vector pACT2 or pACT2-URA or the prokaryotic expression vector pET15b (Novagen, Madison, WI). All cDNA constructs were generated using primers that added appropriate start or stop codons along with restriction sites allowing subcloning in a PCR reaction with proofreading Pfu polymerase (Promega). The keratin constructs used were as follows: K8 consists of residues 1–488, K18 consists of residues 1–430, K5 consists of residues 1–590, K5 headless (HL) encompasses residues 169–590, K5 tailless (TL) encompasses residues 1–477, K14 consists of residues 1–472, K14 HL encompasses residues 115–472, K14 TL encompasses residues 1–421. The chimeric constructs generated were as follows: (1) BP(B)–DP(L) consists of residues 2077–2321 of BP230 in fusion with residues 2445–2627 of DP followed by residues 2457–2462 of BP230; (2) BP(BC)–DP(L) encompasses residues 2077–2321 of BP230 fused to residues 2445–2627 of DP followed by residues 2457–2649 of BP230; and (3) BP(BC)–DP(Ct) consists of residues 2077–2649 of BP230 fused to residues 2821–2871 of DP with or without the S2849G mutation. The correctness of all constructs was verified by sequence analysis. The strategy used for cloning in pET15b vector leads to the replacement of the serine at position 2 by alanine in K8 and K18.
Expression of Keratins in Escherichia coli and Purification
The E. coli strain BL21 (DE3) (F-, ompT, gal [dcm], [lon], hsdSB, DE3 λ prophage [T7 RNA polymerase]) (Novagen) was transformed with recombinant IF expression plasmids, and the colonies obtained were used to inoculate Luria-Bertani medium containing 100 μg/ml ampicillin. Overnight cultures were then diluted 1:20 in fresh medium, grown to an OD600 nm of 0.7 at 30°C, and induced by the addition of isopropyl β-d-thiogalactopyranoside for 16 h. Bacteria were harvested by centrifugation at 4000 × g and frozen at -20°C.
Purification of keratins from total extracts of bacteria was achieved in two steps. First, a keratin-rich fraction was prepared as follows. Frozen bacteria were lysed by sonification in low-salt buffer (50 mM Tris-HCl, pH 6.7, 10 mM EDTA, 50 mM NaCl, 1% vol/vol Triton X-100) until complete disruption of the pellet. After centrifugation at 3000 × g for 20 min, the pellet was washed 5 times with the same buffer using cycles of resuspension and centrifugation as above. The pellet was then washed three times in high-salt buffer (50 mM Tris-HCl, pH 7.2, 2 mM EDTA, 1 M NaCl, 0.5 M KCl, 0.5% vol/vol Triton X-100) as described above. The final pellet was solubilized in urea buffer (50 mM Tris-HCl, pH 8, 8 M urea, 2 mM β-mercaptoethanol), and the proteins were extracted for 16 h at 4°C. After an additional centrifugation at 12,000 × g, the supernatant was collected and further purified using anion exchange chromatography. Keratin fractions were passaged through a 1-ml mono Q column. Keratins were eluted with a 50-ml gradient of 0–200 mM guanidine-HCl. Fractions (1 ml) were collected and analyzed by SDS-PAGE and Coomassie blue staining. For K5 purification, the pH of the urea buffer and of the keratin fraction was adjusted to 10 to ensure a strong binding of the protein to the column. Peak fractions that contained keratins at 80–90% purity were pooled and dialyzed against urea buffer to remove guanidine-HCl. A second round of anion exchange chromatography was performed as described above, and peak fractions containing keratins at nearly 100% purity were pooled, dialyzed against urea buffer, and concentrated to 2 mg/ml using Ultrafree 4–Biomax 10 K filters (Millipore, Bedford, MA), then stored at -20°C. Recombinant human K14 was kindly provided by Dr. H. Herrmann (DKFZ, Heidelberg). Vimentin was from Progen (Heidelberg, Germany).
Metabolic Labeling of c-myc–tagged Proteins
[35S]methionine/cysteine–labeled recombinant forms of BP230 and DP were generated by coupled in vitro transcription/translation of pcDNA3-myc constructs using the Quick TNT coupled reticulocyte lysate system (Promega). Nonincorporated amino acids were removed from the in vitro translation mixture (50 μl) using Ultrafree 0.5–Biomax 5 K (Millipore) filters. The translation mixture was diluted into 4 ml of binding buffer (20 mM HEPES, 10 mM PIPES, pH 7.2, 0.2 mM CaCl2, 2 mM MgCl2, 50 mM KCl) supplemented with 0.1% (wt/vol) of heat-inactivated BSA and 2 mM β-mercaptoethanol.
Overlay Assays
Purified IF proteins were polymerized into filaments by stepwise dialysis as described previously (Coulombe and Fuchs, 1990). The quality of the filaments was verified by electron microscopy after negative staining with uranyl acetate. The polymerization mixture was spotted onto a nitrocellulose sheet using a Dot-Blot apparatus (Bio-Rad). Immobilized monomers of keratins were obtained by spotting purified polypeptides solubilized in urea buffer onto nitrocellulose membranes. Membranes were subsequently washed three times in binding buffer and incubated overnight at 4°C in blocking buffer (binding buffer supplemented with 2% wt/vol of heat-inactivated BSA). Nitrocellulose strips were then incubated overnight at 4°C with [35S]methionine/cysteine–labeled proteins prepared as described above. After five washes with binding buffer, nitrocellulose strips were air-dried and bound proteins were visualized by autoradiography.
RESULTS
Development of the Yeast Three-Hybrid System
Because keratin IFs are obligate heteropolymers (Fuchs, 1994; Parry and Steinert, 1999), it was essential to study the interaction of plakins with heterodimers by using a developed yeast three-hybrid system (Zhang, 2000) that allowed the concomitant expression of three proteins, one plakin and two complementary keratins. Previous studies have demonstrated that IF proteins are capable of strong interaction in yeast (Leung and Liem, 1996; Meng et al., 1996; Schnabel et al., 1998). Therefore, we replaced the LEU2 selective marker in the plasmid pACT2 by URA3 to generate pACT2-URA and to have three plasmids available for interaction assays in yeast: one to express GAL4-BD fusion protein (pAS2–1) and two for the expression of GAL4-AD fusion protein (pACT2 or pACT2-URA), each encoding a different selection marker, TRP1, LEU2, or URA3, respectively. By introducing two complementary type I and type II keratins fused to GAL4-AD, one in pACT2 and the other in pACT2-URA, that are expected to dimerize properly (Leung and Liem, 1996; Meng et al., 1996; Schnabel et al., 1998), the effect of heterodimerization of keratins on their interaction with the IF-binding domain of plakins fused to GAL4-BD in pAS2–1 can be studied in yeast.
Selective Binding of BP230 with Dimeric Epidermal K5/K14 Requires Both the COOH-Terminal B and C Subdomains
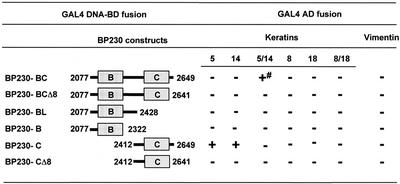
The complete tail of human BP230 (residues 1881–2649) was shown to be codistributed with the epidermal K5/K14 IF network in transfected SCC-13 keratinocytes (Yang et al., 1996). To define the ability of human BP230 to associate with keratins and vimentin, we first used the yeast three-hybrid system. Initially, two large BP230 constructs, encompassing either the entire globular COOH tail domain (residues 1881–2649) or a shortened version (residues 1946–2649), respectively, produced GAL4-AD fusion proteins, which caused autotransactivation of reporter genes, precluding yeast three-hybrid analysis. By contrast, a BP230 fragment encompassing the B and C subdomains, BP230-BC residues 2077–2648 (Figure 1), did not cause autotransactivation and specifically interacted with the K5/K14 heterodimer in yeast three-hybrid assays (Figure 2). This interaction appeared to be weak, because yeast growth was sustained only on plates lacking histidine, not on plates lacking adenine. Indeed, it is known that in the PJ69–4A strain, the ADE2 reporter gene requires more GAL4 activity than the HIS3 reporter gene (Cagney et al., 2000).
