Abstract
Intraflagellar transport (IFT) is a process required for flagella and cilia assembly that describes the dynein and kinesin mediated movement of particles along axonemes that consists of an A and a B complex, defects in which disrupt retrograde and anterograde transport, respectively. Herein, we describe a novel Caenorhabditis elegans gene, xbx-1, that is required for retrograde IFT and shares homology with a mammalian dynein light intermediate chain (D2LIC). xbx-1 expression in ciliated sensory neurons is regulated by the transcription factor DAF-19, as demonstrated previously for genes encoding IFT complex B proteins. XBX-1 localizes to the base of the cilia and undergoes anterograde and retrograde movement along the axoneme. Disruption of xbx-1 results in cilia defects and causes behavioral abnormalities observed in other cilia mutants. Analysis of cilia in xbx-1 mutants reveals that they are shortened and have a bulb like structure in which IFT proteins accumulate. The role of XBX-1 in IFT was further confirmed by analyzing the effect that other IFT mutations have on XBX-1 localization and movement. In contrast to other IFT proteins, retrograde XBX-1 movement was detected in complex A mutants. Our results suggest that the DLIC protein XBX-1 functions together with the CHE-3 dynein in retrograde IFT, downstream of the complex A proteins.
INTRODUCTION
Transport mechanisms that utilize microtubule based kinesin and dynein motor proteins play a variety of important functions in cells such as movement of vesicles and organelles, chromosomal segregation, Golgi organization, and intraflagellar transport (IFT; Mitchell, 1994; Porter, 1996; Vaisberg et al., 1996; Hirokawa, 1998; Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999a; Goldstein, 2001). IFT is required for flagella and cilia assembly that describes the anterograde and retrograde migration of protein particles from the base to the tip of the cilia and back to the base, respectively (Signor et al., 1999a; Rosenbaum, 2002).
IFT appears to be a highly conserved process common to all ciliated eukaryotic organisms (Kozminski et al., 1993; Signor et al., 1999a; Haycraft et al., 2001; Rosenbaum, 2002). Through biochemical approaches in Chlamydomonas, many of the IFT proteins have been assigned to one of several substructures in the IFT particle including the kinesin-II complex, complex A, complex B, and the dynein motor complex (Cole et al., 1998; Pazour et al., 1999; Porter et al., 1999; Iomini et al., 2001). In Caenorhabditis elegans, mutations that disrupt proteins in the kinesin or complex B result in severely stunted cilia (Perkins et al., 1986; Collet et al., 1998; Haycraft et al., 2001). In contrast, mutations in complex A proteins or the dynein CHE-3 result in slightly shortened cilia axonemes with an accumulation of electron dense material along the axoneme compared with wild type (Perkins et al., 1986; Signor et al., 1999a). These data suggest that the kinesin-II and complex B proteins are required for the anterograde directed particle movement, whereas the complex A proteins and the dynein function in retrograde transport (Cole et al., 1998; Piperno et al., 1998; Signor et al., 1999a).
Dyneins are high-molecular-weight motor protein complexes that generate minus end directed movement along microtubules. There are two classes of dyneins: axonemal dyneins that are involved in cilia and flagella beating and cytoplasmic dyneins that are involved in IFT and intracellular trafficking (reviewed in Holzbaur and Vallee, 1994; Gibbons, 1995; Porter, 1996). Most dynein complexes consist of two heavy chains, two or more intermediate chains, several light intermediate chains, and numerous light chains. The heavy chains function as the ATPase and motor component, whereas the other accessory chains are thought to provide diversity through interactions with specific cargo molecules (Holzbaur and Vallee, 1994; Tynan et al., 2000b; Karcher et al., 2002).
Although the full composition of the dynein complex involved in IFT has yet to be determined, the dynein heavy chains have been identified in Chlamydomonas and C. elegans (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999a; Wicks et al., 2000). In Chlamydomonas, mutation of the dynein heavy chain (DHC1B) results in shortened flagella that exhibit IFT particle accumulation at the distal tips indicative of defective retrograde transport (Pazour et al., 1999; Porter et al., 1999). Similar results have been observed in C. elegans due to disruption of the dynein heavy chain CHE-3 (Perkins et al., 1986; Signor et al., 1999a; Wicks et al., 2000).
Herein, we describe a novel gene in C. elegans, xbx-1, that shares significant similarity with the recently identified mammalian dynein light intermediate chain (D2LIC; Grissom et al., 2002) as well as with a Chlamydomonas ortholog that functions in IFT (Perrone et al., 2003). Expression of xbx-1 is regulated by the DAF-19 transcription factor. The cilia of xbx-1(ok279) mutant worms are shortened and terminate in a bulb-like structure at the distal tip where IFT proteins accumulate. As is typical of proteins involved in IFT (Signor et al., 1999a; Haycraft et al., 2001; Qin et al., 2001), XBX-1::YFP localizes to the base of cilia and migrates along the axoneme in both anterograde and retrograde directions. In contrast to results obtained with other IFT proteins, retrograde movement of XBX-1::YFP was normal in complex A mutants. Together, these data suggest that the light intermediate chain subunit of the dynein complex, XBX-1, functions as part of the retrograde motor for IFT.
MATERIALS AND METHODS
General Molecular Biology Methods
General molecular biology procedures were performed according to standard protocols (Sambrook et al., 1989). Cloned worm DNA, total worm DNA, worm cDNA, or single worms were used for PCR amplifications, for direct sequencing, or for subcloning (Sambrook et al., 1989). Clones, primer sequences, and PCR conditions are available on request. DNA sequencing was performed either by MWG Biotech (http://www.mwg-biotech.com/html/index.shtml; Ebersberg, Germany) or the UAB Genomics Core Facility of the Heflin Center for Human Genetics.
DNA Sequence Analyses
Genome sequence information used in this study was obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), from the C. elegans Genome Sequencing Centers (http://www.sanger.ac.uk/Projects/C_elegans/; http://genome.wustl.edu/projects/celegans/; Consortium, 1998) or from the Celera Database (http://www.celera.com/). Gene identities were derived from the C. elegans database WormBase or references therein (http://www.wormbase.org/; Stein et al., 2001). BLAST and visual inspection were used to identify and evaluate orthologues of XBX-1 (http://www.ncbi.nlm.nih.gov/BLAST/; Altschul et al., 1997).
The C. elegans genome sequence wide search for putative transcriptional target genes of the RFX-type transcription factor DAF-19 was conducted using a specially designed computer search algorithm (K. Bubb and P. Swoboda, unpublished information), which searches for the X-box promoter element consensus sequence (Swoboda et al., 2000; Haycraft et al., 2001) upstream of the translational start site (ATG) of genes or predicted genes. F02D8.3 was one in a list of candidate genes that fit the experimental criteria and, when mutated, resulted in phenotypes that had previously been described for direct transcriptional DAF-19 targets (Swoboda et al., 2000; Haycraft et al., 2001). Thus, the gene F02D8.3 was renamed xbx-1 for X-box promoter element regulated gene.
Strains
General growth conditions for C. elegans strains were as described (Brenner, 1974). Strains were grown at 20°C unless stated otherwise. The wild-type strain was N2 Bristol. The following mutations were used: LG (linkage group) I: che-3(e1124), che-13(e1805); LG II: daf-19(m86), dpy-10(e128), unc-52(e444); LG III: dpy-1(e1), unc-32(e189); LG IV: daf-10(e1387), dpy-20(e1282), him-8(e1489), unc-24(e138); LG V: che-11(e1810), dyf-4(m158), dpy-11(e224), osm-6(p811), unc-76(e911), xbx-1(ok279); LG X: lin-15(n765), osm-5(m184). The following extrachromosomal arrays were used: saEx523, yhEx105, yhEx107, and yhEx109 were used for xbx-1::gfp expression experiments; yhEx19 was used for OSM-5::GFP localization analyses (Haycraft et al., 2001); yhEx80 was used for OSM-6::YFP localization studies; yhEx100 was used for XBX-1::YFP localization studies; myEx10 was used for CHE-11::GFP localization studies (Qin et al., 2001). All strains used and strain construction details are available on request.
