Abstract
CLA4, encoding a protein kinase of the PAK type, and CDC11, encoding a septin, were isolated in a screen for synthetic lethality with CHS3, which encodes the chitin synthase III catalytic moiety. Although Ste20p shares some essential function with Cla4p, it did not show synthetic lethality with Chs3p. cla4 and cdc11 mutants exhibited similar morphological and septin localization defects, including aberrant and ectopic septa. Myo1p, which requires septins for localization, formed abnormally wide rings in cla4 mutants. In cultures started with unbudded cells, an inhibitor of Chs3p activity, nikkomycin Z, aggravated the abnormalities of cla4 and cdc11 mutants and gave rise to enlarged necks at the mother-bud junction, leading to cell death. It is concluded that Cla4p is required for the correct localization and/or assembly of the septin ring and that both the septin ring and the Chs3p-requiring chitin ring at the mother-bud neck cooperate in maintaining the neck constricted throughout the cell cycle, a vital function in budding yeast.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, a chitin ring is formed in the cell wall at the base of an emerging bud, in a reaction requiring chitin synthase III, of which Chs3p is the putative catalytic unit (for reviews see Cabib et al., 1996, 2001). After further bud growth and mitosis, a primary septum is constructed between mother and daughter cell, requiring the joint action of three elements: the septin filament ring, which seems to serve as a scaffold for other components of the septation apparatus (for reviews see Longtine et al., 1996; Gladfelter et al., 2001); the actomyosin contractile ring, which invaginates the plasma membrane inward at cytokinesis (Tolliday et al., 2001); and the chitin synthase II system. This system, with Chs2p as the catalytic subunit, acts in the formation and secretion of chitin into the membrane invagination, giving rise to the primary septum and separating mother and daughter cell (Cabib et al., 1996; Schmidt et al., 2002). Finally, secondary septa are formed from both dividing cells and a trilaminar structure is built, with the chitin layer in the middle. At this point, the chitin ring encircles the septum and is contiguous to it. After cell separation, the ring ends up as the rim of the bud scar, which remains on the mother cell and contains most of the septum. We have reported earlier (Slater et al., 1985) and confirmed recently (Roh et al., 2002b) that in septin mutants structures morphologically similar to septa are made at ectopic locations in the cell. This result emphasizes both the scaffold function of the septin ring and the autonomous nature of the septation apparatus. It also highlights the need for cellular mechanisms for the organization and retention of the septins at the mother-bud neck.
In contrast with all the changes taking place at the mother-bud junction, the diameter of the neck, as measured from the inner surface of the cell wall, remains unmodified during septation and indeed throughout the cell cycle. The mechanisms that prevent the neck from being enlarged in the face of adjacent growth of the bud are unknown. We found information on these mechanisms in an unexpected way. In an attempt to identify genes necessary for Chs2p function, we had initiated a genetic screen for mutants that, like chs2, would show synthetic lethality with a chs3 mutation. Two genes isolated in that screen, CLA4 and CDC11, coding for a protein kinase of the PAK type (Cvrcková et al., 1995) and for a septin, respectively, did not seem to have a direct role in Chs2p function. On the other hand, a study of mutants in those genes and of their interaction with the chs3 mutation has shown that septins and the chitin ring formed at bud emergence cooperate in the maintenance of mother-bud neck size. When the function of both systems is compromised simultaneously, the neck enlarges and the cell eventually dies, which shows that neck integrity is essential for viability.
MATERIALS AND METHODS
Strains and Growth Conditions
The yeast strains used in this study are listed in Table 1. Growth media and conditions were as previously described (Schmidt et al., 2002), except that synthetic complete media (SC) were prepared from dropout powders (Qbiogene, Carlsbad, CA).
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| ECY36-3D | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 | Shaw et al. (1991) |
| ECY46-4-1B | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2Δ1 chs3::LEU2 | Crotti et al. (2001) |
| ECY101 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG [pEC28] | This study |
| ECY101-39 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG cla4-39 [pEC28] | This study |
| ECY105 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG ste20::URA3 [pEC28] | This study |
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | Sikorski and Hieter (1989) |
| YMS134 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cla4::LEU2 | This study |
| YMS189 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG cla4-39 swe1::kanMX6 [pEC28] | This study |
| YMS190 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 chs3::LEU2 cla4::URA3 swe1::kanMX6 | This study |
| YMS197 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG cla4::URA3 [pEC28] | This study |
| YMS306 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cla4::TRP1 | This study |
| YMS332 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cla4::LEU2 swe1::kan MX6 | This study |
| 1238 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 swe1::kanMX6 | Research Genetics |
| DHY103-9B | MATaura3-52 lys2 his3-Δ200 leu2 tyr1 cdc3-1 [pMS55] | Roh et al. (2002b) |
| AVY1 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG ade3::hisG | This study |
| AVY2 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG ade3::hisG leu2::TRP1 [pAV1] | This study |
| AVY2-25 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG ade3::hisG leu2::TRP1 cdc11-25 [pAV1] | This study |
| AVY3 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG ade3::hisG leu2::TRP1 cdc11-25 cla4::LEU2 [pAV1] | This study |
| AVY5 | MATaura3-52 leu2-2 trp1-1 chs1-23 chs3-1 ade2::hisG ade3::hisG leu2::TRP1 cdc11-25 | This study |
Strain Construction
General methods of DNA manipulation were as described in Ausubel et al. (1994). Yeast transformation was carried out with the lithium acetate method (Ito et al., 1983). Deletion of ADE2 in ECY36-3D was carried out with a deletion cassette from ATCC vector 99604 (pΔADE2; Aparicio et al., 1991), according to instructions from the supplier. The URA3 initially inserted in the ADE2 gene between two hisG was eliminated by growth on uracil-containing medium and plating on fluoroorotic acid medium. The resulting strain, ECY101, was transformed with pEC28 to generate ECY101[pEC28]. This strain was used for mutagenesis and red-white selection.
STE20 was disrupted in strain ECY101[pEC28] by transformation with a ste20::URA3 cassette cut from plasmid pEL45 (Leberer et al., 1992) with SalI and XbaI, to yield ECY105 (Table 1).
The cla4::LEU2 gene disruption was achieved by transforming yeast with SphI/SmaI-cut plasmid pFD26 (Cvrcková et al., 1995). For disruption of CLA4 with URA3 or TRP1, plasmid pMS17, was cut with MluI/XcmI. The DNA fragment was then blunted and ligated to the reporter genes, prepared as follows. A 1.20-kb URA3 fragment or a 1.0-kb TRP1 fragment was generated by PCR, blunted, and phosphorylated. The blunt-ended ligations yielded plasmid pMS32 (URA3) and pMS46 (TRP1), respectively. The disruption fragments were amplified by PCR from these plasmids, using primers CLA4UP: 5′-AGTAGAGGAGATCTACAAACTTGA-3′ and CLA4DOWN: 5′-GATATGCTTCTAGAAATAGTTGTGTG-3′.
