Abstract
Nuclear imports of uridine-rich small nuclear ribonucleoprotein (U1 snRNP) and proteins with classical nuclear localization signal (cNLS-protein) are mediated by importin β. However, due to the presence of different import signals, the adapter protein of the imported molecules and importin β is different for each pathway. Although the adapter for cNLS-protein is importin α, the adapter for U1 snRNP is snurportin1 (SPN1). Herein, we show that the use of distinct adapters by importin β results in differences at the docking and releasing step for these two import pathways. Nuclear pore complex (NPC) docking of U1 snRNP but not of cNLS-protein was inhibited by an anti-CAN/Nup214 antibody. Thus, the initial NPC-binding site is different for each pathway. Pull-down assays between immobilized SPN1 and two truncated forms of importin β documented that SPN1 and importin α have different binding sites on importin β. Importin β fragment 1–618, which binds to SPN1 but not to importin α, was able to support the nuclear import of U1 snRNPs. After the translocation through the NPC, both import complexes associated with the nuclear side of the NPC. However, we found that the nature of the importin β-binding domain of the adapters influences the release of the cargo into the nucleoplasm.
INTRODUCTION
Active nuclear transport occurs through the nuclear pore complex (NPC) and is a highly selective process that requires a signal residing on the transported molecules or “cargo.” The different signals are recognized by soluble transport receptors shuttling between the cytoplasm and the nucleus (reviewed by Görlich and Kutay, 1999; Kuersten et al., 2001; Macara, 2001). Several distinct signals on different cargo molecules have been identified. The first identified nuclear import signal is characterized by short stretches of basic amino acids called nuclear localization sequences (NLSs). It is now referred to as the classical NLS or cNLS. The receptor for the cNLS import pathway consists of two subunits, importin α and importin β. Importin α harbors an importin β-binding (IBB) domain at its N terminus and acts as an adapter between the cNLS-bearing protein (cNLS-protein) and importin β. Many other (but not all) nuclear transport pathways are mediated by transport receptors that are members of a large importin β-related protein family (reviewed by Görlich and Kutay, 1999; Conti and Izaurralde, 2001).
At the molecular level, nuclear import is a sequential process that starts with the interaction between the targeting signal and soluble cellular receptors. After targeting to the NPC, the cargo–receptor complex crosses the NPC, and the cargo is released into the nucleus. Sequential interactions between the cargo–receptor complex and nucleoporins are thought to be the driving force behind the translocation of the cargo–receptor complex through the NPC (Radu et al., 1995; Rexach and Blobel, 1995). Although there have been several studies documenting in vitro interactions between import receptors and nucleoporins (reviewed by Ryan and Wente 2000), in vivo interactions between the cargo–receptor complex and NPC components are still not well characterized. Nevertheless, by microinjecting gold-labeled nucleoplasmin into the cytoplasm of Xenopus oocytes and following its nuclear import by electron microscopy (EM), it has been possible to depict in vivo interactions between the cargo–receptor complex and the NPC. For example, three different conditions that yield docking of the cNLS–cargo–receptor complex to the nuclear envelope by immunofluorescent microscopy yielded three distinct NPC-arrested intermediates by EM. Gold-labeled nucleoplasmin is arrested: 1) at the terminal end of the cytoplasmic filaments when import is inhibited by wheat germ agglutinin (WGA) (Panté and Aebi, 1996); 2) at the cytoplasmic entrance of the central channel when import is inhibited by low temperature (Panté and Aebi, 1996); and 3) at the nuclear basket when import is followed in the presence of a mutant form of importin β that does not bind Ran (Görlich et al., 1996). By using this methodology, it will also be possible to follow the path of other transport cargo molecules through a single NPC.
Importin β also mediates the nuclear import of spliceosomal uridin-rich small nuclear ribonucleoprotein particles (U snRNPs; Palacios et al., 1997). However, the signal for U1 snRNP nuclear import is not a cNLS. The U1 snRNP nuclear import signal is bipartite, and it is formed by the m3G-cap and the Sm core domain of the Sm proteins (Fischer and Lührmann, 1990; Hamm et al., 1990; Fischer et al., 1991, 1993). Both components of the signal are formed after the assembly of the U snRNPs in the cytoplasm. As a consequence, only fully assembled U1 snRNPs enter the nucleus. The m3G-cap is specifically recognized by the import receptor snurportin1 (SPN1) that functions as an adapter between the m3G-cargo and importin β (Huber et al., 1998). An additional but different receptor seems to interact with the second part of the signal required for nuclear import of U1 snRNPs. However, the identity of this receptor and the molecular nature of the second signal remain to be elucidated.
Despite the fact that the U1 snRNP and the cNLS-protein import pathways use both importin β as an import receptor, there are some differences between these pathways, which indicates that they are driven by different molecular mechanisms. These differences are as follows: 1) the apparent absence of docking of U snRNPs at the NPC under conditions that yield the accumulation of cNLS-proteins at the NPC (Palacios et al., 1996); 2) in contrast to the nuclear import of cNLS-proteins, the nuclear import of U snRNP is independent of Ran (Marshallsay et al., 1996; Huber et al., 2002); and 3) the limited inhibitory effect of WGA on the nuclear import of U snRNPs under conditions that completely inhibit the import of cNLS-proteins (Fischer et al., 1991; Michaud and Goldfarb, 1992; Marshallsay and Lührmann 1994). Because WGA binds to a group of ∼10 nucleoporins that are modified with O-linked N-acetylglucosamine, the latter difference indicates that different nucleoporins are involved in these two nuclear import pathways.
Both importin α and SPN1 contain an IBB domain for importin β binding, located at the N-terminal end of both proteins. The amino acid sequences of importin α and SPN1 are otherwise unrelated. The IBB of importin α binds to the 3/4 C-terminal region of importin β (a region comprising residues 256–876; Kutay et al., 1997b; Cingolani et al., 1999). The region of importin β that binds to the IBB of SPN1 has not yet been mapped. Despite having a similar role as adapters, there are differences between importin α and SPN1. Some of these differences are as follows: 1) the C-terminal m3G-cap–binding region of SPN1 has no structural similarity to the C-terminal region of importin α (Huber et al., 1998); 2) the affinity of SPN1 for importin β is different than the affinity of importin α for importin β (Huber et al., 1998); and 3) although the nuclear export of importin α is mediated by the export receptor CAS (Kutay et al., 1997a), the nuclear export of SPN1 is mediated by CRM1 (Paraskeva et al., 1999). The differences between the molecular mechanism for nuclear import of U snRNPs and cNLS-proteins might be a consequence of the differences between importin α and SPN1.
To determine the molecular basis of the differences between these two import pathways, herein we have studied nuclear import of U1 snRNP and we have compared our results with those of nuclear import of cNLS-proteins. Our data indicate that the adapter proteins for these two nuclear import pathways are involved in the NPC-docking and -releasing steps.