Isolation, Genetic, and Molecular Characterization of the Deletion Allele xbx-1(ok279) V
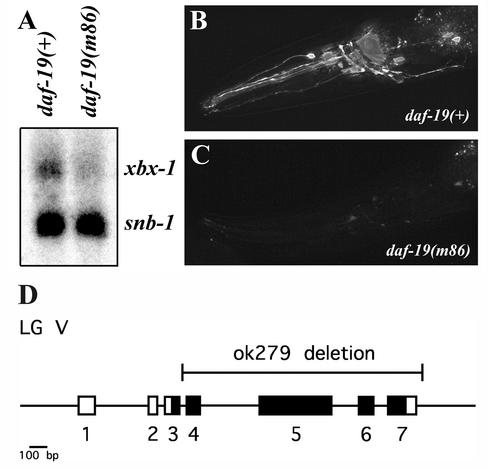
The deletion allele ok279 was generated by the C. elegans Gene Knockout Consortium (http://elegans.bcgsc.bc.ca/knockout.shtml) using publicly available methodology (http://www.mutantfactory.ouhsc.edu/protocols.asp; Anderson, 1995). The original mutated strain carrying the deletion allele ok279 was out-crossed seven times with N2 and CB2065: dpy-11 (e224) unc-76 (e911) V, eventually resulting in the homozygous mutant strain JT11069: xbx-1 (ok279) V, which was then used as the basis for all further analyses. Using standard genetic crossing methods and fluorescent dye filling assays we determined 1) that xbx-1(ok279) V is fully recessive and 2) that xbx-1(ok279) V complements another gene, dyf-4(m158) V, that maps nearby and results in a Dyf phenotype (Starich et al., 1995). In fluorescent dye filling assays the progeny of dyf-4(m158) heterozygous males crossed to xbx-1(ok279) homozygous hermaphrodites behaved similarly to wild-type and other control cross progeny (our unpublished results). Thus, xbx-1 and dyf-4 are different genes. After restriction enzyme mapping the deletion allele ok279 was DNA sequenced directly from purified bulk PCR products that spanned the deletion from both sides. The ok279 deletion extends over 1610 base pairs starting in the intron between exons 3 and 4 and ending 30 base pairs after the STOP codon (cosmid F02D8 base pairs 25954–27563 are deleted; Figure 1D).
Figure 1.
DAF-19 regulation of xbx-1 expression. (A) Northern blot analysis of xbx-1 expression in daf-19(+) and daf-19(m86) worms. xbx-1 expression is reduced in daf-19(m86) worms. The DAF-19 independent gene snb-1 was used as a loading control. (B and C) In vivo analysis of xbx-1 promoter driven GFP expression in (B) daf-19(+) and (C) daf-19(m86) mutant worms. xbx-1 is expressed in the ciliated sensory neurons of the wild-type worms. xbx-1 expression is significantly reduced in the absence of DAF-19. Anterior of the worm is directed toward the left. (D) Genomic organization of the xbx-1 gene (F02D8.3), which maps on linkage group (LG) V. Numbered boxes, exons; black boxes, exons with significant sequence similarities to DLIC proteins from other species (cf. Grissom et al., 2002, and Perrone et al., 2003). The region of the gene deleted in the xbx-1(ok279) mutant allele is shown above the schematic.
Assays
Fluorescent dye-filling assays were performed as described (Malone and Thomas, 1994; Starich et al., 1995; Fujiwara et al., 1999) using FITC (Sigma, St. Louis, MO), DiI-C12 or DiD (Molecular Probes, Inc., Eugene, OR). Adult hermaphrodites were observed at 1000× magnification by conventional fluorescence microscopy (Zeiss Axioplan 2, Carl Zeiss MicroImaging, Thornwood, NY) and at the highest magnification on a standard stereo dissecting microscope (Olympus Optical SZX12, Oympus America, Melville, NY) equipped with a fluorescent light attachment.
Osmotic avoidance assays were performed essentially as described (Culotti and Russell, 1978) by testing whether adult hermaphrodites placed in the center of a ring of high osmotic strength (8 M glycerol) will cross that ring or not during a time period of 10 min.
A semiquantitative assessment of male mating efficiency was made as described by Starich et al. (1995) with slight modification. Five L4 or young adult males were placed on a regular agar plate together with two L4 hermaphrodites, respectively. Hermaphrodites were one of the following two genotypes: 1) JT7273: unc-24(e138) dpy-20(e1282) IV; or 2) SP17: unc-32(e189) dpy-1(e1) III. For the mutations che-13 I, dyf-4 V and xbx-1 V, double mutants were constructed using him-8 IV, which then segregated homozygous mutant male progeny for mating tests. Numbers of cross-progeny were counted for each cross and compared with appropriate controls involving N2 or him-8 males. At least four separate crosses using two different types of marked hermaphrodites were performed for each mutant tested.
Generation and Analyses of the xbx-1::gfp Expression Constructs
A genomic fragment consisting of 2 kb of the promoter region upstream of xbx-1 was amplified from cloned worm DNA by PCR. The primers contained restriction enzyme sites for ligation into the GFP vector pPD95.77 (gift of A. Fire). The promoter, containing the wild-type X-box element, was fused in-frame to the GFP gene at codon ten of xbx-1. This fusion was introduced into worms by germline transformation at 100 ng/μl by using standard methods (Mello et al., 1991) using lin-15(+) at 60 ng/μl as a cotransformation marker DNA (Huang et al., 1994), and expression levels were analyzed as previously described (Swoboda et al., 2000; Haycraft et al., 2001).
Generation of Constructs and Strains Used for Localization Studies
For xbx-1 rescue experiments and analysis of XBX-1::YFP localization, a general YFP expression vector was derived from pPD95.81 vector (gift of A. Fire) by replacing GFP with YFP from pPD132.102 (gift of A. Fire) using the restriction enzymes NcoI and MfeI. The 250-base pair osm-5 promoter was cloned into this vector (Haycraft et al., 2001) along with the 2.2-kb xbx-1 or 3.1-kb osm-6 (Collet et al., 1998) coding region from N2 genomic DNA to create the osm-5::xbx-1::yfp or osm-5::osm-6::yfp expression vector, respectively. Genomic DNA for cloning was amplified using AccuTaq LA Polymerase Mix (Sigma-Aldrich, St. Louis, MO). Germline transformations were carried out as previously described (Mello et al., 1991). Wild-type or xbx-1(ok279) adult hermaphrodites were injected with 1–5 ng/μl test DNA and pRF4, which contains the dominant marker rol-6(su1006) (Mello et al., 1991). Transgenic worms were identified based on the right-handed roller phenotype (Rol) and maintained by picking Rol hermaphrodites.
To obtain transgenic mutant strains used for localization and cilia morphology analyses, adult Rol males carrying the desired extrachromosomal array were mated to homozygous mutant hermaphrodites. F1 hermaphrodites were screened for the Rol phenotype and allowed to self-fertilize. F2 hermaphrodites were screened for the presence of Rol and subsequently screened by fluorescent dye-filling to identify homozygous mutant hermaphrodites.
Imaging
For imaging, adult worms were anesthetized in 10 mM levamisole and mounted on 2% agar. Time-lapse imaging of worms expressing xbx-1::yfp was performed on an Olympus IX70 inverted microscope and captured with a Retiga 1300 cooled CCD camera (Qimaging, Burnaby, BC, Canada). Shutters and filters were computer driven. Images were acquired for at least 15 s at ∼2 frames/s using IPLab Spectrum 3.6 (Scanalytics, Fairfax, VA). Movies were then exported into Quicktime (Adobe Systems, Inc., San Jose, CA) and sequential still frames were taken from Quicktime movies. Localization and morphology images were captured using a Leica Confocal Imaging Spectrophotometer TCS SP unit mounted on a Leica DMIRBE inverted research microscope (Leica Microsystems, Bannockburn, IL). Further processing of images was done using Photoshop 6.0 and AfterEffects 6.0 (Adobe Systems, Inc., San Jose, CA). QuickTime (Adobe Systems, Inc.) movies were created at a rate of two frames per second.
Northern Blot Analysis
RNA for Northern blot analysis was isolated from mixed stage worms by addition of 5 volumes 5 M guanidine isothiocyanate followed by homogenization using a PowerGen 700 (Fisher Scientific, Pittsburgh, PA), centrifugation at 6000 × g to remove particulate matter, and purification over a CsCl cushion. Total RNA was purified over oligo-dT cellulose (Stratagene, La Jolla, CA) to obtain poly-A–enriched RNA. Radiolabeled xbx-1 and snb-1 (Nonet et al., 1998) probes were created using the Random Prime-A-Gene kit (Promega, Madison, WI) according to the manufacturer's instructions. Strains used for Northern analyses contained daf-12(sa204) X, which suppresses the Daf-c phenotype of daf-19(m86) worms.