Disruption of SWE1 with the kanMX6 module was performed by amplifying the swe1::kanMX6 allele from strain 1238 (Invitrogen, Carlsbad, CA) via PCR with primers SWE1UP: 5′-TTGAACATTGGCGTGCCC-3′ and SWE1DOWN: 5′-TTATCTGCTACATCTGTAA-3′.
Disruption of MYO1 was obtained as described by Schmidt et al. (2002).
Deletion of ADE3 in ECY101 was carried out with an ade3::hisG-URA3-hisG deletion cassette amplified by PCR from plasmid pAV4. From the resulting strain, URA3 was eliminated by growth on uracil-containing medium followed by plating on fluoroorotic acid medium. The resulting strain, AVY1, was transformed with pAV1 to generate AVY1[pAV1].
Deletion of LEU2 in AVY1[pAV1] was done with a disruption cassette cut out from pAV10 by HpaI-SphI double digestion, thus generating strain AVY2. This strain was used for mutagenesis and red-white selection.
To check the effect of a CLA4 deletion in the cdc11-25 mutant (AVY2–25), the CLA4 gene was disrupted with a cla4::LEU2 disruption cassette as described above.
All disruptions were confirmed by PCR. Construction of a cdc11-25 chs3-1 double mutant, AVY5, was done by segregating the plasmid pAV1 from the strain AVY2–25. This was achieved by streaking AVY2–25 on YEPD-agar containing 1 M sorbitol at 26°C. Those cells that lost the plasmid formed white sectors or white colonies. Loss of plasmid was confirmed by Calcofluor White staining, growth on Calcofluor White, ability to grow on fluoroorotic acid, and uracil auxotrophy at 26°C.
Plasmid Construction
To overexpress a nonfunctional chs3R995A allele, the mutant allele contained in plasmid pHV7–37 (C. Roncero) was excised with ClaI and SalI and cloned into YEp352 cut with the same enzymes, yielding vector pMS75 (Table 2). To construct pAS8, the CHS2 ORF contained in the multiple cloning site of YEp352 (pEC2, Ford et al., 1996) was cut out with EcoRV and SacI and ligated to SmaI/SacI digested vector pRS314.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| Library in pRS200 | ATCC 77164 | |
| pΔADE2 | p[ade2::hisG::URA3::hisG] | Aparicio et al. (1991) (ATCC 99604) |
| pEC2 | YEp352CHS2 | Ford et al. (1996) |
| pEC28 | pRS412CHS3 | Schmidt et al. (2002) |
| pEL45 | pBluescript KS+ ste20::URA3 | Leberer et al. (1992) |
| pFD26 | cla4::LEU2 | Cvrcková et al. (1995) |
| pFLC1 | pGALISTE20 | S. Davis |
| pHV7-37 | YCp50chs3R995A | C. Roncero |
| pHV8 | YEp352CHS3 | Valdivieso et al. (1991) |
| pLP17 | pRS315CDC12-GFP | Lippincott and Li (1998b) |
| pNKY50 | YEp24 hisG::URA3::hisG | Alani et al. (1987) |
| pSM491 | Contains triple HA tag | S. Michaelis |
| pMS17 | pRS200CLA4 | This study |
| pMS32 | pRS200cla4::URA3 | This study |
| pMS46 | pRS200cla4::TRP1 | This study |
| pMS39 | From pRS200 library | This study |
| pMS55 | pRS316MYO1-GFP | Schmidt et al. (2002) |
| pMS63 | pRS426MYO1-GFP | This study |
| pMS75 | YEp352chs3R995A | This study |
| pMS76 | pRS313CDC12-GFP | This study |
| pMS79 | YEp352chs3R995A::HA | This study |
| pMS80 | YEp352CHS3::HA | This study |
| pRS316CDC3GFP | CDC3-GFP | M. Longtine |
| p366 | YCp50 with URA3 replaced by LEU2 | ATCC 77163 |
| Library in p366 | ATCC 77162 | |
| pFD10 | YCp50ADE3 | A. Bender |
| pAS8 | pRS314CHS2 | This laboratory |
| pLP8 | pRS315MYO1-GFP | Lippincott and Li, (1998a) |
| p19 | From p366 library | This study |
| pAV1 | YCp50CHS3ADE3 | This study |
| pAV4 | YCp50(URA3 cut out) + ade3::hisG-URA3-hisG | This study |
| pAV10 | YEp351leu2::TRP1 | This study |
| pAV11 | p366mog1HOC1CDC11mig1 | This study |
| pAV12 | YEp351CDC11 | This study |
| pAV13 | p366CDC11 | This study |
| pAV17 | YEp351cdc11-25 | This study |
| pAV21 | pRS425MYO1-GFP | This study |
To obtain pMS76, the LEU2-containing PvuI fragment from pLP17 (Lippincott and Li, 1998b) was replaced with the HIS3-containing PvuI fragment from pRS313. For construction of the screening plasmid, pAV1, a 4.6-kb PvuII fragment containing the CHS3 gene was cut out from pHV8 and ligated to pFD10 at the SmaI site.
For the construction of an ADE3 disruption cassette, a 2.5-kb fragment containing the URA3 gene was removed from pFD10 by BamHI digestion. The remaining plasmid was religated and a 1665-base pair AgeI-PvuII fragment was replaced by a 3.8-kb BamHI-BglII fragment of hisG-URA3-hisG from plasmid pNKY50 after Klenow filling-in treatment. This yielded plasmid pAV4.
To construct a leu2::TRP1 disruption cassette, a 5000-base pair AgeI-EcoRV fragment from YEp351 was replaced with a blunt ended 1.0-kb TRP1 fragment amplified by PCR from pRS314, yielding plasmid pAV10.
To clone CDC11, the gene was amplified by PCR from p19 with oligonucleotides CDC11HindIIIUP: 5′-CTGTAAATTAACAAGCTTTTATAAATAT-3′ with an engineered HindIII site and CDC11SphIDN: 5′-CTCATTTGGCATGCCAATTTTGG-3′ with an engineered SphI site. The PCR product was digested with HindI-SphI and ligated to p366 digested with the same enzymes, resulting in pAV13.
To clone CDC11 in YEp351, the CDC11 PCR product from p19 with the above mentioned oligonucleotides was blunt-ended, phosphorylated, and ligated to YEp351 at the SmaI site, yielding plasmid pAV12.
pAV21 was constructed as follows: an ∼8-kb PvuI fragment from pLP8 was ligated to the large PvuI fragment from pRS426 to yield pMS63. From pMS63, the ∼8-kb PvuI fragment was cut and ligated to the 5.2-kb PvuI fragment of pRS425 to yield pAV21.
Mutagenesis
Mutagenesis of strain ECY101 with ethyl methane sulfonate was carried out as described in Ausubel et al. (1994). Mutagenized cells were plated on SD agar containing nutritional requirements and 10 μg/ml adenine. White colonies were transferred to plates with the same medium and cell morphology was checked after growth by phase contrast microscopy. Those colonies showing similarity to chs2 mutants, i.e., clumps of cells with some cells of aberrant shape, were picked for further study.