MATERIALS AND METHODS
Reagents
U1 snRNP, Δ5′ U1 snRNP, and the recombinant proteins SPN1 and β-galactosidase molecules fused to the IBB domains of SPN1 (IBBspn1) or importin α (IBBα) were kindly provided by Drs. Reinhard Lührmann, Jochen Huber, and Achim Dickmanns (Max-Planck-Institut fuer Biophysikalische Chemie, Goettingen, Germany). Bovine serum albumin (BSA) coupled to the peptide CGGGPKKKRKVED (a cNLS) was kindly provided by Dr. Achim Dickmanns. The antibody QE5 was kindly provided by Dr. Brian Burke (Department of Anatomy and Cell Biology, University of Florida, Gainesville, FL). The anti-CRM1 antibody was a kind gift of Dr. Iain Mattaj (European Molecular Biology Laboratory, Heidelberg, Germany).
Recombinant Protein Expression
The clone for full-length SPN1 tagged to two immunoglobulinbinding domains of Staphylococcus aureus protein A (zz-tagged SPN1) was kindly provided by Dr. Dirk Görlich (University of Heidelberg, Germany). Zz-tagged SPN1 was expressed as described in Paraskeva et al. (1999). The three different importin β constructs (1–876, 1–618, and 1–452) and the zz-tagged importin α were expressed as described in Kutay et al. (1997b).
Pull-Down Assays with Biotinylated Proteins
Importin α and SPN1 were biotinylated by incubation for 1 h on ice with stoichiometric amounts of PEO-biotin (Pierce Chemical, Rockford, IL). To remove unincorporated PEO-biotin, reaction mixtures were passed over NAP5 columns (Amersham Pharmacia, Freiburg, Germany) preequilibrated with 50 mM Tris, pH 7.6, 200 mM NaCl, and 4 mM MgCl2. For each binding reaction, 10 μl of streptavidin-agarose beads was presaturated with biotinylated importin α or SPN1 for 1 h at 4°C. The beads were then washed three times with B-buffer (50 mM Tris, pH 7.6, 150 mM potassium acetate, and 4 mM MgCl2). Bound proteins were incubated for 1 h at 4°C in B-buffer supplemented with recombinant importin β to allow complex formation between importin α and importin β or SPN1 and importin β, respectively. After three times washing with B-buffer, the beads were incubated in B-buffer supplemented with 50 μl of Xenopus egg extract in a total volume of 500 μlfor4hat4°C. The beads were then washed extensively with B-buffer and bound proteins were eluted in 30 μl of SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
Pull-Down Assays with zz-tagged Proteins
Recombinant zz-tagged importin α or SPN1 were prebound to IgG-Sepharose beads for 45 min at 4°C. The beads were washed several times with a buffer containing 50 mM Tris, pH 7.5, 500 mM NaCl, and 5 mM MgCl2. Then 250 μl of lysate of Escherichia coli expressing importin β fragments was incubated each with 20 μl of affinity matrix overnight at 4°Cina final volume of 1.5 ml of binding buffer (50 mM HEPES-KOH, pH 7.5, 225 mM NaCl, 2 mM MgCl2, and 0.005% digitonin). The beads were then washed three times with binding buffer. Bound proteins were eluted from the beads with 100 μl of MgCl2 buffer (1.5 M MgCl2, 50 mM Tris, pH 7.5), and eluted proteins were precipitated with 1 ml of 100% isopropanol. The precipitated proteins were dissolved in SDS sample buffer and analyzed by 8% SDS-PAGE.
Gold Conjugation of U1 snRNP, Proteins, and Import Complexes
Colloidal gold particles (6 and 8 nm) were prepared by reduction of tetrachloroauric acid with sodium citrate in the presence of tannic acid (Slot and Geuze, 1985). U1 snRNP, BSA coupled with a cNLS (cNLS-BSA), SPN1, IBBspn1, and IBBα were directly conjugated to the colloidal gold particles as described by Baschong and Wrigley (1990). After conjugation, the complexes were centrifuged at 32,000 × g for 15 min. The soft pellet was taken for microinjection into Xenopus oocytes.
Complexes between SPN1 and gold-labeled U1 snRNPs were formed by mixing both solutions in a 1:1 M ratio followed by incubation of 20 min at room temperature. The formation of complexes of importin β with IBBspn1 or IBBα was performed by mixing gold-labeled IBBspn1 or gold-labeled IBBα with a 1:1 M ratio of importin β, following by incubation at room temperature for 20 min.
Microinjection of Xenopus Oocytes
Mature oocytes were removed from female Xenopus laevis as described previously (Reichelt et al., 1990) and stored in modified Barth's saline (MBS) containing 88 mM NaCl, 1 mM KCl, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, and 10 mM HEPES, pH 7.5. Oocytes were defolliculated by treatment with 5 mg/ml collagenase in calcium-free MBS for 1 h. After intensive washing with MBS the oocytes were used within the next 2 d for microinjection.
Xenopus oocytes were injected into their cytoplasm with 50–100 nl of gold-conjugated molecules, and the injected oocytes were incubated in MBS at room temperature for the time indicated in the figure legends and prepared for EM as indicated below. The inhibition studies with the antibody QE5 were performed by cytoplasmic injecting QE5 at a 1:10 dilution. After incubation for 2 h at room temperature, gold-U1 snRNP, gold-cNLS-BSA, or gold-U1 snRNP-SPN1 was injected into the cytoplasm of different QE5-preinjected oocytes. After incubation for further 2 h, the oocytes were prepared for EM as indicated below.
Preparation of Oocytes for Electron Microscopy
After incubation of injected oocytes as indicated above, the oocytes were fixed overnight at 4°C with 2% glutaraldehyde in MBS. The oocytes were then washed three times with MBS, and the animal pole of the oocytes (including the nucleus) was dissected and fixed again with 2% glutaraldehyde in MBS for 30 min at room temperature. Then the dissected oocytes were washed three times in MBS and embedded in 2% agar. After a postfixation with 1% OsO4 in MBS for 1 h the samples were dehydrated and embedded in Epon 812 (Fluka, Buchs, Switzerland) by standard procedures (Jarnik and Aebi, 1991).