RESULTS
Transcriptional Regulation of xbx-1
Several genes involved in IFT and ciliogenesis in C. elegans have been identified (Cole et al., 1998; Collet et al., 1998; Signor et al., 1999b; Wicks et al., 2000; Haycraft et al., 2001; Qin et al., 2001). In the case of the IFT complex B genes, all are regulated by the DAF-19 transcription factor (Swoboda et al., 2000; Haycraft et al., 2001). This regulation occurs through an X-box promoter element generally located within the first 150 nucleotides upstream of the translational start site (ATG). In a previous genome sequence–based search, the gene xbx-1 (F02D8.3) was identified as a candidate DAF-19 target because of the presence of an X-box sequence located at position –79 upstream of the predicted ATG (Swoboda et al., 2000).
XBX-1 shares homology with a recently identified mammalian dynein light intermediate chain (DLIC) known as D2LIC (Grissom et al., 2002). Putative orthologs are also present in Chlamydomonas, Drosophila, and Caenorhabditis briggsae (Perrone et al., 2003, and our unpublished results). Interestingly, a near consensus X-box sequence was also detected in the promoter region of the putative xbx-1 orthologs in human, C. briggsae, and Drosophila (Table 1). These data suggest that xbx-1 and its orthologs in higher eukaryotes are regulated though a common transcriptional mechanism. Intriguingly, cross-species comparisons have also identified similar promoter regions in genes encoding several other IFT complex B proteins (B.K. Yoder and P. Swoboda, unpublished results).
Table 1.
X-box sequence in the promoter region of xbx-1 and its putative orthologs
| Species | Locationa | X-box Sequenceb | |
|---|---|---|---|
| C. elegans | -79 to -66 ATG | G T T T C C AT | G G T A A C |
| C. briggsae | -93 to -80 ATG | G T T T C C AT | G G T T A C |
| Drosophila | -68 to -54 ATG | G T T G C T AGT | A G C A A C |
| Human | -67 to -54 ATG | G C T C C C AT | G G C A A C |
| Consensus X-box | G T N R C C N (0-3) | R G Y A A C | |
Note: The sequence of the five prime end of the mouse ortholog has not yet been completed and thus the presence of an X-box could not be evaluated.
ATG denotes translational start site.
R = G/A; Y = C/T; N = G/A/T/C; bold denotes a match to the mammalian X-box consensus sequence (Emery et al., 1996).
To determine whether xbx-1 is regulated by DAF-19, we compared the level of xbx-1 expression in wild-type and daf-19 mutant worms. By Northern blot analysis, we determined that the expression of xbx-1 was nearly abolished in the absence of DAF-19 (Figure 1A). To further establish the importance of DAF-19 in xbx-1 expression, we measured GFP expression in wild-type (daf-19(+)) and daf-19 mutant strains (daf-19(m86)) carrying the xbx-1 promoter region fused to the GFP gene (see MATERIALS AND METHODS). In wild-type worms, xbx-1::gfp expression is detected in ciliated sensory neurons and like the results obtained on the Northern blot, mutation of daf-19 caused a significant reduction in the level of xbx-1::gfp expression compared with wild-type N2 background (Figure 1, B and C, and Table 2).
Table 2.
xbx-1 expression analyses using an in vivo GFP expression assay
| Genotype | Wild type
|
daf-19(m86)
|
||||
|---|---|---|---|---|---|---|
| Strength of Expression | Strong | Weak | Absent | Strong | Weak | Absent |
| xbx-1::gfp | ||||||
| line 1 | 86 | 14 | 0 | 1 | 37 | 62 |
| line 2 | 66 | 31 | 3 | 1 | 33 | 66 |
| line 3 | 61 | 33 | 6 | 0 | 22 | 78 |
| line 4 | 51 | 41 | 8 | 0 | 3 | 97 |
Data are given as percent expression in ciliated sensory neurons in the head of adult animals. Only the ciliated sensory neurons adjacent to the nerve ring were examined. To classify expression as “strong,” “weak,” or “absent,” the number of neurons expressing xbx-1::gfp as well as the visibility of GFP in neuronal cell bodies and in processes extending to the tip of the nose were taken into consideration. Four independent transgenic lines were analyzed. Each row represents the data for one transgenic line. For each transgenic line (containing the wild-type X-box promoter element sequence) the transgene was also moved from wild type into a daf-19 mutant background. Eighty to 120 animals were analyzed for each transgenic line.
Generation and Characterization of the xbx-1 Deletion Mutant
To begin analyzing the role of xbx-1, the C. elegans knockout consortium screened a mutant library for worm strains carrying a deletion in xbx-1 (see MATERIALS AND METHODS). One allele, xbx-1(ok279), was identified that carried a 1610-base pair deletion starting in the middle of the third intron and ending 30 base pairs after the translational termination codon (base pairs 25,954–27,563 are deleted in the cosmid F02D8 sequence). The ok279 deletion would truncate the 369 amino acid XBX-1 protein at position 87 and therefore most likely represents a functional null allele (Figure 1D).
Ciliary and Sensory Defects in xbx-1 Mutants
Because mutations in several other X-box containing and DAF-19 regulated genes have been shown to result in ciliary and sensory defects (Collet et al., 1998; Swoboda et al., 2000; Haycraft et al., 2001), we analyzed xbx-1 mutant worms for such phenotypes. A first approach often used to evaluate whether sensory cilia are correctly formed is the fluorescent dye-filling assay. In wild-type worms, some of the sensory neurons that extend cilia through the cuticle into the environment allow fluorescent dye uptake into these sensory neurons. As is seen in other IFT mutants (Perkins et al., 1986; Collet et al., 1998; Haycraft et al., 2001), xbx-1(ok279) worms failed to absorb fluorescent dye (Dyf phenotype; Figure 2B and Table 3), suggesting that cilia are either malformed in xbx-1(ok279) worms or that the cilia do not extend through the cuticle.
Figure 2.
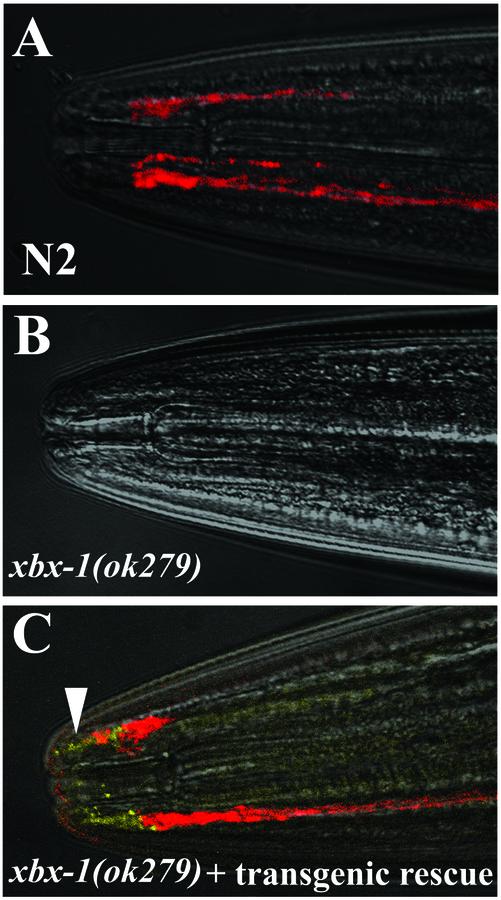
Restoration of cilia in xbx-1 mutants by transgenic rescue. All panels are bright field images overlaid with fluorescent images to show dye-filling (red) and XBX-1:: YFP localization (yellow). (A) Six pairs of amphid neurons of wild-type N2 worms fill with the fluorescent dye DiD. (B) In contrast, xbx-1(ok279) mutant worms fail to take up the dye. (C) Transgenic expression of xbx-1::yfp in xbx-1(ok279) mutant worms restores the ability to uptake fluorescent dye (red). The XBX-1::YFP protein is detected at the transition zones and within cilia on the ends of the amphid neurons (yellow, arrowhead). Anterior is toward the left in all panels.
Table 3.