Figure 2.
Survey of the sites of interaction between the COOH-terminal domain of BP230 fused to GAL4-BD and IF proteins fused to GAL4-AD: + and - indicate growth or no growth, respectively, on selective media. The PJ69–4A yeast strain was cotransformed (triple transformation) with pACT2, pACT2-URA plasmids encoding IF proteins in fusion with GAL4-AD and pAS2–1 plasmids encoding BP230 fused at its NH2-terminus with GAL4-DNA-BD. To test the interaction with either vimentin or monomeric keratins, triple transformations were performed with either pACT2 or pACT2-URA without insert. After selection on SC-LWUra agar plates, eight colonies of each transformation were arrayed in 96-well microtiter plates and then transferred onto agar SC-LWUra (positive control), SC-LWUra without adenine, and SC-LWUra without histidine and supplemented with 2 mM 3-amino 1,2,3-triazole. Growth was estimated after 5 d of incubation at 30°C. Transactivation controls were performed systematically for each construct with the opposite vectors without insert. # indicates no growth on medium lacking adenine. Each experiment was repeated at least twice.
Neither the single keratins K5, K14, K8, and K18 nor the heterodimeric keratin K8/K18 or vimentin, all fused to the GAL4-AD, bound to BP230-BC (Figure 2). To further map the sequences essential for binding to K5/K14, we tested a series of deletion mutants of BP230. A construct encompassing the B subdomain and the linker, BP230-BL, or the B subdomain, BP230-B, did not interact with any of the IF proteins tested (Figure 2). In contrast, the C subdomain, BP230-C, did bind to monomeric K5 and K14 but neither to K8 or K18 nor to dimeric K5/K14, K8/K18, or vimentin (see below). Finally, we deleted a motif situated at the extremity of BP230 2641GXXSXYXXS2649 (where X is not defined), which is also conserved in DP and PL (Figure 1B). Removal of these last eight residues of BP230 from previously active deletion mutants completely abrogated their binding to IF proteins, indicating that the COOH extremity of BP230 is crucial for binding (Figure 2).
The B and C Subdomains of the BP230 Tail Are Essential for Its Selective Coalignment with K5/K14 IFs
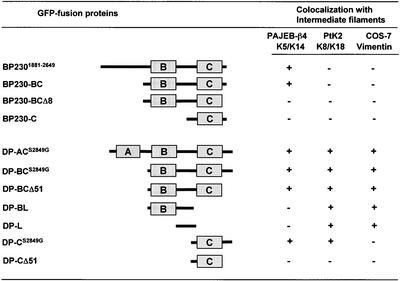
To further investigate whether specific sequences of BP230 are responsible for its association with various types of IFs, we generated expression vectors encoding deletion mutants of the BP230 tail GFP-tagged at the NH2-terminus. These hybrid proteins, like other members of the plakin family, were found to be more stable and easier to analyze than c-myc–tagged deletion mutants (not shown) (Smith and Fuchs, 1998). To determine whether deletion mutants of BP230 coaligned with different IF networks, we transiently transfected the above constructs into various cell lines, including PA-JEB/β4 keratinocytes, which contain a K5/K14 keratin network, PtK2 cells containing K8/K18 IFs, and COS-7 cells for vimentin IFs.
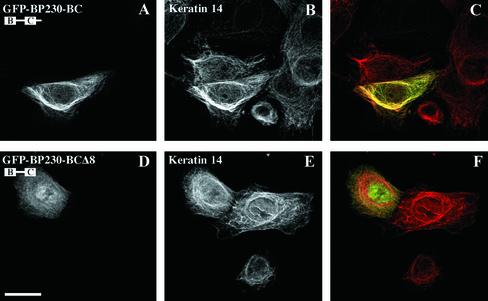
Immunofluorescence microscopy studies of transfected PA-JEB/β4 keratinocytes demonstrated that BP2301881–2649, encompassing the entire tail, decorated the epidermal K5/K14 IF network (not shown, and Figure 3). In up to 10% of the transfected keratinocytes, the epidermal keratin network had partially collapsed. In contrast, when expressed in PtK2 cells, the BP230 tail was not codistributed with the K8/K18 network but rather remained diffusely distributed in the cytoplasm. The same distribution pattern was observed in transfected COS-7 cells, in which no obvious colocalization with vimentin was seen (not shown, and Figure 3). The deletion mutant BP230-BC also coaligned with and, rarely, disrupted the K5/K14 networks of transfected PA-JEB/β4 cells (Figures 3 and 4). BP230-BC was not codistributed with either K8/K18 or vimentin IFs but rather was found primarily diffusely distributed in the cytoplasm of PtK2 and COS-7 cells, respectively (not shown, and Figure 3). The deletion of the ultimate eight residues from the COOH terminus of BP230-BC abrogated the coalignment of the recombinant BP230-BCΔ8 with K5/K14 in transfected PA-JEB/β4 cells (Figures 3 and 4). Finally, in all transfected cell lines, BP230-C was present in small cytoplasmic aggregates, which were not coaligned with and did not disrupt the K5/K14, K8/K18, and vimentin IF networks (Figure 3). When expressed by itself in the various cell lines tested, GFP was found either diffusely distributed over the cytoplasm or in the nucleus without obvious staining of the IF network (not shown).
Figure 3.
Summary of the data obtained in cell transfection experiments in PA-JEB/β4, PtK2, and COS-7 cell lines: + and - indicate colocalization or not between GFP-tagged BP230 or DP fusion proteins and the network of IFs consisting of K5/K14 in PAJEB-β4 cells, K8/K18 in PtK 2 cells, and vimentin in COS-7 cells.
Figure 4.
The codistribution of BP230 with the epidermal keratin network is mediated by sequences contained within the B and C subdomains and the most COOH-terminal extension. Ec-topic expression of the COOH-terminal domain of BP230 fused to GFP in PA-JEB/β4 keratinocytes. Cells were transiently transfected with either GFP-BP230-BC (A–C) or GFP-BP230-BCΔ8 cDNA (D–F). After 48 h, cells were stained with anti-GFP (A and D) and anti-keratin 14 antibodies (B and E). Overlays are shown in C and F. The deletion of the eight COOH-terminal amino acid residues impairs the ability of the BP230 tail to codistribute with epidermal keratins. Bar, 25 μm
Consistent with the results in yeast, the transfection studies indicate that (1) the BP230 tail contains sequences conferring binding specificity for the epidermal K5 and K14 keratins and (2) a region of BP230 encompassing the B and C subdomains, including the intervening linker region, as well as the COOH-terminal extremity is essential for its interaction with K5/K14 filaments. Finally, (3) the observation that BP230-C is unable to become colocalized with the K5/K14 IF network correlates with its failure to interact with dimeric K5/K14 in yeast, suggesting that its binding activity with monomeric K5 and K14 is artificial, most likely as a result of the exposure of cryptic binding sites by the N-terminal truncation.