Electron Microscopy Import Assay in HeLa Cells
SPN1 labeled with 8-nm gold particles was preincubated with a equimolar amount of importin β fragment 1–618 at room temperature for 20 min. HeLa cells were grown as monolayers on thermanox plastic coverslips (Nalge Nunc International, Naperville, IL) to 80–90% confluence in DMEM (Hyclone, Logan, UT) supplemented with 10% fetal calf serum and penicillin/streptomycin at 37°C. Cells were permeabilized with 50 μg/ml digitonin for 5 min at room temperature (Adam et al., 1990). Coverslips with attached permeabilized cells were incubated for 30 min at room temperature with transport buffer (40 mM HEPES-KOH, pH 7.5, 110 mM potassium acetate, 4 mM magnesium acetate, 1 mM dithiothreitol, and 1:1000 of the following protease inhibitor mix: 10 mg/ml chymostatin, 10 mg/ml leupeptin, 10 mg/ml antipain, and 10 mg/ml pepstatin in dimethyl sulfoxide; Bastos et al., 1996) containing 25 μl of ∼1.3 μM SPN1-importin β (1–618) preformed complex, 0.2 mg/ml tRNA, and 4 mg/ml BSA. After incubation, the coverslips were fixed with 2% glutaraldehyde in phosphate-buffered saline for 1 h at room temperature and postfixed for 1 h with 1% OsO4 in phosphate-buffered saline at room temperature. Coverslips with fixed cells were then sequentially dehydrated in 30, 70, and 90% ethanol each for 10 min; followed by three times 100% ethanol (each for 10 min); and finally with 100% acetone for 10 min. Coverslips were then infiltrated with mixtures of Epon 812 (Fluka) and acetone 1:1 for 2 h and 2:1 for 2 h, and finally in pure Epon 812 for 3 h. Gelatin capsules filled with fresh pure Epon resin were place on top of coverslips (with the layer of cells facing the gelatin capsule) and polymerized at 60°C for at least 24 h.
Electron Microscopy and Quantitation of Gold Labeling
Thin sections were cut on a Reichert Ultracut ultramicrotome by using a diamond knife. Ultrathin sections were collected on phalloidin/carbon-coated cooper grids, stained with 2% uranyl acetate for 30 min, and poststained with 2% lead citrate for 5 min. Micrographs were digitally recorded in an H-7000 transmission electron microscope (Hitachi, Tokyo, Japan) operated at an acceleration voltage of 100 kV.
The position of gold particles associated with NPCs was determined from digital electron micrographs of cross sections along the nuclear envelope. Distances of gold particles were measured from the central plane of the NPC.
RESULTS
Gold-labeled U1 snRNPs Are Imported into Xenopus Oocyte Nuclei
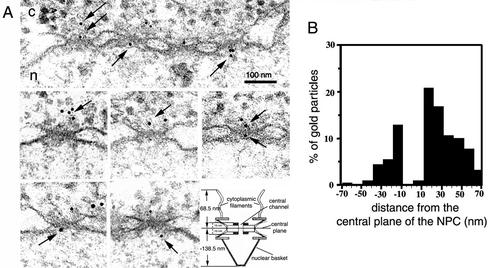
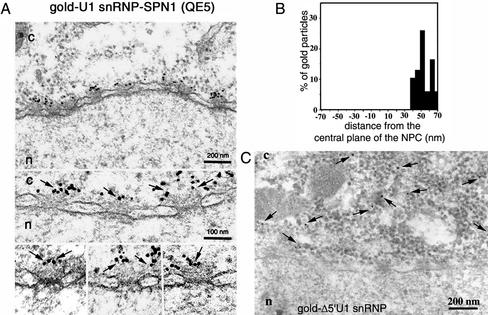
To characterize the nuclear import of U1 snRNP at the ultrastructural level, we labeled U1 snRNP with colloidal gold and microinjected these gold complexes into the cytoplasm of Xenopus oocytes (a system well suited for the study of nuclear transport that yields structurally well-preserved NPCs). Visualization of the U1 snRNP–gold complexes in the EM after negative staining revealed that the labeling was very efficient with all gold particles surrounded by light areas of protein (our unpublished data). Injected oocytes were then processed for embedding and thin sectioning EM, as described under MATERIALS AND METHODS, and the position and distribution of gold particles were determined on cross-sectioned nuclear envelopes. As shown in Figure 1A, gold-U1 snRNP was imported into the nucleus of Xenopus oocyte and was often associated with the NPC. Thus, gold labeling did not interfere with the nuclear import of U1 snRNP.
Figure 1.
The nuclear import of gold-labeled U1 snRNP. (A) Nuclear envelope cross sections from Xenopus oocytes that have undergone cytoplasmic injection with gold-U1 snRNPs (gold diameter, 6 nm). After injection, oocytes were incubated for 2 h at room temperature and then processed for embedding and thin section EM, as indicated under MATERIALS AND METHODS. Cytoplasm and nucleus are indicated with c and n, respectively. The arrows indicate gold-U1 snRNP interacting with the NPC. Bar, 100 nm. Also shown is a schematic diagram of the NPC, indicating some NPC components and their sizes measured from the central plane of the NPC. Negative values represent the nuclear side of the NPC (this diagram was adapted from Fahrenkrog et al., 1998). (B) Quantitative analysis of gold particles associated with the NPC from experiments performed under conditions indicated in A. Nuclear envelopes from three different experiments were analyzed, which yielded 178 gold particles.
The route that the gold–U1 snRNP complexes took through the NPC is as follows: first, they bound to the cytoplasmic filaments (gold particles located at 30–65 nm from the NPC central plane), and then to the cytoplasmic entrance of the central channel (gold particles detected at ∼20 nm from the NPC central plane). Second, after the translocation through the central channel, a process that is obviously too fast to be detected in our embedded/thin-sectioned oocytes, the gold–U1 snRNP import complex interacted with the nuclear entrance/exit of the central channel (gold particles at ∼20 nm from the NPC central plane). Finally, the gold–U1 snRNP import complex bound to the nuclear basket (gold particles at ∼30–120 nm from the NPC central plane). The release from the nuclear basket into the nucleus and the subsequent movement away from the NPC occurred very quickly, because gold particles inside the nucleus were always found far away from the NPCs.
