Abstract
A well-established function of centrosomes is their role in accomplishing a successful mitosis that gives rise to a pair of identical daughter cells. We recently showed that DNA replication defects and DNA damage in Drosophila embryos trigger centrosomal changes, but it remained unclear whether comparable centrosomal responses can be provoked in somatic mammalian cells. To investigate the centrosomal organization in the presence of impaired DNA integrity, live and ultrastructural analysis was performed on γ-tubulin–GFP and EGFP–α-tubulin–expressing Chinese hamster ovary cells. We have shown that during mitosis in the presence of incompletely replicated or damaged DNA, centrosomes split into fractions containing only one centriole. This results in the formation of multipolar spindles with extra centrosome-like structures. Despite the extra centrosomes and the multipolarity of the spindles, cells do exit from mitosis, resulting in severe division errors. Our data provide evidence of a novel mechanism showing how numerous centrosomes and spindle defects can arise and how this can lead to the formation of aneuploid cells.
INTRODUCTION
When normal diploid somatic mammalian cells undergo successful mitosis, a pair of genetically identical daughter cells are generated. A successful mitosis requires the presence of a bipolar spindle organized by two centrosomes, and in normal cells the centrosome numbers are regulated by strict control mechanisms (Hinchcliffe and Sluder, 2001; Rieder et al., 2001; Stearns, 2001). Initially, cells start in G1 with one centrosome that contains two barrel-shaped centrioles surrounded by the pericentriolar matrix. Around the G1/S transition, a procentriole (or daughter centriole) forms adjacent to each of the two parental centrioles. By late G2, the cell has two centrosomes, each consisting of two doublets of centrioles. Shortly before the onset of mitosis, the centrosomes migrate in opposite directions to play a key role in organizing the mitotic spindle that pulls the duplicated chromosomes away from each other during cell division. After cell division, each daughter cell starts again with one centrosome. Centrosomes are not only involved in achieving a successful mitosis but have also been shown to be required for somatic cells to progress through G1 into S phase (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001). In addition to a role in cytokinesis and cell-cycle progression, centrosomes may also be involved in cellular responses to impaired DNA integrity. We have shown that DNA replication defects and DNA damage in Drosophila embryos trigger the disappearance of a set of centrosomal proteins at the spindle poles (Sibon et al., 2000). As a result of this, anastral nonfunctional spindles arise that are unable to segregate the chromosomes.
In normal mammalian cells, there are S-phase and G2/M checkpoint mechanisms that prevent those cells with incompletely replicated or damaged DNA from entering mitosis (Hartwell and Weinert, 1989; Elledge, 1996; Smits and Medema, 2001). However, it is unclear what happens when mammalian cells with DNA replication defects (for example, if the G2/M checkpoint is defective or is being overruled) enter mitosis. In the present study, we aimed to explore the possibility that in mammalian somatic cells, centrosomes are being altered when DNA replication defects or DNA damage persists in mitosis. To do this, we took advantage of the fact that Chinese hamster ovary (CHO) cells progress through mitosis in the presence of DNA damage induced by the interstrand cross-linker mitomycin-C (MMC) or can be forced into mitosis with DNA replication defects by use of hydroxyurea (HU) and caffeine (Schlegel and Pardee, 1986; Balczon et al., 1995; Balczon et al., 1999). We were able to demonstrate the modification of centrosomes during mitosis when the integrity of DNA was impaired. This led to extra centrosome-like structures, multipolar spindles, and division errors. Extra centrosomes, multipolar spindles, and aneuploidy have been observed in numerous tumor cells (Fukasawa et al., 1996; Lingle et al., 1998, 2002; Xu et al., 1999; Brinkley, 2001; Marx, 2001). Despite the strong correlation between the occurrence of aneuploidy, extra centrosomes, and genetic instability, the question remains which of these characteristics is a cause or a consequence of the others. Our live and ultrastructural analysis provides evidence of one simple order of events. In the presence of incompletely replicated DNA or damaged DNA, centrosomes break up during mitosis, multipolar spindles arise, and aneuploid cells may be formed. Centrosome and spindle abnormalities may therefore be a general response of mammalian cells when DNA defects persist in mitosis. This may explain the presence of extra centrosome-like structures and multipolar spindles in a large variety of cancer cells.
MATERIALS AND METHODS
Cell Lines and Culturing
CHO cells and irs1SF cells (kindly provided by Prof. M.Z. Zdzienicka, Leiden, The Netherlands) were cultured in HAM's F-12 medium. V79 and O23 cells were cultured in DMEM (high glucose). The media was supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin purchased from GIBCO (Paisley, UK). All cultures were maintained at 5% CO2 in a humidified 37°C incubator.
Stable cell lines expressing γ-tubulin–GFP or enhanced green fluorescent protein (EGFP)–α-tubulin were generated in CHO cells and irs1SF cells by standard techniques. The pcDNA3-γTGFP that encodes the fusion protein γ-tubulin–GFP was kindly provided by A. Khodjakov (Albany, NY) (Khodjakov and Rieder, 1999). The pEGFP-Tub that encodes the fusion protein EGFP–α-tubulin was purchased from Clontech (Palo Alto, CA). From ∼30 single-cell colonies, one clone or a subsequent subclone was chosen that showed moderate expression that did not affect progression through mitosis and showed a normal organization of α-tubulin– and γ-tubulin–containing structures. A selective marker, 400 μg/ml geneticin (GIBCO) was added to the medium.
Treatments
All our standard laboratory chemicals were purchased from Sigma (St. Louis, MO). Cells were treated with 2.5 mM HU or 5 μg/ml aphidicolin in the medium for variable times to inhibit DNA synthesis. Caffeine 5 mM or UCN-01 0.3 μM was added to the medium for 4 h in addition to the HU to force cells with incompletely replicated DNA into mitosis. When indicated, 0.5 μg/ml colcemid or 1 μg/ml cytochalasin D was added to the medium in addition to the HU treatment. To induce DNA damage via interstrand DNA cross-links, 25 μM MMC (Christiaens BV, Breda, The Netherlands) was added to the medium for 1 h.
Immunofluorescence Analysis
Cells were grown for 2 d on coverslips, washed twice with PBS, and fixed with methanol:acetone 3:1 for 10 min. Coverslips were washed three times for 10 min with PBS, permeabilized with 0.2% Triton-X100 in PBS for 15 min, and incubated with 50 mM glycine in PBS for 10 min. After 30 min of blocking with PBG (0.5% BSA and 0.1% glycine in PBS), cells were labeled with polyclonal anti–γ-tubulin (Sigma T3559) or anti-pericentrin (Babco, Richmond, CA) in combination with anti-rabbit FITC (DAKO, Glostrup, Denmark) to visualize the centrosomes. To visualize the mitotic spindles, a monoclonal anti–α-tubulin (Sigma T5168) was used in combination with anti-mouse CY3 (Amersham, Piscataway, NJ). To visualize the DNA, cells were stained for 10 min with 0.2 μg/ml DAPI. To visualize mitotic cells, Phospho-Histone 3 (PH3) mAb (Cell Signaling Technology, Beverly, MA) was used in combination with antimouse CY3 (Amersham). Pictures were taken with a confocal laser-scanning microscope (Leica TCS SP2, Leica Microsystems, Heidelberg, Germany) with 351/364-, 488-, and 543-nm lasers. Mitotic indexes were obtained by counting the number of mitotic spindles in >1000 cells. The percentages of normal and abnormal spindles were obtained by analyzing >100 spindles per condition. The number of centrosomes in interphase cells was scored in 500 interphase cells per condition. For this purpose, only interphase cells with one nucleus were scored.