The same method was used with strain AVY2, except that red colonies were selected.
Cloning and Sequencing of CLA4 and cla4-39
Mutant strain ECY101-39 was transformed with a genomic library contained in vector pRS200 (ATCC 77164). Transformants were screened for their ability to segregate the ADE2-containing plasmid pEC28, leading to the appearance of red sectors, and for the correction of the mutant morphology. Plasmid pMS39 was isolated from two of the transformants. pMS39 induced sectoring of transformant colonies and corrected the mutant morphology after retransformation into the original mutant. The DNA contained in the cloning site of pMS39 was sequenced using T3 and T7 primers, the ABI prism sequencing kit, and the ABI-prism 310 sequencer according to manufacturer's (Perkin Elmer-Cetus, Oak Ridge, TN) instructions. The insertion in the cloning site was found to originate from chromosome XIV, position 71828–63727. Removing a SgrAI/EcoRI fragment from this vector left CLA4 as the only intact reading frame in the insert. This truncated plasmid pMS17 still complemented both morphology and sectoring phenotype of ECY101-39; therefore, the mutant allele was designated cla4-39. To determine the nature of the cla4-39 mutation, the allele was sequenced from ECY101-39 chromosomal DNA as described above.
Cloning and Sequencing of cdc11-25
The mutant strain AVY2-25 was transformed with a genomic library in the vector p366 (ATCC 77162). The transformants were screened for their ability to segregate the ADE3 CHS3 containing plasmid pAV1, leading to the formation of white sectors and for the correction of the mutant morphology. From three of the transformants, plasmid p19 was isolated, which was able to induce sectoring of the transformant colonies and complement the mutant morphology when transformed into the original mutant AVY2-25. The DNA insert in p366 cloning site was sequenced (Seqwright DNA Sequencing) with oligos: 5′-GCCACTATCGACTACGCGATC-3′ and 5′-GTGGCGCCGGTGATGCCGCT-3′. The insert in p19 was found to be from chromosome X at position 572600–583595. A 7-kb HindIII fragment of p19 (pAV11) was able to induce sectoring and complement mutant morphology of AVY2-25. The ORFs in this fragment were further subcloned. It was found that CDC11 was the only ORF able to induce the sectoring phenotype and complementation of the mutant morphology.
To determine the position of the cdc11-25 mutation, the mutant DNA allele was recovered from AVY2-25 by gap repair (Rothstein, 1991). Gap-repaired plasmid pAV17 was isolated from the colonies, which showed a restriction pattern identical to pAV12 but was unable to complement the mutant phenotype of cdc11-25 mutant and did not induce sectoring. The insert was sequenced with several oligonucleotides from the flanking regions of CDC11. The sequences revealed a G-A change at position 95, changing the GGA triplet codon for glycine to glutamic acid (G32E).
Quantifying Chs3R995A Expression
To quantify the expression of the mutant Chs3p, we introduced a triple-HA tag into the N-terminus of the protein. The tag was amplified from pSM491 with primers CHS3HAUP: 5′-GGCCGCACCGGTTACCCATACGATGTTCCT-3′ and CHS3HADOWN: 5′-GCGGCCACCGGTAGCAGCGTAATCTGGAAC-3′ (engineered AgeI sites in bold). Plasmids pHV8 and pMS75, encoding the wild-type and mutant Chs3p, respectively, were linearized with AgeI and ligated to the PCR fragments, yielding plasmids pMS79 (chs3R995A::HA) and pMS80 (CHS3::HA). Both plasmids were then introduced into strain ECY46-4-1B (chs3::LEU2). Calcofluor White staining of transformants showed that pMS80 gave rise to wild-type chitin synthase III activity, whereas pMS79 did not. Sample preparation and Western blotting were as described previously for Spa2HA (Schmidt et al., 2002).
Electron Microscopy
Electron microscopy was carried out as previously described (Schmidt et al., 2002).
Fluorescence Microscopy
Fluorescence of GFP-tagged proteins was observed as already reported (Roh et al., 2002b). Septin rings showing breaks in continuity or an asymmetric distribution of septin-GFP were scored as abnormal.
Isolation of Single, Unbudded Cells
Single, unbudded cells were isolated by centrifugation on sucrose gradients as already described (Drgonová et al., 1999), except that methylmannoside was not added to the cell suspension before sonication.
RESULTS
Isolation of CLA4 in a Screen for Synthetic Lethality with CHS3
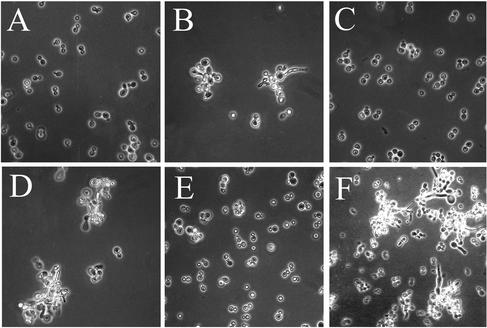
A genetic screen for mutants that would be inviable in a chs3 background was conducted with a modification of the red-white selection (Bender and Pringle, 1991; see MATERIALS AND METHODS). In this modification, an ade2Δ chs3-1 strain that can lose a plasmid containing ADE2 and CHS3 (ECY101[pEC28]) gives rise to white colonies with red sectors, whereas strains containing a mutation that is synthetically lethal with chs3-1 cannot lose the plasmid and therefore remain white. To increase the probability of selecting mutants compromised in septation, only white colonies whose cells showed some clumping were chosen for further study. One such strain was mutant ECY101-39 (Figure 1B).
Figure 1.
Morphology of different strains related to the isolation o'f a cla4 mutant. (A) Wild-type (ECY101). (B) ECY101-39 (cla4-39 mutant). (C) ECY101-39 transformed with pMS17 (pRS200CLA4). (D) YMS197 (ECY101 in which CLA4 was disrupted). (E) ECY105 (ECY101 in which STE20 was disrupted). (F) YMS197[pFLC1] (cla4:: URA3 with STE20 overexpressed by growth in 2% galactose medium).
After transformation of ECY101-39 with a genomic DNA library, one plasmid was found to restore the sectoring. The insert was subcloned and it was found that a fragment containing as the only ORF CLA4, which encodes a protein kinase of the PAK type (Cvrcková et al., 1995), was able to complement both the morphology and the sectoring defect of ECY101-39 (Figure 1C); therefore, the mutation was designated as cla4-39. By appropriate crosses it was determined that ECY101-39 carries a recessive mutation in the CLA4 locus. Sequencing of the allele detected a G-A transition at position 200, turning the TGG triplet coding for Trp67 into a TAG stop codon and leaving a 66-amino acid Cla4p fragment that is not functional (Benton et al., 1997). In fact, a null mutant of CLA4 behaved as cla4-39 with respect both to lack of sectoring and morphology (Figure 1D).