Nucleoporin CAN/Nup214 Is the First NPC Binding Site for the U1 snRNP Import Pathway
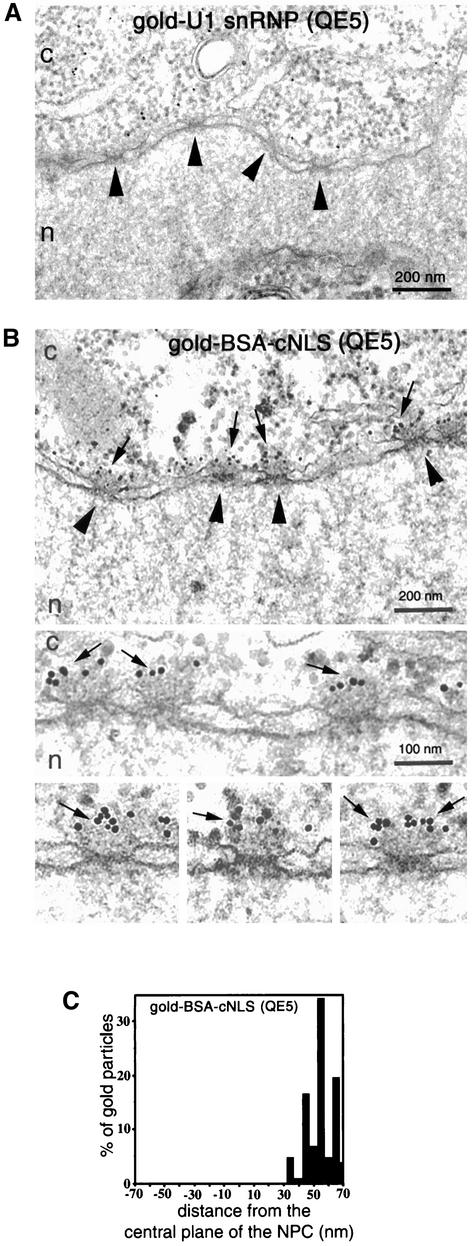
To identify the nucleoporin(s) involved in the docking step of U1 snRNP to the NPC, we used the antibody QE5 that has an epitope at the cytoplasmic filaments corresponding to the nucleoporin CAN/Nup214 (Panté et al., 1994). To guarantee that CAN/Nup214 is completely blocked by the antibody, QE5 was preinjected into the cytoplasm of the oocytes and the oocytes were incubated at room temperature for 2 h. Next, gold-labeled U1 snRNP (or gold-cNLS-BSA) was injected into the oocytes. After further incubation at room temperature, the oocytes were fixed, prepared for EM, and gold distribution was determined in EM cross sections. We found that QE5 differentially blocked each import pathway. Whereas gold-U1 snRNP was found throughout the cytoplasm and was not associated with the NPC of oocytes preinjected with QE5 (Figure 2A), gold-cNLS-BSA was found associated with the distal part of the NPC cytoplasmic filaments under the same conditions (Figure 2B). However, gold-cNLS-BSA remained at the cytoplasmic filament and did not associate with the cytoplasmic entrance/exit of the central channel. Our explanation for this result is that the antibody QE5, which also recognizes nucleoporin p62 (located near or at the cytoplasmic entrance/exit of the central channel; Guan et al., 1995), had blocked this NPC-binding site.
Figure 2.
An anti-CAN/Nup214 antibody inhibits NPC docking of U1 snRNP but not NPC docking of cNLS-proteins. (A and B) Nuclear envelope cross sections of Xenopus oocytes that have undergone cytoplasmic injection first with the antibody QE5 and then with 8-nm gold-U1 snRNP (A), or with 8-nm gold cNLS-BSA (B). QE5 was preinjected into the cytoplasm of oocytes, and the oocytes were incubated for 2 h at room temperature followed by the second injection and further incubation for 2 h at room temperature. Cytoplasm and nucleus are indicated with c and n, respectively. Bars, 200 nm (overview) and 100 nm (gallery). Arrowheads point to NPC, and arrows indicate gold-cNLS-BSA associated with the NPC. (C) Quantitative analysis of gold particles associated with the NPC from oocytes injected with QE5 and then with gold-cNLS-BSA. Nuclear envelopes from three different experiments were analyzed, which yielded 102 gold particles. In comparison with the cNLS-BSA import complex (B and C), which is able to bind to the NPC cytoplasmic filaments (distance 30–70 nm from the NPC central plane) after the preinjection of QE5, the U1 snRNP import complex (A) is not able to bind to the NPC.
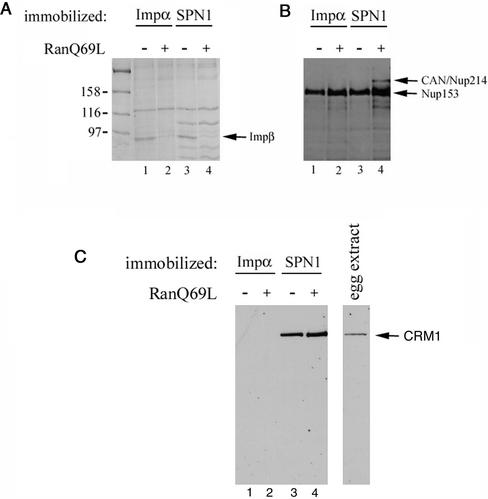
To demonstrate biochemically the different association between the two cargo–receptor complexes with CAN/Nup214, we performed pull-down experiments with immobilized adapters and proteins from a Xenopus egg extract. For this purpose, biotinylated SPN1 and importin α were immobilized on streptavidin-agarose beads, and the immobilized proteins were bound to importin β. Then, the beads were incubated with Xenopus egg extracts in the absence or presence of RanQ69L, a mutant of Ran, which is insensitive to RanGAP, and which persists in the GTP bound state. As documented in Figure 3, A and B, when proteins from the egg extract that bound to the beads were eluted and analyzed by immunoblots using the antibody mAb414 against several nucleoporins, we found that nucleoporin CAN/Nup214 interacted with the immobilized SPN1 only in the presence of RanQ69L. In contrast, CAN/Nup214 was not present on the immunoblots when importin α was immobilized. The Coomassie-stained gel (Figure 3A) shows that importin β was indeed dissociated from SPN1 (and from importin α) by RanQ69L. Thus, the interaction of gold-U1 snRNP with CAN/Nup214 occurs when SPN1 is not loaded with importin β (in the presence of Ran-GTP, SPN1 does not interact with importin β; Paraskeva et al., 1999). As shown in Figure 3C, CRM1 (the export factor for SPN1) was present in the pull-down experiments when SPN1 was immobilized, but not when importin α was immobilized. The CRM1 band was more intense in the presence of RanQ69L (Figure 3C, lane 4). This could be explained by the interaction of CRM1 with CAN/Nup214, which is present in the pull-down experiment in the presence of RanQ69L (Figure 3B, lane 4).
Figure 3.
Nucleoporin CAN/Nup214 is the first NPC binding site for the U1 snRNP import pathway. (A) Xenopus egg extracts were incubated with immobilized importin α (lanes 1 and 2) or SPN1 (lanes 3 and 4) precomplexed with importin β. Where indicated 1 μM RanQ69L was also added. Bound proteins were eluted, separated by SDS-PAGE, and detected by Coomassie staining (A) or immunoblotting with either the antibody mAb414 that recognizes several nucleoporins (B) or an anti-CRM1 antibody (C).
The U1 snRNP Import Pathway Has a Second NPC Binding Site at the Cytoplasmic Filament
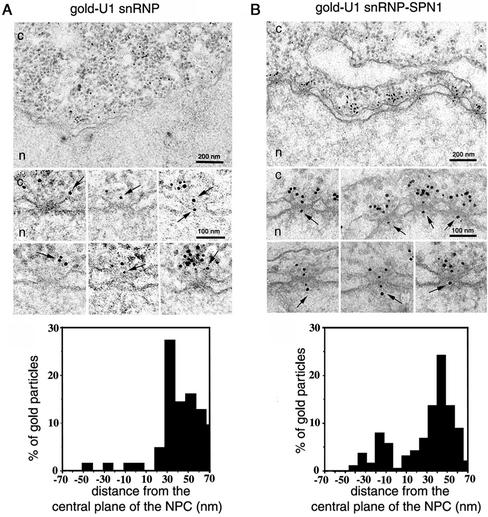
Our results with the QE5 antibody indicate that U1 snRNP binds its adapter at CAN/Nup214. To test this implication, we performed EM import experiments with the in vitroformed U1 snRNP–SPN1 complex. This complex was formed by mixing gold-labeled U1 snRNP with recombinant SPN1 at a 1:1 M ratio, followed by incubation of the solution at room temperature for 20 min. We first tested whether this complex was import competent by injecting them into the cytoplasm of Xenopus oocytes and followed their fate by EM. We found that at any given time of incubation there were more gold particles targeted to the NPC, in transit through the NPC and imported into the nucleus for the gold-U1 snRNP-SPN1 in vitro-formed complex than for gold-U1 snRNP (Figure 4, A and B). These results indicate that SPN1 accelerates the nuclear import of U1 snRNP by enhancing the targeting of U1 snRNP to the NPC and by increasing the interaction of the cargo–receptor complex with the NPC.