Time-Lapse Imaging
Images were made with a confocal laser-scanning microscope (Zeiss LSM510NLO, Carl Zeiss, Jena, Germany). To keep cells alive during the experiment (17 h), cells were cultured on a round cover glass mounted in a special chamber with 2 ml of medium. The chamber was placed on a heated microscope stage that was kept at 37°C. A mixture of air with 5% CO2 was blown into the heated stage. The 63× oil immersion lens was also heated to 37°C. Cells were imaged in six optical planes (thickness <1 μm, 1-μm distance). A macro was developed (Carl Zeiss) to move the electronic XY stage to different locations. In this way, 10 different locations could be imaged sequentially. To correct for focal drift, an autofocus routine was implemented in the time-lapse macro (Carl Zeiss). During the autofocus procedure, a Z-scan of one line was made with a 633 laser (XZ plane). The reflection of the laser on the cover glass was measured by removing the filter in front of the detector. An offset from the highest-intensity line was used in the macro to start scanning at the same distance from the cover glass. All 10 locations were imaged in a cycle of 10 min. This cycle was repeated for 17 h. Cells expressing the EGFP or GFP fusion proteins were excited with a 488 laser, and emission was measured with a BP 500–550 filter. Together with the fluorescent image, a transmitted light (differential interference contrast optics) image was made. At the end of the 17-h period, the cells were still dividing. Data were analyzed using the LSM510 software (Carl Zeiss).
Electron Microscopy
Cells were grown on Thermanox coverslips and treated with HU and caffeine or left untreated (control cells) as described above. Cells were immunolabeled with monoclonal anti–α-tubulin (Sigma T5168) and goat anti-mouse ultrasmall gold particles (Aurion, Wageningen, The Netherlands) followed by silver enhancement (British BioCell International, Cardiff, UK) as previously described (Macville et al., 1995). Cells were preselected by light microscopy before consecutive sections were made for electron microscopic (Philips 201, CM100, Philips, Eindhoven, The Netherlands) analysis.
RESULTS
Abnormal Spindles with Extra Centrosome-like Structures Are Formed When CHO Cells Enter Mitosis in the Presence of Incompletely Replicated DNA
To investigate whether centrosomal changes occur in the presence of impaired DNA integrity in mammalian cells, we first attempted to reproduce the experiments described for CHO cells and forced them into mitosis in the presence of incompletely replicated DNA (Schlegel and Pardee, 1986; Balczon et al., 1995; Wise and Brinkley, 1997). CHO cells were incubated with 2.5 mM of HU for up to 18 h, inhibiting DNA synthesis by >98% (Kung et al., 1993). In control cells, the mitotic index was 2%, and after HU treatment, the mitotic index was 0% (Table 1). When 5 mM caffeine was added for 4 h in addition to the HU treatment, a mitotic index of 3% was found. These results show that caffeine overrides the checkpoint induced by HU, leading to entrance into mitosis in the presence of incompletely replicated DNA. These results are in agreement with earlier studies (Schlegel and Pardee, 1986; Balczon et al., 1995; Wise and Brinkley, 1997).
Table 1.
CHO cells forced into mitosis in the presence of incompletely replicated DNA show a high percentage of abnormal mitotic spindles
| Percentage of cells in mitosis | Percentage of mitotic cells that are abnormal | Percentage of interphase cells with more than two centrosomes | |
|---|---|---|---|
| Control | 2% | 1% | 0% |
| 18 h HU | 0% | 1% | |
| 18 h HU + 4 h HU and caffeine | 3% | 70% | 2% |
| 4 h caffeine | 2% | 2% | 2% |
| 6 h HU | 0% | 2% | |
| 6 h HU + 4 h HU and caffeine | 1% | 31% | 2% |
| 6 h HU and 8 h recovery | 2% | 1% | 2% |
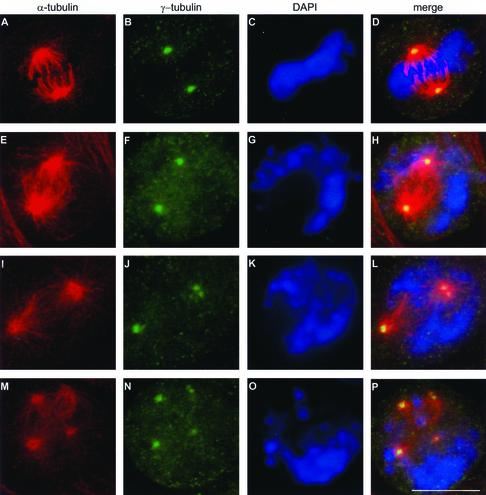
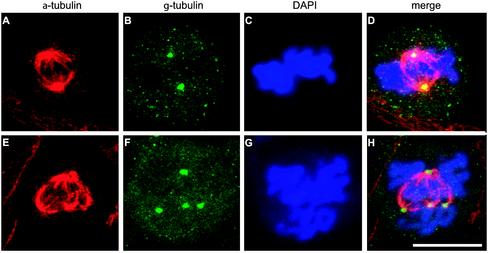
We then analyzed the spindle and centrosome morphology under these conditions. Control cells and cells treated with HU and caffeine were fixed and labeled with an antibody specific to α-tubulin and an antibody specific to γ-tubulin (centrosomal protein) (Oakley, 2000). The vast majority of spindles in control cells were bipolar, with γ-tubulin localized exclusively at the spindle poles and the chromosomes aligned in the metaphase plate, as previously described in numerous other reports (for review, see Karsenti and Vernos, 2001) (Figure 1, A–D). Cells treated with HU and caffeine showed three types of spindles: (1) bipolar spindles with γ-tubulin localized in one spot at each pole of the spindle (Figure 1, E–H), (2) bipolar spindles but with an abnormal localization of γ-tubulin detected in more than one spot at the spindle poles or localized anywhere along the spindle (Figure 1, I–L), and (3) multipolar spindles with γ-tubulin localized at every pole of the spindle (Figure 1, M–P). In all three types of spindles, DNA was never aligned properly in the metaphase plate. The same results were obtained when an anti-pericentrin antibody (Doxsey et al., 1994) was used as a centrosomal marker (our unpublished results). This showed not only that the localization of γ-tubulin was affected but that the localization of at least one other centrosomal protein was also influenced. These results indicated that a centrosome-like structure is present at sites at which the γ-tubulin was detected. Comparable results were found when aphidicolin was substituted for HU or when UCN-01 (a drug that is a potent overruler of the G2/M checkpoint) (Wang et al., 1996; Yu et al., 1998) was used instead of caffeine. Caffeine treatment alone had no effect on the mitotic spindles (Table 1). When a lower concentration of HU (0.5 mM) was used, a small population of cells entered mitosis without the use of caffeine (mitotic index, 0.8%). This suggests that low concentrations of HU are not sufficient to completely block cell-cycle arrest and that cells can enter mitosis in the presence of incompletely replicated DNA, even without the addition of caffeine. Under these conditions, the same types of abnormal spindles were observed. The results were also similar when other hamster cell lines (V79 or O23 cells) were used (our unpublished results). To summarize: these results indicated that the abnormal mitotic spindles were not the result of caffeine or UCN-01 but merely of DNA synthesis inhibitors.