Analysis of xbx-1(ok279) mutant phenotypes
| Genotype | Fluorescent dye-fillinga | Osmotic avoidanceb | Male mating efficiency (%)c |
|---|---|---|---|
| N2 (wild type) | + | + | 100 |
| che-13(e1805) | - | - | 0 |
| dyf-4(m158) | - | - | ≥100 |
| xbx-1(ok279) | - | - | 8 |
Using the fluorescent dyes FITC and DiI-C12, 100% of wild-type animals exhibited staining of 6 amphid neurons (occasionally an animal stained only 4 or 5) and 2 phasmid neurons, whereas all three mutant strains tested did not exhibit any staining, either in amphid or in phasmid neurons. At least 3 separate experiments were performed using a total of >50 adult hermaphrodites per strain.
Less than 5% of wild-type animals and >80% of all three mutant strains tested crossed a ring of high osmotic strength (8 M glycerol) during a time period of 10 min. At least 4 separate experiments were performed using a total of 55 to 90 adult hermaphrodites per strain.
Data are given as percent mating efficiency (ME) compared with N2 wild type (set to 100%). Combined data from two independent, representative crosses are shown. Percentages as shown translate to the standard C. elegans ME scale (cf. Starich et al., 1995) as follows: ME = 4 (30-100% as efficient mating as N2 wild type), ME = 3 (10-30% as efficient), ME = 2 (1-10% as efficient), ME = 1 (<1% as efficient), ME = 0 (no detectable mating). In repeated crosses xbx-1 mutant males displayed an ME = 2-3.
Defects in the cilia of the sensory neurons in C. elegans are associated with characteristic changes in behavior, including the inability to avoid substances of high osmotic strength (Osm phenotype), defects in chemotaxis (Che phenotype) toward attractants as well as a reduction in mating efficiency (Starich et al., 1995). To determine if the deletion of xbx-1 resulted in behavioral defects similar to that seen for other ciliogenic mutants, we conducted an osmotic avoidance assay. In this assay, we measured the frequency with which xbx-1(ok279) worms crossed a ring of 8 M glycerol on agar plates. Wild-type control N2 worms were found to cross the osmotic barrier at a rate of <5%. In contrast, worms lacking xbx-1 were found to cross the barrier at a rate >80%, a result similar to that seen for other mutants lacking sensory ciliary function such as che-13(e1805) and dyf-4(m158) (Table 3; Starich et al., 1995).
To further confirm the ciliary defects, we analyzed male xbx-1(ok279) worms for their mating efficiency. Male mating behavior, in particular the ability to locate the hermaphrodite vulva is mediated through cilia that extend off sensory neurons located in specialized rays in the male tail (Liu and Sternberg, 1995). Loss of cilia function has been shown to cause a significant reduction in mating efficiency (Starich et al., 1995). Our analysis of xbx-1 mutants revealed a mating efficiency significantly <30% as efficient as wild type (Table 3). For comparison, che-13(e1805) has a mating efficiency of 0%, and both dyf-4(m158) and wild-type N2 have mating efficiencies of 100%. The defects in xbx-1(ok279) mating efficiency are milder than in che-13(e1805), but more severe than in dyf-4(m158). Thus, xbx-1 mutants have a measurable sensory defect with regard to male mating.
Transgenic Rescue of xbx-1(ok279) Ciliary Defects
To confirm that the ok279 deletion is responsible for the defects in xbx-1 mutants, we constructed xbx-1 mutant lines that express the wild-type xbx-1 gene fused to the yellow fluorescent protein (YFP) as a transgene. Expression in these transgenic strains was under control of the DAF-19 regulated osm-5 promoter (Haycraft et al., 2001), which, like the xbx-1 promoter, drives expression in many ciliated sensory neurons. The effect on the cilia phenotype was evaluated using the fluorescent dye-filling assay. As indicated previously, xbx-1 mutants were not able to absorb dye (Figure 2B), however, all of the rescued lines tested (n = 4) were able to absorb dye, indicating the restoration of normal cilia (Figure 2, A and C). These data verify that the ok279 deletion is responsible for the xbx-1 phenotype.
Localization and Movement of XBX-1 Protein within Cilia
To analyze the possible role of XBX-1 in the IFT process, we utilized transgenic worm strains expressing XBX-1 protein tagged with YFP to evaluate XBX-1 localization and movement within sensory neurons. The XBX-1::YFP protein was detected specifically at the transition zone at the base of the cilia and in the axoneme (Figure 2C), consistent with a role in IFT. Furthermore, using time-lapse fluorescence microscopy, XBX-1::YFP particles were detected migrating along the cilium axoneme in both anterograde and retrograde directions similar to known IFT proteins (Figure 3; Signor et al., 1999a; Haycraft et al., 2001; Qin et al., 2001). These results, along with the ciliary defects observed in xbx-1 mutants, demonstrate a role for XBX-1 in the IFT process.
Figure 3.
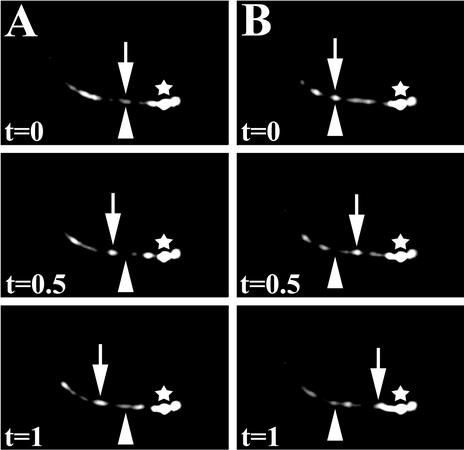
Movement of XBX-1::YFP along the cilia axoneme in phasmid neurons of a wild-type hermaphrodite. Using time-lapse fluorescence image analysis, XBX-1::YFP movement was observed in both (A) anterograde and (B) retrograde directions similar to that seen for other IFT proteins (Signor et al., 1999a; Haycraft et al., 2001; Qin et al., 2001). The transition zone is marked (*). The arrowhead indicates the initial position of the particle at time zero (t = 0). The arrows indicate the same particle at subsequent times (t = 0.5 s, and t = 1 s). The distal tips of the phasmid cilia are directed toward the left. Three sequential frames from Movie 1 are shown for each.
xbx-1 Mutants Display Specific Ciliary Defects
There is a large collection of mutations that disrupt cilia formation in C. elegans, some of which impinge directly on the IFT process. The ciliary phenotype in many of these IFT mutants has been extensively analyzed by morphological description at the level of serial section transmission electron microscopy. Interestingly, the differences in the cilia morphology observed in the IFT mutants correlate nicely with whether the mutation disrupts a protein that functions as part of complex A, complex B, or the dynein motor complex. For example, in osm-1, osm-5, osm-6, and che-13 mutants, the cilia are severely stunted and have ectopic posteriorly directed cilia-like projections (Perkins et al., 1986). The analysis of the corresponding proteins affected in these mutants revealed that they are all orthologs of IFT complex B proteins identified biochemically in Chlamydomonas (Cole et al., 1998; Qin et al., 2001). The cilia phenotype exhibited by complex B mutants is supportive of their function in anterograde transport because the IFT particles fail to enter and migrate from the base to the distal tip of the cilia but rather accumulate at the transition zones. In the daf-10 and che-11 complex A mutants, cilia were slightly stunted relative to wild type, and contained dense material interspersed throughout swollen axonemes (Perkins et al., 1986; Qin et al., 2001). In the che-3 dynein heavy chain mutant, the cilia axoneme is shorter than that of the complex A mutants with a similar accumulation of dense material in the swollen tip (Perkins et al., 1986; Signor et al., 1999a). In both the che-3 and complex A mutant worms as well as in complex A mutants in Chlamydomonas the phenotype has been attributed to defects in retrograde transport (Piperno et al., 1998; Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999a).