Figure 4.
The nuclear import of the preformed U1 snRNP–SPN1 complex. Nuclear envelope cross sections of Xenopus oocytes that have undergone cytoplasmic injection with 8-nm gold-U1 snRNP (A) or with 8-nm gold-U1 snRNP-SPN1 (B) and kept at room temperature for 20 min. Complexes between gold-U1 snRNP and SPN1 were preformed by mixing both solution in a 1:1 M ratio, followed by incubation at room temperature for 20 min. Cytoplasm and nucleus are indicated with c and n, respectively. Bars, 200 nm (overview) and 100 nm (galleries). Arrows indicate gold particles associated with the NPC. Quantitative analysis of gold particles associated with the NPC for the two different experiments are indicated at the bottom of each panel. For each histogram, nuclear envelopes from three different experiments were analyzed, which yielded 65 and 190 gold particles for A and B, respectively.
We then performed EM import experiments with the gold-U1 snRNP-SPN1 in vitro-formed complex in oocytes that had been preinjected with the antibody QE5 (exactly as in the experiments performed with gold-U1 snRNP; Figure 2A). Surprisingly, we found that in contrast to gold-U1 snRNP (Figure 2A), the gold-U1 snRNP-SPN1 in vitroformed complex could still bind to the distal part of the cytoplasmic filaments in the presence of the QE5 antibody against the nucleoporin CAN/Nup214 (Figure 5A).
Figure 5.
An anti-CAN/Nup214 antibody does not inhibit NPC docking of the preformed U1 snRNP–SPN1 complex. (A) Nuclear envelope cross sections of Xenopus oocytes that have undergone cytoplasmic injection first with the antibody QE5 and then with 8-nm gold-U1 snRNP-SPN1. Complexes between gold-U1 snRNP and SPN1 were preformed, as described in Figure 4. Cytoplasm and nucleus are indicated with c and n, respectively. Bars, 200 nm (overview) and 100 nm (gallery). Arrows indicate gold particles associated with the NPC. (B) Quantitative analysis of gold particles associated with the NPC for experiments performed under the conditions indicated in (A). Nuclear envelopes from three different experiments were analyzed that yielded 115 gold particles. (C) Cross section of a Xenopus oocyte nuclear envelope after cytoplasmic injection of gold-Δ 5′ U1 snRNP, a mutant U1 snRNP that does not have the m3G-cap. Δ 5′ U1 snRNP was conjugated with 8-nm gold particles and injected into the cytoplasm of Xenopus oocytes. The oocytes were then incubated for 2 h at room temperature and prepared for EM. Gold-Δ 5′ U1 snRNP remained in the cytoplasm and did not interact with the NPC (arrows). c, cytoplasmic side; n, nuclear side of the nuclear envelope. Bar, 200 nm.
Because the nuclear import of U1 snRNP is also mediated by a second import signal located in the core domain of the Sm protein, this receptor and not SPN1 might be involved in the interaction of the cargo–receptor complex with the NPC. To test this hypothesis, we performed EM import experiments with gold-Δ 5′ U1 snRNP, a mutant form that does not have the m3G-cap. As shown in Figure 5C, 2 h after the cytoplasmic injection, gold-Δ 5′ U1 snRNP remained in the cytoplasm and did not interact with the NPC. This data suggests a strong m3G-cap-dependent interaction of U1 snRNP with CAN/Nup214, and demonstrates that the interaction of the cargo–receptor complex with the NPC depends on the presence of SPN1 in the import complex.
Together, our results with the QE5 antibody indicate that there are two binding sites for the U1 snRNP import complex at the cytoplasmic filaments: one at the nucleoporin CAN/Nup214 that involves the SPN1 export complex, and a second that requires the previous formation of the U1 snRNP import complex.
Importin α and SPN1 Bind to Different Regions of Importin β
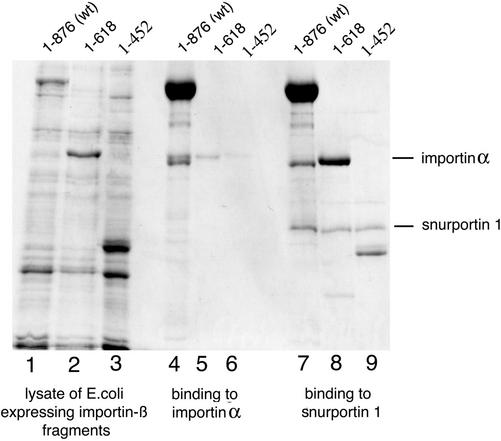
Next, we performed experiments to address the question of whether molecular differences between SPN1 and importin α explain the differences between the U1 snRNP and the cNLS import pathways. Our hypothesis was that SPN1 and importin α have different binding sites on importin β that allow different interactions of the cargo–receptor complex to distinct nucleoporins. To test this hypothesis, pull-down assays between zz-tagged SPN1 and two truncated forms of importin β (1–618 and 1–452) were performed, as it has been done for importin α (Kutay et al., 1997b). As a control, parallel experiments were done with the full-length importin β (1–876) and with zz-tagged importin α. As expected and as described previously (Kutay et al., 1997b), importin β 1–618 and importin β 1–452 did not bind to importin α (Figure 6, lanes 5 and 6). Surprisingly, importin β 1–618 and importin β 1–452 bound to SPN1 (Figure 6, lanes 8 and 9). The two fragments, however, bound less efficiently than the wild-type protein, and fragment 1–452 bound less efficiently than fragment 1–618. These results indicate that the binding of importin α and SPN1 to importin β differs. Moreover, SPN1 and importin α cannot bind simultaneously to importin β (our unpublished data).
Figure 6.
Importin α and SPN1 bind to different regions of importin β. Full-length importin β (1–876) and two shorter constructs (1–618, 1–452) were tested for their ability to bind to immobilized importin α and snurportin1. Importin α and SPN1 were tagged with two immunoglobulin-binding domains of protein A from S. aureus (zz-tag), and zz-tagged proteins were immobilized to IgG-Sepharose beads. These beads were then incubated with the different importin β constructs. The eluted proteins were precipitated with isopropanol, dissolved in SDS-sample buffer and detected by Coomassie staining after 8% SDS-PAGE. The gel shows E. coli lysates of the different importin β constructs (lanes 1–3), as well as the bound fractions to importin α (lanes 4–6) and snurportin1 (lanes 7–9).