Although by the sectoring assay cla4Δ and chs3-1 behaved as synthetically lethal, we were able to obtain a cla4Δ chs3Δ double mutant by deleting CLA4 in a chs3::LEU2 strain. The double mutant, however, grew very slowly, giving rise to large aggregates and was temperature sensitive (our unpublished results). Even at 30°C, the cells showed a wide neck and many cells lysed, which may explain the slow growth. Furthermore, null mutations of Bni4p, which is needed for binding of Chs3p to the septin ring (DeMarini et al., 1997), and of Chs4p, which is required both for that binding (DeMarini et al., 1997) and for enzymatic activity of Chs3p (Choi et al., 1994a; Trilla et al., 1997), clearly aggravated the morphological defect of a cla4Δ strain, especially when the double mutants were shifted from minimal medium to YEPD (our unpublished results).
STE20 Does Not Complement the Observed cla4 Defect
CLA4 and STE20, which encodes another kinase of the PAK type (Leberer et al., 1992) have been found to be synthetically lethal (Cvrcková et al., 1995), therefore it has been postulated that they have some function(s) in common. To explore whether any such function was involved in the defects observed in the cla4 mutants, we deleted STE20 in the strain used for mutagenesis (ECY101[pEC28]). The resulting strain showed normal sectoring and wild-type morphology (Figure 1E). Moreover, when STE20 was overexpressed in a cla4Δ strain by placing it under the control of the GAL1 promoter, the morphological defects of the strain were unchanged (Figure 1F). We conclude that the defects observed in the cla4 mutants reveal function(s) of Cla4p that are not shared with Ste20p.
CDC11 Shows Synthetic Lethality with CHS3
A screen similar to that outlined above but done exactly as described by Bender and Pringle (1991) was carried out. In this case, the original strain, AVY2, contained both ade2 and ade3 deletions, as well as a chs3-1 mutation and a plasmid carrying CHS3 and ADE3. With this approach, the colonies that cannot lose the plasmid are red and those that can lose it show white sectors. After mutagenesis, a red colony was isolated that showed cell clumps and elongated cells when the culture was grown at 30°C (Figure 2, C and D). The morphology was closer to that of wild-type at 26°C (Figure 2, A and B).
Figure 2.
Morphology of the cdc11-25 mutant isolated in the screen for synthetic lethality with chs3-1 and of related strains. (A and B) AVY2–25 (cdc11-25) at 26°C in minimal and YEPD medium, respectively. (C and D) AVY2–25 at 30°C in minimal and YEPD medium, respectively. (E and F) AVY5 (cdc11-25 chs3-1) at 26°C in minimal and YEPD medium, respectively. (G) AVY2–25 transformed with pAV13 (YEp351CDC11) at 30°C in minimal medium. (H) AVY3 (cdc11-25 cla4:: LEU2) at 26°C in minimal medium.
After transformation with a genomic library, one plasmid was found to complement the defect. The insert was subcloned and a plasmid containing CDC11, which codes for a septin, as the only ORF was able to complement both the morphological and the sectoring defect of strain AVY2–25 (Figure 2G). Crosses with cla4Δ and chs3Δ strains showed that the mutant does not harbor a mutation in CLA4 and that the mutation is recessive. Sequencing located the cdc11-25 mutation at position 32, changing a GGA triplet codon for glycine to GAA for glutamic acid.
As in the case of the cla4 mutant, it was possible to obtain a cdc11-25 chs3-1 double mutant (see MATERIALS AND METHODS). The double mutant, although viable, showed a more aberrant morphology than cdc11-25 both at 26 and 30°C in minimal medium (Figure 2, E and F) and did not grow in YEPD at 30°C. We also disrupted CLA4 in the cdc11-25 mutant, which resulted in a much more abnormal morphology (Figure 2H).
Both cla4 and cdc11 Mutants Show Septin Defects That are Exacerbated by a chs3 Mutation
Because both a cla4 strain and a septin mutant, cdc11-25, showed synthetic lethality in a red-white screen and because some septin defects were previously observed in cla4 strains (Cvrcková et al., 1995; Longtine et al., 2000), we decided to examine in detail the septin organization and localization in both cla4Δ and cdc11-25 strains. Septins fused to GFP were used for visualization of the septin rings. A cla4::TRP1 mutant showed many more aberrant rings than wild-type as well as some mislocalized patches (Figure 3B and Table 3). A manifestation of the cla4 phenotype is the formation of elongated buds (see above and Cvrcková et al., 1995; Longtine et al., 2000). To find out whether this modality of growth had given rise to the septin defects, we introduced in a cla4Δ strain a swe1 deletion that eliminates the elongation (Longtine et al., 2000). The double mutant, although clumpy, did not show elongated buds (Figure 3E), as expected, but the septin defect was similar to that of a single cla4Δ mutant (Figure 3C and Table 3). Although a swe1 deletion suppresses lethality of a double cla4 ncs1 mutant (Mitchell and Sprague, 2001) a cla4 swe1 double mutant, either in the ECY101-39 background (YMS189) or in the YPH499 background (YMS190), still showed synthetic lethality with chs3, as shown by lack of sectoring (see also below).
Figure 3.
Abnormalities in septin ring organization in cla4Δ, cla4Δ swe1Δ and chs3Δ mutants. All four strains shown were transformed with plasmid pMS76 (pRS313CDC12-GFP). (A) Wild-type (YPH499). (B) cla4Δ (YMS306). (C) cla4Δ swe1Δ (YMS332). (D) chs3Δ (ECY46–4-1B). (E) Morphology of cla4Δ swe1Δ double mutant. Introduction of the swe1 mutation eliminated bud elongation (compare with Figure 1, B or D), although not cell aggregation of a cla4 strain.
Table 3.
Septin localization in different strains
| Straina | Genotype | Normal rings (%)b | Abnormal rings (%)b | Mislocalized patches (%)b | Total rings and patches analyzed |
|---|---|---|---|---|---|
| YPH499 | Wild type | 84 | 16 | 0 | 43 |
| YMS134 | cla4Δ | 39 | 50 | 11 | 62 |
| ECY46-4-1B | chs3Δ | 55 | 43 | 2 | 56 |
| YMS332 | cla4Δ swe1Δ | 32 | 44 | 24 | 72 |
Strains were transformed with pRS316CDC3-GFP.
Percentages are relative to the total number of rings and patches observed. For criteria in scoring of abnormal rings, see MATERIALS AND METHODS.
We wanted to determine the effect of a Chs3p defect on the septin organization of cla4Δ mutants. Because the double mutant cla4Δ chs3Δ grows very poorly, an alternative procedure to abolish Chs3p activity in a cla4 strain was used, by adding to the cultures nikkomycin Z, a specific inhibitor of chitin synthase III (Choi et al., 1994b; Gaughran et al., 1994). To observe septin rings during budding, when they are formed, we isolated single, unbudded cells by centrifugation on sucrose gradients and incubated them in the absence or presence of the inhibitor. Addition of nikkomycin Z to wild-type already caused the appearance of abnormal rings (Table 4) in a proportion similar to that found in a random culture of a chs3Δ mutant (Table 3 and Figure 3D). In a cla4 null mutant, nikkomycin Z greatly increased the proportion of aberrant rings and mislocalized patches (Table 4). Finally, a cla4Δ swe1Δ double mutant showed more aberrant rings than the single cla4Δ mutant, and nikkomycin addition resulted in many more mislocalized patches (Table 4), thus confirming the above-mentioned sectoring results. The septin abnormalities produced by the addition of nikkomycin Z, although more numerous, were similar in aspect to those shown in Figure 3.