Figure 1.
Treatment with HU and caffeine induces the formation of abnormal spindles. Control cells (A–D) and cells treated with HU for 18 h and subsequently with HU and caffeine for 4 h (E–P) were fixed and labeled with an antibody against α-tubulin (red) and an antibody against γ-tubulin (green). DNA was stained with DAPI (blue). Right, overlay of the three signals. A–D, Control cells showed bipolar spindles with γ-tubulin localized at the spindle poles and the DNA aligned in the metaphase plate. After HU and caffeine treatment, three types of abnormal spindles were observed: (1) bipolar spindles with one γ-tubulin–containing spot localized at each pole of the spindle poles (E–H); (2) bipolar spindles containing more than 2 γ-tubulin–containing spots (I–L); and (3) multipolar spindles with γ-tubulin–containing spots localized at every pole of the spindle (M–P). Bar, 10 μm.
Several reports have already demonstrated that a prolonged S-phase, induced by HU over a period of >40 h, allows multiple rounds of centrosome duplication and that after subsequent caffeine treatment, multipolar spindles and extra centrosome-like structures are observed (Balczon et al., 1995, 1999). These results indicate that the most plausible hypothesis is that multipolar spindles are the result of over-duplication of centrosomes in interphase. However, careful examination of the abnormal spindles, observed in Figure 1, leads us to suggest an alternative, although less obvious, sequence of events. The three types of observed spindles suggest that spindles may start as bipolar (Figure 1, E–H), progress toward bipolar with too many centrosome-like structures (Figure 1, I–L), and finally result in the formation of multipolar spindles (Figure 1, M–P). We therefore propose that the centrosomes do not necessarily have to duplicate in the preceding interphase but that centrosomes may break up in response to the presence of incompletely replicated DNA during mitosis. Subsequently, the fragmented centrosomes may give rise to the multipolar spindles.
Formation of Extra Centrosome-like Structures Can Occur during Mitosis
To test the theory that extra centrosome-like structures can develop during mitosis, we chose conditions in which centrosome reduplication did not occur in the preceding interphase, using short (6-h) incubation times with HU. After this incubation time with HU, the mitotic index was 0%, indicating that DNA synthesis was inhibited and that cell-cycle progression had been arrested. Such short HU incubations did not cause the formation of extra centrosomes during interphase. This was determined by antibody labeling of centrosomes (Table 1). It may be possible that the centrosomes duplicate during 6 h of HU treatment but that the centrosomes remain in close proximity and extra centrosomes become detectable only after the cells have been forced into mitosis. To exclude this possibility, we designed the following experiment: CHO cells were treated with HU for 6 h, followed by a recovery period without HU and caffeine. After 8 h of recovery, the cells started to enter mitosis, and almost no multipolar spindles or extra centrosome-like structures were observed. The spindles were almost exclusively bipolar (Table 1). These results show that a short HU treatment in itself does not alter centrosomes during interphase and does not induce the formation of multipolar spindles. Only when this short HU treatment was immediately followed by incubation with HU and caffeine for an additional 4 h was a high percentage of abnormal spindles with extra centrosome-like structures observed (Table 1, Figure 1). Together, these results strongly suggest that aberrant spindles, as observed under these conditions, are not the result of multiple centrosomes accumulated in interphase but that the entry into mitosis in the presence of incompletely replicated DNA may act as a trigger for the formation of multiple centrosome-like structures and multipolar spindles.
In Response to Incompletely Replicated DNA during Mitosis, Centrosomes Split into Fragments Containing Only One Centriole
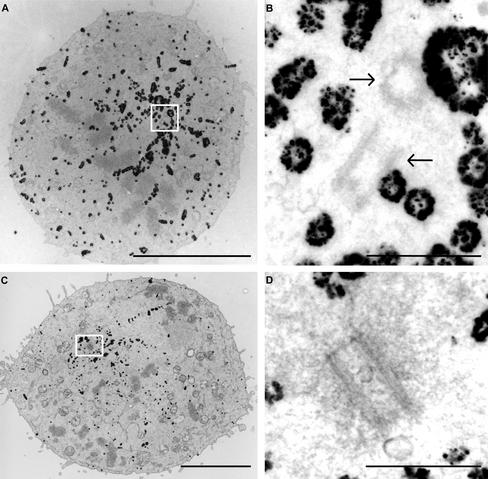
Centrosomes normally duplicate once per cell cycle around the G1/S transition. Our results demonstrate that centrosome numbers in interphase cells remain normal after 6 h of HU treatment. However, when cells are forced into mitosis, extra centrosomal structures arise. The extra centrosomal structures could be the result of centrosome duplication or fragmenting in response to the presence of impaired DNA integrity. Mitotic cells were examined at the ultrastructural level to investigate these possibilities further. Control cells and cells after treatment with HU and caffeine were fixed and labeled with α-tubulin antibodies, followed by immunogold labeling and silver enhancement. Immunogold labeling of the tubulin filaments allows preselection of mitotic cells at the light microscopic level. In addition, the prelabeling of the tubulin filaments allows fast determination of the position of the spindle poles at low magnifications at the electron microscopic level (Figure 2, A and C), and thus, centrosomes can be traced and analyzed relatively quickly. Serial sections were made of the selected cells, and centrosomal structures were examined. In almost every spindle pole of the control cells (8/9), two centrioles were detected in the same section (Figure 2, A and B) or two centrioles were visible in consecutive sections, indicating that centrioles are in close proximity under control conditions in most mitotic cells, as previously described (for review, see Rieder et al., 2001). In contrast, in most (18/23) spindle poles of multipolar spindles after HU and caffeine treatment (Figure 2, C and D), only one centriole could be found, and no other centrioles were observed in consecutive sections. In addition, centriole numbers of centrosomes in interphase cells containing one nucleus (see Discussion) in HU- and caffeine-treated samples were also analyzed, and none (0/43) of these interphase cells showed more than four centrioles or more than two centrosomes.
Figure 2.
When mitosis is initiated in the presence of incompletely replicated DNA, centrosomes split into fragments containing only one centriole. Electron microscopic analysis of mitotic cells under control conditions (A and B) and after HU and caffeine treatment (C and D). Fixed cells were labeled with an α-tubulin antibody, followed by immunogold labeling and silver enhancement. Mitotic cells were preselected by light microscopy and subsequently processed for ultrastructural analysis. The gold particles decorating the tubulin filaments are clearly visible at low magnifications of the sections (A and C) and allow fast identification of the spindle poles. Control mitotic cells show two centrioles (arrows) per spindle pole (A and B), whereas most mitotic cells after HU and caffeine treatment show only one centriole per pole (C and D). Bars: (A and C) 5 μm and (B and D) 500 nm.