Importin β Fragment 1–618 That Does Not Bind to Importin α Is Able to Import SPN1
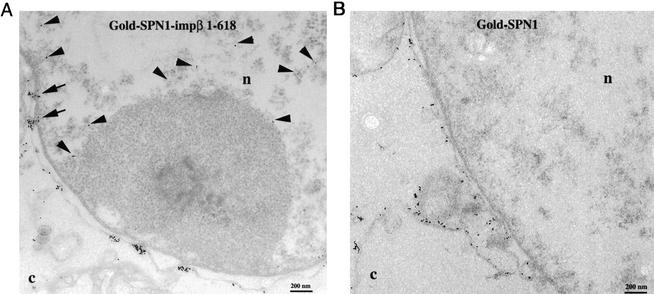
To obtain more insight on functional differences caused by molecular differences between SPN1 and importin α, we tested whether importin β fragment 1–618, which binds with high affinity to SPN1 but not to importin α, is able to support the nuclear import of U1 snRNPs (as it has been done with importin β fragments for the cNLS pathway; Görlich et al., 1996; Kutay et al., 1997b). To avoid competition with endogenous importin β, digitonin-permeabilized HeLa cells were used for these experiments instead of Xenopus oocytes. Recombinant SPN1 labeled with colloidal gold was used as import substrate. Gold-SPN1 was preincubated in vitro with recombinant importin β 1–618, and this complex was tested in our EM import assay in HeLa cells (see MATERIALS AND METHODS). Similarly to the nuclear import of histone H1 by importin β 1–618 (Jäkel et al., 1999), we found that this fragment crossed the NPC and carried SPN1 into the nucleus of permeabilized HeLa cells (Figure 7A). However, compared with control experiments using full-length importin β, the amount of gold-SPN1 found in the nucleus was reduced to ∼40% for importin β 1–618. In contrast, when the experiment was done with gold-SPN1 alone, the gold particles were not found in the nucleus of permeabilized HeLa cells (Figure 7B). Consistent with results from Huber et al. (2002), nuclear import of gold-SPN1-importin β 1–618 occurred in the absence of energy and Ran. The release of the gold-SPN1-importin β 1–618 complex from the nuclear side of the NPC into the nucleoplasm and its subsequent movement into the nucleus must have been very fast, because gold particles were found inside the nucleus but away from the NPC.
Figure 7.
Importin β 1–618 is able to import SPN1. Electron micrographs of cross-sectioned nuclear envelopes from digitonin-permeabilized HeLa cells that have been incubated with the gold-SPN1-importin β 1–618 complex (A) or with gold-SPN1 (B). Import was performed as described under MATERIALS AND METHODS. c, cytoplasm; n, nucleus. Bars, 200 nm.
The IBB Domain of Importin α and the IBB Domain of SPN1 Interact Differently with the NPC
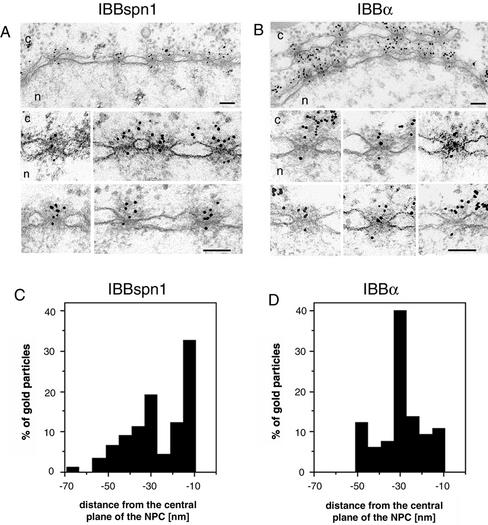
To investigate whether the differences in the nuclear import of cNLS-protein and U1 snRNPs is due to the IBB of the adapters, we performed import experiments with β-galactosidase molecules fused to the IBB domains of SPN1 (IBBspn1) or importin α (IBBα). Both IBBspn1 and IBBα molecules were conjugated to colloidal gold, and the IBB–importin β complexes were formed in vitro by incubating gold-IBBspn1 or gold-IBBα with importin β at a 1:1 M ratio. These complexes were then microinjected into the cytoplasm of Xenopus oocytes, and their nuclear import was followed by EM. As documented in Figure 8, A and B, gold particles from both gold-IBBspn1 and gold-IBBα, were found associated with both sides of the NPCs. Some gold particles were also found within the nucleus, indicating that the nuclear import had taken place. Quantitative analysis revealed that the distribution of gold particles associated with the nuclear side of the NPC was significantly different for the two gold-conjugated IBB molecules. As shown in Figure 8, C and D, the amount of gold particles accumulated at -30 nm was higher for IBBα than for IBBspn1, and at -10 nm it was lower for IBBα than for IBBspn1. Thus, the IBBα–import complex remains associated to the nuclear basket for longer time than the IBBspn1–import complex.
Figure 8.
The IBB domain of importin α and SPN1 interact differently with the NPC. Nuclear envelope cross sections with adjacent cytoplasm (c) and nucleoplasm (n) and a gallery of selected NPC cross sections from Xenopus oocytes that have been microinjected into their cytoplasm with in vitro-formed gold-IBBspn1-importin β (A) and in vitro-formed gold-IBBα-importin β complexes (B). The gold particles were 8 nm in diameter and were coupled with IBBspn1 or IBBα before the formation of the IBB–importin β complexes. After microinjection, oocytes were incubated at room temperature for 1 h and prepared for thin section electron microscopy as described under MATERIALS AND METHODS. Bars, 100 nm. (C and D) Quantitative analysis of gold particles associated with the nuclear face of the NPC for the experiments described in A and B, respectively. Nuclear envelopes from two different experiments were analyzed, which yielded 89 and 65 gold particles for C and D, respectively.
DISCUSSION
Despite recent advances in the understanding of how import signals are recognized by import receptors and in characterizing in vitro interactions between import receptors and nucleoporins, the molecular mechanism by which the cargo–receptor complex crosses the NPC has remained elusive. It is thought that while crossing the NPC the cargo–receptor complex has multiple interactions with many components of the NPC. However, it is not yet clear whether the different import substrates that cross the NPC share the same NPC docking, translocation, and releasing sites. To answer these questions, this article structurally analyzes the U1 snRNPs import pathway and compares its results with those of the cNLS import pathway. As it is discussed below, we have detected differences between the intermediate states for these two import pathways, which are a consequence of using different adapters between the transported molecule and importin β.