The Interaction of the DP Tail with IFs Depends on the Heterodimerization of Type I and Type II Keratins in Yeast and Is Regulated by Ser 2849
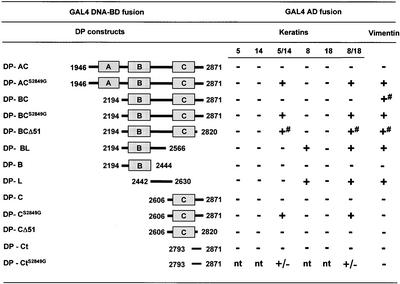
Because previous studies have demonstrated that the DP tail associates with epidermal and nonepidermal keratins (Stappenbeck et al., 1993; Kouklis et al., 1994; Meng et al., 1997), we investigated the ability of two DP constructs containing its entire tail or the B and C regions, DP-AC and DP-BC, respectively, to bind to monomeric or dimeric keratins K5, K14, K8, K18, or vimentin in yeast. Surprisingly, there was no binding with any of the keratins tested, and only DP-BC bound weakly to vimentin (Figure 5). It has been observed that only DP mutants carrying the amino acid substitution S2849G were coaligned with keratin IF networks in certain cell lines and showed increased binding activity with vimentin and K8/K18 in yeast (Stappenbeck et al., 1994; Meng et al., 1997). Therefore, we next assessed the effect of a mutated DP construct in which Ser 2849 was substituted by Gly (S2849G), thereby disrupting a potential protein kinase A consensus phosphorylation site. As shown in Figure 5, DP-ACS2849G and DP-BCS2849G bound to dimeric K5/K14, K8/K18, and vimentin in yeast, whereas there was no interaction with monomeric K5, K14, K8, or K18.
Figure 5.
Yeast three-hybrid analysis of the interaction between various subdomains of the COOH-terminal tail of DP fused to GAL4-BD and IF proteins fused to GAL4-AD: +, +/- and - indicate growth, slow growth, or no growth, respectively, on selective media, tested as described under MATERIALS AND METHODS and in Figure 2. # indicates slow growth on medium lacking adenine; nt, not tested.
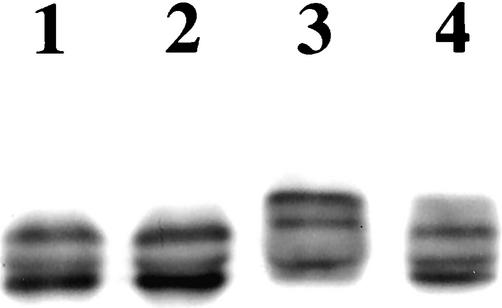
Because Western blot analysis of yeast protein extracts showed that the electrophoretic mobility of DP-C was lower than that of DP-CS2849G, we tested the possibility that phosphorylation of Ser 2849 in the DP tail was responsible for this mobility change. Therefore, the DP-C and DP-CS2849G recombinant proteins were immunoprecipitated from yeast extracts, subjected to dephosphorylation by calf intestine alkaline phosphatase (CIAP), and immunoblotted. As shown in Figure 6, phosphatase treatment results in a shift in the electrophoretic mobility of DP-C, making it indistinguishable from that of DP-CS2849G. Consistent with previous reports that Ser 2849 is phosphorylated in mammalian cells (Stappenbeck et al., 1994), these findings suggest that Ser 2849 is phosphorylated in yeast as well. In this context, we tested the possibility that phosphorylation of Ser 2849 modulates the folding of DP tail by regulating intramolecular interactions within this region. However, no evidence was found for a direct association between the extremity of the COOH terminus and the B and C subdomains or between the C and B subdomains in yeast (not shown).
Figure 6.
Ser 2849 in the DP tail is phosphorylated in yeast. GAL4-recombinant proteins DP-CS2849G (lanes 1 and 2) and DP-C (lanes 3 and 4) were immunoprecipitated from yeast extracts and subjected (lanes 2 and 4) or not (lanes 1 and 3) to dephosphorylation by CIAP and immunoblotted with mouse anti-GAL4 antibody. DP-CS2849G and DP-C produce three distinct bands presumably as a result of a proteolytic activity in yeast cells and extracts. DP-C has a lower electrophoretic mobility than DP-CS2849G. Treatment with CIAP has no impact on the electrophoretic mobility of recombinant protein DP-CS2849G (lanes 1 and 2), whereas it affects recombinant protein DP-C (lane 3), the electrophoretic mobility of which (lane 4) becomes indistinguishable from that of recombinant protein DP-CS2849G (lanes 1 and 2).
These results (1) imply that the presence of either K5/K14 or K8/K18 in the form of a heterodimer made an interaction of DP with these keratins possible and (2) suggest that phosphorylation of Ser 2849 in yeast negatively affects the ability of DP to interact with keratins. Nevertheless, the binding activity of the DP constructs, in which this Ser residue was replaced by Asp to try mimicking phosphorylated Ser (DP-ACS2849D and DP-BCS2849D), was unaffected (not shown).
Identification of Sequences within the DP Tail Interacting Specifically with Various IF Proteins
Next, we assessed the requirement of specific sequences within the DP tail important for its binding to various types of IFs using yeast three-hybrid analysis. A construct encompassing the C subdomain and the COOH terminus of DP with the S2849G substitution, DP-CS2849G, was able to bind to K5/K14 and K8/K18 but no longer interacted with vimentin (Figure 5). Deletion of an additional stretch of 51 residues, comprising the GSRS/T repeats (DP-CΔ51), abolished the interaction with K5/K14 and K8/K18. In contrast, deletion of the same 51 residues from the construct DP-BC (DP-BCΔ51) did not prevent binding to any of the dimeric IF proteins, including keratins. However, the interaction appeared to be weaker than that of DP-BC, as inferred from the delayed growth of yeast cells on medium without adenine. This finding suggested that the COOH-terminal extremity of DP contains residues that contribute to IF binding (Figure 5). Consistent with this idea, we found that a recombinant protein encompassing the last 79 residues of DP, DP-CtS2849G, showed some binding activity with both K5/K14 and K8/K18.
In contrast, a construct encompassing the B subdomain and a large portion of the linker, DP-BL, did not interact with K5 and K14 either as single or heterodimeric proteins but did interact with K8 and heterodimeric K8/K18 and vimentin. To better define which portion of DP-BL is implicated in binding, we tested a construct containing either the linker region of DP, DP-L, or the B subdomain, DP-B. DP-L was sufficient to mediate binding to K8, K8/K18, and vimentin but not to K18 or to K5 and K14 as either single or heterodimeric proteins. By contrast, DP-B was unable to interact with any of the IF proteins tested. These results indicate that (1) a region of DP encompassing the C subdomain is essential but not sufficient for binding to the epidermal K5/K14 keratin pairs, because sequences contained within either the B and the linker region or the 51 COOH-terminal have an impact on the ability of the C subdomain to interact with K5/K14 (see DP-BCΔ51 and DP-CΔ51); (2) the linker region of DP contains recognition sites for simple epithelial keratins and vimentin; and (3) the COOH extremity displays binding activity with both simple and epidermal keratins.