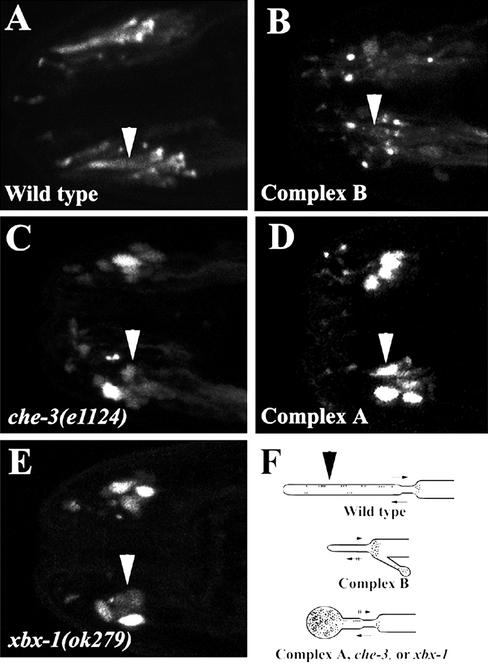
The correlation between the cilia morphology and the function of the protein affected by the mutation suggests that we may be able to glean insight into the role of XBX-1 by comparing its ciliary phenotype to that seen in other known mutants that affect complex A, complex B, or the CHE-3 dynein. To conduct this analysis, we used fluorescence-tagged OSM-5 or OSM-6 (complex B) proteins to visualize the cilia morphology in wild-type, xbx-1(ok279), che-11(e1810) (complex A mutant), osm-5(m184) (complex B mutant), and che-3(e1124) (dynein heavy chain mutant) worms. In wild-type worms, OSM-5::GFP localizes to the base of cilia and within cilia (Figure 4A). OSM-6::YFP shows an identical localization in wild-type worms (Collet et al., 1998; Signor et al., 1999a; and our unpublished results). Similar to the analysis described at the electron microscopy level, we could clearly distinguish between cilia morphology in mutants affecting complex B relative to that in complex A or the CHE-3 dynein using this G/YFP fusion protein approach (Figure 4). However, it was difficult to distinguish between the cilia morphology seen in complex A and the che-3 mutants that both affect retrograde transport (Figure 4, C and D). Interestingly, our analysis of the xbx-1(ok279) worms indicated that the cilia morphology did not resemble those typified by complex B mutants. Rather, the cilia morphology in xbx-1(ok279) worms was indistinguishable from that seen in the complex A or che-3 mutants (Figure 4, C–E). The cilia were stunted and contained massive accumulation of OSM-5::GFP in a bulb at the distal cilia tip. These results were surprising since there is an X-box sequence in the promoter of xbx-1. To date, X-box sequences have been identified in the promoter regions of complex B genes but not in the promoter regions of complex A genes or the IFT dynein (Swoboda et al., 2000; Haycraft et al., 2001). However, the ciliary phenotype of the xbx-1 mutant supports a role for XBX-1 in retrograde IFT similar to that of the complex A proteins and CHE-3, but not in anterograde transport as proposed for the complex B proteins. Furthermore, the strong sequence similarity between XBX-1 and the DLIC protein D2LIC (Grissom et al., 2002) is also suggestive of a role for XBX-1 in conjunction with CHE-3 in retrograde movement.
Figure 4.
Analysis of cilia morphology in wild-type and IFT mutants using fluorescence-tagged IFT proteins. (A) OSM-5::GFP in wild-type worms shows full-length cilia with OSM-5::GFP localized at the transition zones and within cilia. (B) Complex B mutants (osm-5 (m184)) exhibit severely truncated cilia axonemes as visualized by localization of OSM-6::YFP to the base of the truncated cilia. (C) Because of loss of retrograde IFT movement in che-3 mutants, the cilia are severely shortened and exhibit a significant accumulation of OSM-5::GFP along the enlarged cilia axonemes. (D) The cilia in complex A mutants (che-11(e1810)) as determined by using OSM-5:: GFP are similar to those seen in the che-3 mutants. The cilia are truncated and show significant accumulation of OSM-5::GFP along the swollen cilia axonemes. (E) Analysis of the cilia morphology in xbx-1 mutants using OSM-5::GFP. Similar to that seen in both che-3 and complex A mutants, the cilia of xbx-1 mutants were truncated and OSM-5::GFP concentrated along the enlarged axonemes. (F) Schematic diagram depicting the cilia morphology as determined using the IFT::G/YFP fusion proteins. Arrows pointing right indicate retrograde movement while arrows pointing left indicate anterograde movement. Two lines drawn through the arrows indicate defects in transport. Images A–E are 2-D confocal projections from the amphid region of the worm. In all panels, arrowheads point to the cilia axonemes of one bundle of amphid neurons. The distal end of the cilium is directed toward the left in all panels.
Effect of xbx-1 Mutation on the Localization of IFT Proteins
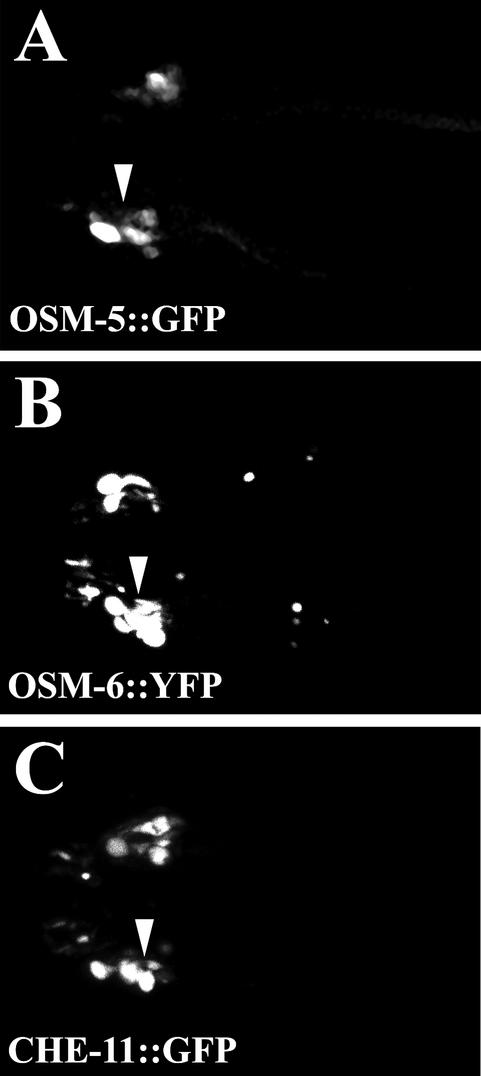
To evaluate what effect the loss of XBX-1 protein has on localization and movement of other proteins in the IFT particle, we moved extrachromosomal arrays that express known IFT proteins tagged with GFP or YFP into the xbx-1 mutant background. Included in this analysis were two complex B proteins, OSM-5 and OSM-6, and a complex A protein, CHE-11. Because of the large size of che-3 (∼12 .4 kb transcript) we were unable to generate a GFP fusion protein to include in the analyses. As seen for OSM-5::GFP in che-3 mutants (see Figure 4C), all the IFT proteins analyzed were found to accumulate in the bulb structure at the distal end of the cilia in xbx-1 mutants, further supporting a role for XBX-1 in retrograde movement (Figure 5). Because all of the IFT proteins analyzed here enter the cilia efficiently, the data suggest that anterograde movement in the xbx-1 mutants is not overtly affected and that XBX-1 function is not required for their assembly into the IFT particle at the base of cilia.
Figure 5.
Localization of IFT proteins in xbx-1 mutants. To evaluate the effect that the xbx-1(ok279) mutation has on the localization of IFT proteins, we crossed transgenic lines expressing either complex B or complex A IFT proteins fused to either GFP or YFP into the xbx-1 mutant background. The loss of XBX-1 results in accumulation of both complex A and B proteins along the axoneme as shown using (A) OSM-5::GFP (complex B), (B) OSM-6::YFP (complex B), and (C) CHE-11::GFP (complex A). Arrowheads indicate the swollen cilia axonemes. Anterior of the worm and the distal tips of the cilia are directed toward the left. All panels are 2-D confocal projections.