Different Initial NPC Binding Sites for the U1 snRNP and the cNLS–Protein Import Complex
Because both U1 snRNP and cNLS-proteins use importin β, it is considered that the interactions of the cargo–receptor complex with the NPC are the same in both cases. However, we found by both EM and pull-down assays that the nucleoporin CAN/Nup214 is involved in the U1 snRNP import pathway but not in the cNLS import pathway. This is in agreement with the recent finding by Walther et al. (2002) that this nucleoporin is not involved in nuclear import of cNLS-protein. Thus, the U1 snRNP and the cNLS import pathways have different initial NPC-binding sites at the cytoplasmic filaments, even though they both use importin β.
A second binding site at the cytoplasmic filament different from CAN/Nup214 was also observed for the U1 snRNP import pathway. We found that whereas U1 snRNP did not bind to the NPC, the in vitro-formed U1 snRNP–SPN1 complex bound to the NPC-cytoplasmic filaments in the presence of the anti-CAN/Nup214 antibody. From our data, we cannot distinguish whether this second cytoplasmic filament-binding site is the same as the first docking site for the cNLS import pathway (which is not inhibited by QE5).
The U1 snRNP Import Complex Is Formed at the Nucleoporin CAN/Nup214
We found that the cytoplasmic preinjection of QE5 did not prevent the interaction of the preformed U1 snRNP–SPN1 complex with the NPC cytoplasmic filaments. Similarly to the result with the cNLS-protein (Figure 2B), the interaction of the in vitro-formed U1 snRNP–SPN1complex with the cytoplasmic side of the central channel was inhibited (because QE5 recognizes p62). It seems that if SPN1 is present in the import complex, it can skip the first binding site at the nucleoporin CAN/Nup214. These results support the conclusions that U1 snRNP binds to CAN/Nup214 without forming an import complex with SPN1 in the cytoplasm.
We were surprised to find two different binding sites at the cytoplasmic filament for the U1 snRNP import pathway. These results raise the question of why the U1 snRNP pathway requires two binding sites at the cytoplasmic filaments, whereas the cNLS pathway requires only one. Our explanation for this difference is that, most probably, the first binding of U1 snRNP to CAN/Nup214 is mediated by its interaction with SPN1, which is already bound to CAN/Nup214 via CRM1 when SPN1 is exported (i.e., the SPN1 export complex). Thus, the first NPC interaction of U1 snRNP via CAN/Nup214 enables the U1 snRNP to interact with SPN1 and to form the U1 snRNP import complex directly at the NPC. This is in contrast to the cNLS pathway where the cargo–receptor complex is formed before the interaction with the NPC (Görlich et al., 1995).
Importin β Is a Carrier That Can Interact with Cargo in Three Different Modes
Two different importin β-binding domains for cargo have been previously identified. The first one involves the 3/4 C-terminal region of importin β (residues 256–876; Kutay et al., 1997b; Cingolani et al., 1999), which binds to the IBB domain of importin α. The crystal structure of importin β bound to the IBB domain of importin α shows that when the two molecules interact, the IBB of importin α fits within the helical structure of the snail-shaped molecule of importin β (Cingolani et al., 1999). Thus, the IBB is embedded within the structure of importin β. The second cargo-binding domain of importin β comprises residues 286–462, which binds to the BIB (β-like import receptor binding) domain of the ribosomal protein L23a (Jäkel and Görlich, 1998). This article reports a third mode of cargo-binding to importin β. Although the N-terminal 618 residues of importin β are able to bind to SPN1, the same importin β fragment does not interact with importin α. This result was surprising because of the high degree of homology between the IBB of SPN1 and the IBB of importin α (31% identity, 62% similarity; Huber et al., 1998). Most probably, the IBB of SPN1 folds in a conformation that cannot fit within the helical structure of importin β like the IBB of importin α does (Cingolani et al., 1999). SPN1 binding to importin β also seems to be different from the BIB binding, because transport receptors such as transportin, importin 5 and importin 7, which greatly stimulate nuclear uptake of the BIB domain of L23a, do not promote the efficient nuclear import of SPN1 (our unpublished data).
The SPN1-binding domain on importin β might partially overlap with the Ran-binding domain (residues 1–342; Kutay et al., 1997b; Vetter et al., 1999). Crystallographic studies have shown that the Ran-binding domain is a loop within the importin β molecule, which corresponds to the small tail of the snail-like molecule (Vetter et al., 1999). If the IBB of SPN1 fits into this loop, this could explain that Ran is not required for the nuclear import of U1 snRNPs (Huber et al., 2002). Alternatively, the binding of SPN1 to importin β might modify the conformation of this loop in such a way that RanGTP cannot bind to importin β.
Importin β seems to be a highly flexible molecule that can adopt a number of different conformations (Cingolani et al., 1999; Vetter et al., 1999; Bayliss et al., 2000). The mode of interaction with the import cargo may force importin β into different conformations, which, in turn, determine the Ran requirement of the NPC passage. This leads to the assumption that importin β, when bound to different cargo, changes its way of interaction with the NPC.
The Last Step of Nuclear Import at the NPC Depends on the Nature of the Adapters
After the translocation through the central channel of the NPC, the cNLS–receptor complex binds to the nuclear basket of the NPC. From this last NPC-binding site the cargo– receptor complex is then released into the nucleus, a step that requires the binding of nuclear RanGTP to importin β (Görlich et al., 1996). For the U1 snRNP import pathway we have also observed interaction of the cargo–receptor complex with the nuclear baskets. Using complexes of importin β with the IBB domain of SPN1 or with the IBB domain of importin α we found that the nature of the IBB domain of the adapters influences the association of the gold-IBB with the nuclear basket. There were more gold particles associated with the nuclear baskets for gold-IBBα than for IBBspn1-gold. An interpretation of this result is that the dissociation of the import cargo from the NPC and its delivery into the nucleus is faster for the U1 snRNP import pathway than for the cNLS import pathway. Alternatively, because different regions of importin β are engaged in the binding with the different adapters, depending on whether SPN1 or importin α is bound to importin β, it will interact with different nucleoporins at the release site. Our data support both interpretations. However, because RanGTP is not required for SPN1-mediated nuclear import (Huber et al., 2002), one can speculate that the different gold distribution at the nuclear basket is because the IBBspn1 import complex does not have to wait for nuclear RanGTP to bind to importin β. Thereby, IBBspn1-gold released from the nuclear basket faster than gold-IBBα.
In summary, our structural analysis of the U1 snRNP import pathway is consistent with a model in which the U1 snRNP binds to SPN1 in its export complex at CAN/Nup214. The U1 snRNP-SPN1 import complex then incorporates importin β to advance the translocation through the NPC. Similarly to the cNLS import complex, after the translocation through the central channel, the U1 snRNP–receptor complex binds to the nuclear basket. This interaction might involve the binding of importin β to a region of Nup153 (or other nucleoporins located at the nuclear baskets) that is different from the one used by importin β when it is loaded with a cNLS-cargo. This importin β–nucleoporin interaction might involve different regions of importin β, depending on the import cargo. From this final NPC-binding site, the import complex dissociates and the cargo is released into the nucleus. However, because different regions of importin β are involved in the interaction with SPN1 and importin α, RanGTP is not required for the release of the m3G-cap-cargo, and the kinetics of this step is faster than the release of the cNLS-cargo. Thus, our comparative analysis of the U1 snRNP and the cNLS import pathways points to the differences in the NPC docking and releasing steps of each pathway. It will be interesting to investigate whether this is also true for other nuclear import pathways.