Table 4.
Changes in septin localization in different strains upon inhibition of Chs3p with nikkomycin Za
| Strainb | Genotype | Nikkomycin Zc | Normal rings (%) | Abnormal rings (%) | Mislocalized patches (%) | Total rings and patches analyzed |
|---|---|---|---|---|---|---|
| YPH499 | wt | - | 93 | 7 | 0 | 56 |
| + | 55 | 45 | 0 | 53 | ||
| YMS134 | cla4Δ | - | 60 | 32 | 8 | 53 |
| + | 4 | 69 | 27 | 134 | ||
| YMS322 | cla4Δ swe1Δ | - | 32 | 58 | 8 | 52 |
| + | 0 | 46 | 54 | 60 |
Single, unbudded cells were isolated in sucrose gradients as indicated in MATERIALS AND METHODS. Cells were incubated in SC medium lacking uracil, until most cells had well-developed buds (2.5-6 h, depending on the strain). The septin distribution was determined by fluorescence microscopy as outlined in MATERIALS AND METHODS. Percentages are referred to the total number of rings and patches observed.
All strains were transformed with pRS316-CDC3-GFP.
When added, the final concentration of nikkomycin Z was 1 mM.
Because the localization of Myo1p, a component of the actomyosin contractile ring, depends on septins (Lippincott and Li, 1998a), we examined the distribution of a Myo1p-GFP construct in a cla4Δ mutant (strain YMS134[pMS55]). The Myo1-GFP rings in the mutant were often somewhat blurred and almost twice the diameter of the wild-type rings (Figure 4, A and B) a finding consistent with a septin ring defect.
Figure 4.
cla4Δ mutants have wider Myo1p-GFP rings than wild-type. Wild-type is YPH499[pMS55]; the cla4Δ strain is YMS134[pMS55]. At left, fluorescence images superimposed to Nomarski images. Panel at right, statistical measurements of ring diameter: circles, wild-type; squares, cla4 mutant.
Finally, because of the separation defect in cla4 strains, which results in clumping (Figure 1B), mutant cells were observed by electron microscopy (Figure 5). Some septa were essentially normal, with a typical trilaminar structure (Shaw et al., 1991; Schmidt et al., 2002). Many others, however, had an aberrant structure, with frequent lacunae and multiple intersecting chitin layers (Figure 5, C and D). Multiple septa were also observed in some cases (Figure 5B). A few septa were completely delocalized (Figure 5A), like those found in septin mutant at a nonpermissive temperature (Slater et al.,1985; Roh et al., 2002b), again pointing to a septin defect.
Figure 5.
Electron microscopy of abnormal and delocalized septa in a cla4Δ mutant (strain YMS134). In A, a delocalized septum is shown (cf. Slater et al., 1985; Roh et al., 2002b). In B, the multiple septa may have resulted from repeated “landings” of septins unable to bind well to their normal locations. Note in C and D the meandering chitin lines and, in D, a lacuna. Bars, 1 μm.
Not unexpectedly, the cdc11-25 strain showed abnormal and delocalized septin rings when grown at 30°C, a defect that was aggravated by growth in rich medium (Table 5). The defect was further exacerbated by the presence of a chs3-1 mutation (Figure 6 and Table 5). In accordance with the high proportion of mislocalized septins in the mutant, ectopic septa were seen at high frequency by electron microscopy (Figure 7, A and C). The septa with normal localization had structures resembling those shown in Figure 5 of the cla4Δ septa (Figure 7A), including the presence of multiple septa (Figure 7B).
Table 5.
Septin localization in cdc11-25 strains
| Strain | Temp (°C) | Medium | Normal rings (%)a | Abnormal rings (%)a | Mislocalized patches (%)a | Total rings and patches observed |
|---|---|---|---|---|---|---|
| AVY2-25[pLP17]b | 26 | Minimal | 89 | 8 | 3 | 63 |
| 30 | Minimal | 62 | 32 | 6 | 82 | |
| 26 | YEPD | 81 | 13 | 6 | 126 | |
| 30 | YEPD | 14 | 43 | 43 | 103 | |
| AVY5c-[pRS316-CDC3-GFP] | 26 | Minimal | 53 | 41 | 6 | 80 |
| 30 | Minimal | 32 | 44 | 24 | 72 | |
| 26 | YEPD | 0 | 32 | 68 | 28 |
Percentages are relative to the total number of rings and patches observed. For criteria in scoring of abnormal rings, see MATERIALS AND METHODS.
Strain containing cdc11-25 and CDC12-GFP in plasmid.
Strain containing cdc11-25 and chs3-1. This strain does not grow in YEPD at 30°C.
Figure 6.
Septin defects in cdc11-25. Strains were grown in minimal medium at 30°C. A–D, fluorescence images; E–H, Nomarski images. (A, B, E, and F) Strain AVY2–25[pLP17] (cdc11-25); (C, D, G, and H) strain AVY5[pRS316-CDC3-GFP] (cdc11-25 chs3-1).
Figure 7.
Electron microscopy of abnormal and delocalized septa in cdc11-25. In A and C, delocalized septa are shown. See also in A and B, septa with lacunae. In B, the double septum resembles the triple one in Figure 5B. Bars, 1 μm.
A Simultaneous Defect in Chitin Synthase III and Septins Causes Enlargement of the Mother-Bud Neck
Why is the defect of the cla4 or the cdc11-25 mutant much aggravated by a CHS3 deletion? One way to look into this problem is to ask another question, i.e., whether the mere presence of the Chs3 protein is sufficient to prevent the defect seen in the double mutant or whether the enzymatic activity of the protein is also required. This question arises because Chs3p is known to be connected to a septin, Cdc10p, through Chs4p and Bni4p (DeMarini et al., 1997). This linkage is necessary for the appropriate localization of Chs3p, but it might reciprocally help to retain the septins in their location. We approached this question in two ways. One was to express, in a cla4Δ chs3Δ strain or in a cdc11-25 chs3-1 strain, a CHS3 gene containing a single mutation in the putative active site, chs3R993A. The mutation causes loss of Chs3p activity in vitro and of function in vivo (Cos et al., 1998). If only the presence of the protein were sufficient, expression of this protein in a plasmid should suppress the defect of the double mutants. However, the neck widening, clumping, and lysis of the original cla4Δ chs3Δ or the cdc11-25 chs3-1 strain were not affected by the presence of chs3R993A, either on a centromeric (pHV7–37) or a high-copy (pMS75) plasmid (unpublished results). Western blots of extracts from cells containing a chs3 null mutation and a high-copy plasmid expressing HA-fusions with wild-type or mutated CHS3 (see MATERIALS AND METHODS) showed similar levels of both Chs3 proteins (unpublished results). Furthermore, the wild-type HA-CHS3 conferred Calcofluor White sensitivity to the chs3-1 strain, but the mutated gene did not (unpublished results).