Distinct Regions within the DP Tail Are Required for Its Coalignment with Various IF Networks
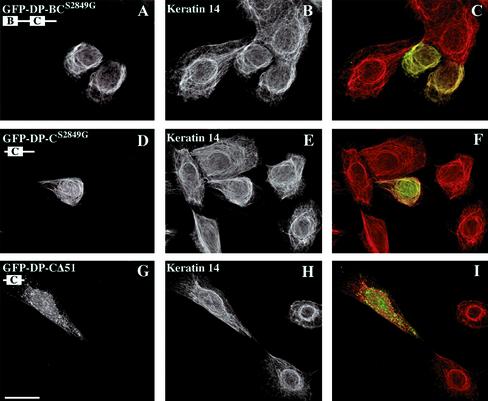
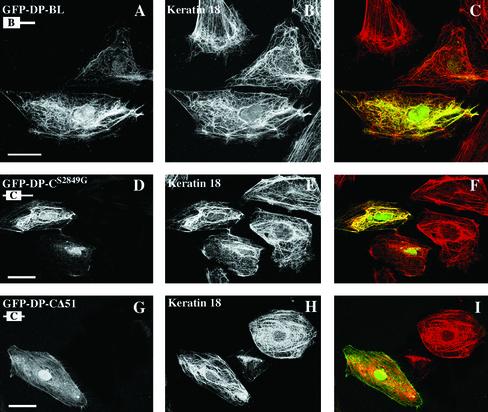
We next performed transfection studies using expression vectors encoding fusion proteins encompassing portions of the DP tail linked to GFP at the NH2-terminus. To ensure the best codistribution potential of these hybrid proteins with various IF networks (Stappenbeck et al., 1994), we used DP mutants in which Ser 2849 was replaced by Gly. We first tested the behavior in transfected cells of two deletion mutants, which encompassed the entire tail of DP or the tail domain without the A subdomain, DP-ACS2849G and DP-BCS2849G, respectively. When expressed in PA-JEB/β4 keratinocytes, PtK2 cells, and COS-7 cells, these DP mutants were able to decorate epidermal K5/K14 and simple K8/K18 keratins and vimentin IF networks, respectively (Figures 3 and 7, A–C). In addition, partial disruption and collapse of these networks was observed occasionally, particularly in cells with high DP fusion protein expression levels (not shown). Additional deletion of 51-amino-acid residues from the COOH-terminal, DP-BCΔ51, had no effect on the codistribution of DP with IFs (Figure 3). The deletion mutant DP-CS2849G coaligned with epidermal and simple keratin IF networks, although occasionally, it was also distributed in the cytoplasm in cells with high levels of expression (Figures 3, 7, and 8, D–F). This DP mutant protein did not decorate the vimentin network in transfected COS-7 cells but remained diffusely distributed over the cytoplasm (Figure 3). Notably, the mutant DP-CΔ51 did not become codistributed with any of the IF networks but instead was diffusely distributed over the cytoplasm (Figures 3, 7, and 8, G–I). In contrast, a DP mutant containing the B subdomain and the linker, DP-BL, became colocalized with both the simple keratin and vimentin networks (Figures 3 and 8, A–C) but not with epidermal keratins (Figure 3). Finally, a DP-L recombinant protein encompassing the linker alone also exhibited this codistribution potential but was more frequently diffusely distributed in the cytoplasm than the recombinant protein DP-BL and even more than DP-BC, which contain one and two modules, respectively (Figure 3). In agreement with the results in yeast and in extension to results of previous studies, our findings clearly indicate that (1) a region encompassing the C subdomain and the COOH extremity contains sequences sufficient for the interaction of DP with epidermal and simple keratin filaments; (2) the presence of the B subdomain and the linker region or the COOH-terminal stretch of 51 residues has an important impact on the potential of the C subdomain and COOH-terminal region to become coaligned with keratin networks; and (3) sequences within the linker region confer additional binding activity for the vimentin IF network and are thus essential for such an interaction.
Figure 7.
Distinct subdomains affect the coalignment potential of DP tail with the epidermal K5/K14 keratin network. Representatives of double immunofluorescence microscopy analyses of PA-JEB/β4 keratinocytes transiently transfected with cDNAs encoding deletion mutants of the DP tail fused to GFP. Cells were transfected with the cDNA constructs GFP-DP-BCS2849G (A–C), GFP-DP-CS2849G (D–F), and GFP-DP-CΔ51 (G–I), respectively, and stained with anti-GFP (A, D, and G) and anti-keratin 14 antibodies (B, E, and H). Overlays are shown in the right panel. Bar, 25 μm
Figure 8.
The ability of the COOH-terminal tail of DP to codistribute with the K8/K18 IF network is affected by sequences contained within either the B subdomain and the linker or the C subdomain and the COOH-terminal extension. PtK2 cells were transiently transfected with the cDNA construct GFP-DP-BL (A–C), GFP-DP-CS2849G (D–F), or GFP-DP-CΔ51 (G–I), respectively. After 48 h, cells were double-stained with anti-GFP (A, D, and G) and anti-keratin 18 antibodies (B, E, and H). Overlays are shown in the right panel. Bar, 25 μm
The DP Tail Preferentially Associates with Polymeric IF Proteins in In Vitro Binding Assays
Because our data suggest that binding of DP and BP230 to epidermal and simple cytokeratins requires that the keratins are in the form of heterodimers, we tested the ability of DP and BP230 tails to associate with monomeric and polymeric IF proteins in in vitro binding assays. K5, K14, K8, and K18 monomers and keratin and vimentin filaments were immobilized on nitrocellulose membranes and overlaid with recombinant DP and BP230 proteins that were in vitro transcribed and translated and used as fluid-phase ligands. In agreement with our yeast two-hybrid results, the recombinant DP-BCS2849G interacted strongly with filaments of K5/K14, K8/K18, and vimentin but not with K5, K8, and K18 monomers (Figure 9), whereas weak associations with K14 were observed. Identical results were found using the recombinant protein DP-BC without the Ser substitution, most likely because the in vitro translated product was not phosphorylated (not shown). Furthermore, the recombinant protein DP-C bound to both K5/K14 and K8/K18, although less efficiently than DP-BCS2849G. Finally, deletion of the last 51 residues from the tail of DP-C, DP-CΔ51, abrogated these interactions. These results provide strong support for the contention that DP associates preferentially with heterodimers of keratin. Attempts to demonstrate an association of the BP230 tail with keratin filaments in overlay binding assays failed (not shown). It is possible that the interaction of BP230 with keratin filaments is too weak to be detected in our in vitro binding assay. Alternatively, the in vitro translated recombinant form of BP230 used may fold incorrectly or lack posttranslational modifications essential for binding to IFs. Such modifications were recently shown to be critical for the association of PL with IF proteins (Janda et al., 2001)
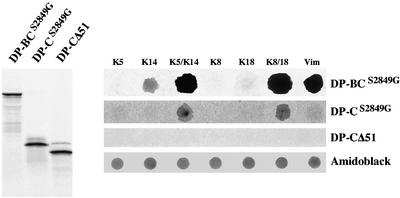
Figure 9.
Dot-blot overlay assays of recombinant purified monomeric or polymerized IF proteins with radiolabeled fragments of the tail of DP. Left, 35[S]-radiolabeled c-myc–tagged recombinant forms of DP, DP-BCS2849G, DP-CS2849G, or DP-CΔ51 were generated by coupled in vitro transcription/translation and analyzed by SDS-PAGE and autoradiography. Right, identical amounts (1 μg) of monomeric or polymerized IF proteins were immobilized on a nitrocellulose membrane by dot blotting and incubated with 35[S]-radiolabeled DP-BCS2849G, DP-CS2849G, or DP-CΔ51. Protein loading was verified by amino-black staining of the spotted proteins. Overlay assays using recombinant protein DP-BC yielded results identical to those with DP-BCS2849G (not shown).