This ultrastructural analysis confirmed that 6-h HU treatment does not alter interphase centriole and centrosome numbers. However, when entrance into mitosis is initiated in the presence of incompletely replicated DNA, extra centrosome-like structures are formed. These extra centrosome-like structures are not the result of centrosome duplication but arise because of the splitting of the centrosomes in fragments containing only one centriole.
In Vivo Analysis Shows That Bipolar Spindles Convert into Multipolar Spindles Early in Mitosis When Incompletely Replicated DNA Is Present
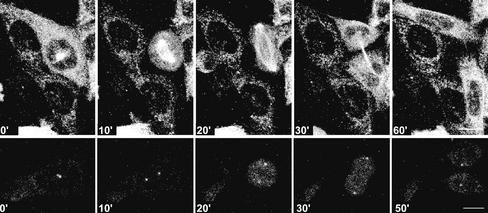
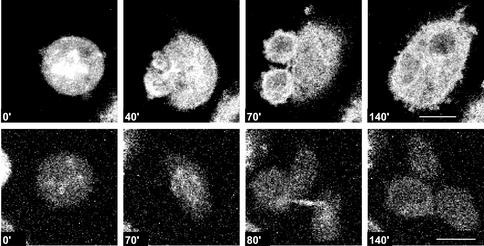
It has been demonstrated previously that a severe mitotic delay can induce centrosome splitting in monocentriolar centrosomes (Keryer et al., 1984; Gallant and Nigg, 1992; Hinchcliffe et al., 1998). It may therefore be possible that the presence of incompletely replicated DNA induces a mitotic delay that in turn is the trigger for centrosomal splitting. However, the alternative may also be possible, that impaired DNA integrity itself elicits centrosome splitting. To distinguish between these two possibilities, CHO cell lines were generated that stably express EGFP–α-tubulin or γ-tubulin–GFP, and live analyses were performed. Using these cell lines, every centrosome-like structure or γ-tubulin–containing spot and α-tubulin–containing structure were followed in time, in parallel, with the corresponding cell morphology. Mitosis was defined as the time that a cell was rounded up and the nuclear envelope was absent. The presence in interphase cells and absence in mitotic cells of the nuclear envelope can be followed in the live recordings by the exclusion of the GFP signal from the nucleus (interphase) or the presence of the GFP signal in the nucleus (mitosis). A normal mitosis of an EGFP–α-tubulin–expressing cell was defined as the “rounding up” of the cells, followed by the formation of a bipolar mitotic spindle. Subsequently, a midbody develops, and two equal-size “rounded-up” daughter cells appear that reflatten and remain separated (Figure 3) (http://coo.med.rug.nl/sscb/film/film.htm, Figure 1). The average time from the appearance of a mitotic spindle to the formation of two daughter cells was 17 min (n = 82). In γ-tubulin–GFP–expressing control cells, centrosomes were clearly visible as one, or two in close proximity, γ-tubulin–GFP–containing spots. On nuclear envelope breakdown, they migrate away from each other and remain visible as two defined diametrically opposed spots during mitosis (Figure 3). In control cells, 82 mitotic cells were observed, 81 of which proceeded normally through mitosis. The cell divisions observed in the control situation were highly comparable to our observations in fixed samples and to the results described by others using both live recordings (Khodjakov and Rieder, 1999; Sibon et al., 2000; Rusan et al., 2001) and fixed samples (Zheng et al., 1991; Karsenti and Vernos, 2001, and references therein). This implies that the intensity of the laser beam was not causing significant cell-cycle arrest and predominantly allowed mitoses to occur normally. Higher intensities of the laser beam resulted in improved quality of the live images. However, these higher laser intensities caused a cell-cycle arrest in most of the recorded cells.
Figure 3.
Time-lapse confocal microscopy allows normal cell-cycle progression of CHO cells. Time-lapse confocal analysis of CHO cells expressing EGFP–α-tubulin (upper row) or γ-tubulin–GFP (lower row) under control conditions. Time progression is shown in minutes. Bar, 10 μm.
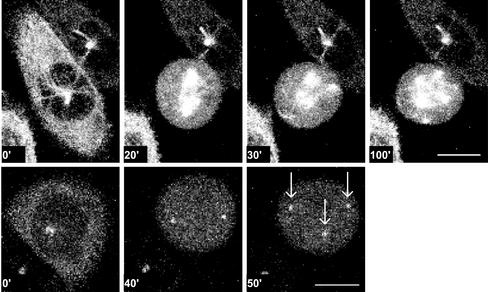
After HU and caffeine treatment, 55 mitoses were analyzed; 22 mitoses of EGFP–α-tubulin–expressing cells and 33 mitoses of γ-tubulin–GFP–expressing cells. Live recordings performed with the EGFP–α-tubulin–expressing CHO cells revealed that in most cases (18/22), a bipolar spindle was formed initially that subsequently developed into a multipolar spindle during mitosis (Figure 4). In some mitotic cells (3/22), a multipolar spindle was observed without the previous detection of a bipolar spindle. It was unclear whether the bipolar spindle phase occurred in the 10-min interval between two recorded images or whether a bipolar spindle was never really formed. Live recordings using the γ-tubulin–GFP–expressing CHO cells revealed that in many cases (17/33), two centrosomes were visible early in mitosis and extra centrosome-like structures were formed during this mitosis (Figure 4). In some cells (12/33), two centrosome-like structures were visible throughout the entire mitosis, and in some cells (4/33), extra centrosome-like structures were already observed in the first recording in mitosis, whereas only two centrosome-like structures were observed in the preceding interphase. These data confirm our idea that when extra centrosome-like structures appear, they appear during mitosis rather than in the preceding interphase.
Figure 4.
In cells treated with HU and caffeine, extra centrosome-like structures and multipolar spindles arise early in mitosis. Time-lapse confocal analysis of CHO cells expressing EGFP–α-tubulin or γ-tubulin–GFP after HU and caffeine treatment. (upper row) EGFP–α-tubulin–expressing cells after HU and caffeine treatment showed that several mitoses started with a bipolar spindle that converted into a multipolar spindle. (lower row) γ-tubulin–GFP–expressing cells after HU and caffeine treatment showed that several mitoses started with two centrosomes, progressing to the formation of extra centrosome-like structures (arrows). Time points are indicated in minutes. Bar, 10 μm.
The formation of these extra centrosome-like structures after HU and caffeine treatment already became visible, on average, within the first 18 min after entrance into mitosis, showing that these structures appeared within the time frame of a normal mitosis (17 min, n = 82). Live recordings also revealed that cells with multipolar spindles and extra centrosome-like structures showed a severe mitotic delay. Most mitoses (52/55) lasted >60 min. The observation that extra centrosome-like structures had already appeared early during mitosis excludes the theory that the mitotic delay is the trigger for the centrosome splitting. A more plausible hypothesis is that multipolar spindles or the presence of DNA replication defects causes a mitotic delay.