The Effect of Mutations in Other IFT Proteins on Localization of XBX-1::YFP
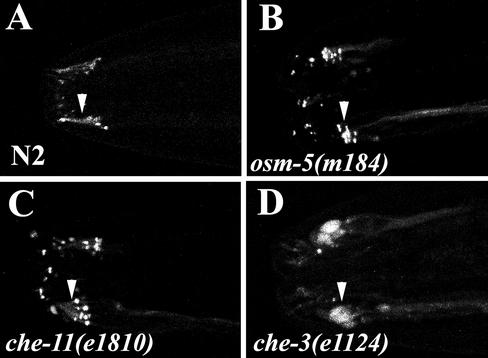
The IFT particle is a complex structure containing 17 or more proteins (Piperno and Mead, 1997; Cole et al., 1998). The hierarchy with which these proteins assemble into the two complexes A and B and how these proteins interact to form the IFT particle is currently unknown. To determine where XBX-1 function is required during particle assembly or transport, we expressed XBX-1::YFP protein in the context of other ciliogenic mutant backgrounds that disrupt proteins in either complex B, complex A, or the dynein motor CHE-3. In complex B mutants such as osm-5(m184), XBX-1::YFP was retained at the base of the stunted cilia and in the severely stunted axonemes (Figure 6B), suggesting a defect in anterograde movement. Similarly, in complex A mutants, such as che-11(e1810), XBX-1::YFP was positioned at the cilia base and could also be detected in the shortened cilia axoneme extending off these transition zones (Figure 6C). These data suggest that the complex A and complex B proteins tested here are not critical for normal targeting or retention of XBX-1 to the transition zones at the base of the cilia. An interesting observation was that XBX-1::YFP does not accumulate in the axonemes of the complex A mutants (Figure 6C) as seen with IFT complex B proteins analyzed in complex A mutant backgrounds (Piperno et al., 1998, and Figure 4D). These data suggest that retrograde transport of XBX-1 is still active in the absence of complex A. The transport of XBX-1 was further examined in the che-3 mutants that lack a functional dynein that acts downstream of the complex A proteins during retrograde IFT. Unlike the result in the complex A mutants, our analysis of XBX-1::YFP in the che-3(e1124) strain revealed a significant accumulation of XBX-1::YFP in the massive bulb like structures at the distal tip of cilia as seen for all other IFT proteins analyzed in this mutant background (Figure 6D and our unpublished results).
Figure 6.
The effect of IFT mutations on XBX-1 localization. Localization of XBX-1::YFP was analyzed by moving the XBX-1::YFP transgene into the indicated IFT mutant backgrounds. (A) XBX-1::YFP localizes to the base of cilia and within cilia in wild-type N2 worms. (B) XBX-1::YFP localizes to the transition zone at the base of cilia in the osm-5 complex B mutant, with little if any fluorescence detected along the cilia axoneme. (C) XBX-1::YFP is localized to the base of cilia in the che-11 complex A mutant. Unlike other IFT proteins analyzed in complex A mutants, there is no significant accumulation of XBX-1::YFP within cilia (see Figure 4D). (D) XBX-1::YFP accumulates in large bulb-like endings in the che-3 IFT dynein mutant. This is similar to that reported for other IFT proteins (see Figure 4C). Anterior of the worm and distal tip of the cilium are toward the left. Arrowheads indicate the position of the cilia axonemes on one bundle of amphid neurons.
Retrograde Movement of XBX-1::YFP in the Context of Complex A Mutants
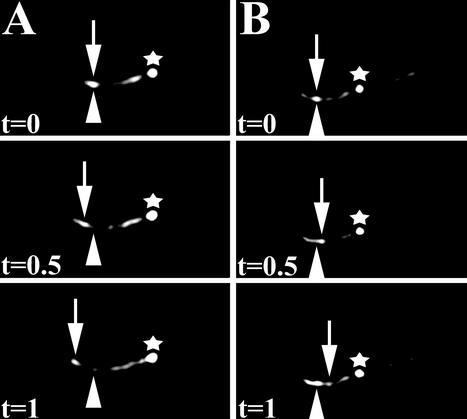
The fact that XBX-1 does not accumulate in the cilia of complex A mutants raised the possibility that XBX-1 retrograde transport is not inhibited by loss of the complex A proteins. To test this possibility, we conducted time-lapse fluorescence imaging of XBX-1::YFP in che-11 and daf-10 complex A mutant backgrounds (Figure 7, and our unpublished results). Because of the low expression level of this fusion protein and the limiting sensitivity of the camera, it was difficult to obtain high-quality time-lapse images in these mutants. However, under visual inspection through the microscope, both anterograde and retrograde movement of XBX-1::YFP were clearly evident in the stunted cilia. This is further supported by the lack of accumulation of XBX-1 in the complex A mutants, which is distinct from complex A and complex B proteins analyzed in this background (Figure 4D and our unpublished results). Thus, retrograde transport of XBX1::YFP is still active in complex A mutants (Figure 7). Together with the accumulation of XBX-1::YFP in che-3 mutants, these data support a role for XBX-1 and the CHE-3 dynein heavy chain in the retrograde transport of the IFT particle.
Figure 7.
Movement of XBX-1::YFP along the cilia axonemes on one pair of phasmid neurons of a che-11 complex A mutant. Using time-lapse fluorescence image analysis, XBX-1::YFP movement was observed in the (A) anterograde direction along the stunted cilia axoneme of the che-11 mutants. In contrast to other IFT proteins, XBX-1::YFP transport was also detected in the (B) retrograde direction. This continued retrograde trafficking of XBX-1 in che-11 mutants is consistent with the failure of XBX-1 to accumulate in the cilia axoneme. Images were captured at a rate of two frames per second. The distal tips of the phasmid cilia are directed toward the left. The transition zone is marked (*). The arrowhead indicates the initial IFT particle at time zero (t = 0). The arrows indicate the same particle at the subsequent times indicated (t = 0.5 s, and t = 1 s). Three frames representative of the movement seen in Movie 2 are shown for each direction.
DISCUSSION
IFT was first observed in Chlamydomonas as a process required for flagella assembly (Kozminski et al., 1993; Piperno and Mead, 1997). Subsequently, IFT has been reported in cilia assembly in C. elegans (Orozco et al., 1999; Signor et al., 1999a), and it is likely involved in flagella and cilia construction in higher eukaryotes as well (Pazour et al., 2000, 2002; Yoder et al., 2002). IFT is a process that describes the assembly of ciliary and flagellar proteins into large particles at the base of the organelle, transport of the particle toward the ciliary and flagellar tip through the action of the heterotrimeric kinesin-II complex, and return of the particle to the base utilizing a cytoplasmic dynein (Cole et al., 1998; Piperno et al., 1998; Orozco et al., 1999; Signor et al., 1999a, 1999b). Biochemical analysis of the IFT particle in Chlamydomonas indicates that the IFT particle consists of more than 17 peptides that form A and B complexes (Piperno and Mead, 1997; Cole et al., 1998). How these complexes assemble, how they interact with one another in the IFT particle, and how they associate with the IFT motor proteins is still poorly understood.
Herein, we describe a novel gene, xbx-1, which shares strong sequence similarity with a DLIC protein recently identified in mammalian systems (Grissom et al., 2002). Our analysis of xbx-1 in C. elegans reveals that its function is required for IFT and cilia formation in sensory neurons. xbx-1 was originally identified based on the presence of an X-box that is also present in the promoters of several ciliogenic genes encoding complex B IFT proteins (Swoboda et al., 2000; Haycraft et al., 2001). Our data confirm that xbx-1 is regulated by DAF-19, that the XBX-1 protein localizes to the cilia base and moves along the axoneme typical of an IFT protein (Signor et al., 1999a; Haycraft et al., 2001; Qin et al., 2001), and that disruption of xbx-1 results in cilia defects causing sensory behavior defects common to other ciliogenic mutants (Starich et al., 1995).
Surprisingly, despite the presence of an X-box sequence in the xbx-1 promoter, the cilia morphology in xbx-1 mutant worms did not resemble those typically seen in complex B mutants (Perkins et al., 1986; Cole et al., 1998; Haycraft et al., 2001). Rather, the morphology is similar to that seen in mutants affecting retrograde transport such as complex A mutants or the CHE-3 IFT dynein mutant (Perkins et al., 1986; Signor et al., 1999a; Qin et al., 2001). Typical of retrograde defects, the cilia show a bulb like structure at the distal tip with extensive accumulation of IFT proteins along the axoneme. These data suggest that in xbx-1 mutants, IFT particles enter the cilium axoneme where they are transported in anterograde direction but then fail to return to the transition zones.