Evidence That Sequences within the DP Tail Confer Binding Specificities to Distinct IF Proteins
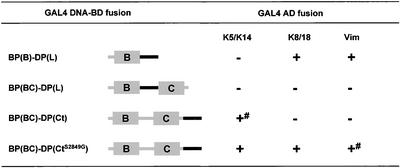
The assumption that distinct subdomains of the plakin tails are responsible for their selective binding to IF proteins was tested further in yeast by generation of chimeric constructs in which distinct regions of BP230 were swapped with the equivalent sequences in DP (Figure 10). A chimeric construct consisting of the B subdomain of BP230 fused at its carboxyl extremity with the linker of DP, BP(B)–DP(L), had binding activities similar to those of the linker of DP (Figure 10). Complementary to this, we swapped the linker region of BP230 with the corresponding region of DP in BP230-BC to obtain BP(BC)–DP(L). Despite this swapping, this latter construct was unable to interact with any of the IF proteins tested (Figure 10). Finally, a chimeric protein consisting of BP230-BC fused at its COOH extremity with the last 51 residues of DP, BP(BC)–DP(Ct) with or without the S2849G substitution, was able to bind to K5/K14. Most importantly, the chimeric construct carrying the S2849G acquired the ability to interact with K8/K18 and weakly with vimentin. Overall, these observations reveal that the linker region and/or the COOH extremity of DP contain sequences critical for the association with K8/K18 and vimentin. Proper folding and exposure of the recognition sites within the linker appear to be affected by the context of the flanking sequences such as the B and/or C subdomains, as inferred from the behavior of BP(BC)–DP(L).
Figure 10.
Yeast three-hybrid analysis of the interaction between IF proteins and chimeric proteins BP230 and DP. + and - indicate growth or no growth, respectively, on selective media, tested as described under MATERIALS AND METHODS and in Figure 2. # indicates no growth on medium lacking adenine.
The Head and Tail Domains of K5 and K14 Are Not Needed for Their Interaction with BP230 and DP
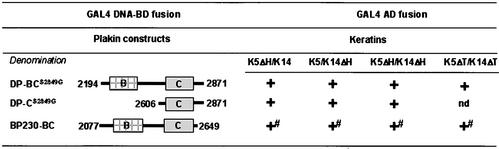
Finally, because previous studies have provided strong evidence that the head domain of K1 and K5 contains sequences that mediate binding to DP (Kouklis et al., 1994; Meng et al., 1997), we studied the effect of the truncation of the head or the tail domain of K5 (K5ΔH and K5ΔT, respectively) and of K14 (K14ΔH and K14ΔT, respectively) on the binding of DP and BP230 to IF proteins. In yeast two-hybrid assays, we first verified that the deletion of the head or tail domains did not impair the ability of K5 and K14 to associate with each other (data not shown). In accordance with previous studies using other combinations of keratin pairs (Schnabel et al., 1998), we found that the head or tail domains of K5 and K14 are unnecessary for dimerization. We then tested the binding activity of DP-BCS2849G, DP-CS2849G, and BP230-BC with various combinations of K5 and K14 with or without their head and tail domains (Figure 11). The results of all experiments were positive. In contrast, DP-CΔ51 did not bind to the various keratin pair combinations (not shown). These findings strongly suggest that the keratin rod domain is sufficient for binding.
Figure 11.
Yeast three-hybrid analysis of the interaction between the COOH terminus of either DP or BP230 and K5/K14 keratins, from which the head or tail domains were removed. + and - indicate growth or no growth, respectively, on selective media, tested as described under MATERIALS AND METHODS and in Figure 2. # indicates no growth on medium lacking adenine.
DISCUSSION
We have performed a comparative investigation of the regions within the tail of BP230 and DP involved in their binding to distinct IF proteins. Our results indicate that these plakins display different binding specificities for the various IF types. These specificities seem to depend, at least for DP, on the sequences within the linker region between the B and C subdomains and/or within its COOH-terminal extension. Furthermore, the association of BP230 and DP with epidermal and simple keratins is critically affected by the tertiary structure induced by heterodimerization of these IF proteins and most likely involves recognition sites located primarily in the rod domain of these keratins.
Identification of Distinct Sequences within the COOH-Terminal Regions of BP230 and DP Required for Their Association with IFs
Our results demonstrate that a region encompassing the B and C subdomains of BP230 contains the minimal sequences required for its specific interaction with the epidermal K5/K14 keratins (Figure 12). It does not bind to the simple epithelial keratins or, in line with a recent study (Leung et al., 1999), vimentin. These findings further support the role of BP230 in tethering the keratin network in basal keratinocytes, in which BP230 and the keratin pair K5/K14 are coexpressed (Arnemann et al., 1993; Guo et al., 1995; Fuchs and Cleveland, 1998).
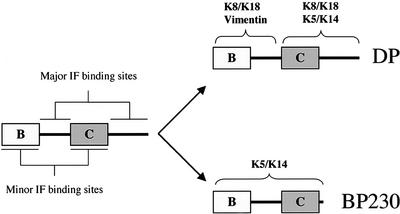
Figure 12.
Binding sites for IF proteins on plakin family members. The linker region between the B and C subdomains and the COOH extremity contain the major IF binding sites and ensure binding specificity. The B and C subdomains contribute to the association with IF proteins by providing additional binding sites and/or by affecting the proper folding of the linker region and the COOH extremity. DP binds to vimentin and K8/K18 via the B subdomain and the linker, whereas the C subdomain and the COOH extremity mediate the association with K8/K18 and K5/K14. In contrast, the ability of BP230 to associate with K5/K14 depends on a region encompassing the B and C subdomains and the COOH extremity. Although there is no direct evidence that they contain a recognition site, the linker region and the COOH extremity are essential for IF binding.
Our data demonstrate that DP has a broader binding capacity than BP230, because it interacts with dimeric K5/K14, K8/K18, and vimentin, as well as with desmin (Meng et al., 1997). This versatility is most likely a prerequisite for the association of DP with distinct IF networks in several cell types, including meningeal cells, dendritic reticulum cells, and cardiac muscle cells, in which DP is thought to link nonkeratin IFs to desmosomes (Kartenbeck et al., 1983, 1984; Meng et al., 1997). Different regions within the DP tail are important for its interaction with vimentin or keratins (Figure 12). The linker region is a specific binding domain for vimentin and simple epithelial keratins but not for epidermal keratins. Furthermore, the C subdomain and the COOH extremity are implicated in its interaction with keratins (see Figures 3 and 5). The association of the C subdomain with simple epithelial and epidermal keratin filaments depends on the presence of a stretch of 51 amino acids at the COOH extremity of DP, because its deletion abrogated binding. However, additional sequences contributing to binding are also contained in a region encompassing the B subdomain and the linker, as evidenced by the fact that construct DP-BCΔ51 but not DP-CΔ51 interacts with K5/K14 keratins. Our findings pose a disparity with the reported inability of the C subdomain and the COOH extremity of DP to become coaligned with the keratin network (Stappenbeck et al., 1993). A possible explanation is that, whereas our construct was GFP-tagged at its NH2-terminus, the DP mutant previously used had a c-myc tag at its COOH extremity, which might interfere with its function.
Importance of the Head and Tail Domains of Cytokeratins and of Their Heterodimerization on Their Interaction Potential with BP230 and DP
As judged by the consistent findings obtained in yeast three-hybrid assays with keratin pairs and the cell transfection studies, both DP and BP230 preferentially interact with K5/K14 and K8/K18 heterodimers. This contention is also supported by the overlay binding assays, which confirmed a significant association of DP with keratin filaments but not with monomeric keratins. These observations suggest that in our yeast assays, the binding activity of certain plakin deletion mutants with monomeric keratins (see BP230-C), which was not observed with larger constructs, is artifactual and results from the exposure of cryptic binding sites by the truncation procedure (see BP230-C).
Our results seem to be at variance with two previous studies, which indicated that DP is able to interact with various monomeric type II keratins, including K5 and K1, as demonstrated by solution binding assays or yeast two-hybrid analysis (Kouklis et al., 1994; Meng et al., 1997).