Cell-Cycle Progression Does Occur in Mitotic Cells with Abnormal Spindles
We then performed experiments in an attempt to address the question of whether mitotic cells with abnormal spindles progress into anaphase and proceed through cytokinesis or whether an internal checkpoint mechanism prevents cell-cycle progression in the presence of aberrant spindles. Live recordings of CHO cells incubated with HU and caffeine showed that the abnormal multipolar spindles cause a severe mitotic delay. Most mitoses (52/55) lasted >60 min, but none of the cells arrested completely in mitosis. While cells progressed through mitosis, multiple lobes were formed, followed by a complete or incomplete cytokinesis (Figure 5). Forty-two mitotic cells with multipolar spindles (using the EGFP–α-tubulin clone) or with extra centrosome-like structures (using the γ-tubulin–GFP clone) were followed in time. Eleven mitoses resulted in the collapse of the daughter cells after cytokinesis, and one cell with two or multiple nuclei was formed (Figure 5). Eighteen mitoses resulted in the formation of two daughter cells of equal or unequal size (http://coo.med.rug.nl/sscb/film/film.htm, Figure 2). Thirteen mitoses resulted in the formation of more than two daughter cells (Figure 5 and for additional information, http://coo.med.rug.nl/sscb/film/film.htm, Figure 3). Especially the finding that more than two daughter cells can arise provides evidence that aneuploid cells must have been formed. Live recordings performed with low concentrations of HU and in the absence of caffeine showed similar results (our unpublished data), indicating that the progression of the cell cycle in the presence of aberrant spindles and the possible outcomes are not influenced or forced by caffeine.
Figure 5.
Cells with multipolar spindles and extra centrosome-like structures do progress through the cell cycle and can form aneuploid cells. Time-lapse confocal analysis of CHO cells expressing EGFP–α-tubulin after HU and caffeine treatment. Cells were treated with HU and caffeine, and time-lapse recordings were made of mitotic cells with multipolar spindles. After a delayed mitosis and cytokinesis, some of the daughter cells collapsed back to one cell (upper row). Several of the cytokineses resulted in the formation of more than two daughter cells (lower row). Time points are indicated in minutes. Bar, 10 μm.
To summarize, cells with multipolar spindles show a mitotic delay; however, cell-cycle progression is not blocked. Cytokinesis can occur that can finally result in the formation of cells with multiple nuclei or can result in the formation of aneuploid cells.
Formation of Extra Centrosome-like Structures Requires Microtubulin Filaments but Is Independent of Actin Filaments
We then went on to test whether the formation of extra centrosome-like structures was dependent on the presence of microtubules. To do this, cells were forced into mitosis in the presence of incompletely replicated DNA with treatment with HU for 6 h and caffeine and colcemid for 4 h. The PH3 antibody was used as a marker for mitotic cells, and a pericentrin antibody was used as a centrosomal marker. None (0/100) of the PH3-positive cells showed more than two centrosomes or centrosome-like structures. When cytochalasin D was used instead of colcemid, multipolar spindles with extra centrosome-like structures were observed.
These results show that in the presence of incompletely replicated DNA, centrosome splitting depends on the presence of microtubulin filaments but is not dependent on the presence of actin filaments.
Formation of Extra Centrosome-like Structures and Multipolar Spindles Also Occurs When MMC–Treated Cells Enter Mitosis
To examine whether centrosomal changes would also occur in the presence of other types of DNA damage, CHO cells were treated with the interstrand DNA cross-linking compound MMC for 1 h. The cells were then fixed at various time points after treatment, and spindle morphology was analyzed. Mitotic cells in samples fixed and labeled with α-tubulin and γ-tubulin antibodies immediately after the MMC treatment showed a normal morphology indistinguishable from those observed in Figure 1, A–D (Figure 6, A–D). This indicates that MMC itself does not alter microtubulin filaments or spindle morphology. MMC did not provoke a cell-cycle arrest in CHO cells, and as time progressed, the number of multipolar spindles increased after the MMC treatment even in the absence of caffeine or UCN-01 (Figure 6, E–H). In all multipolar spindles (50/50), chromosome-congression defects were observed (Figure 6G), whereas in a bipolar spindle, in most cases (29/50), the DNA was not aligned. Live analysis of MMC-treated cells confirmed that cells entered mitosis predominantly with bipolar spindles. In a small percentage of mitotic cells observed (4/65), the bipolar spindles converted into multipolar spindles with extra centrosome-like structures early in mitosis. In some mitotic cells (6/65), a multipolar spindle was formed without the detection of a previous bipolar spindle. It is again unclear whether the bipolar spindle phase occurred in the 10-min interval between the two recorded images or whether the bipolar spindle was never really formed. Progression through mitosis subsequently occurred, and mitotic exit was comparable to that with HU- and caffeine-treated cells, although the formation of more than two daughter cells and the collapse of the daughter cells was observed in a smaller percentage of the cells. Mitosis of cells with multipolar spindles lasted, on average, 32 min (n = 10). These results show that splitting of centrosomes also occurs when DNA damage persists in mitosis. These data further support our theory that centrosome splitting is not the result of a mitotic delay but rather is merely the consequence of impaired DNA integrity present during mitosis.
Figure 6.
Treatment with MMC induces the formation of abnormal spindles. Cells were treated with MMC and fixed, and spindle morphology was analyzed by use of α-tubulin (A) and γ-tubulin (B) antibodies immediately after the MMC treatment. Under these conditions, only normal spindles were observed. Abnormal spindles were observed in mitotic cells fixed after a 24-h recovery after MMC treatment and visualized with α-tubulin (E) and γ-tubulin (F) labeling. In addition, cells were labeled with DAPI (C and G). (D and H) Overlay of the three signals. Bar, 10 μm.
A Mutant Defective in Homologous Recombination Shows the Formation of Comparable Extra Centrosome-like Structures and Aberrant Spindles in Mitosis
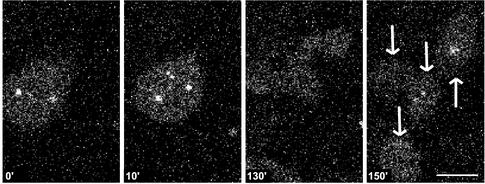
Finally, we speculated that if impaired integrity of the DNA coincides with the formation of aberrant spindles, mutant cell lines carrying a mutation in one of the DNA repair genes might spontaneously show an increased percentage of these abnormal spindles compared with normal cells. To test this hypothesis, irs1SF cells carrying a mutation in the XRCC3 gene that is involved in homologous recombination were used. It had been reported previously that in this cell line, the percentage of mitotic cells but not interphase cells with too many centrosomes is higher than in control cells (Griffin et al., 2000). Irs1SF cells were generated that express EGFP–α-tubulin or γ-tubulin–GFP, and these were analyzed in vivo. In nontreated cells, we found that in 10 of 123 cases, extra centrosome-like structures recorded in γ-tubulin–GFP–expressing cells or multipolar spindles recorded in EGFP–α-tubulin–expressing cells were formed during mitosis. In some mitotic cells (10/123), a multipolar spindle was formed without the detection of a previous bipolar spindle. Compared with the results obtained in CHO cells after HU and caffeine treatment, the extra centrosome-like structures appeared within the time frame of a normal mitosis. A number of these abnormal mitoses (4/20) resulted in the formation of more than two daughter cells (Figure 7 and http://coo.med.rug.nl/sscb/film/film.htm, Figure 4).