Acknowledgments
We thank Drs. Reinhard Lührmann, Jochen Huber, and Achim Dickmanns for helpful advice and discussion during the course of this work, and for the gifts of reagents. We thank Drs. Brian Burke and Iain Mattaj for providing antibodies. This work was supported by a grant from the Swiss National Science Foundation (3100-053034) and by a grant from the Natural Sciences and Engineering Research Council of Canada.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-06-0372. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-06-0372.
Abbreviations used: BSA, bovine serum albumin; cNLS, classical nuclear localization sequence; cNLS-protein, cNLS-bearing protein; EM, electron microscopy; IBB, importin β-binding; IBBα, recombinant β-galactosidase molecules fused to the IBB domain of importin α; IBBspn1, recombinant β-galactosidase molecules fused to the IBB domain of snurportin1; NPC, nuclear pore complex; SPN1, snurportin1; WGA, wheat germ agglutinin; zz-tag, two immunoglobulin-binding domains of Staphylococcus aureus protein A.
References
- Adam, S.A., Sterne-Marr, R., and Gerace, L. (1990). Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschong, W., and Wrigley, N.G. (1990). Small colloidal gold conjugated to Fab fragments or to immunoglobulin G as high-resolution labels for electron microscopy: a technical overview. J. Electron. Microsc. Tech. 14, 313–323. [DOI] [PubMed] [Google Scholar]
- Bastos, R., Lin, A., Enarson, M., and Burke, B. (1996). Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss, R., Littlewood, T., and Stewart, M. (2000). Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 102, 99–108. [DOI] [PubMed] [Google Scholar]
- Cingolani, G., Petosa, C., Weis, K., and Müller, C.W. (1999). Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229. [DOI] [PubMed] [Google Scholar]
- Conti, E., and Izaurralde, E. (2001). Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., Hurt, E., Aebi, U., and Panté, N. (1998). Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol. 143, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, U., Darzynkiewicz, E., Tahara, S.M., Dathan, N.A., Lührmann, R., and Mattaj, I.W. (1991). Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J. Cell Biol. 113, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, U., and Lührmann, R. (1990). An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science 249, 786–790. [DOI] [PubMed] [Google Scholar]
- Fischer, U., Sumpter, V., Sekine, M., Satoh, T., and Lührmann, R. (1993). Nucleo-cytoplasmic transport of U1 snRNPs: definition of a nuclear location signal in the SM core domain that binds a transport receptor independently of the m3G-cap. EMBO J. 12, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–60. [DOI] [PubMed] [Google Scholar]
- Görlich, D., Panté, N., Kutay, U., Aebi, U., and Bischoff, F.R. (1996). Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., Vogel, F., Mills, A.D., Hartmann, E., and Laskey, R.A. (1995). Distinct functions for the two importin subunits in nuclear protein import. Nature 377, 246–248. [DOI] [PubMed] [Google Scholar]
- Guan, T., Müller, S., Kleir, G., Panté, N.,. Blevitt, J.M, Häner, M., Paschal, B., Aebi, U., and Gerace, L. (1995). Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell 6, 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm, J., Darzynkiewicz, E., Tahara, S.M., and Mattaj, I.W. (1990). The trimethylguamosin cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell 62, 569–577. [DOI] [PubMed] [Google Scholar]
- Huber, J., Cronshagen, U., Kadokura, M., Marshallsay, C., Wada, T., Sekine, M., and Lührmann, R. (1998). Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17, 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, J., Dickmanns, A., and Lührmann, R. (2002). The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 156, 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel, S., Albig, W., Kutay, U., Bischoff, F.R., Schwamborn, K., Doenecke, D., and Görlich, D. (1999). The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel, S., and Görlich, D. (1998). Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnik, M., and Aebi, U. (1991). Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 107, 291–308. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Ohno, M., and Mattaj, I.W. (2001). Nucleocytoplasmic transport. Ran, β and beyond. Trends Cell Biol. 11, 497–503. [DOI] [PubMed] [Google Scholar]
- Kutay, U., Bischoff, F.R., Kostka, S., Kraft, R., and Gorlich, D. (1997a). Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay, U., Izaurralde, E., Bischoff, F.R., Mattaj, I.W., and Görlich, D. (1997b). Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I.G. (2001). Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshallsay, C., and Lührmann, R. (1994). In vitro nuclear import of snRNPs: cytosolic factors mediate m3G-cap dependence of U1 and U2 snRNP transport. EMBO J. 13, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshallsay, C., Dickmanns, A., Bischoff, F.R., Ponstingl, H., Fanning, E., and Luhrmann, R. (1996). In vitro and in vivo evidence that protein and U1 snRNP nuclear import in somatic cells differ in their requirement for GTP-hydrolysis, Ran/TC4 and RCC1. Nucleic Acids Res. 24, 1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, N., and Goldfarb, D.S. (1992). Microinjected U snRNAs are imported to oocyte nuclei via the nuclear pore complex by three distinguishable targeting pathways. J. Cell Biol. 116, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I., Hetzer, M., Adam, S.A., and Mattaj, I.W. (1997). Nuclear import of U snRNPs requires importin β. EMBO J. 16, 6783–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I., Weis, K., Klebe, C., Mattaj, I.W., and Dingwall, C. (1996). Ran/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J. Cell Biol. 133, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté, N., and Aebi, U. (1996). Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science 273, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Panté, N., Bastos, R., Mc Morrow, I., Burke, B., and Aebi, U. (1994). Interactions and three-dimensional localization of a group of pore complex proteins. J. Cell Biol. 126, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva, E., Izaurralde, E., Bischoff, F.R., Huber, J., Kutay, U., Hartmann, E., Lührmann, R., and Görlich, D. (1999). CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu, A., Blobel, G., and Moore, M.S. (1995). Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA 92, 1769–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt, R., Holzenburg, A., Buhle, E.L., Jr., Jarnik, M., Engel, A., and Aebi, U. (1990). Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 110, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach, M., and Blobel, G. (1995). Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Ryan, K.J., and Wente, S.R. (2000). The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12, 361–371. [DOI] [PubMed] [Google Scholar]
- Slot, J.W., and Geuze, H.J. (1985). A new method of preparing gold probes for multiple-labeling cytochemistry. Eur. J. Cell Biol. 38, 87–93. [PubMed] [Google Scholar]
- Vetter, I.R., Arndt, A., Kutay, U., Görlich, D., and Wittinghofer, A. (1999). Structural view of the Ran-importin β interaction at 2.3 A resolution. Cell 97, 635–646. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Pickersgill, H.S., Cordes, V.C., Goldberg, M.W., Allen, T.D., Mattaj, I.W., and Fornerod, M. (2002). The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]