To determine at which step XBX-1 function is required during ciliogenesis, we analyzed how mutations in IFT complex A or B proteins affect the localization of XBX-1::YFP in sensory cilia. In a recent study, we demonstrated that mutations in one complex B protein could differentially affect the localization of other complex B proteins thought to be within the same complex (Haycraft et al., 2003). These data suggest that particle assembly at the transition zone occurs in an ordered process determined by specific protein interactions within the complex. Thus, by placing fluorescence-tagged IFT proteins in the context of different IFT mutants (complex A or B), it may be possible to dissect the hierarchy with which the proteins function in IFT particle assembly and how the A and B complexes interact with each other and the motor proteins. Herein, we observed that none of the complex B mutants analyzed disrupted the ability of XBX-1 to concentrate at the base of the cilia, suggesting that XBX-1 localization and assembly into the particle is independent of the complex B proteins tested. Similarly, our analysis of XBX-1 in complex A mutants revealed that XBX-1::YFP effectively enters the axoneme and is transported to the distal tip of the cilia. Thus, complex A protein function is not required for XBX-1 anterograde IFT movement. Additionally, there was no significant accumulation of XBX-1::YFP in the swollen axonemes of the complex A mutants, unlike that observed with the complex B proteins. This is surprising considering the proposed retrograde transport role for complex A proteins. Because XBX-1::YFP fails to accumulate in the cilia of the complex A mutants, we speculated that XBX-1 retrograde transport was not disrupted by the loss of complex A. To test this possibility, we used time-lapse image analysis of XBX-1::YFP in two complex A mutant backgrounds. In contrast to what was seen for other IFT proteins analyzed, XBX-1::YFP retrograde transport was still detected in both complex A mutants. Furthermore, when XBX-1::YFP was analyzed in the che-3 dynein mutant, XBX-1 retrograde transport was abolished. Collectively, these data suggest that XBX-1 function is required for retrograde IFT in parallel with CHE-3. Moreover, it suggests that XBX-1 activity is required subsequent to proteins in complex A, because the complex A proteins accumulate in xbx-1 mutants but XBX-1 does not accumulate in the complex A mutants. These data raise the possibility that XBX-1 functions to connect the IFT particle and CHE-3 motor. This possibility is further supported by biochemical evidence in other species showing that the orthologs of XBX-1 both copurify and coimmuno-precipitate with the IFT dynein complex (Grissom et al., 2002; Perrone et al., 2003). Further biochemical and genetic analyses of XBX-1 will be required to confirm whether XBX-1 directly associates with a protein in the IFT particle and/or the CHE-3 dynein motor.
In contrast to the localization of XBX-1 in che-3(e1124) mutants reported here, in the Chlamydomonas cDhc1b mutants the LIC (the XBX-1 ortholog) is not detected in the stunted flagella (Perrone et al., 2003). However, the complex A and B proteins do accumulate similarly to that seen in the cilia of che-3 mutant worms (Pazour et al., 1999; Porter et al., 1999). These apparently conflicting effects may be explained by the differences in the dynein mutations. The che-3(e1124) strain used in this study has a nonsense mutation located in the middle of CHE-3 and truncates the proteins before the motor domain (Wicks et al., 2000). Although the che-3 mutation may be a hypomorphic allele, the Chlamydomonas cDhc1b (che-3 ortholog) mutation analyzed by Perrone et al. (2003) is a deletion and presumably produces no protein. Because the interaction between the dynein heavy chain and its LIC is thought to occur through the N-terminal region of the dynein (Tynan et al., 2000a), it is likely that XBX-1 and the mutant form of CHE-3 used in our analysis still associate and are transported into the axoneme, where it would then accumulate because of loss of the retrograde motor activity. However, in the Chlamydomonas cDhc1b mutants, the putative LIC binding site is lost, thus preventing LIC's entry into the cilium and subsequent movement along the axoneme. Furthermore, these data suggest that because the A and B complexes accumulate in both dynein mutants, their entry into the cilia does not appear to be dependent on the function of the dynein heavy chain/LIC complex.
Dyneins are high-molecular-weight, multisubunit complexes that play diverse roles in the cell (reviewed in Porter and Johnson, 1989; Holzbaur and Vallee, 1994; Hirokawa, 1998). Two classes of dyneins have been identified, including the extensive family of axonemal dyneins required for cilia and flagella beating and the cytoplasmic dyneins that play a role in intracellular vesicle transport, organelle movement, mitotic spindle assembly, chromosomal segregation, axonal transport, and retrograde movement in IFT (Mitchell, 1994; Porter, 1996; Vaisberg et al., 1996; Hirokawa, 1998; Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999a; Goldstein, 2001). Diversification of dynein function appears to be regulated by the accessory proteins associated with the dynein heavy chains that provide motive force driving movement along the microtubules (Karcher et al., 2002). As discussed above, our data support a role for XBX-1 as an accessory protein to the CHE-3 IFT dynein. This prediction is further corroborated by the recent analysis of orthologs of XBX-1 in both the mouse (D2LIC) and in Chlamydomonas (Perrone et al., 2003). In its initial characterization, the mammalian D2LIC was identified as a DLIC that associates with the dynein heavy chain DHC2 (Grissom et al., 2002). Interestingly, DHC2 is the ortholog of the worm CHE-3 and Chlamydomonas DHC1b IFT dynein. In mammalian systems, D2LIC and DHC2 were found to localize to the Golgi apparatus and centrosomes of nonpolarized cells where they were thought to be involved in intracellular trafficking and Golgi organization (Grissom et al., 2002). Although the initial report on D2LIC demonstrated no role in ciliogenesis (Grissom et al., 2002), data provided by Perrone et al. (2003) do show localization of D2LIC and DHC2 in the cilium of polarized epithelia in vivo and in vitro in MDCK cells. We believe that the different results with regard to D2LIC/XBX-1 protein localization are a direct result of the respective cell types analyzed and the conditions in which these experiments were conducted. The study performed by Grissom et al. (2002) was done in COS-7 cells under relatively nonpolarized conditions where cilia are not likely to be present. Furthermore, their expression analysis of D2LIC in various mammalian organs at the level of Western blot shows a good correlation between expression levels and the extent of ciliated cells present in the respective organ. The highest levels of expression were seen in the testis, lung, brain, and kidney, with little expression seen in the heart, liver, and spleen (Grissom et al., 2002). These data parallel the expression profiles seen for polaris, which is known to be a cilia protein in the mouse (Taulman et al., 2001).
In contrast to what was suggested for mammalian D2LIC, we do not believe that either XBX-1 or CHE-3 has a significant role in Golgi organization in C. elegans. This is supported by several lines of evidence. First xbx-1 expression is restricted to a small number of cells, all of which are ciliated. Second, the protein is prominently localized to cilia and third, mutations in xbx-1 and che-3 result in cilia specific defects (Perkins et al., 1986; Signor et al., 1999a; Wicks et al., 2000). Although we cannot unequivocally exclude a role for XBX-1 in Golgi, we would predict that disruption of this organelle would have a more profound effect than simply the loss of cilia. An additional possibility is that these proteins have acquired multiple functions in mammals both in the Golgi as seen in COS-7 cells (Grissom et al., 2002) and in cilia as shown in Perrone et al. (2003), while retaining a single function in IFT in C. elegans. Finally, recent work has shown DHC2/dynein 2 and D2LIC/LIC3 both localize to connecting cilia of photoreceptor cells and primary cilia of cultured kidney epithelial cells (Mikami et al., 2002). Furthermore, no colocalization of either DHC2/dynein 2 or D2LIC/ LIC3 with the Golgi was seen (Mikami et al., 2002). This work also notes that although XBX-1 (F02D8.3 protein) shares homology with D2LIC/LIC3, XBX-1 does not contain the same P-loop motif, suggesting that it may have distinct functions in mammals (Mikami et al., 2002).
Cilia assembly is a highly coordinated process in which as many as 200 distinct proteins need to be transported from their site of synthesis in the cytoplasm to their site of function in the axoneme. Proper localization and function of these proteins appear to be regulated by balancing the activity of the IFT kinesin and dynein (Cole et al., 1998; Pazour et al., 1999; Signor et al., 1999a). Thus, to understand how the IFT particle is transported and how cargo specificity is determined will require more detailed analysis of the kinesin and dynein accessory proteins, such as XBX-1. Disruption of this process, as seen in mice with mutations in the Tg737 gene (complex B), demonstrates the importance of elucidating the mechanism of IFT and cilia function as these mice exhibit major pathologies such as random left-right axis specification, skeletal pattering defects, cystic kidney disease, biliary and bile ductule hyperplasia, hydrocephalus, pancreatic acinar and ductule abnormalities, retinal degeneration, and sterility (Moyer et al., 1994; Murcia et al., 2000; Pazour et al., 2000, 2002; Taulman et al., 2001; Yoder et al., 2002).