Figure 7.
A DNA repair-defective mutant (irs1SF) shows extra centrosome-like structures spontaneously during mitosis that can result in the formation of aneuploid cells. Time-lapse confocal analysis of irs1SF cells expressing γ-tubulin–GFP. Several of the mitoses recorded spontaneously showed the formation of extra centrosome-like structures during mitosis, and some of the cells with extra centrosome-like structures finally formed more than two daughter cells (arrows) as shown here. Time points are indicated in minutes. Bar, 10 μm.
These results show that during mitosis, multipolar spindles or extra centrosome-like structures were formed spontaneously in a repair-defective cell line and that the aberrant centrosome-like structures are not induced by a mitotic delay in this cell line. In addition to this, a fraction of the aberrant mitoses resulted in the formation of aneuploid cells.
DISCUSSION
In this study, we have shown that impaired DNA integrity coincides with centrosomal changes leading to extra centrosome-like structures and division errors. The changes in centrosomal organization occurred only in mitotic cells and not in interphase cells. However, we cannot exclude the possibility that the protein constitution of interphase centrosomes could change after HU or MMC treatment. Even if this occurs, our results show that this does not result in increased centrosome numbers, changes in morphology, or the behavior of the centrosomes during interphase. On the contrary, when mitosis is initiated in the presence of incompletely replicated or damaged DNA, early in mitosis centrosomes split into fragments containing one centriole, and multipolar spindles are formed. The centrosomal alterations in mammalian somatic cells we describe here differ from what was observed in Drosophila embryos, in which DNA replication defects occur consecutively with the disappearance of centrosomal proteins, for example, γ-tubulin, resulting in nonfunctional anastral spindles (Sibon et al., 2000). Although we currently have no explanation for this, it is not surprising that rapidly cycling syncytial Drosophila embryos (each cell-cycle time is ∼10 min, and the cell cycle consists of only S- and M-phase) show different responses to impaired DNA integrity compared with somatic mammalian cells with relatively long cell cycles (20 h). Despite the differences between the two systems, centrosomes of both organisms do change in the presence of DNA replication defects and DNA damage. Both centrosomal alterations can lead to the formation of collapsed nuclei and cells with extra centrosomal structures.
After HU or MMC treatment and spontaneously in the irs1SF cell line (our unpublished results and Griffin et al., 2000), multipolar spindles always coincide with chromosome-congression defects. Comparable chromosome-congression defects were observed in embryos of the Drosophila mutant grapes. Grapes mutant embryos enter mitosis prematurely and most likely in the presence of incompletely replicated DNA (Sibon et al., 2000). It may be possible that impaired DNA integrity interferes with DNA alignment at the metaphase plate, or it may be that multipolar spindles with split centrosomes are unable to align the DNA properly. We prefer the first explanation because in all bipolar spindles after HU treatment and in most bipolar spindles after MMC treatment, the chromosomes do not form a compact mass at the metaphase plate. These results suggest that chromosome-congression defects occur independently of centrosome splitting. In addition, it may be that chromosome-congression defects precede the splitting of centrosomes and precede the formation of multipolar spindles. To examine DNA-congression defects in relation to spindle abnormalities in more detail, live recordings are necessary to visualize the DNA and spindle/centrosomes simultaneously.
Our data suggest the existence of a signaling system between impaired DNA integrity and centrosomal organization in mitotic cells. The mechanism of this sensing system, however, remains elusive. Our data show that actin filaments are not involved but that microtubules may play a role. It may also be possible that microtubules are not required in the sensing mechanism per se but rather are involved only in splitting the centrosomes. The latter, however, seems unlikely, because it has been demonstrated by others that during interphase, centrosomal splitting induced by several protein kinases does occur in the absence of microtubules (Meraldi and Nigg, 2001).
Under control conditions, centrioles are kept together by a proteinaceous link (Paintrand et al., 1992). The tightness of this connection seems to be flexible, because the mother and daughter centriole separation occurs at specific points during the cell cycle. In Drosophila embryos, centriole splitting happens during mitotic exit, and interphase cells start with two centrosomes, each containing one centriole (Callaini et al., 1997). The anaphase-promoting complex activator Cdc20fizzy is either directly or indirectly required for this timely disengagement of centrioles in Drosophila embryos (Vidwans et al., 1999). In mammalian cells, mother and daughter centrioles move away from each other twice during the cell cycle. The first time occurs immediately after mitosis when the centriole pair splits. Before cytokinesis is completed and when daughter cells are still linked by a cytoplasmic bridge, the mother centriole transiently leaves the daughter centriole and moves to the intercellular bridge (Piel et al., 2000). Evidence has shown that proper cytokinesis depends on these movements of the mother centriole (Piel et al., 2001). A second splitting event of centrioles occurs later in G1, when a slight separation marks the first identifiable event of centrosome duplication (Hinchcliffe et al., 1999). Using Xenopus embryo extracts, several factors have been identified that play a role in centriole separation, such as Cul1, Skp1, and Cdk2 (Lacey et al., 1999; Freed et al., 1999).
Separation of the duplicated centrosomes and thus of the parental centrioles in G2/M is controlled by a balance between phosphatases and kinases, such as Nek2, PP1, and Cdc14A (Fry et al., 1998; Helps et al., 2000; Mailand et al., 2002; Meraldi and Nigg, 2002). Together, these results show that centriole and centrosome separation are under tight control, and factors have been identified that play a role in these timely events. Whether one of these factors is involved in centriole splitting early in mitosis in the presence of impaired DNA integrity still remains to be tested.
Cells with aberrant spindles somehow manage to progress through mitosis, indicating that there is no evidence of a checkpoint pathway that arrests progression through mitosis in the presence of aberrant spindles. These results are in agreement with an earlier report demonstrating that aberrant spindles do not themselves cause an arrest, whereas unattached kinetochores, for example, do lead to a delay in anaphase onset (Sluder et al., 1997). In contrast to the latter report, our results show that in the presence of multipolar spindles, mitosis is severely delayed. This is most likely a specific response to the presence of incompletely replicated or damaged DNA. DNA damage and DNA replication defects that persist in mitosis also coincide with a severe mitotic delay in Drosophila embryos (Sibon et al., 2000).
Mitotic exit of cells with abnormal spindles results in different outcomes, such as the formation of two daughter cells, two or more daughter cells that collapse, or the formation of more than two daughter cells that remain separated. The collapsed cells contain two or more nuclei and four centrioles. This will probably give rise to more than two centrosome-like structures in the next interphase. In fixed samples after HU and caffeine, it was indeed the case that an increased fraction of cells with more than one nucleus and more than two centrosomes was observed (our unpublished results). This corresponds with an earlier report that demonstrated that abortive cell division results in cells with two or more nuclei and more than two centrosome-like structures. This has been shown to be a major route to multiple centrosomes (Meraldi et al., 2002). The failed cytokinesis in the latter was caused by overexpression of Polo-like kinase 1 and Aurora-B, and in our report, the failed cytokinesis is induced by centrosome abnormalities observed in the presence of impaired DNA integrity.