Nevertheless, in line with our findings, Meng et al. (1997) showed in yeast two-hybrid assays that the association of DP with K8/K18, in contrast to that with the epidermal keratin K1, required the presence of both keratin partners. Together, the latter observation and our data strongly suggest that the association of DP and BP230 with epidermal and simple keratins is favored by the tertiary structure associated with the keratin α-helical coiled-coil and heterodimers. In this context, it is of interest to note that Kouklis et al. (1994) reported that a region encompassing the KSIS motif within the head domain of K1 and K5 was crucial for the interaction of DP with epidermal keratins, whereas Meng et al. (1997) found that the head domain of K1 contributed to the interaction of K1 with DP in yeast and peptide competition assays. Although our findings do not exclude a role of the head domain for binding to DP and BP230, they strongly suggest that there are additional, critical interaction sites in the keratin α-helical coiled-coil of K5 and K14, because deletion of their head or tail domains does not abrogate their binding with either DP or BP230 in yeast. It is likely that the association of DP with K8/K18 involves similar IF domains. In fact, Meng et al. (1997) showed that the association of DP with K8/K18 heterodimer was not dependent on the head domain of K8 and K18. However, additional experiments are needed to clarify whether the mechanisms by which simple epithelial and epidermal keratins bind to plakins are comparable.
Potential Role of the Phosphorylation of the DP and BP230 Tail for Binding with IFs
Previous studies have shown that only DP mutants carrying the amino acid substitution S2849G were coaligned with keratin IF networks in certain cell lines and showed increased binding activity with vimentin and K8/K18 in yeast (Stappenbeck et al., 1994; Meng et al., 1997). These observations suggested that phosphorylation of Ser 2849 in the DP COOH terminus critically affects its association with IF proteins. In line with this idea, we found here that none of the DP GAL4-fusion proteins containing the wild-type COOH-terminal tail were able to interact with epidermal and simple keratins or vimentin in yeast three-hybrid assays. However, whereas the replacement of Ser 2849 by Asp to mimic phosphorylation did not inhibit the interaction of the DP tail with IF proteins, substitution of Ser 2849 by Gly resulted in significant binding activity in yeast. Our findings obtained with alkaline phosphatase–treated yeast extracts provide indirect evidence that Ser 2849 is phosphorylated in yeast, probably by a cAMP-dependent protein kinase, an enzyme that is highly conserved in all eukaryote cells (Taylor et al., 1990).
It is possible that the potential to phosphorylate Ser 2849 in yeast varies among the strains used. This would provide a plausible explanation for the observation that Meng et al. (1997), using the PCY2 strain, found, in apparent contrast to our results, a weak interaction between DP-AC and K8/K18. An alternative and more likely explanation is that the LacZ reporter gene of the PCY2 strain is more sensitive than the HIS3 and ADE2 reporter genes of the PJ69 strain used in this study and also allowed detection of less robust interactions.
Regulation of the IF-binding properties of BP230 by its phosphorylation has not been reported as yet. However, our finding that the deletion of the last stretch of eight amino acids from the BP230 tail containing Tyr and Ser residues that can be phosphorylated had a dramatic effect on its interaction with K5/K14 raises the possibility that posttranslational modifications of the COOH extremity of BP230 regulate its function.
Elucidating the Function of the Linker Region of Plakins for IF Binding
Previous in vitro binding studies (Nikolic et al., 1996; Steinbock et al., 2000) have demonstrated that the IF-binding activity of PL with vimentin and cytokeratins depends on an ∼50-amino-acid stretch within its linker. The latter is highly conserved in DP and PL (70% identity) but not in BP230 (35% identity) (Figure 1A) (Nikolic et al., 1996; Steinbock et al., 2000). The four basic residues that were shown to be crucial for the association of PL with vimentin are present in DP but not in BP230. Hence, we argued that the linker of DP contains sequences sufficient for IF binding. In line with this idea, we found that the linker of DP binds to K8/K18 keratins and vimentin in yeast and is colocalized with these IF networks when expressed in transfected cells. However, it remains unclear whether the binding sites for K8/K18 and vimentin overlap or are located on distinct sequences within the linker.
The fact that the linker region of BP230 is different from that in DP and PL can explain the inability of BP230 to associate with either vimentin or K8/K18. BP230 lacks not only the crucial short stretch of basic residues but also a series of Ser residues between the B and C repeats (Figure 1A). Notably, the swapping of the linker of BP230 with that of DP in the construct BP(BC)–DP(L) abolished the ability of the BP230 tail to associate with K5/K14. This result suggests that the proper folding and alignment of the linker region and/or of the B and C subdomains are essential for binding to IF proteins.
Finally, the idea that the linker region contains sequences critical for the codistribution of plakins with distinct IF proteins is further supported by recent studies of periplakin. The coalignment with IFs of this protein and its binding ability to simple keratins and vimentin in yeast appeared to depend on sequences within the linker at its COOH extremity (DiColandrea et al., 2000; Karashima and Watt, 2002; Kazerounian et al., 2002).
An Increased Importance of the COOH Extremity of Plakins
The importance of the COOH extremity of DP is supported by the observation that patients with mutations in the DP gene leading to a deletion of the C subdomain have abnormalities of the heart and the skin with a disorganized cytokeratin network (Norgett et al., 2000). Furthermore, it has been shown that a stretch encompassing residues 48–68 from the extremity of the carboxyl-terminus of DP was necessary for the coalignment of the DP tail with the keratin network (Stappenbeck et al., 1993). As an extension of these findings, we demonstrate here that the COOH extremity of DP is critical for its association with both K5/K14 and K8/K18, because its deletion, as in DP-CΔ51, abrogates all binding activities in yeast and biochemical assays. This stretch of residues may contribute to the proper folding of the recognition sites for IFs and/or participate directly in the binding to IFs, as inferred from the ability of the last 79 amino acid residues of DP to weakly bind to keratins in yeast.
The COOH-terminal extremity of DP seems to contribute not only to keratin but also to vimentin binding, a role hitherto unappreciated. In fact, the presence of the terminal 51-amino-acid stretch of the COOH extremity of DP confers upon the BP230 tail the ability to bind to simple epithelial keratins and vimentin [see chimeric protein BP(BC)–DP(Ct) in Figure 10]. An alternative explanation for this finding is that the BP230 tail has the potential to interact, as previously suggested (Yang et al., 1996), to vimentin. The presence of the terminal 51-amino-acid stretch of the DP may contribute to increase or stabilize this interaction. However, the function of this portion of DP appears to be regulated by phosphorylation, because no interaction with either vimentin or K8/K18 was observed when a similar chimeric construct without the S2849G substitution was used (see Figure 10), whereas binding to K5/K14 remained largely unaffected.
Finally, it should be noted that the region of residues 51–68 from the carboxyl extremity of DP contains a motif, GXXSXYXXS (where X is not defined), that is conserved in DP, BP230, and PL. Removal of this motif from the BP230 tail as in construct BP230-BCΔ8 abrogated its binding to K5/K14, an observation underlining a key role of this sequence for keratin binding (see Figures 2 and 3).
Contribution of the B and C Subdomains to IF Binding
On the basis of their sequence homology, the B and C subdomains in DP, PL, and BP230 have been thought to represent the major IF binding modules, the available number of which determines the binding affinity for IFs (Stappenbeck et al., 1993). However, using different approaches, we found here no compelling evidence for this model for either DP or BP230. Nevertheless, these subdomains most likely contribute to IF binding, as inferred from the reduced ability of DP-L compared with that of DP-BL and DP-BC to become codistributed with the IF network in transfected cells. These results suggest that the single B and C subdomains of DP contain recognition sites for IFs, but their affinity is too low to detect interactions in transfection studies and yeast assays. Notably, crystallographic studies have shown that the B and C subdomains are globular and exhibit a conserved basic groove that may serve as recognition site for the rod domain of vimentin (Choi et al., 2002). Alternatively, it is possible, as previously proposed for PL (Steinbock et al., 2000), that these subdomains are involved in the proper exposure and folding of linker sequences containing the actual IF binding sites. In line with this idea, in vitro binding assays have demonstrated that a recombinant protein of DP encompassing both the B and C subdomains showed better binding to vimentin than the isolated B or C subdomain, the interaction potential of which was very weak and occurred only when a large molar excess of DP recombinant proteins was used (Choi et al., 2002).