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. S. Nozell and M. Porter for critical reading of this manuscript. We thank Drs. A. Coulson and A. Fire for the gifts of C. elegans clones and expression vectors and A. Tousson and S. Williams of the UAB Imaging Facility for assistance in fluorescence microscopy. We thank Dr. T. Hays and S. Mische for generous use of the UltraVIEW LCI and assistance with time-lapse imaging. We thank Thomas Bürglin for transiently providing laboratory space. We thank Dr. M. Barr for the CHE-11::GFP transgenic strain used for localization studies and Kerry Bubb for help with computer programming and algorithm design. The C. elegans Genome Sequencing Consortium provided sequence information and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, provided some of the C. elegans strains used in this study. We thank the C. elegans Knockout Consortium for providing the xbx-1(ok279) deletion mutant. This work was supported in part by a Public Health Service grant to J.H.T. (R01 GM48700), by a grant to P.S. from the Swedish Foundation for Strategic Research (SSF) in Stockholm, Sweden, and by Grant RO1-DK-62758 to B.K.Y. from the National Institute of Diabetes and Digestive and Kidney Diseases, and the March of Dimes Research Grant FY01-105.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-10-0677. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-10-0677.
Online version of this article contains video materials. Online version is available at www.molbiolcell.org.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. (1995). Mutagenesis. Methods Cell Biol. 48, 31–58. [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D.G., Diener, D.R., Himelblau, A.L., Beech, P.L., Fuster, J.C., and Rosenbaum, J.L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet, J., Spike, C.A., Lundquist, E.A., Shaw, J.E., and Herman, R.K. (1998). Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, C.e.S. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- Culotti, J.G., and Russell, R.L. (1978). Osmotic avoidance defective mutants of the nematode Caenorhabditis elegans. Genetics 90, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, P., Strubin, M., Hofmann, K., Bucher, P., Mach, B., and Reith, W. (1996). A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol. Cell. Biol. 16, 4486–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, M., Ishihara, T., and Katsura, I. (1999). A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126, 4839–4848. [DOI] [PubMed] [Google Scholar]
- Gibbons, I.R. (1995). Dynein family of motor proteins: present status and future questions. Cell Motil. Cytoskel. 32, 136–144. [DOI] [PubMed] [Google Scholar]
- Goldstein, L.S. (2001). Kinesin molecular motors: transport pathways, receptors, and human disease. Proc. Natl. Acad. Sci. USA 98, 6999–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom, P.M., Vaisberg, E.A., and McIntosh, J.R. (2002). Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol. Biol. Cell 13, 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft, C.J., Schafer, J.C., Zhang, Q., Taulman, P.D., and Yoder, B.K. (2003). Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 284, 249–261. [DOI] [PubMed] [Google Scholar]
- Haycraft, C.J., Swoboda, P., Taulman, P.D., Thomas, J.H., and Yoder, B.K. (2001). The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development 128, 1493–1505. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N. (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279, 519–526. [DOI] [PubMed] [Google Scholar]
- Holzbaur, E.L., and Vallee, R.B. (1994). DYNEINS: molecular structure and cellular function. Annu. Rev. Cell Biol. 10, 339–372. [DOI] [PubMed] [Google Scholar]
- Huang, L.S., Tzou, P., and Sternberg, P.W. (1994). The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5, 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini, C., Babaev-Khaimov, V., Sassaroli, M., and Piperno, G. (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher, R.L., Deacon, S.W., and Gelfand, V.I. (2002). Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 12, 21–27. [DOI] [PubMed] [Google Scholar]
- Kozminski, K.G., Johnson, K.A., Forscher, P., and Rosenbaum, J.L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K.S., and Sternberg, P.W. (1995). Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron 14, 79–89. [DOI] [PubMed] [Google Scholar]
- Malone, E.A., and Thomas, J.H. (1994). A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics 136, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C.C., Kramer, J.M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami, A., Tynan, S., Hama, T., Luby-Phelps, K., Saito, T., Crandall, J., Besharse, J., and Vallee, R. (2002). Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 115, 4801–4808. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R. (1994). Cell and molecular biology of flagellar dyneins. Int. Rev. Cytol. 155, 141–180. [DOI] [PubMed] [Google Scholar]
- Moyer, J.H. et al. (1994). Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science 264, 1329–1333. [DOI] [PubMed] [Google Scholar]
- Murcia, N.S., Richards, W.G., Yoder, B.K., Mucenski, M.L., Dunlap, J.R., and Woychik, R.P. (2000). The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development 127, 2347–2355. [DOI] [PubMed] [Google Scholar]
- Nonet, M.L., Saifee, O., Zhao, H., Rand, J.B., and Wei, L. (1998). Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 18, 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, J.T., Wedaman, K.P., Signor, D., Brown, H., Rose, L., and Scholey, J.M. (1999). Movement of motor and cargo along cilia. Nature 398, 674. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., Baker, S.A., Deane, J.A., Cole, D.G., Dickert, B.L., Rosenbaum, J.L., Witman, G.B., and Besharse, J.C. (2002). The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., Vucica, Y., Seeley, E.S., Rosenbaum, J.L., Witman, G.B., and Cole, D.G. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., and Witman, G.B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L.A., Hedgecock, E.M., Thomson, J.N., and Culotti, J.G. (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487. [DOI] [PubMed] [Google Scholar]
- Perrone, C.A., Tritschler, D., Taulman, P., Bower, R., Yoder, B.K., and Porter, M.E. (2003). Mol. Biol. Cell 14, 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., and Mead, K. (1997). Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 94, 4457–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Siuda, E., Henderson, S., Segil, M., Vaananen, H., and Sassaroli, M. (1998). Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143, 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E. (1996). Axonemal dyneins: assembly, organization, and regulation. Curr. Opin. Cell Biol. 8, 10–17. [DOI] [PubMed] [Google Scholar]
- Porter, M.E., Bower, R., Knott, J.A., Byrd, P., and Dentler, W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E., and Johnson, K.A. (1989). Dynein structure and function. Annu. Rev. Cell Biol. 5, 119–151. [DOI] [PubMed] [Google Scholar]
- Qin, H.M., Rosenbaum, J.L., and Barr, M.M. (2001). An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C-elegans ciliated sensory neurons. Curr. Biol. 11, 457–461. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, J. (2002). Intraflagellar transport. Curr. Biol. 12, R125. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Signor, D., Wedaman, K.P., Orozco, J.T., Dwyer, N.D., Bargmann, C.I., Rose, L.S., and Scholey, J.M. (1999a). Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 147, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor, D., Wedaman, K.P., Rose, L.S., and Scholey, J.M. (1999b). Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol. Biol. Cell 10, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich, T.A., Herman, R.K., Kari, C.K., Yeh, W.H., Schackwitz, W.S., Schuyler, M.W., Collet, J., Thomas, J.H., and Riddle, D.L. (1995). Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139, 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L., Sternberg, P., Durbin, R., Thierry-Mieg, J., and Spieth, J. (2001). WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda, P., Adler, H.T., and Thomas, J.H. (2000). The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5, 411–421. [DOI] [PubMed] [Google Scholar]
- Taulman, P.D., Haycraft, C.J., Balkovetz, D.F., and Yoder, B.K. (2001). Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol. Biol. Cell 12, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan, S.H., Gee, M.A., and Vallee, R.B. (2000a). Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J. Biol. Chem. 275, 32769–32774. [DOI] [PubMed] [Google Scholar]
- Tynan, S.H., Purohit, A., Doxsey, S.J., and Vallee, R.B. (2000b). Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 275, 32763–32768. [DOI] [PubMed] [Google Scholar]
- Vaisberg, E.A., Grissom, P.M., and McIntosh, J.R. (1996). Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J. Cell Biol. 133, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S.R., de Vries, C.J., van Luenen, H.G., and Plasterk, R.H. (2000). CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 221, 295–307. [DOI] [PubMed] [Google Scholar]
- Yoder, B.K., Tousson, A., Millican, L., Wu, J.H., Bugg, C.E., Jr., Schafer, J.A., and Balkovetz, D.F. (2002). Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am. J. Physiol. Renal Fluid Electrolyte Physiol. 282, F541–F552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.