The percentages of multipolar spindles and subsequently the formation of collapsed cells or aneuploid cells differ between the HU-treated cells and the MMC-treated cells and irs1SF cells. One possible explanation may be that 6 h of HU followed by caffeine treatment produces DNA defects in a population of partly synchronized cells. This results in a more severe phenotype and more pronounced centrosomal alterations compared with what occurs spontaneously in the repair-deficient cell line or what is induced after MMC treatment for 1 h. It might also be possible that there are differences in centrosomal responses between replication inhibition and DNA damage.
Our live analyses demonstrate that aneuploid cells can arise from cytokinesis of cells with multipolar spindles. Our observations did not allow us to follow the path of the aneuploid cells over more generations. However, it is likely that a small percentage of the aneuploid cells were able to survive and divide. In the irs1SF cell line, in which multiple centrosome-like structures arise spontaneously during mitosis and can therefore lead to the formation of more than two daughter cells, aneuploidy is indeed enhanced (Griffin et al., 2000). Although we cannot exclude the possibility that in this cell line, aneuploidy may be caused by additional mechanisms, the order of events as we describe them here might be a major contributor to the formation of viable aneuploid cells.
Extra centrosomes, multipolar spindles, and aneuploidy as we have reported here are three characteristics also observed in numerous tumor cells (Fukasawa et al., 1996; Lingle et al., 1998; Xu et al., 1999; Brinkley, 2001; Marx, 2001). However, the origin of multiple centrosomes was not captured by “live analysis” in any of the above-mentioned studies, and none of the data provided evidence as to whether multiple centrosomes are a cause or a consequence of aneuploidy. However, it is highly plausible that mutations in genes that normally regulate centrosome duplication, such as p53, lead to the formation of multiple centrosomes in interphase and that extra centrosomes in turn lead to the formation of multipolar spindles and subsequently aneuploidy (Fukasawa et al., 1996; Tarapore and Fukasawa, 2000; Tarapore et al., 2001). In addition to this, our studies suggest that extra centrosome-like structures and multipolar spindles can arise independently from mutations in genes that regulate centrosome duplication (see also Figure 8).
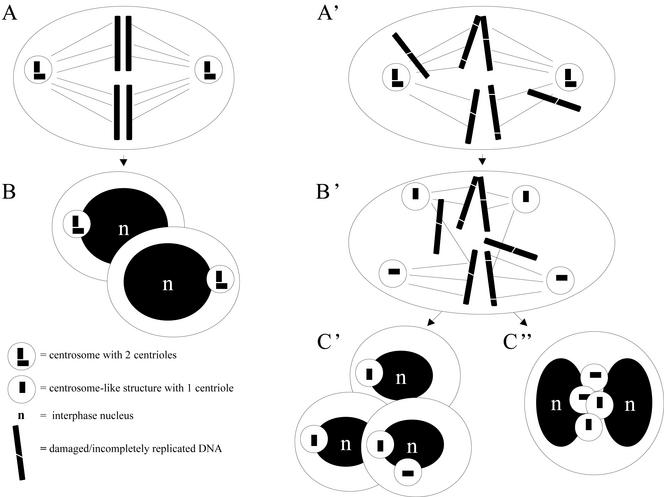
Figure 8.
Centriole splitting occurs in the presence of impaired DNA integrity during mitosis and may lead to aneuploidy. (A) Under control conditions, at mitosis, a bipolar spindle is formed containing two centrosomes located at the spindle poles, and the DNA is aligned in the metaphase plate. (B) Mitotic exit results in the formation of two identical daughter cells containing one centrosome each consisting of two centrioles. (A') In the presence of damaged or incompletely replicated DNA, mitosis starts with a bipolar spindle containing two centrosomes, and the chromosomes are not aligned in the metaphase plate. (B') Centrioles split early in mitosis, extra centrosome-like structures arise, and multipolar spindles are being formed. (C′) After a severe mitotic delay, cytokinesis can result in the formation of more than two daughter cells, and aneuploid cells are being formed, or (C”) a failed cytokinesis results in the formation of one cell with multiple nuclei and extra centrosome-like structures. In the absence of a functional checkpoint (e.g., in a P53-/- background), these polyploid cells may progress through the cell cycle (see also Borel et al., 2002; Meraldi et al., 2002), and after DNA replication and centrosome duplication, multipolar spindles will arise in the next mitosis, resulting in chromosome instability and aneuploidy.
We demonstrated that during mitosis, centrosomes can split into monocentriolar fragments and multipolar spindles can arise in the presence of impaired DNA integrity. This shows that not only interphase can be an initial source for the formation of extra centrosome-like structures. Despite the presence of multipolar spindles, cells do exit mitosis, and aneuploid cells and cells with multiple nuclei can arise. Our data provide a simple model for a possible order of events as to how impaired DNA integrity, numerous centrosomes, aberrant spindles, and aneuploidy are linked.
Supplementary Material
Acknowledgments
We thank A. Khodjakov for the γ-tubulin–GFP construct, M. Zdzienicka for the irs1SF cell line, and B.J.L. Eggen, W.E. Theurkauf, and B. Humbel for critical reading and helpful comments on the manuscript. The work was supported by grants from The Netherlands Organization for Scientific Research: NWO (901–01-221), and from the Interuniversitairy Institute for Radiopathology and Radiation Protection: IRS (7.2.6).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-08-0510. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-08-0510.
Online version of this article contains video material. Online version of this article is available at www.molbiolcell.org.