The other experiment consisted in inhibiting the activity of chitin synthase III in vivo with the competitive inhibitor nikkomycin Z, as described above. In wild-type, nikkomycin Z does not affect growth, although it largely abolishes Calcofluor staining, because the chitin that requires Chs3p for synthesis is not made (see also Figure 8B). In these experiments, we again started from single, unbudded cells. When cla4Δ cells were incubated without inhibitor, they budded in a fairly normal way, with an occasional elongated bud, as seen in random culture (Figure 8A). The formation of the chitin ring at the mother cell-bud neck and hence the localization of Chs3p appeared also to be normal in these cells, as detected with Calcofluor White (Figure 8B). In the presence of nikkomycin Z, however, the buds became extremely long and the neck between mother cell and bud widened, so that many cell pairs looked like long tubes, with little constriction between the two cells (Figure 8, A and B). Staining with Calcofluor White showed that nikkomycin Z prevented deposition of chitin at the neck both in wild-type and in the cla4Δ strain, although some diffuse fluorescence of unknown origin sometimes remained in the mutant (Figure 8B). The rapid inhibition of chitin ring formation upon the addition of nikkomycin Z before budding strongly supports the notion that the compound acts as an inhibitor of chitin synthase III in vivo, rather than interfering with expression of Chs3p. Furthermore, in previous work we observed that turning off the CHS3 gene did not result in a decrease of enzyme activity for many hours (Choi et al., 1994b). Accordingly, we found that turning off CHS3 (under a MET3 promoter) in a cla4Δ strain required overnight incubation to detect changes in septins or morphology (our unpublished results).
Figure 8.
Effect of nikkomycin Z on morphology of wild-type (YPH499) and a cla4Δ mutant (YMS306). The cultures were started with single unbudded cells (see MATERIALS AND METHODS) in synthetic complete medium with or without 1 mM nikkomycin Z and incubated at 30°C. (A) Phase contrast images of the cultures of YMS306 at different times. Note in the control a few elongated cells at 5 h. Bar, 10 μm. (B) Fluorescence images of cells of wild-type and different mutants after 4-h incubation without or with nikkomycin Z, followed by staining with Calcofluor White. Note in wild-type cells the absence of a fluorescent chitin ring, when incubated in the presence of nikkomycin Z. Some residual and largely delocalized fluorescence in the cla4Δ mutant under the same conditions remains unexplained.
Neck diameters were measured in the Calcofluor-stained cells as photographed with fluorescence filters, because the cell contour was easier to see under those conditions. After 4 h of incubation at 30°C, the average diameter of the necks of cla4Δ in the presence of nikkomycin Z was almost double of that in the control (Figure 8B and Table 6). No such increase was seen in a wild-type strain (Figure 8B and Table 6). The possibility that in the cla4Δ strain the necks formed in the presence of nikkomycin Z were already wide at bud emergence was explored by measuring neck diameters of very small buds. The results showed only a small increase in nikkomycin Z–treated cells, ascribable to enlargement after budding (Table 6). This clearly indicates that the neck widens during bud growth. The cla4Δ cells incubated in the presence of the inhibitor appeared to bud only once, although branches sometimes appeared on the long tubes. Again, the results were not changed appreciably by the presence of a swe1 deletion in addition to the cla4 deletion (Table 6), except that the buds formed in the presence of nikkomycin Z were less elongated in this case.
Table 6.
Effect of nikkomycin Z on mother-bud neck diameter in different strainsa
| Neck diameter (μm)
|
|||||
|---|---|---|---|---|---|
| Strain | Genotype | Incubation time (h) | Temp (°C) | Control | +Nikkomycin Z |
| YPH499 | wild type | 4 | 30 | 1.1 ± 0.1 (28) | 1.16 ± 0.25 (22) |
| YMS306b | cla4Δ | 2 | 30 | 1.35 ± 0.14 (49) | 1.65 ± 0.2 (53) |
| YMS306 | cla4Δ | 4.5 | 30 | 1.4 ± 0.24 (64) | 2.6 ± 0.3 (68) |
| YMS332 | cla4Δswe1Δ | 6.5 | 30 | 0.97 ± 0.25 (41) | 2.6 ± 0.45 (32) |
| AVY2-25 | cdc11-25 | 6.5 | 26 | 1.1 ± 0.1 (42) | 1.5 ± 0.18 (36) |
| AVY2-25 | cdc11-25 | 4 | 30 | 1.14 ± 0.15 (32) | 1.7 ± 0.2 (38) |
| DHY103-9B | cdc3-1 | 4 | 26 | 0.96 ± 0.16 (36) | 1.3 ± 0.2 (33) |
| DHY103-9B | cdc3-1 | 4 | 30 | 1.1 ± 0.15 (51) | 1.6 ± 0.2 (55) |
| DHY103-9B | cdc3-1 | 5.5 | 30 | 1.16 ± 0.18 (61) | 1.9 ± 0.4 (50) |
Data are given as averages plus/minus the standard deviation, which indicates the variability of the neck diameter. Values in parentheses indicate the number of necks measured. The final concentration of nikkomycin Z, when added, was 1 mM. The difference between the values with or without nikkomycin Z, as measured by the z score, was at a significance level of <0.001 in all cases, except for the wild-type strain.
Only the neck diameters of small buds were measured here.
Chitin synthase III is responsible for the formation of the chitin ring that is laid down at the onset of budding. If the effect of nikkomycin Z is due only to inhibition of ring formation, the results should not be changed by withdrawal of the inhibitor after the time for ring deposition has elapsed. This was indeed the case. In one experiment, cells were incubated with nikkomycin Z until small buds appeared in most of them, then centrifuged, washed, and suspended in fresh medium. On further incubation the cells showed the same morphological changes as those that were constantly incubated with nikkomycin Z (unpublished results).
The results with the cdc11-25 strain were similar to those with the cla4Δ mutant (Figure 8B), although the widening of the neck was less pronounced. The diameter increased ∼50% in the presence of nikkomycin Z (Figure 8B and Table 6). Similar results were obtained with another septin mutant, cdc3-1 (Figure 8B and Table 6).
DISCUSSION
cla4 and cdc11 Mutants Show Similar Septin Abnormalities
Two genes, CLA4 and CDC11, were isolated in our screen for synthetic lethality with CHS3. The appearance of CDC11, coding for a septin, in the screen was not unexpected, because a similar finding was briefly mentioned by Osmond et al. (1999). On the other hand, the isolation of CLA4 was surprising. Cla4p is a protein kinase of the PAK type (Cvrcková et al.1995). cla4 mutants have elongated buds and some cytokinesis defect (Cvrcková et al., 1995; Holly and Blumer, 1999; Longtine et al., 2000; Weiss et al., 2000), but the mechanism of action of Cla4p, an effector of Cdc42p (Benton et al., 1997) has not been determined. Cla4p appears to be functionally redundant with another kinase, Ste20p, based on the finding that CLA4 and STE20 are synthetically lethal (Cvrcková et al., 1995). The function described here, however, is not shared with Ste20p, because deletion of STE20 did not result in the same defect and overexpression of Ste20p did not correct the morphology of the cla4 mutant. In previous work Cla4p was usually studied in a ste20 background; therefore, the specific functions of Cla4p and those shared with Ste20p were observed together (see, e.g., Holly and Blumer, 1999; Weiss et al., 2000).