Together, on the basis of our and previous studies, the following model is proposed (see Figure 12). Sequences between the highly homologous B and C subdomains and within the COOH extremity of DP and BP230 have a critical impact on their function: the linker and the COOH extremity of plakins contain recognition sites crucial for IF binding and confer specificity for various IF proteins. In this context, the B and C subdomain contribute to efficient binding by either providing additional interaction sites or ensuring the proper conformation and folding of the linker and COOH extremity. Furthermore, the latter region is subject to phosphorylation events that profoundly affect the ability of plakins to interact with various and distinct IF networks. Finally, although epidermal and simple keratins may associate with DP or BP230 via different binding sites located on either their rod or globular end-domains, in both cases the association is favored by the tertiary structure induced by heterodimerization.
Acknowledgments
The authors are indebted to Drs. J. Stanley (Philadelphia), T. Hashimoto (Kurume Fukuoka), and K. Owaribe (Nagoya), who generously provided antibodies used in this study; to Dr. E.B. Lane (Dundee), who kindly provided cDNAs for human K5 and K14; and to Dr. H. Herrmann (Heidelberg), who kindly provided recombinant human K14. This work was supported by grants from the Swiss National Foundation for Scientific Research (32–51083.97 and 32–56727.99 to L.B.), from the Office fédéral de l'éducation et de la science (no. 010113, to L.B.), the Commission of the European Communities (CT-2001–02007 to L.B.), Telethon Action Suisse (Aubonne, to L.B.), the Dutch Cancer Society (NKI 99–2039 to A.S.), and NIH grant RO1-AR-43380 to K.G.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-08-0548. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-08-0548.
Abbreviations used: mAb, monoclonal antibody; AD, transcription-activation domain; BD, DNA-binding domain; BP230, bullous pemphigoid antigen 1; DP, desmoplakin; EGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; IF, intermediate filament; PL, plectin
References
- Andrä, K., Lassmann, H., Bittner, R., Shorny, S., Fässler, R., Propst, F., and Wiche, G. (1997). Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev. 11, 3143–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst, B.D., Nilles, L.A., and Green, K.J. (1990). Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97, 247–257. [DOI] [PubMed] [Google Scholar]
- Arnemann, J., Sullivan, K.H., Magee, A.I., King, I.A., and Buxton, R.S. (1993). Stratification-related expression of isoforms of the desmosomal cadherins in human epidermis. J. Cell Sci. 104, 741–750. [DOI] [PubMed] [Google Scholar]
- Borradori, L., and Sonnenberg, A. (1999). Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Invest. Dermatol. 112, 411–418. [DOI] [PubMed] [Google Scholar]
- Brown, A., Bernier, G., Mathieu, M., Rossant, J., and Kothary, R. (1995). The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat. Genet. 10, 301–306. [DOI] [PubMed] [Google Scholar]
- Cagney, G., Uetz, P., and Field, S. (2000). High-throughput screening for protein-protein interaction using the two-hybrid assay. Methods Enzymol. 328, 3–14. [DOI] [PubMed] [Google Scholar]
- Choi, H.J., Park-Snyder, S., Pascoe, L.T., Green, K.J., and Weis, W.I. (2002). Structure of two intermediate filaments-binding fragments of desmoplakin reveal a unique repeat motif structure. Nat. Struct. Biol. 9, 612–620. [DOI] [PubMed] [Google Scholar]
- Coulombe, P.A., and Fuchs, E. (1990). Elucidating the early stages of keratin filament assembly. J. Cell Biol. 111, 153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiColandrea, T., Karashima, T., Määttä, A., and Watt, F.M. (2000). Subcellular distribution of envoplakin and periplakin: insights into their role as precursors of the epidermal cornified envelope. J. Cell Biol. 151, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, J., Yang, Y., Wollmann, R., Reichardt, L.F., and Fuchs, E. (1997). Developmental expression of BPAG1-n: insights into the spastic ataxia and gross neurologic degeneration in dystonia musculorum mice. Dev. Biol. 187, 131–142. [DOI] [PubMed] [Google Scholar]
- Favre, B., Fontao, L., Koster, J., Shafaatian, R., Jaunin, F., Saurat, J.H., Sonnenberg, A., and Borradori, L. (2001). The hemidesmosomal proteins bullous pemphigoid antigen 1 and the integrin β4 subunit bind to ERBIN. J. Biol. Chem. 276, 32427–32436. [DOI] [PubMed] [Google Scholar]
- Foisner, R., Leichtfried, F.E., Herrmann, H., Small, J.V., Lawson, D., and Wiche, G. (1988). Cytoskeleton-associated plectin: in situ localization, in vitro reconstitution, and binding to immobilized intermediate filament proteins. J. Cell Biol. 106, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, E. (1994). Intermediate filaments and disease: mutations that cripple cell strength. J. Cell Biol. 125, 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, E., and Cleveland, D.W. (1998). A structural scaffolding of intermediate filaments in health and disease. Science. 279, 514–519. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and Weber, K. (1994). Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 63, 345–382. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and Yang, Y. (1999). Crossroads on cytoskeletal highways. Cell. 98, 547–550. [DOI] [PubMed] [Google Scholar]
- Gallicano, G.I., Bauer, C., and Fuchs, E. (2001). Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development 128, 929–941. [DOI] [PubMed] [Google Scholar]
- Gallicano, G.I., Kouklis, P., Bauer, C., Yin, M., Vasioukhin, V., Degenstein, L., and Fuchs, E. (1998). Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143, 2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, K.J., Virata, M.L., Elgart, G.W., Stanley, J.R., and Parry, D.A. (1992). Comparative structural analysis of desmoplakin, bullous pemphigoid antigen and plectin: members of a new gene family involved in organization of intermediate filaments. Int. J. Biol. Macromol. 14, 145–153. [DOI] [PubMed] [Google Scholar]
- Guo, L., Degenstein, L., Dowling, J., Yu, Q.C., Wollmann, R., Perman, B., and Fuchs, E. (1995). Gene targeting of BPAG1: abnormalities in mechanical strength and cell migration in stratified epithelia and neurologic degeneration. Cell. 81, 233–243. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T., Amagai, M., Ebihara, T., Gamou, S., Shimizu, N., Tsubata, T., Hasegawa, A., Miki, K., and Nishikawa, T. (1993). Further analyses of epitopes for human monoclonal anti-basement membrane zone antibodies produced by stable human hybridoma cell lines constructed with Epstein-Barr virus transformants. J. Invest. Dermatol. 100, 310–315. [DOI] [PubMed] [Google Scholar]
- Herrmann, H., and Aebi, U. (2000). Intermediate filaments and their associates: multi-talented structural elements specifying cytoarchitecture and cytodynamics. Curr. Opin. Cell Biol. 12, 79–90. [DOI] [PubMed] [Google Scholar]
- Hopkinson, S.B., and Jones, J.C. (2000). The N-terminus of the transmembrane protein BP180 interacts with the N-terminal domain of BP230, thereby mediating keratin cytoskeleton anchorage to the cell surface at the site of the hemidesmosome. Mol. Biol. Cell 11, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, L., Damborsky, J., Rezniczek, G., and Wiche, G. (2001). Plectin repeats, and modules: strategic cysteines and their presumed impact on cytolinker functions. Bioassays 23, 1064–1069. [DOI] [PubMed] [Google Scholar]