References
- Balczon, R., Bao, L., Zimmer, W.E., Brown, K., Zinkowski, R.P., and Brinkley, B.R. (1995). Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon, R., Varden, C.E., and Schroer, T.A. (1999). Role for microtubules in centrosome doubling in Chinese hamster ovary cells. Cell Motil. Cytoskeleton 42, 60–72. [DOI] [PubMed] [Google Scholar]
- Borel, F., Lohez, O.D., Lacroix, F.B., and Margolis, R.L. (2002). Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA 99, 9819–9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley, B.R. (2001). Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 11, 18–21. [DOI] [PubMed] [Google Scholar]
- Callaini, G., Whitfield, W.G., and Riparbelli, M.G. (1997). Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp. Cell Res. 234, 183–190. [DOI] [PubMed] [Google Scholar]
- Doxsey, S.J., Stein, P., Evans, L., Calarco, P.D., and Kirschner, M. (1994). Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639–650. [DOI] [PubMed] [Google Scholar]
- Elledge, S.J. (1996). Cell cycle checkpoints: preventing an identity crisis. Science 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Freed, E., Lacey, K.R., Huie, P., Lyapina, S.A., Deshaies, R.J., Stearns, T., and Jackson, P.K. (1999). Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13, 2242–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, A.M., Meraldi, P., and Nigg, E.A. (1998). A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa, K., Choi, T., Kuriyama, R., Rulong, S., and Vande Woude, G.F. (1996). Abnormal centrosome amplification in the absence of p53. Science 271, 1744–1747. [DOI] [PubMed] [Google Scholar]
- Gallant, P., and Nigg, E.A. (1992). Cyclin B2 undergoes cell cycle-dependent nuclear translocation and, when expressed as a non-destructible mutant, causes mitotic arrest in HeLa cells. J. Cell Biol. 117, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, C.S., Simpson, P.J., Wilson, C.R., and Thacker, J. (2000). Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2, 757–761. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H., and Weinert, T.A. (1989). Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634. [DOI] [PubMed] [Google Scholar]
- Helps, N.R., Luo, X., Barker, H.M., and Cohen, P.T. (2000). NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Bio-chem. J. 349, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., Cassels, G.O., Rieder, C.L., and Sluder, G. (1998). The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J. Cell Biol. 140, 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., Li, C., Thompson, E.A., Maller, J.L., and Sluder, G. (1999). Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283, 851–854. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., Miller, F.J., Cham, M., Khodjakov, A., and Sluder, G. (2001). Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science 291, 1547–1550. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., and Sluder, G. (2001). “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle: a self-made machine. Science 294, 543–547. [DOI] [PubMed] [Google Scholar]
- Keryer, G., Ris, H., and Borisy, G.G. (1984). Centriole distribution during tripolar mitosis in Chinese hamster ovary cells. J. Cell Biol. 98, 2222–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and Rieder, C.L. (1999). The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle do not require microtubules. J. Cell Biol. 146, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and Rieder, C.L. (2001). Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, A.L., Sherwood, S.W., and Schimke, R.T. (1993). Differences in the regulation of protein synthesis, cyclin B accumulation, and cellular growth in response to the inhibition of DNA synthesis in Chinese hamster ovary and HeLa S3 cells. J. Biol. Chem. 268, 23072–23080. [PubMed] [Google Scholar]
- Lacey, K.R., Jackson, P.K., and Stearns, T. (1999). Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 96, 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle, W.L., Barrett, S.L., Negron, V.C., D'Assoro, A.B., Boeneman, K., Liu, W., Whitehead, C.M., Reynolds, C., Salisbury, J.L. (2002). Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. USA 99, 1978–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle, W.L., Lutz, W.H., Ingle, J.N., Maihle, N.J., and Salisbury, J.L. (1998). Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA 95, 2950–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macville, M.V., Wiesmeijer, K.C., Dirks, R.W., Fransen, J.A., Raap, A.K. (1995). Saponin pre-treatment in pre-embedding electron microscopic in situ hybridization for detection of specific RNA sequences in cultured cells: a methodological study. J. Histochem. Cytochem. 43, 1005–1018. [DOI] [PubMed] [Google Scholar]
- Mailand, N., Lukas, C., Kaiser, B.K., Jackson, P.K., Bartek, J., and Lukas, J. (2002). Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 4, 317–322. [DOI] [PubMed] [Google Scholar]
- Marx, J. (2001). Cell biology: do centrosome abnormalities lead to cancer? Science 292, 426–429. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Honda, R., and Nigg, E.A. (2002). Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53(-/-) cells. EMBO J. 21, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., and Nigg, E.A. (2001). Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci. 114, 3749–3757. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., and Nigg, E.A. (2002). The centrosome cycle. FEBS Lett. 521, 9–13. [DOI] [PubMed] [Google Scholar]
- Oakley, B.R. (2000). γ-Tubulin. Curr. Top. Dev. Biol. 49, 27–54. [DOI] [PubMed] [Google Scholar]
- Paintrand, M., Moudjou, M., Delacroix, H., and Bornens, M. (1992). Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108, 107–128. [DOI] [PubMed] [Google Scholar]
- Piel, M., Meyer, P., Khodjakov, A., Rieder, C.L., and Bornens, M. (2000). The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, M., Nordberg, J., Euteneuer, U., and Bornens, M. (2001). Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550–1553. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., Faruki, S., and Khodjakov, A. (2001). The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 11, 413–419. [DOI] [PubMed] [Google Scholar]
- Rusan, N.M., Fagerstrom, C.J., Yvon, A.M., and Wadsworth, P. (2001). Cell cycle–dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell 12, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel, R., and Pardee, A.B. (1986). Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science 232, 1264–1266. [DOI] [PubMed] [Google Scholar]
- Sibon, O.C., Kelkar, A., Lemstra, W., and Theurkauf, W.E. (2000). DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2, 90–95. [DOI] [PubMed] [Google Scholar]
- Sluder, G., Thompson, E.A., Miller, F.J., Hayes, J., and Rieder, C.L. (1997). The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J. Cell Sci. 110, 421–429. [DOI] [PubMed] [Google Scholar]
- Smits, V.A., and Medema, R.H. (2001). Checking out the G2/M transition. Biochim. Biophys. Acta 1519, 1–12. [DOI] [PubMed] [Google Scholar]
- Stearns, T. (2001). Centrosome duplication: a centriolar pas de deux. Cell 105, 417–420. [DOI] [PubMed] [Google Scholar]
- Tarapore, P., and Fukasawa, K. (2000). p53 mutation and mitotic infidelity. Cancer Invest. 18, 148–155. [DOI] [PubMed] [Google Scholar]
- Tarapore, P., Horn, H.F., Tokuyama, Y., and Fukasawa, K. (2001). Direct regulation of the centrosome duplication cycle by the p53–p21Waf1/Cip1 pathway. Oncogene 20, 3173–3184. [DOI] [PubMed] [Google Scholar]
- Vidwans, S.J., Wong, M.L., and O'Farrell, P.H. (1999). Mitotic regulators govern progress through steps in the centrosome duplication cycle. J. Cell Biol. 147, 1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Fan, S., Eastman, A., Worland, P.J., Sausville, E.A., and O'Connor, P.M. (1996). UCN-01: a potent abrogator of G2 check-point function in cancer cells with disrupted p53. J. Natl. Cancer Inst. 88, 956–965. [DOI] [PubMed] [Google Scholar]
- Wise, D.A., and Brinkley, B.R. (1997). Mitosis in cells with unreplicated genomes (MUGs): spindle assembly and behavior of centromere fragments. Cell Motil. Cytoskeleton 36, 291–302. [DOI] [PubMed] [Google Scholar]
- Xu, X., Weaver, Z., Linke, S.P., Li, C., Gotay, J., Wang, X.W., Harris, C.C., Ried, T., and Deng, C.X. (1999). Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3, 389–395. [DOI] [PubMed] [Google Scholar]
- Yu, L., Orlandi, L., Wang, P., Orr, M.S., Senderowicz, A.M., Sausville, E.A., Silvestrini, R., Watanabe, N., Piwnica-Worms, H., and O'Connor, P.M. (1998). UCN-01 abrogates G2 arrest through a Cdc2-dependent pathway that is associated with inactivation of the Wee1Hu kinase and activation of the Cdc25C phosphatase. J. Biol. Chem. 273, 33455–33464. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Jung, M.K., and Oakley, B.R. (1991). Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell 65, 817–823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.