The finding that both CLA4 and CDC11, which codes for a septin, show synthetic lethality with CHS3 suggested that Cla4p may be involved in septin function. Cvrcková et al. (1995) and Longtine et al. (2000) observed abnormalities in septin organization in a cla4 mutant. We confirmed and extended their results (Figure 3, Tables 3 and 4). Both cla4 and cdc11-25 mutants showed frequent and very similar abnormalities in septa as well as delocalized septal structures (Figures 5 and 7). The aberrant actomyosin contractile rings found in the cla4Δ strain may result from the septin defect, because Myo1p localization depends on septins (Lippincott and Li, 1998a). In turn, the mislocalization of the contractile ring may lead to the abnormal septa as a result of an irregular binding to the plasma membrane, resulting in erratic invagination and abnormal deposition of primary septum chitin (Roh et al., 2002b; Schmidt et al., 2002). As for the ectopic septa, we previously found them in cdc3, cdc10, cdc11, and cdc12 mutants and concluded that they resulted from attachment of defective septin rings to aberrant sites, where they served as scaffolds for an improperly placed septum (Slater et al., 1985; Roh et al., 2002b). From all the above we conclude that Cla4p is necessary for normal septin function, either for attachment of the septin filaments to the plasma membrane, or for organization of the filaments, or both. An attractive hypothesis is that Cla4p, directly or indirectly, causes a modification of some protein in the plasma membrane at the bud neck region, which enables it to bind the septin filaments and/or to organize the septin ring. This would provide an explanation for the more aberrant phenotype of cla4Δ cdc11-25 double mutants compared with the single mutants, because in the double mutant both the septins and their receptor would be defective.
Septins and the Chitin Ring Cooperate in Maintaining the Mother-Bud Neck Size
Our results implied an involvement of Cla4p in septin function but did not explain the puzzling interaction between Cla4p or Cdc11p on one side and Chs3p on the other. Because septins retain Chs3p at the correct localization through a Bni4p-Chs4p bridge, it seemed possible that Chs3p, a plasma membrane protein, might exert a reciprocal action and tether the septin ring to the membrane. If so, mere presence of Chs3p, even in an enzymatically inactive form, could be sufficient to stabilize the septin ring. However, either a centromeric or a high-copy plasmid carrying a mutated form of CHS3 was unable to suppress the defect of a cla4Δ chs3Δ mutant, although the protein was expressed at normal levels. The Chs3 protein used had a single point mutation, resulting in the change of one of three adjacent arginine residues into alanine in the putative catalytic site of the protein, which eliminates the enzymatic activity (Cos et al., 1998). Therefore, little change would be expected in charge, general conformation, and ability to bind other proteins. On the other hand, inhibition of chitin synthase III activity by nikkomycin Z, a competitive inhibitor that is supposed to act exclusively by displacing the substrate (the structural analog UDP-GlcNAc), not only aggravated the septin abnormalities, but also the morphological defect of a cla4 mutant. The unbudded cells used in this experiment budded only once and gave rise to very elongated buds, with a much widened neck at the junction with the mother cell (Figure 8B and Table 6). A similar widening of the neck as well as cell lysis were observed in a cla4Δ chs3Δ double mutant (see RESULTS), although this strain was able to grow slowly, possibly because of some suppressor. Neck widening, albeit less pronounced, was detected in septin mutants, cdc11-25 and cdc3-1, upon treatment with nikkomycin Z (Figure 8B and Table 6). Thus, under conditions in which attachment to the membrane and/or organization of the septins is compromised by a cla4 or a septin mutation, a defect in Chs3p enzymatic activity results in abnormal growth and expansion of the neck region, which normally is unchanged throughout the cell cycle. This expansion ultimately leads to cell death, which explains the synthetic lethality between cla4 or cdc11 and chs3.
How does the chitin ring, which requires Chs3p for its synthesis, contribute to prevent growth in the neck region? A reasonable explanation is suggested by what we know about cell wall structure and the order of assembly of cell wall components. As previously reported (Kollár et al., 1995, 1997), chitin is bound to the cell wall structure in two types of linkages: to side chains of β(1→6)glucan and to the nonreducing end of β(1→3)glucan. The latter position is the same where, in other chains, β(1→6)glucan is attached to β(1→3)glucan. Mannoproteins, in turn, are linked to β(1→6)glucan. The order of addition in vivo is β(1→3)glucan, β(1→6)glucan, mannoprotein (Roh et al., 2002a). Because of its high concentration in the neck region, chitin may cap most of the β(1→3)glucan nonreducing ends, making them unavailable for addition of β(1→6)glucan, hence of mannoprotein. Thus, cell wall growth and consequent neck expansion would be prevented in that area.
Our conclusions are summarized in Scheme 1. Cla4p has a role in septin localization and/or assembly. This scheme does not postulate a mechanism for Cla4p action and it would be valid even if Cla4p acted through Cdc42p, as recently postulated by Gladfelter et al. (2002), so long as the ultimate targets are the septins. Septins, in turn, determine the localization of the chitin synthase III system, which is necessary for the formation of a chitin ring at the neck, at bud emergence (Shaw et al., 1991). The chitin ring prevents growth at the neck, probably also blocked by the presence of the septins, which appear to act as a barrier between mother cell and bud, impeding the movement of proteins on the plasma membrane (Gladfelter et al., 2001). When either the septins or the chitin synthase system are functional, relatively little change is seen at the neck. When both are defective, growth in that region is not controlled and the neck enlarges. Thus, in the same way that septins and Chs3p normally cooperate with each other, a defect in one aggravates that of the other. A septin defect leads to partial delocalization of chitin synthesis, whereas a Chs3p defect causes enlargement of the neck with reduced binding of the already abnormal septins.
Scheme 1.
Proposed effect of Cla4p and Chs3p on septin anchoring and organization. Cla4p helps anchoring of the septin ring at the neck. The septin ring, in turn, localizes Chs3p at the neck through binding of Bni4p and Chs4p as intermediaries (DeMarini et al., 1997). Both septins and the chitin ring, dependent on Chs3p activity, cooperate in preventing growth at the neck. When either the septins or Chs3p are defective, not much change in neck growth is noted. If both are faulty, however, neck growth becomes unrestrained, which contributes further to septin disorganization (curved arrow).
These results show that two stabilizing systems, the septins and the chitin ring, are required to ensure the constancy of diameter and structure of the bud neck. When both fail, the neck enlarges and the cell dies, an outcome that underlines the importance of neck integrity.