- Karashima, T., and Watt, F.M. (2002). Interaction of periplakin and envoplakin with intermediate filaments. J. Cell Sci. 115, 5027–5037. [DOI] [PubMed] [Google Scholar]
- Kartenbeck, J., Franke, W.W., Moser, J.G., and Stoffels, U. (1983). Specific attachment of desmin filaments to desmosomal plaques in cardiac myocytes. EMBO J. 2, 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck, J., Schwechheimer, K., Moll, R., and Franke, W.W. (1984). Attachment of vimentin filaments to desmosomal plaques in human meningiomal cells and arachnoidal tissue. J. Cell Biol. 98, 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazerounian, S., Uitto, J., and Aho, S. (2002). Unique role for the periplakin tail in intermediate filament association: specific binding to keratin 8 and vimentin. Exp. Dermatol. 11, 428–438. [DOI] [PubMed] [Google Scholar]
- Kouklis, P.D., Hutton, E., and Fuchs, E. (1994). Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J. Cell Biol. 127, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk, A.P., Bornslaeger, E.A., Borgwardt, J.E., Palka, H.L., Dhaliwal, A.S., Corcoran, C.M., Denning, M.F., and Green, K.J. (1997). The amino-terminal domain of desmoplakin binds to plakoglobin and clusters desmosomal cadherin-plakoglobin complexes. J. Cell Biol. 139, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, C.L., Green, K.J., and Liem, R.K. (2002). Plakins: a family of versatile cytolinker proteins. Trends Cell Biol. 12, 37–45. [DOI] [PubMed] [Google Scholar]
- Leung, C.L., and Liem, R.K. (1996). Characterization of interactions between the neurofilament triplet proteins by the yeast two-hybrid system. J. Biol. Chem. 271, 14041–14044. [PubMed] [Google Scholar]
- Leung, C.L., Liem, R.K., Parry, D.A., Green, K.J. (2001b). The plakin family. J. Cell Sci. 114, 3409–3410. [DOI] [PubMed] [Google Scholar]
- Leung, C.L., Sun, D., and Liem, R.K. (1999). The intermediate filament protein peripherin is the specific interaction partner of mouse BPAG1-n (dystonin) in neurons. J. Cell Biol. 144, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, C.L., Zheng, M., Prater, S.M., Liem, R.K. (2001a). The B.P.A.G1 locus: alternative splicing produces multiple isoforms with distinct cytoskeletal linker domains, including predominant isoforms in neurons and muscles. J. Cell Biol. 154, 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, J.J., Bornslaeger, E.A., Green, K.J., Steinert, P.M., and Ip, W. (1997). Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J. Biol. Chem. 272, 21495–21503. [DOI] [PubMed] [Google Scholar]
- Meng, J.J., Khan, S., and Ip, W. (1996). Intermediate filament protein domain interactions as revealed by two-hybrid screens. J. Biol. Chem. 271, 1599–1604. [DOI] [PubMed] [Google Scholar]
- Nikolic, B., Mac Nulty, E., Mir, B., and Wiche, G. (1996). Basic amino acid residue cluster within nuclear targeting sequence motif is essential for cytoplasmic plectin-vimentin network junctions. J. Cell Biol. 134, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgett, E.E., Hatsell, S.J., Carvajal-Huerta, L., Cabezas, J.C., Common, J., Purkis, P.E., Whittock, N., Leigh, I.M., Stevens, H.P., and Kelsell, D.P. (2000). Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 9, 2761–2766. [DOI] [PubMed] [Google Scholar]
- Parry, D.A., and Steinert, P.M. (1999). Intermediate filaments: molecular architecture, assembly, dynamics and polymorphism. Q. Rev. Biophys. 32, 99–187. [DOI] [PubMed] [Google Scholar]
- Ruhrberg, C., and Watt, F.M. (1997). The plakin family: versatile organizers of cytoskeletal architecture. Curr. Opin. Genet. Dev. 7, 392–397. [DOI] [PubMed] [Google Scholar]
- Schnabel, J., Weber, K., and Hatzfeld, M. (1998). Protein-protein interactions between keratin polypeptides expressed in the yeast two-hybrid system. Biochim. Biophys. Acta 1403, 158–168. [DOI] [PubMed] [Google Scholar]
- Skaria, M., Jaunin, F., Hunziker, T., Riou, S., Schumann, H., Bruckner-Tuderman, L., Hertl, M., Bernard, P., Saurat, J.H., Favre, B., and Borradori, L. (2000). IgG autoantibodies from bullous pemphigoid patients recognize multiple antigenic reactive sites located predominantly within the B and C subdomains of the COOH-terminus of BP230. J. Invest. Dermatol. 114, 998–1004. [DOI] [PubMed] [Google Scholar]
- Smith, E.A., and Fuchs, E. (1998). Defining the interactions between intermediate filaments and desmosomes. J. Cell Biol. 141, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck, T.S., Bornslaeger, E.A., Corcoran, C.M., Luu, H.H., Virata, M.L., and Green, K.J. (1993). Functional analysis of desmoplakin domains: specification of the interaction with keratin versus vimentin intermediate filament networks. J. Cell Biol. 123, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck, T.S., Lamb, J.A., Corcoran, C.M., and Green, K.J. (1994). Phosphorylation of the desmoplakin COOH-terminus negatively regulates its interaction with keratin intermediate filament networks. J. Biol. Chem. 269, 29351–29354. [PubMed] [Google Scholar]
- Steinbock, F.A., Nikolic, B., Coulombe, P.A., Fuchs, E., Traub, P., and Wiche, G. (2000). Dose-dependent linkage, assembly inhibition and disassembly of vimentin and cytokeratin 5/14 filaments through plectin's intermediate filament-binding domain. J. Cell Sci. 113, 483–491. [DOI] [PubMed] [Google Scholar]
- Steinbock, F.A., and Wiche, G. (1999). Plectin: a cytolinker by design. Biol. Chem. 380, 151–158. [DOI] [PubMed] [Google Scholar]
- Steinert, P.M., Chou, Y.H., Prahlad, V., Parry, D.A., Marekov, L.N., Wu, K.C., Jang, S.I., and Goldman, R.D. (1999). A high molecular weight intermediate filament-associated protein in BHK-21 cells is nestin, a type VI intermediate filament protein: limited co-assembly in vitro to form heteropolymers with type III vimentin and type IV alpha-internexin. J. Biol. Chem. 274, 9881–9890. [DOI] [PubMed] [Google Scholar]
- Sterk, L.M., Geuijen, C.A., Oomen, L.C., Calafat, J., Janssen, H., and Sonnenberg, A. (2000). The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 149, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.S., Buechler, J.A., and Yonemoto, W. (1990). cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 59, 971–1005. [DOI] [PubMed] [Google Scholar]
- Wiche, G., Becker, B., Luber, K., Weitzer, G., Castanon, M.J., Hauptmann, R., Stratowa, C., and Stewart, M. (1991). Cloning and sequencing of rat plectin indicates a 466-kD polypeptide chain with a three-domain structure based on a central alpha-helical coiled coil. J. Cell Biol. 114, 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche, G., Gromov, D., Donovan, A., Castanon, M.J., and Fuchs, E. (1993). Expression of plectin mutant cDNA in cultured cells indicates a role of COOH-terminal domain in intermediate filament association. J. Cell Biol. 121, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Dowling, J., Yu, Q.C., Kouklis, P., Cleveland, D.W., and Fuchs, E. (1996). An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell 86, 655–665. [DOI] [PubMed] [Google Scholar]
- Zhang, J. (2000). Use of yeast three-hybrid system to clone bridging proteins. Methods Enzymol. 328, 103–110. [DOI] [PubMed] [Google Scholar]