It is interesting to note that, although the existence of the chitin ring has been known for 30 years (Hayashibe and Katohda, 1973), its function remained unknown until now. Retrospectively, this is understandable, because a chitin ring defect has relatively little consequence (Shaw et al., 1991) in the absence of a concomitant septin abnormality.
Supplementary Material
Acknowledgments
We thank A. Bender, F. Cvrcková, N. Kleckner, E. Leberer, J. Lippincott, M. Longtine, T. Roberts, C. Roncero, S. Michaelis, and M. Thomas for plasmids and strains. We are also indebted to J. Hanover for the use of the fluorescence microscope and to O. Cohen-Fix and W. Prinz for a critical reading of the manuscript.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-08-0547. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-08-0547.
References
- Alani, E., Cao, L., and Kleckner, N. (1987). A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, O.M., Billington, B.L., and Gottschling, D.E. (1991). Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66, 1279–1287. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, G.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. New York: John Wiley & Sons.
- Bender, A., and Pringle, J.R. (1991). Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton, B.K., Tinkelenberg, A., Gonzalez, I., and Cross, F.R. (1997). Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 17, 5067–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib, E., Roh, D.-H., Schmidt, M., Crotti, L.B., and Varma, A. (2001). The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276, 19679–19682. [DOI] [PubMed] [Google Scholar]
- Cabib, E., Shaw, J.A., Mol, P.C., Bowers, B., and Choi, W.-J. (1996). Chitin biosynthesis and morphogenetic processes. In: The Mycota, vol. III, ed. R. Bramble and G.A. Marzluf, Berlin: Springer-Verlag, 243–267. [Google Scholar]
- Choi, W.-J., Sburlati, A., and Cabib, E. (1994a). Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc. Natl. Acad. Sci. USA 91, 4727–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W.-J., Santos, B., Durán, A., and Cabib, E. (1994b). Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol. Cell. Biol. 14, 7685–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cos, T., Ford, R.A., Trilla, J.A., Durán, A., Cabib, E., and Roncero, C. (1998). Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 256, 419–426. [DOI] [PubMed] [Google Scholar]
- Crotti, L.B., Drgon, T., and Cabib, E. (2001). Yeast cell permeabilization by osmotic shock allows determination of enzymatic activities in situ. Anal. Biochem. 292, 8–16. [DOI] [PubMed] [Google Scholar]
- Cvrcková, F., De Virgilio, C., Manser, E., Pringle, J.R., and Nasmyth, K. (1995). Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9, 1817–1830. [DOI] [PubMed] [Google Scholar]
- DeMarini, D.J., Adams, A.E.M., Fares, H., De Virgilio, C., Valle, G., Chuang, J.S., and Pringle, J.R. (1997). A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonová, J., Drgon, T., Roh, D.-H., and Cabib, E. (1999). The GTP-binding protein Rho1p is required for cell cycle progression and polarization of the yeast cell. J. Cell Biol. 146, 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, R.A., Shaw, J.A., and Cabib, E. (1996). Yeast chitin synthases 1 and 2 consist of a non-homologous and dispensable N-terminal region and of a homologous moiety essential for activity. Mol. Gen. Genet. 252, 420–428. [DOI] [PubMed] [Google Scholar]
- Gaughran, J.P., Lai, M.H., Kirsch, D.R., and Silverman, S.J. (1994). Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J. Bacteriol. 176, 5857–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A.S., Pringle, J.R., and Lew, D.J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681–689. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., Bose, I., Zyla, T.R., Bardes, E.S.G., and Lew, D.J. (2002). Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashibe, M., and Katohda, S. (1973). Initiation of budding and chitin-ring. J. Gen. Appl. Microbiol. 19, 23–39. [Google Scholar]
- Holly, S.P., and Blumer, K.J. (1999). PAK-Family kinases regulate cell and actin polymerization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollár, R., Petráková, E., Ashwell, G., Robbins, P.W., and Cabib, E. (1995). Architecture of the yeast cell wall. The linkage between chitin and β(1→3)glucan. J. Biol. Chem 270, 1170–1178. [DOI] [PubMed] [Google Scholar]
- Kollár, R., Reinhold, B.B., Petráková, E., Yeh, H.J.C., Ashwell, G., Drgonová, J., Kapteyn, J.C., Klis, F.M., and Cabib, E. (1997). Architecture of the yeast cell wall. β(1→6)glucan interconnects mannoprotein, β(1→3)glucan, and chitin. J. Biol. Chem. 272, 17762–17775. [DOI] [PubMed] [Google Scholar]
- Leberer, E., Dignard, D., Harcus, D., Thomas, D.Y., and Whiteway, M. (1992). The protein kinase homologue Ste20 is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signaling components. EMBO J. 11, 4815–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998a). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998b). Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143, 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., DeMarini, D.J., Valencik, M.L., Al-Awar, O.S., Fares, H., De Virgilio, C., and Pringle, J.R. (1996). The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8, 106–119. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., Theesfeld, C.L., McMillan, J.N., Weaver, E., Pringle, J.R., and Lew, D. (2000). Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.A., and Sprague, G.F. (2001). The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G2/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond, B.C., Specht, C.A., and Robbins, P.W. (1999). Chitin synthase III synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA 96, 11206–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh, D.-H., Bowers, B., Riezman, H., and Cabib, E. (2002a). Rho1p mutations specific for regulation of β(1→3)glucan synthesis and the order of assembly of the yeast cell wall. Mol. Microbiol. 44, 1167–1184. [DOI] [PubMed] [Google Scholar]
- Roh, D.-H., Bowers, B., Schmidt, M., and Cabib, E. (2002b). The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell, 13, 2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., Bowers, B., Varma, A., Roh, D.-H., and Cabib, E. (2002). In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115, 293–302. [DOI] [PubMed] [Google Scholar]
- Shaw, J.A., Mol, P.C., Bowers, B., Silverman, S.J., Valdivieso, M.H., Durán, A., and Cabib, E. (1991). The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R., and Hieter, P. (1989). A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, M.L., Bowers, B., and Cabib, E. (1985). Formation of septumlike structures at locations remote from the budding sites in cytokinesis-defective mutants of Saccharomyces cerevisiae. J. Bacteriol. 162, 763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday, N., Bouquin, N., and Li, R. (2001). Assembly and regulation of the cytokinetic apparatus in budding yeast. Curr. Opin. Microbiol. 4, 690–695. [DOI] [PubMed] [Google Scholar]
- Trilla, J.A., Cos, T., Durán, A., and Roncero, C. (1997). Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and. CSD4. Yeast 13, 795–807. [DOI] [PubMed] [Google Scholar]
- Valdivieso, M.H., Mol, P.C., Shaw, J.A., Cabib, E., and Durán, A. (1991). CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J. Cell Biol. 114, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E.L., Bishop, A.C., Shokat, K.M., and Drubin, D.G. (2000). Chemical genetic analysis of the budding yeast-p21-activated kinase Cla4p. Nat. Cell Biol. 2, 677–685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.